During corneal stromal wound healing, quiescent stromal keratocytes are activated to myofibroblasts with changed phenotypic characteristics, which are responsible for the formation of nontransparent scar tissue. The present study demonstrates that ethyl pyruvate (EP) modulates phenotypic changes during TGF-β1–induced activation of keratocytes to myofibroblasts in vitro, and EP may be a potential therapeutic agent in corneal wound healing.
Abstract
Purpose.
Ethyl pyruvate (EP) has pharmacologic effects that remediate cellular stress. In the organ-cultured murine lens, EP ameliorates oxidative stress, and in a rat cataract model, it attenuates cataract formation. However, corneal responses to EP have not been elucidated. In this study, the potential of EP as a therapeutic agent in corneal wound healing was determined by examining its effects on the transition of quiescent corneal stromal keratocytes into contractile myofibroblasts.
Methods.
Three independent preparations of cultured human keratocytes were treated with TGF-β1, to elicit a phenotypic transition to myofibroblasts in the presence or absence of 10 or 15 mM EP. Gene expression profiles of the 12 samples (keratocytes ± EP ± TGF-β1 for three preparations) were produced by using gene microarrays.
Results.
TGF-β1–driven twofold changes in at least two of three experiments defined a group of 1961 genes. Genes showing twofold modulation by EP in at least two experiments appeared exclusively in myofibroblasts (857 genes), exclusively in keratocytes (409 genes), or in both phenotypes (252 genes). Analysis of these three EP-modulated groups showed that EP (1) inhibited myofibroblast proliferation with concomitant modulation of some cell cycle genes, (2) augmented the NRF2-mediated antioxidant response in both keratocytes and myofibroblasts, and (3) modified the TGF-β1–driven transition of keratocytes to myofibroblasts by inhibiting the upregulation of a subset of profibrotic genes.
Conclusions.
These EP-induced phenotypic changes in myofibroblasts indicate the potential of EP as a therapeutic agent in corneal wound healing.
Pyruvic acid is the final product of the glycolytic pathway, the starting substrate for the tricarboxylic acid (TCA) cycle, and a scavenger of reactive oxygen species (ROS).1,2 Ethyl pyruvate (EP) is a membrane-permeant ester of pyruvate, and exogenous EP has the potential to augment intracellular pyruvate levels. In hypoxia, elevated intracellular pyruvate enables the cell to protect itself from ROS-mediated damage and to slough off excess reducing equivalents (by converting pyruvate to lactate). However, intracellular hydrolysis of EP is relatively slow, and several studies (for a review, see Fink 3) have shown that the intact ester also has direct pharmacologic effects. Using murine lens in organ culture, Varma et al.4 showed that EP ameliorates oxidative stress when present concurrently and can partly reverse deleterious effects when 2 hours are added to the stress period.5 Moreover, in intact rats fed a 30% galactose diet (a model for the development of sugar cataract) the concurrent application of EP eye drops attenuated cataract development up to 40 days.6 These authors point out that the reaction of ROS with glycated lens proteins is a major contributor to cataract formation, and so EP very likely protects against cataract development by decreasing ROS levels.
Apart from the work of Varma et al.,4 the potential therapeutic effects of EP have been investigated predominantly in splanchnic systems (for a review, see Fink3). These studies focused mainly on rodent models of endotoxin (bacterial lipopolysaccharide [LPS]) induced damage (e.g., LPS infusion, bacterial peritonitis, or acute endotoxemia). The NF-κB pathway is prominent in mediating the proinflammatory effects seen in these models, and EP inhibits NF-κB-dependent signaling by directly targeting p65.7 Therefore EP is of obvious interest in the corneal response to bacterial infection. However, a separate clinical concern is corneal scarring absent infection. This scarring is largely driven by the TGF-β-mediated conversion of quiescent stromal keratocytes to myofibroblasts.
Although TGF-β isoforms are absent from the corneal stroma in the normal human eye,8 increased local TGF-β2 is seen in patients with superior limbic keratoconjunctivitis.9 In the rabbit, antibodies against TGF-β1 decrease subepithelial collagen deposition (corneal haze) after excimer laser photorefractive keratectomy (PRK),10 and antibodies against TGF-β2 reduce subconjunctival scarring after glaucoma filtration surgery.11 In the rat, antibodies against TGF-β1 inhibit the increase in the number of stromal cells in the laser-ablated area 5 days after PRK,12 including the recruitment of highly reflective activated keratocytes. Myofibroblast transformation and consequent stromal fibrosis also are inhibited. Experiments in vitro suggest that in the cornea, stromal-to-epithelial signaling predominantly involves HGF and KGF (FGF7),13 whereas epithelial-to-stromal signaling is predominantly by TGF-β1, bFGF (FGF2), and EGF.14 Cultured corneal keratocytes undergo phenotype shifts to fibroblasts and myofibroblasts in response to FGF2 and TGFβ, respectively.15 In corneal fibroblasts expression of TGF-β1 and TGF-βRI (but not TGF-βRII or -RIII) is upregulated by exogenous TGF-β1.16 Exogenous FGF-2 decreases TGF-β1 mRNA levels,17 but TGF-β1 has no reciprocal effect on FGF-2. Relative to keratocytes, myofibroblasts show upregulation of α-smooth muscle actin and TGF-β receptors, and downregulation of connexin 43,18,19 they stain positively for integrin α5β1, myosin, and α-actinin, and form f-actin microfilament bundles (stress fibers) that co-localize with fibronectin.
The previous reports in splanchnic models mentioned earlier suggest that EP should have therapeutic anti-inflammatory effects in the cornea; and in the intact rat, long-term (40 day) instillation of EP eye drops is well tolerated.6 However, before human trials are conducted, it is important to survey the responses to EP of both normal and activated human corneal stromal cells. In the present study, we use cultured human keratocytes to model the effects of EP on normal stroma, and TGF-β1–activated keratocytes to model the effects of EP on the TGF-β–driven scarring process that can occur in vivo. The present microarray analyses provide global surveillance of the resultant gene expression changes and an encouraging picture of the potential therapeutic uses of EP.
Materials and Methods
Isolation, Culture, and Treatment of Human Keratocytes and Myofibroblasts
Keratocytes were isolated from single corneas from three human donors according to the method of Guerriero et al.20 Briefly, corneas were cut in half, and the endothelium, along with Descemet's membrane, and the epithelium, along with a thin layer of underlying stroma, were removed. The remaining stromal pieces were digested in 0.25% collagenase (Sigma-Aldrich, Inc., St. Louis, MO) at 37°C for 16 to 18 hours. After centrifugation at 1200 rpm for 7 minutes, the pellets containing keratocytes were resuspended in DMEM/F12 with 0.021% of a dipeptide l-glutamine (Glutamax), 0.011% pyruvate, and penicillin/streptomycin (all from Invitrogen/Gibco, Carlsbad, CA), and the suspension was filtered through a cell strainer (70 μm; BD-Falcon, Bedford, MA). Keratocytes from a single cornea were plated onto four 35-mm dishes (Falcon Primaria; BD Biosciences, Lincoln Park, NY) in serum-free (SF) medium, to maintain the keratocyte phenotype. The keratocytes were then maintained in serum-free DMEM/F12 containing 50 mM HEPES and 0.1 mM l-ascorbic acid 2-phosphate. Addition of 8 ng/mL TGF-plus 0.1% fetal bovine serum (FBS) was used to activate keratocyte transition into myofibroblasts, and 10 or 15 mM EP was added when appropriate. Treatment continued for 2 days, with the medium replaced every 12 hours. The cells were then processed for microarray analysis.
Proliferation Analysis
The effect of EP on human corneal stromal fibroblasts was determined by activating keratocytes to fibroblasts with DMEM/F12 with 10% FBS and then subculturing them in the same medium. Fibroblasts in passages 1 or 2 were subcultured into the desired number of 35-mm tissue culture dishes at a density of 5 × 104 cells/per dish. After 24 hours of incubation, the medium were replaced with DMEM/F12 containing 8 ng/mL TGF-β1 and 0.1% FBS. The medium also contained (final concentrations) 50 mM HEPES and 0.1 mM l-ascorbic acid 2-phosphate, with or without 10 or 15 mM EP. The medium was replaced every 24 hours. Several regions were marked on the bottoms of the dishes, and the cells in marked regions were counted at 24-hour intervals for 5 days from the start of the treatment. Phase-contrast digital images of the cells in the marked fields were captured, and the cells in the images were counted with image-analysis software (MetaMorph; Molecular Devices, Sunnyvale, CA). The rate of proliferation was evaluated by determining the number of cells at specific time points after treatment in relation to the number at the start of the treatment.
After 2 days of treatment with EP, one set of cells was fixed; permeabilized; stained for Ki67 with a monoclonal anti-Ki67 antibody (Zymed, South San Francisco, CA), followed by Alexa 488-conjugated goat anti-mouse IgG (1:2500; Molecular Probes, Inc.-Invitrogen), and for phalloidin with Alexa Fluor 546 phalloidin (1:50; Molecular Probes-Invitrogen); and counterstained with DAPI, as described previously.21 The total number of cells (DAPI stained) and Ki67-positive cells were then counted (MetaMorph imaging software; Molecular Devices).
Gene Microarray Analysis of Keratocyte and Myofibroblasts
Total RNA (3.6 ± 1.7 μg, mean ± SD; n = 12) was extracted from 12 distinct samples (RNeasy and QiaShredder kits; Qiagen, Valencia, CA). The samples were processed and analyzed by using appropriate gene microarray products (Affymetrix Inc., Santa Clara, CA: cited as catalog numbers). Briefly, Eukaryote Poly A RNA internal standards (cat no. 900433) were added to the samples, and the mRNA component of the total RNA was reverse transcribed in the presence of a T7-(dT)24 primer (900431). The resulting cDNA was extracted (900371) and transcribed in vitro in the presence of biotin-labeled ribonucleotides (900449). The biotinylated cRNA product (41.7 ± 25.0 μg, mean ± SD, n = 12) was extracted (900371), and 20 μg was fragmented (900371) for 35 minutes at 94°C. Hybridization controls (900454) were added, and each sample was hybridized overnight to a gene chip (U133 Plus 2.0 GeneChip [900466]; Affymetrix). The chips were then washed, developed, and scanned (ChipScanner; Agilent, Palo Alto, CA). Raw data were processed and analyzed (GeneChip Operating System [GCOS], ver. 1.4; Affymetrix) with default statistical settings. Processed data were sorted and inspected (Excel; Microsoft, Redmond, WA). The gene microarray used (HG-U133 Plus 2.0; Affymetrix) contains 54,675 panels, each targeting a specific transcript sequence. Approximately 20,550 transcripts identified by Entrez Gene symbols (National Center for Biotechnology Information, Bethesda, MD) are redundantly targeted by 46,000 panels; the remaining panels target transcripts that are less well characterized. Unscaled mean expression levels were 136 ± 25 (mean ± SD, n = 11) with one outlier value of 26. Expression levels were scaled using the software default (2% trimmed mean scaled to 500). Five redundant panels measured the transcript for the housekeeping gene GAPDH, yielding a total of 5 × 3 (sample pairs) = 15 measurements of GAPDH change. These changes were (mean ± SD) 1.04 ± 0.12-fold for TGF-β, 1.11 ± 0.13-fold for EP in myofibroblasts, and 1.03 ± 0.12-fold for EP in keratocytes. Therefore, GAPDH expression is unchanged, and the SD is consistently 12% to 13% of the mean.
We showed previously that in cultured human corneal stromal cells, donor-to-donor variation is greater than chip-to-chip variation.22 Therefore, transcripts were selected on the basis of more than twofold changes between each treated sample and its matched, untreated control in two of three preparations (see Table 2). Panels showing consistent changes were subjected to a Web-based pathways analysis program (Ingenuity Pathways Analysis ver.7.6; IPA 7.6: https://analysis.ingenuity.com/pa/login/applet.jsp/ Ingenuity Systems, Redwood, CA). This software classifies genes according to ontology groups, functional interactive networks, or canonical pathways. For the present data, we found the third, canonical, classification to be the simplest and most instructive.
Table 2.
Canonical Pathways Significantly Enriched with EP-Modulated Genes
| Canonical Pathways (Total Number of Genes in Pathway) | Number of Genes in Each Phenotype (P) |
||
|---|---|---|---|
| Myofibroblast | Keratocyte | Both Phenotypes | |
| NRF2-mediated oxidative stress response (185) | 16 (3.0E-03) | 9 (9.3E-03) | 10 (6.8E-05) |
| Hepatic fibrosis/hepatic stellate cell activation (135) | 18 (9.3E-06) | 9 (3.7E-05) | |
| Cell Cycle: G2/M DNA damage checkpoint regulation (43) | 6 (5.3E-03) | 6 (7.4E-06) | |
| p53 signaling (89) | 11 (1.5E-03) | 6 (9.6E-04) | |
| ATM signaling (52) | 9 (3.5E-04) | 5 (5.1E-04) | |
| C21-steroid hormone metabolism (71) | 4 (8.5E-03) | ||
| Molecular mechanisms of cancer (372) | 25 (6.8E-03) | ||
| Biosynthesis of steroids (128) | 8 (2.6E-05) | ||
| Metabolism of xenobiotics by cytochrome P450 (210) | 10 (3.8E-03) | ||
| Factors promoting cardiogenesis in vertebrates (89) | 10 (2.9E-03) | ||
| Pancreatic adenocarcinoma signaling (117) | 12 (1.8E-03) | ||
| Cell cycle: G1/S checkpoint regulation (59) | 6 (3.8E-03) | ||
| Role of BRCA1 in DNA damage response (53) | 9 (3.0E-04) | ||
| Aryl hydrocarbon receptor signaling (157) | 20 (7E-05) | ||
| Phospholipid degradation (106) | 6 (8.3E-03) | ||
| 14-3-3-Mediated signaling (114) | 7 (6.6E-03) | ||
| Glutathione metabolism (98) | 5 (5.6E-03) | ||
| Glycerophospholipid metabolism (193) | 8 (5.1E-03) | ||
| Macropinocytosis signaling | 6 (2.3E-03) | ||
| Starch and sucrose metabolism (72) | 7 (8.7E-04) | ||
| RAN signaling (23) | 4, 3.0E-04) | ||
| Thyroid cancer signaling (41) | 6 (1.4E-04) | ||
| Colorectal cancer metastasis signaling (245) | 8 (8.91E-03) | ||
| Glioma signaling (112) | 5 (7.2E-03) | ||
| IL-8 signaling (187) | 8 (1.4E-03) | ||
| Role of CHK proteins in cell cycle checkpoint control (34) | 4 (8.5E-04) | ||
| Mitotic roles of polo-like kinase (62) | 7 (7.8E-06) | ||
Canonical pathways significantly enriched (P < 0.01) in the myofibroblast group contained 88 unique, characterized genes making 167 appearances, with 77 (87%) appearing in three pathways or fewer. The equivalent keratocyte data show 42 genes making 58 appearances, with 41 (98%) appearing in three pathways or fewer. Forty-four genes made 68 appearances and are shown in the “both phenotypes” column, with 40 (91%) appearing in three pathways or fewer. The aggregate data contained 163 unique genes, whereas (88 + 42 + 44) = 172 were expected from the component groups (i.e., there are nine redundancies). Redundancies among groups were due to multiple panels that may assign a given gene to more than one group. The following genes: BIRC5, COL1A1, FAS, IGF1, IGFBP5, MAF, GSR, SOD2, SQSTM1 and RRAS2 were redundant among groups. Each of these genes appears in Tables 3, 4, and 5.
Results and Discussion
Effect of EP on Proliferation of Corneal Stromal Myofibroblasts
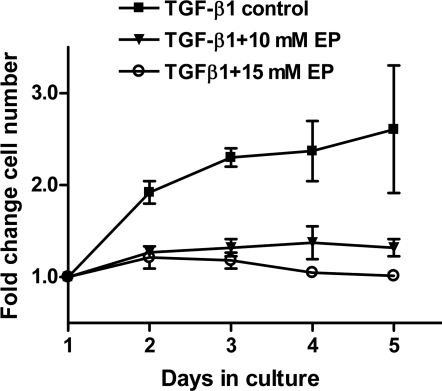
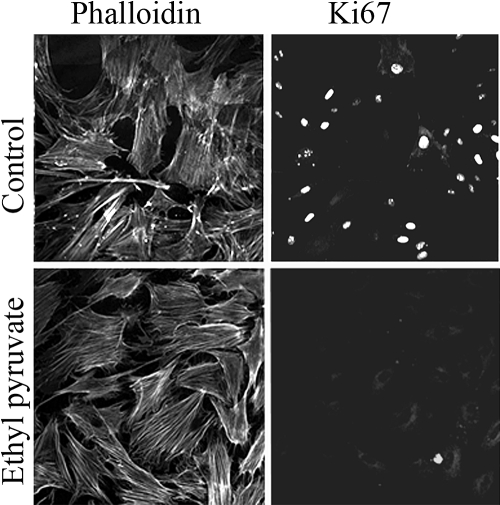
Hypercellularity is an undesirable characteristic of fibrotic tissue that develops after an injury to the corneal stroma. Quiescent, nonproliferative keratocytes can be activated to convert into proliferative fibroblast or myofibroblast phenotypes by FGF2 or TGF-β, respectively, both in vivo and in vitro. We measured the effect of EP on the proliferation of cells cultured in medium with TGF-β1+1% FBS. These myofibroblasts showed a twofold or greater increase in the number of cells in the absence of EP, but only a marginal increase (<1.2 fold) when EP was present (Fig. 1). The number of cells expressing the proliferative nuclear antigen Ki67 was measured (Fig. 2) to determine the fraction of cells in the G1/S and S phases of the cell cycle. Control cells were 30% ± 5% positive for Ki67, whereas <1% of EP-treated cells were positive (representative staining patterns are shown in Figure 2). Microarray determination of MKI67 (the transcript encoding Ki67) in myofibroblasts showed that EP caused a 3.4-fold decrease relative to the control cells, as opposed to the 30-fold decrease in protein. This finding resulted from the GCOS software's calculating a positive numerical value even for undetectable transcripts, thereby underestimating the changes if the transcript was absent in one of the samples being compared. In control myofibroblasts, MKI67 was detected by 12 panels (four redundant panels for MKI67 in each experiments × three experiments), whereas in EP-treated myofibroblasts, only one of the 12 panels showed detectable MKI67. Thus, EP essentially ablates the measurable MKI67 transcript, consistent with the very low protein level seen.
Figure 1.
EP-induced changes in cell proliferation. Effect of EP on the growth of human corneal stromal cells. Corneal stromal cells in P2 growing in DMEM/F12 with 10% FBS (i.e., fibroblasts) were subcultured into the desired number of 35-mm tissue culture dishes. After 24 hours of incubation, the medium was replaced with DMEM/F12 medium containing TGF-β1+1% FBS (i.e., conversion to myofibroblast). The cells were then incubated in this medium, with or without EP.
Figure 2.
EP-induced changes in the expression of Ki67. Human corneal stromal cells were doubled stained with phalloidin and Ki67 nuclear proliferative antigen. The cells were cultured in medium containing 8 ng/mL of TGF-β1 and 1% FBS, with or without 15 mM EP for 2 days. The cells were then double stained with anti-Ki67 antibodies (right) and phalloidin (left). There were only a few Ki67-positive cells in the EP-treated cultures.
TGF-β–Driven Changes: Comparing Keratocytes and Myofibroblasts
We examined all the gene expression changes associated with the phenotypic shift from keratocyte to myofibroblast in the absence of EP. Among the present TGF-β data, 650 panels (440 unique characterized genes) showed consistent twofold changes in all three preparations, whereas 2890 panels (1961 genes) showed twofold changes in at least two preparations. In these two data groups, canonical pathways analysis (IPA, ver. 7.6; Ingenuity Systems) identified six pathways that were significantly (P < 0.01) enriched in both groups (Table 1). The second most populated pathway was hepatic fibrosis/hepatic stellate cell activation. Hepatic fibrosis is largely driven by the responses to TGF-β of hepatic stellate cells, relatively quiescent cells of dendritic morphology that can be activated to a myofibroblast (α-actin expressing) phenotype. Therefore, this canonical pathway is a close hepatic analogue of the present TGF-β–driven response.
Table 1.
Canonical Pathways That Are Significantly (P < 0.01) Enriched with TGF-β1–Modulated Genes
| Canonical Pathways (Total Number of Genes in Pathway) | Number of Genes Modulated in (% of Pathway Genes, P) |
|
|---|---|---|
| All 3 Experiments | At Least 2 of 3 Experiments | |
| Hepatic fibrosis/hepatic stellate cell activation (135) | 13 (7.1E-06) | 27 (20%) (1.0E-04) |
| Biosynthesis of steroids (128) | 9 (1.1E-08) | 14 (11%) (1.7E-07) |
| Bladder cancer signaling (90) | 7 (3.2E-03) | 19 (21%) (5.1E-04) |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis (341) | 15 (3.2E-03) | 46 (14%) (1.1E-03) |
| Butanoate metabolism (133) | 6 (2.3E-03) | 13 (10%) (4.8E-03) |
| Oncostatin M signaling (35) | 4 (8.91E-03) | 12 (6.3E-05) |
These six canonical pathways are populated by 131 instances of 106 unique, characterized genes. Redundant appearances are MMP1 (in four pathways); IL8, FGF2, NRAS, RRAS, HRAS, KRAS, and MAPK1 (in 3 pathways); and MYC, FN1, IL6R, IL6ST, CCND1, VCAM1, TLR4, and TNFRSF11 (in 2 pathways).
Downregulation of keratan sulfate proteoglycans (lumican and keratocan) is one of the reported undesirable changes associated with activation of keratocytes in vivo and in vitro.20,23–27 Lumican and keratocan are critical in the regulation of collagen fibrillogenesis and in maintaining corneal transparency. All 12 samples had detectable levels of lumican and keratocan, which are known to be highly expressed in keratocytes. TGF-β1 treatment decreased expression of lumican by 21%, 30%, and 26%, respectively, in the three preparations, which is not enough to fulfill our twofold requirement. However, keratocan expression fulfilled the requirement with decreases of 60%, 54%, and 57%, respectively. Interestingly, 15 mM EP also decreased keratocan expression both in keratocytes (by 49% and 45% respectively, narrowly missing the twofold requirement) and in myofibroblasts (64% and 54%, attaining the twofold requirement). Again, lumican expression showed similar trends, but the effects were less marked.
Distribution of EP Effects between Phenotypes
Within each experiment, EP modulated more genes in myofibroblasts than in keratocytes, suggesting that the two phenotypes were differently affected. Across all experiments, the lowest number of genes modulated in either phenotype was associated with 10 mM EP, suggesting a modest dosage effect and necessitating a two-of-three selection to avoid false negatives. Accordingly, we selected genes that showed at least a two twofold change in myofibroblasts and those that showed at least two twofold change in keratocytes. Genes appearing in both groups were abstracted into a third group, yielding exclusively myofibroblast changes (1095 panels, 857 genes), exclusively keratocyte changes (462 panels, 409 genes), and changes in both phenotypes (305 panels, 252 genes). Canonical pathways significantly (P < 0.01) enriched in any of these groups are listed in Table 2. The most highly populated pathway and the only one enriched in all three groups, was the NRF2-mediated oxidative stress response. This finding is consistent with those of Varma et al.4–6 that EP enables ocular systems to resist oxidative stress.
Cell Cycle Control Genes Modulated by EP
EP inhibited proliferation in myofibroblasts (Fig. 1), and Table 2 contains five canonical pathways associated with control of the cell cycle (Table 2: G2/M DNA damage checkpoint regulation, ATM signaling, role of CHK proteins in cell cycle checkpoint control, role of BRCA1 in DNA damage response, and mitotic role of polo-like kinase). These pathways are specific to myofibroblasts or to the combined (myofibroblast plus keratocyte) group. Of the aggregate 22 members of these pathways (Table 3), 15 were downregulated. The seven transcripts upregulated all were p53-inducible and also appeared in the p53 canonical pathway (Table 2, row 4). Prominent was MDM2, which is a potent homeostatic controller of p53 effects.28 These p53 effects include arrest of proliferation at the G1/S interface, predominantly via CDKN1A,29 which encodes WAF1/Cip 1/p21, a potent suppressor of G1 cyclin-dependent kinases.30 The CDKN1A product also upregulates the NRF2 response by binding to and stabilizing the Nrf2 protein.31 Once cell proliferation has ceased, GADD45A plays a key role in nucleotide-excision repair of DNA damage,32 as does the ribonucleotide reductase subunit RRM2B.33 RRM2B has been reported to have a catalase-like activity and to lower ROS levels directly.34
Table 3.
EP Modulation of Cell-Cycle Control Signaling
| Panel | Gene Title | Gene Symbol | EP Effect | K, M |
|---|---|---|---|---|
| 204859_s_at | Apoptotic peptidase activating factor 1 | APAF1 | −2.51 | 1, 2 |
| 212672_at | Ataxia telangiectasia mutated (includes complementation groups A, C and D) | ATM | −2.39 | 1, 2 |
| 202095_s_at | Baculoviral IAP repeat-containing 5 (survivin) (also 202094_at, 210334_x_at) | BIRC5 | −6.38 | 2, 2 |
| 204531_s_at | Breast cancer 1, early onset | BRCA1 | −3.13 | 2, 3 |
| 219099_at | Chromosome 12 open reading frame 5 | C12orf5 | 3.24 | 2, 3 |
| 214710_s_at | Cyclin B1 (also 228729_at) | CCNB1 | −5.43 | 2, 3 |
| 202705_at | Cyclin B2 | CCNB2 | −5.17 | 0, 2 |
| 203213_at | Cell division cycle 2, G1 to S and G2 to M (also 203214_x_at, 210559_s_at) | CDC2 | −10.45 | 1, 2 |
| 205167_s_at | Cell division cycle 25 homolog C (S. pombe) | CDC25C | −3.97 | 2, 2 |
| 204252_at | Cyclin-dependent kinase 2 | CDK2 | −2.40 | 2, 2 |
| 202284_s_at | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CDKN1A | 3.96 | 2, 1 |
| 205394_at | CHK1 checkpoint homolog (S. pombe) also 205393_s_at | CHEK1 | −3.12 | 0, 2 |
| 242560_at | Fanconi anemia, complementation group D2 | FANCD2 | −6.66 | 0, 2 |
| 203725_at | Growth arrest and DNA-damage-inducible, alpha | GADD45A | 4.87 | 0, 2 |
| 217373_x_at | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) (also 205386_s_at) | MDM2 | 3.50 | 1, 2 |
| 205024_s_at | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) | RAD51 | −4.61 | 1, 2 |
| 223342_at | Ribonucleotide reductase M2 B (TP53 inducible) | RRM2B | 3.07 | 1, 2 |
| 213253_at | Structural maintenance of chromosome 2 | SMC2 | −3.01 | 2, 3 |
| 235086_at | Thrombospondin 1 (also 201107_s_at) | THBS1 | −4.33 | 1, 3 |
| 209294_x_at | Tumor necrosis factor receptor superfamily, member 10b (also 209295_at, 210405_x_at) | TNFRSF10B | 2.33 | 2, 3 |
| 201291_s_at | Topoisomerase (DNA) II alpha 170kDa (also 201292_at) | TOP2A | −10.46 | 0, 2 |
| 225912_at | Tumor protein p53 inducible nuclear protein 1 | TP53INP1 | 3.34 | 1, 2 |
The “EP effect” is the mean of the valid expression changes only. The number of valid expression changes in keratocytes (K) and myofibroblasts (M) are given in the column headed K, M (e.g., 1,2 denotes one valid keratocyte change and two valid myofibroblast changes).
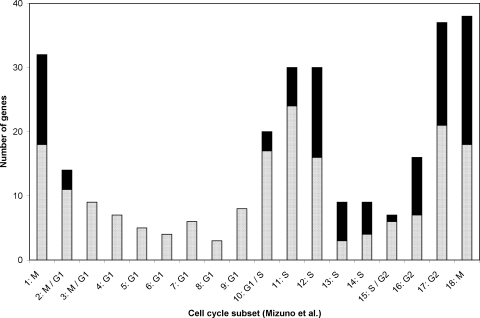
TP53INP1 encodes tumor protein 53–induced nuclear protein 1, a major mediator of p53's antioxidant (as opposed to apoptotic) function,35 whereas C12ORF5 encodes the TP53-induced glycolysis and apoptosis regulator (TIGAR), which functions as a fructose-2,6-bisphosphatase, redirecting glycolytic flux to the pentose phosphate pathway and thereby lowering intracellular ROS levels.36 Although TNFRSF10B encodes a death receptor gene that is transactivated by p53 (TRAIL, tumor necrosis factor-related apoptosis-inducing ligand), overall the data in Table 3 reflect a well-moderated p53 response, causing growth arrest and mitigating ROS damage rather than inducing apoptosis. Analysis of cell cycle control was extended by reference to the work of Mizuno et al.,37 who used both their own and previously published microarray data to place 284 occurrences of 252 signature cell cycle genes into 18 subsets (Fig. 3). We compared all EP-modulated genes with the data of Mizuno et al., and found 97 occurrences of 82 genes in common between the two data sets. The distribution of this overlapping group is strongly biased toward S, G2, and especially M at the expense of G1, which is unaffected by EP (Fig. 3). This result is consistent with CDKN1A/WAF1/Cip 1/p21 inhibition of entry into the S phase; that is, progression through G1 occurs normally with subsequent decreases in transcripts specific for late S, G2, and M. Blockade of the G2/M transition by p53 activation is known38 to involve downregulation of Cdc2, cyclin B1, and topoisomerase 2, which is confirmed in Table 3.
Figure 3.
Signature gene distribution within subsets of the cell cycle: Genes from the data of Mizuno et al.37which are not EP modulated ( : 187 occurrences of 170 genes) and those that are EP modulated (■): 97 occurrences of 82 genes). G1 class genes showed no EP modulation except at the M/G1 or G1/S interfaces.
: 187 occurrences of 170 genes) and those that are EP modulated (■): 97 occurrences of 82 genes). G1 class genes showed no EP modulation except at the M/G1 or G1/S interfaces.
Effect of EP on the NRF2-Mediated Oxidative Stress Response
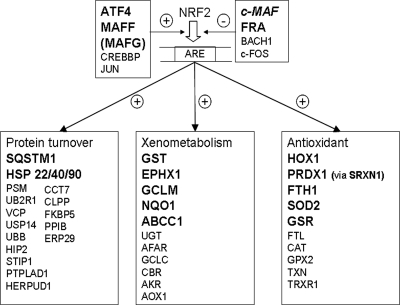
In the nucleus, Nrf2 binds to the antioxidant response element (ARE) and upregulates the expression of three functional clusters of ARE-dependent genes (Fig. 4). Nrf2 is normally retained in the cytoplasm by its interaction with KEAP1, an ROS-sensitive protein. Increased intracellular ROS concentrations modify KEAP1, disrupt the Nrf2-KEAP1 interaction, and permit Nrf2 to enter the nucleus. A recent review39 addresses some possible variations from the canonical Nrf2–KEAP1 interaction. EP modulation of the NRF2 pathway occurred in 36 panels with 27 unique characterized genes (Table 4). Overall, these genes represent only 15% of the 185 members of the pathway. However, the pathway includes 20 cytoplasmic kinase signaling components (not shown in Fig. 4) which can activate NRF2 without increased transcription, and only one of those kinases (MAP3K1) was upregulated. In contrast, of nine entities (Fig. 3) that are direct regulators of NRF2, four (ATF4, FRA1, MAFF, and probably MAFG) were upregulated, whereas c-MAF, which downregulates the NRF2 response40 was downregulated. Not included in Table 4 is a panel for MAFG which had two increases (2.17, 1.99) in myofibroblasts, just missing inclusion at the twofold threshold. MAFF and MAFG are small MAFs that may either activate or repress transcription (for a review, see Ref. 41) but appear to maintain NRF2 activity.42 In the antioxidant stress cluster, peroxiredoxin 1 (PRDX1) was highly expressed but showed no consistent robust response to either EP or TGF-β1. However, sulfiredoxin 1 (SRXN1) showed increases of 4.9- and 3.1-fold in keratocytes, and 3.1-, 4.6- and 3.2-fold increases in myofibroblasts. SRXN1 is induced by activation of NRF243 and is critical to maintaining the antioxidant properties of peroxiredoxin.44 By ensuring optimal peroxiredoxin 1 activity within the cell, increased SRXN1 expression very likely contributes to the function of the NRF2-mediated oxidative stress response, wherein 5 of 10 genes (including SRXN1 with a modulator of PRDX1), or 50%, were upregulated. In the xenometabolism cluster, 5 (33%) of 12 were upregulated. This is an underestimate, since the generic GST (glutathione S-transferase) entry actually contains four modulated genes: GSTA4, GSTM1, GSTO1, and MGST1. Similarly, in the protein turnover (mainly ubiquitination/chaperonin) cluster only two entities (SQSTM1 and heat shock protein-22 -40, and -90) were modulated, but the latter comprised five DNAJ-class genes as well as HSP8 (Table 4).
Figure 4.
EP induced changes in the NRF2 pathway. Nuclear effects of the NRF2 transcription factor, which upregulates ARE-dependent genes in three functional groups. All genes in large, bold letters were upregulated by EP treatment (Table 4), except c-MAP (bold italics), which was downregulated.
Table 4.
EP Modulation of the NRF2-Mediated Oxidative Stress Response
| Panel | Gene Title | Gene Symbol | EP Effect | K, M |
|---|---|---|---|---|
| 202804_at | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | ABCC1 | 2.85 | 1, 2 |
| 200779_at | activating transcription factor 4 (tax-responsive enhancer element B67) | ATF4 | 2.09 | 2, 0 |
| 230893_at | DnaJ homology subfamily A member 5 | DNAJA5 | −2.52 | 2, 2 |
| 223054_at | DnaJ (Hsp40) homolog, subfamily B, member 11 | DNAJB11 | 2.06 | 2, 0 |
| 202500_at | DnaJ (Hsp40) homolog, subfamily B, member 2 | DNAJB2 | 2.64 | 0, 2 |
| 229588_at | DnaJ (Hsp40) homolog, subfamily C, member 10 | DNAJC10 | −2.13 | 1, 2 |
| 213092_x_at | DnaJ (Hsp40) homolog, subfamily C, member 9 | DNAJC9 | −2.47 | 1, 3 |
| 202017_at | Epoxide hydrolase 1, microsomal (xenobiotic) | EPHX1 | 4.98 | 1, 3 |
| 204420_at | FOS-like antigen 1 | FOSL1 | 5.46 | 2, 3 |
| 214211_at | Ferritin, heavy polypeptide 1 | FTH1 | 5.18 | 2, 2 |
| 236140_at | glutamate-cysteine ligase, modifier subunit (also 203925_a) | GCLM | 3.45 | 2, 3 |
| 225609_at | Glutathione reductase (also 205770_at) | GSR | 2.50 | 2, 2 |
| 202967_at | Glutathione S-transferase A4 | GSTA4 | −2.30 | 2, 1 |
| 204550_x_at | Glutathione S-transferase M1 also 215333_x_at | GSTM1 | −2.60 | 2, 0 |
| 201470_at | Glutathione S-transferase omega 1 also 1557915_s_at | GSTO1 | 2.23 | 1, 2 |
| 203665_at | Heme oxygenase (decycling) 1 | HMOX1 | 9.41 | 3, 3 |
| 221667_s_at | Heat shock 22kDa protein 8 | HSPB8 | 2.99 | 3, 0 |
| 209348_s_at | v-Maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) | MAF | −2.73 | 0, 2 |
| 36711_at | v-Maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) 205193_at | MAFF | 4.42 | 2, 2 |
| 225927_at | Mitogen-activated protein kinase kinase kinase 1 | MAP3K1 | 2.89 | 0, 2 |
| 224918_x_at | Microsomal glutathione S-transferase 1 also 1565162_s_at, 231736_x_at | MGST1 | 3.06 | 1, 2 |
| 201467_s_at | NAD(P)H dehydrogenase, quinone 1 | NQO1 | 2.99 | 0, 2 |
| 212647_at | Related RAS viral (r-ras) oncogene homolog | RRAS | −2.48 | 1, 2 |
| 208456_s_at | Related RAS viral (r-ras) oncogene homolog 2 also 212589_at, 212590_at | RRAS2 | 3.00 | 2, 2 |
| 215223_s_at | Superoxide dismutase 2, mitochondrial also 1566342_at, 216841_s_at | SOD2 | 3.12 | 2, 2 |
| 213112_s_at | Sequestosome also 1201471_s_at, 244804_at | SQSTM1 | 6.32 | 2, 3 |
| 201266_at | Thioredoxin reductase 1 | TXNRD1 | 3.16 | 1, 2 |
See Table 3 for an explanation of the EP effect and K, M, values.
Of the 28 genes in Table 4, 21 (75%) were upregulated, confirming the activation of this pathway. Seven genes showed decreased transcription but these most likely represent shifts in emphasis: as noted earlier, MAF was downregulated, whereas MAFF and MAFG were upregulated. The heat shock protein genes DNAJA5, DNAJC10, and DNAJC9 were downregulated but family members DNAJB11 and DNAJB2 were upregulated. Decreased expression of the glutathione-S-transferases GSTM1 and GSTA4 is likely to increase available glutathione for the upregulated GSTO1. Similarly, ras signaling was modulated by decreased expression of the RRAS transcript with a concomitant increase in RRAS2.
Interaction of TGF-β and EP: Modulation of Fibrosis
TGF-β1 modulated 440 genes in all three experiments: of these, 198 (45%) were co-modulated by EP for at least one phenotype in at least two experiments, and 42 (9.5%) were co-modulated by EP for at least one phenotype in all three experiments. For the 1961 genes which responded to TGF-β1 in at least two experiments, equivalent co-modulation values were 755 (39%) and 187 (9.5%) genes, respectively. Table 5 shows 20 genes from the 135-member hepatic fibrosis/hepatic stellate cell activation canonical pathway that were EP-modulated, with suppression of extracellular matrix (ECM) components (COL1A1, COL1A2, COL3A1, and FN1) and of some autocrine signaling contributors (TGF-B2, TGF-B3, and TGF-BR1). Osteoprotegerin (TNFRSF11B) is known to increase in the corneal stroma after epithelial scrape injury and to contribute to monocyte infiltration of the cornea,45 and so its downregulation is a therapeutic advantage. However, other EP-evoked changes would not be advantageous (i.e., upregulation of IL6R and IL8 suggests increased inflammatory activity, whereas increased expression of FAS is proapoptotic46). Elevated levels of TGF-B1 partially repletes the autocrine signaling, and upregulation of LAMA1 suggests increased basement membrane formation. The ambiguous nature of these canonical pathway changes led us to examine the larger group of 42 co-modulated genes referred to earlier. These appear in Table 6, together with genes (bold entries) that showed at least two EP changes in each phenotype (i.e., at least four changes of six). Table 6 is divided into four groups, according to the effects of TGF-β1 and EP. In group 1, six known transcripts (and three additional panels) were consistently decreased by TGF-β and this decrease was inhibited by EP. In this group ALDH3A1 and NR0B1 are of particular interest because they are more highly expressed in keratocytes than in either myofibroblasts or fibroblasts.47 In our hands, ALDH3A1 was the sixth most highly expressed gene in the keratocytes and the only gene expressed at this level that was EP-modulated. ALDH3A1 is a corneal crystallin (structural component) with enzymatic activity that protects against oxidative damage.48 It may also play a role in suppressing keratocyte proliferation.49 The protein encoded by NR0B1 (DAX-1) is a powerful transcriptional repressor,50 which may also contribute to the keratocytes' quiescent state. In group 2, 28 genes were upregulated by TGF-β, an increase attenuated by EP. This group contained a robust subset (COL3A1, CSRP2, LOX, MXRA5, SPARC, ST6GAL2, TNC, and VCAN) that confirms the conclusion drawn from Table 5 that EP modifies the myofibroblast's ability to synthesize ECM. Groups 3 and 4 contain genes that were modulated in the same direction by both EP and TGF-β. In group 3, the transcription factor BNC1 (basonuclin 1) is of interest because it is diagnostically low in keratocytes relative to either myofibroblasts or (not shown) fibroblasts. Although the EP-induced upregulation of NR0B1 may preserve the keratocytes' quiescent phenotype, an EP-induced upregulation of BNC1 apparently acts in the opposite direction. Another gene of interest in group 3 is HBEGF, a candidate for stromal-to-epithelial signaling during wound healing.51,52 EP increased HBEGF expression in keratocytes but markedly inhibited its upregulation by TGF-β1. Our group 4 exemplar is EDNRA. We have reported in another study, without enumeration, that the expression of EDNRA is lower in cultured corneal stromal myofibroblasts than in the equivalent fibroblasts (Table 3 in Ref. 22). The related transcript EDNRB was also downregulated by TGF-β1, with respective responses of 0.14- and 0.20-fold (not shown). The present data yield similar values, with a 1/8.82 = 0.11-fold change for EDNRA from Table 6, and a mean change of 0.21-fold for EDNRB. These transcripts encode, respectively, ETA (endothelin-1 specific) and ETB (endothelin-1 and -3 binding) receptors, which mediate the angiogenic effects of endothelin in the cornea.53 Although both were downregulated by TGF-β1, EDNRA is further downregulated by EP (Table 6), whereas EP upregulated EDNRB in myofibroblasts (Table 5). The physiological significance of these different responses to EP merit further investigation.
Table 5.
EP Modulation of Hepatic Fibrosis/Hepatic Stellate Cell Activation
| Panel | Gene Title | Gene Symbol | EP Effect | K, M |
|---|---|---|---|---|
| 202310_s_at | Collagen, type 1, alpha 1 (also 202311_s_at, 217430_x_at) | COL1A1 | −3.74 | 1, 2 |
| 202404_s_at | Collagen, type I, alpha 2 (also 229218_at) | COL1A2 | −2.85 | 2, 2 |
| 211161_s_at | Collagen, type III, alpha 1 (also 201852_x_at, 215076_s_at) | COL3A1 | −5.62 | 2, 3 |
| 206701_x_at | Endothelin receptor type B (also 204271_s_at, 204273_at) | EDNRB | 4.23 | 1, 2 |
| 215719_x_at | Fas (TNF receptor superfamily, member 6) (also 204780_s_at, 204781_s_at, 216252_x_at) | FAS | 3.65 | 2, 2 |
| 208228_s_at | Fibroblast growth factor receptor 2 (also 203638_s_at, 203639_s_at) | FGFR2 | −3.45 | 2, 2 |
| 222033_s_at | Fms-related tyrosine kinase 1 | FLT1 | −4.91 | 0, 2 |
| 214701_s_at | Fibronectin 1 (also 1558199_at) | FN1 | −4.78 | 0, 2 |
| 209541_at | Insulin-like growth factor 1 (somatomedin C) (also 209540_at) | IGF1 | −11.83 | 1, 2 |
| 210095_s_at | Insulin-like growth factor binding protein 3 (also 212143_s_at) | IGFBP3 | −7.95 | 0, 2 |
| 211959_at | Insulin-like growth factor binding protein 5 (also 211958_at, 1555997_s_at) | IGFBP5 | −3.19 | 2, 3 |
| 205945_at | Interleukin 6 receptor | IL6R | 2.41 | 0, 2 |
| 202859_x_at | Interleukin 8 (also 211506_s_at) | IL8 | 6.07 | 1, 3 |
| 227048_at | Laminin, alpha 1 | LAMA1 | 5.52 | 2, 3 |
| 206584_at | Lymphocyte antigen 96 | LY96 | 3.65 | 0, 2 |
| 203085_s_at | Transforming growth factor, beta 1 | TGF-B1 | 2.22 | 1, 2 |
| 209908_s_at | Transforming growth factor, beta 2 (also 220407_s_at) | TGF-B2 | −3.84 | 0, 2 |
| 209747_at | Transforming growth factor, beta 3 | TGF-B3 | −3.14 | 1, 2 |
| 206943_at | Transforming growth factor, beta receptor 1 (activin A receptor type II-like kinase, 53 kDa) | TGF-BR1 | −2.67 | 0, 2 |
| 204933_s_at | Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | TNFRSF11B | −3.68 | 0, 2 |
The EP effect is the mean of the valid changes shown in the K, M column. For example, COL1A1 shows a mean 3.74-fold decrease for one valid keratocyte (K) change and two valid myofibroblast (M) changes (K, M = 1, 2). Additional information for FGFR2 (bacteria-expressed kinase, keratinocyte growth factor receptor, craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer syndrome, Jackson-Weiss syndrome); FLT1 (vascular endothelial growth factor/vascular permeability factor receptor); and COL3A1 (Ehlers-Danlos syndrome type IV, autosomal dominant). Gene symbols in bold appear in the present TGF-β–modulated set.
Table 6.
Genes Modulated by both EP and TGF-β1
| Panel | Gene Title | Gene Symbol | Normalized Expression Level (Mean) |
|||
|---|---|---|---|---|---|---|
| K | M | K + EP | M + EP | |||
| Group 1: Decreased by TGF-β1, Decrease Partly Inhibited by EP | ||||||
| 205623_at | Aldehyde dehydrogenase 3 family, member A1 | ALDH3A1 | 62.09 | 1.20 | 48.23 | 8.51 |
| 203657_s_at | Cathepsin F | CTSF | 2.89 | 0.46 | 2.01 | 1.22 |
| 209774_x_at | Chemokine (C-X-C motif) ligand 2 | CXCL2 | 2.07 | 0.48 | 4.46 | 3.71 |
| 217966_s_at | Family with sequence similarity 129, member A | FAM129A | 6.05 | 1.32 | 3.06 | 3.53 |
| 207813_s_at | Ferredoxin reductase | FDXR | 0.94 | 0.29 | 1.34 | 1.42 |
| 206645_s_at | Nuclear receptor subfamily 0, group B, member 1 | NR0B1 | 0.51 | 0.07 | 1.34 | 0.69 |
| Group 2: Increased by TGF-β1, Increase Inhibited by EP | ||||||
| 209424_s_at | Alpha-methylacyl-CoA Racemase | AMACR | 0.60 | 2.31 | 0.47 | 0.45 |
| 209425_at | 0.56 | 1.37 | 0.35 | 0.25 | ||
| 209426_s_at | 0.59 | 1.96 | 0.43 | 0.41 | ||
| 205020_s_at | ADP-ribosylation factor-like 4A | ARL4A | 0.32 | 1.79 | 0.61 | 0.58 |
| 201242_s_at | ATPase, Na+/K+ transporting, beta 1 polypeptide | ATP1B1 | 0.32 | 1.91 | 0.44 | 0.71 |
| 236984_at | Chromosome 4 open reading frame 26 | C4orf26 | 0.09 | 1.37 | 0.24 | 0.47 |
| 203967_at | Cell division cycle 6 Homolog (S. cerevisiae) | CDC6 | 0.33 | 1.48 | 0.13 | 0.35 |
| 203968_s_at | 0.32 | 1.32 | 0.22 | 0.42 | ||
| 203440_at | Cadherin 2, type 1, N-cadherin (neuronal) | CDH2 | 0.13 | 1.67 | 0.07 | 0.32 |
| 201852_x_at | Collagen, type III, alpha 1 | COL3A1 | 4.17 | 16.45 | 1.88 | 2.19 |
| 215076_s_at | 9.16 | 27.57 | 3.87 | 4.23 | ||
| 207030_s_at | Cysteine and glycine-rich protein 2 | CSRP2 | 2.48 | 15.23 | 2.93 | 3.86 |
| 208937_s_at | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | ID1 | 0.24 | 2.84 | 0.58 | 0.48 |
| 210095_s_at | Insulin-like growth factor binding protein 3 | IGFBP3 | 0.22 | 5.29 | 0.17 | 1.62 |
| 204679_at | Potassium channel, subfamily K, member 1 | KCNK1 | 0.38 | 2.69 | 0.38 | 0.34 |
| 203276_at | Lamin B1 | LMNB1 | 0.39 | 1.27 | 0.16 | 0.17 |
| 215446_s_at | Lysyl oxidase | LOX | 4.97 | 16.55 | 2.58 | 4.59 |
| 204298_s_at | 3.67 | 11.87 | 1.80 | 3.09 | ||
| 209596_at | Matrix-remodelling associated 5 | MXRA5 | 1.55 | 5.92 | 0.67 | 0.52 |
| 202149_at | Neural precursor cell expressed, developmentally down-regulated 9 | NEDD9 | 0.57 | 2.71 | 0.33 | 0.59 |
| 229461_x_at | Neuronal growth regulator 1 | NEGR1 | 0.20 | 1.11 | 0.09 | 0.18 |
| 211162_x_at | Stearoyl-CoA desaturase (delta-9-desaturase) | SCD | 0.09 | 5.11 | 0.45 | 1.85 |
| 211708_s_at | 0.11 | 6.81 | 0.67 | 2.35 | ||
| 210738_s_at | Solute carrier family 4, sodium bicarbonate cotransporter, member 4 | SLC4A4 | 0.43 | 1.19 | 0.31 | 0.22 |
| 212667_at | Secreted protein, acidic, cysteine-rich (osteonectin) | SPARC | 7.04 | 20.79 | 3.77 | 4.75 |
| 228821_at | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | ST6GAL2 | 0.25 | 0.97 | 0.21 | 0.13 |
| 209277_at | Tissue factor pathway inhibitor 2 | TFP12 | 0.53 | 8.19 | 0.47 | 1.76 |
| 215008_at | Tolloid-like 2 | TLL2 | 0.05 | 0.87 | 0.05 | 0.12 |
| 219410_at | Transmembrane protein 45A | TMEM45A | 6.13 | 19.17 | 4.50 | 4.27 |
| 209753_s_at | Thymopoietin | TMPO | 0.31 | 1.31 | 0.40 | 0.41 |
| 209754_s_at | 0.52 | 1.75 | 0.40 | 0.41 | ||
| 201645_at | Tenascin C (hexabrachion) | TNC | 0.58 | 4.45 | 0.33 | 0.79 |
| 201689_s_at | Tumor protein D52 | TPD52 | 0.13 | 0.62 | 0.13 | 0.17 |
| 211571_s_at | Versican | VCAN | 0.11 | 0.58 | 0.04 | 0.19 |
| 215646_s_at | 0.13 | 0.85 | 0.13 | 0.28 | ||
| 218349_s_at | Zwilch, kinetochore associated, homolog (Drosophila) | ZWILCH | 0.39 | 1.55 | 0.36 | 0.54 |
| Group 3: Increased by Both TGF-β1 and EP with Cumulative Effects or (*) Noncumulative Effects | ||||||
| 205047_s_at | Asparagine synthetase | ASNS | 1.11 | 4.54 | 4.61 | 12.06 |
| 1552487_a_at | Basonuclin 1 | BNC1 | 0.12 | 0.50 | 0.25 | 1.09 |
| 221667_s_at | Heat shock 22kDa protein 8* | HSPB8 | 1.25 | 5.34 | 3.84 | 5.72 |
| 203821_at | Heparin-binding EGF-like growth factor* | HBEGF | 0.52 | 16.32 | 2.11 | 6.03 |
| 38037_at | 0.33 | 8.80 | 0.84 | 3.77 | ||
| 223062_s_at | Phosphoserine aminotransferase 1 | PSAT1 | 1.65 | 8.43 | 7.61 | 18.88 |
| 208456_s_at | Related RAS viral (r-ras) oncogene homolog 2 | RRAS2 | 0.60 | 1.98 | 1.70 | 3.71 |
| 202628_s_at | Serpin peptidase inhibitor, clade E, member 1* | SERPINE1 | 0.63 | 25.86 | 3.14 | 19.72 |
| 222450_at | Transmembrane, prostate, androgen-induced RNA* | TMEPA1 | 0.55 | 9.23 | 2.01 | 6.09 |
| 218368_s_at | Tumor necrosis factor receptor superfamily, member 12A* | TNFRSF12A | 0.96 | 10.86 | 4.50 | 12.71 |
| Group 4: Decreased by Both TGF-β1 and EP with Cumulative Effects | ||||||
| 226665_at | AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) | AHSA2 | 2.73 | 0.96 | 0.69 | 0.39 |
| 239349_at | C1q and tumor necrosis factor related protein 7 | C1QTNF7 | 4.80 | 1.60 | 1.95 | 0.47 |
| 203498_at | Down syndrome critical region gene 1-like 1 | DSCR1L1 | 11.53 | 3.44 | 5.80 | 2.10 |
| 204463_s_at | Endothelin receptor type A | EDNRA | 8.82 | 1.00 | 2.78 | 0.55 |
| 227803_at | Ectonucleotide pyrophosphatase/phosphodiesterase 5 | ENPP5 | 3.44 | 0.43 | 1.60 | 0.43 |
Data are normalized expression levels, not ratios. The mean expression level for all genes across all 12 samples is set to unity: in keratocytes (K), for example ALDH3A1 is expressed at 62.09 times this level. If the K+EP value is in bold, then it changed more than twofold relative to the K value. If the myofibroblast (M)+EP value is in bold, then it changed more than twofold relative to the M value. Gene names in bold indicate two valid changes in each phenotype. Uncharacterized panels not shown are 1555854_at, 238733_at, and 225328_at (group 1); 214078_at and 236277_at, which target the same sequence (group 2); and 1557326_at and 1556185_a_at (group 4).
Focus Genes That Were Modulated by EP in Every Sample, but Were Unaffected by TGF-β1
Eight transcripts were modulated by EP in all six sample pairs (i.e., responded to the lower concentration of EP). This sensitivity led us to examine these genes individually. Six were increased: HMOX1 is a member of the NRF2-mediated oxidative stress response (Table 4), and GDF15 (growth and differentiation factor 15/macrophage inhibitory cytokine-1) is a member of the TGF-β superfamily. GDF15 is p53 inducible,54 antiapoptotic, antiproliferative, and (in the heart) cardioprotective after ischemia/reperfusion.55 Present microarray data showed that cells expressed 9 of the 10 receptors for the TGF-β superfamily, and so an autocrine effect is plausible. C13ORF15 is the response gene to complement 32 (RGC-32, a p53-inducible gene), a substrate and regulator of CDC2, which itself is downregulated (Table 3). In aortic smooth muscle cells, RGC-32 increases p34CDC2 kinase activity and entry into the S-phase,56 whereas overexpression of RGC32 occurs in many tumors,57 suggesting a positive effect on proliferation. However, a more recent report58 shows that in glioma cells, RGC32 is located on centrosomes during mitosis and most likely causes G2/M arrest. It seems that RGC32 can modulate proliferation in either direction, depending on the cell's status. SPON2 is an ECM protein that is expressed in noncancerous but not cancerous lung cells.59 This suggests that elevated SPON2 is associated with decreased proliferation, which would be consistent with the antiproliferative effects of EP. Finally, neuromedin B (NMB) is mitogenic in colon epithelial cells60 but present data show that the NMB receptor is absent from corneal stromal cells, so the autocrine mitogen effect found in other systems61 most likely does not occur here. Interleukin-1 receptor-activated kinases (IRAKs) are key mediators in the signaling pathways of TLRs/IL-1Rs, critical components of the innate immune system (see review in Ref. 62). IRAK1 was increased by EP in all samples and was the only gene in this group to respond to TGF-β1, with valid increases of 1.46-, 2.10-. and 2.92-fold.
The remaining two focus genes were downregulated by EP. The C1QTNF/CTRP (C1q and tumor necrosis factor related protein) family has innate immune functions, and the inhibition of C1QTNF2 expression here is underscored by EP inhibition of C1QTNF7 (Table 6). The Entrez Gene database shows that 332 human genes comprise the TMEM family, of which little is known. One member, TMEM114, has been identified (along with PAX6, PITX2, FOXC1, MAF, SOX2, OTX2, and BMP4) as a gene necessary for orderly development of the human eye; however, there are currently no identified functions of TMEM107.
Summary
Corneal stromal cells are activated after a wound by growth factors and cytokines derived from the corneal epithelium and tear film. TGF-β1–induced activation of keratocytes plays a major role in the formation of scar tissue and tissue contraction. The undesirable phenotypic changes in the activated stromal cells include (1) hyperproliferation, (2) downregulation of the expression of normal stromal proteins that are essential for maintaining corneal transparency, (3) expression of proteins (e.g., tenascin, type III collagen, and fibronectin) that are either not expressed or expressed at low levels in the corneal stroma, and (4) overexpression of normal stromal ECM components including type I collagen. In the present study, EP unequivocally and greatly slowed the proliferation of cultured myofibroblasts, probably by activation of p53 and subsequent blockade of the cell cycle at the transition from the G1 to the S phase. There was also convincing evidence that NRF2-ARE–driven gene activation occurs in both keratocytes and myofibroblasts, with an emphasis on increased protection against ROS and enhanced xenometabolism, rather than mechanisms that affect protein turnover. This evidence is consistent with that in previous reports4–6 that EP treatment increases the resistance of ocular systems to oxidative stress.
EP modified 39% and 45% of keratocyte responses to TGF-β1, without substantially affecting the phenotypic transition from keratocyte to myofibroblast, as determined by microscopy (Fig. 2). In most cases EP counteracted the TGF-β1 effect: the upregulation of some ECM components including tenascin, fibronectin, and type III collagen was attenuated by EP, as was the downregulation of crystallin ALDH3A1. In the intact cornea, these effects would be expected to reduce scarring. However, the expression of some genes is altered in the same direction by TGF-β1 and EP. Notable in this group is keratocan (and possibly lumican), which is required for the regulation of collagen fibril thickness and hydration of corneal stroma. These effects, which include groups 3 and 4 in Table 6, may be countertherapeutic and so require further investigation. However, on balance, the results in the present study suggest that EP has encouraging therapeutic potential that warrants investigation in the intact, wounded cornea. In rats, instillation of 5% EP (50 g/L or 430 mM) is well tolerated over a 40-day experimental period, and penetration of EP through the cornea is rapid, with pyruvate concentration in the aqueous humor reaching 7 mM after 15 minutes.6 Therefore, in the rat model, EP instillation achieves corneal stromal concentrations that are comparable to those reached in the present study.
Footnotes
Supported by National Institutes of Health Grants EY03263 (NS) and EY09098 (core grant); Research to Prevent Blindness, New York, NY; and The Eye and Ear Foundation, Pittsburgh, PA.
Disclosure: S.A.K. Harvey, None; E. Guerriero, None; N. Charukamnoetkanok, None; J. Piluek, None; J.S. Schuman, None; N. SundarRaj, None
References
- 1.Brand K. Aerobic glycolysis by proliferating cells: protection against oxidative stress at the expense of energy yield. J Bioenerg Biomembr 1997;29:355–364 [DOI] [PubMed] [Google Scholar]
- 2.O'Donnell-Tormey J, Nathan CF, Lanks K, DeBoer CJ, de la Harpe J. Secretion of pyruvate: an antioxidant defense of mammalian cells. J Exp Med 1987;165:500–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink MP. Ethyl pyruvate. Curr Opin Anaesthesiol 2008;21:160–167 [DOI] [PubMed] [Google Scholar]
- 4.Varma SD, Devamanoharan PS, Ali AH. Prevention of intracellular oxidative stress to lens by pyruvate and its ester. Free Radic Res 1998;28:131–135 [DOI] [PubMed] [Google Scholar]
- 5.Varma SD, Hegde KR, Kovtun S. Oxidative damage to lens in culture: reversibility by pyruvate and ethyl pyruvate. Ophthalmologica 2006;220:52–57 [DOI] [PubMed] [Google Scholar]
- 6.Devamanoharan PS, Henein M, Ali AH, Varma SD. Attenuation of sugar cataract by ethyl pyruvate. Mol Cell Biochem 1999;200:103–109 [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Englert JA, Yang R, Delude RL, Fink MP. Ethyl pyruvate inhibits nuclear factor-kappaB-dependent signaling by directly targeting p65. J Pharmacol Exp Ther 2005;312:1097–1105 [DOI] [PubMed] [Google Scholar]
- 8.Pasquale LR, Dorman-Pease ME, Lutty GA, Quigley HA, Jampel HD. Immunolocalization of TGF-beta 1, TGF-beta 2, and TGF-beta 3 in the anterior segment of the human eye. Invest Ophthalmol Vis Sci 1993;34:23–30 [PubMed] [Google Scholar]
- 9.Matsuda A, Tagawa Y, Matsuda H. TGF-beta2, tenascin, and integrin beta1 expression in superior limbic keratoconjunctivitis. Jpn J Ophthalmol 1999;43:251–256 [DOI] [PubMed] [Google Scholar]
- 10.Thom SB, Myers JS, Rapuano CJ, Eagle RC, Jr, Siepser SB, Gomes JA. Effect of topical anti-transforming growth factor-beta on corneal stromal haze after photorefractive keratectomy in rabbits. J Cataract Refract Surg 1997;23:1324–1330 [DOI] [PubMed] [Google Scholar]
- 11.Mead AL, Wong TT, Cordeiro MF, Anderson IK, Khaw PT. Evaluation of anti-TGF-beta2 antibody as a new postoperative anti-scarring agent in glaucoma surgery. Invest Ophthalmol Vis Sci 2003;44:3394–3401 [DOI] [PubMed] [Google Scholar]
- 12.Mita T, Yamashita H, Kaji Y, et al. Effects of transforming growth factor beta on corneal epithelial and stromal cell function in a rat wound healing model after excimer laser keratectomy. Graefes Arch Clin Exp Ophthalmol 1998;236:834–843 [DOI] [PubMed] [Google Scholar]
- 13.Wilson SE, Liu JJ, Mohan RR. Stromal-epithelial interactions in the cornea. Prog Retin Eye Res 1999;18:293–309 [DOI] [PubMed] [Google Scholar]
- 14.Wilson SE, Lloyd SA, He YG. EGF, basic FGF, and TGF beta-1 messenger RNA production in rabbit corneal epithelial cells. Invest Ophthalmol Vis Sci 1992;33:1987–1995 [PubMed] [Google Scholar]
- 15.Maltseva O, Folger P, Zekaria D, Petridou S, Masur SK. Fibroblast growth factor reversal of the corneal myofibroblast phenotype. Invest Ophthalmol Vis Sci 2001;42:2490–2495 [PubMed] [Google Scholar]
- 16.Li DQ, Lee SB, Tseng SC. Differential expression and regulation of TGF-beta1, TGF-beta2, TGF-beta3, TGF-betaRI, TGF-betaRII and TGF-betaRIII in cultured human corneal, limbal, and conjunctival fibroblasts. Curr Eye Res 1999;19:154–161 [DOI] [PubMed] [Google Scholar]
- 17.Song QH, Klepeis VE, Nugent MA, Trinkaus-Randall V. TGF-beta1 regulates TGF-beta1 and FGF-2 mRNA expression during fibroblast wound healing. Mol Pathol 2002;55:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petridou S, Maltseva O, Spanakis S, Masur SK. TGF-beta receptor expression and smad2 localization are cell density dependent in fibroblasts. Invest Ophthalmol Vis Sci 2000;41:89–95 [PubMed] [Google Scholar]
- 19.Petridou S, Masur SK. Immunodetection of connexins and cadherins in corneal fibroblasts and myofibroblasts. Invest Ophthalmol Vis Sci 1996;37:1740–1748 [PubMed] [Google Scholar]
- 20.Guerriero E, Chen J, Sado Y, et al. Loss of alpha3(IV) collagen expression associated with corneal keratocyte activation. Invest Ophthalmol Vis Sci 2007;48:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson SC, SundarRaj N. Regulation of a Rho-associated kinase expression during the corneal epithelial cell cycle. Invest Ophthalmol Vis Sci 2001;42:933–940 [PubMed] [Google Scholar]
- 22.Harvey SA, Anderson SC, SundarRaj N. Downstream effects of ROCK signaling in cultured human corneal stromal cells: microarray analysis of gene expression. Invest Ophthalmol Vis Sci 2004;45:2168–2176 [DOI] [PubMed] [Google Scholar]
- 23.Funderburgh JL, Funderburgh ML, Mann MM, Conrad GW. Physical and biological properties of keratan sulphate proteoglycan. Biochem Soc Trans 1991;19:871–876 [DOI] [PubMed] [Google Scholar]
- 24.Beales MP, Funderburgh JL, Jester JV, Hassell JR. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: maintenance of the keratocyte phenotype in culture. Invest Ophthalmol Vis Sci 1999;40:1658–1663 [PubMed] [Google Scholar]
- 25.Funderburgh JL, Funderburgh ML, Mann MM, Corpuz L, Roth MR. Proteoglycan expression during transforming growth factor beta-induced keratocyte-myofibroblast transdifferentiation. J Biol Chem 2001;276:44173–44178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson EC, Wang IJ, Liu CY, Brannan P, Kao CW, Kao WW. Altered KSPG expression by keratocytes following corneal injury. Mol Vis 2003;9:615–623 [PubMed] [Google Scholar]
- 27.Chen J, Guerriero E, Sado Y, SundarRaj N. Rho-mediated regulation of TGF-beta1- and FGF-2-induced activation of corneal stromal keratocytes. Invest Ophthalmol Vis Sci 2009;50:3662–3670 [DOI] [PubMed] [Google Scholar]
- 28.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene 2005;24:2899–2908 [DOI] [PubMed] [Google Scholar]
- 29.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993;75:817–825 [DOI] [PubMed] [Google Scholar]
- 30.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993;75:805–816 [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Sun Z, Wang XJ, et al. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell 2009;34:663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith ML, Seo YR. p53 regulation of DNA excision repair pathways. Mutagenesis 2002;17:149–156 [DOI] [PubMed] [Google Scholar]
- 33.Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet 2003;34:440–445 [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Xue L, Yen Y. Redox property of ribonucleotide reductase small subunit M2 and p53R2. Methods Mol Biol 2008;477:195–206 [DOI] [PubMed] [Google Scholar]
- 35.Cano CE, Gommeaux J, Pietri S, et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res 2009;69:219–226 [DOI] [PubMed] [Google Scholar]
- 36.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006;126:107–120 [DOI] [PubMed] [Google Scholar]
- 37.Mizuno H, Nakanishi Y, Ishii N, Sarai A, Kitada K. A signature-based method for indexing cell cycle phase distribution from microarray profiles. BMC Genomics 2009;10:137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene 2001;20:1803–1815 [DOI] [PubMed] [Google Scholar]
- 39.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 2009;284:13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhakshinamoorthy S, Jaiswal AK. c-Maf negatively regulates ARE-mediated detoxifying enzyme genes expression and anti-oxidant induction. Oncogene 2002;21:5301–5312 [DOI] [PubMed] [Google Scholar]
- 41.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol 2008;376:913–925 [DOI] [PubMed] [Google Scholar]
- 42.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol 2005;25:8044–8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells 2009;27:279–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl 2007;S3–S8 [DOI] [PubMed] [Google Scholar]
- 45.Wilson SE, Mohan RR, Netto M, et al. RANK, RANKL, OPG, and M-CSF expression in stromal cells during corneal wound healing. Invest Ophthalmol Vis Sci 2004;45:2201–2211 [DOI] [PubMed] [Google Scholar]
- 46.Wilson SE, Li Q, Weng J, et al. The Fas-Fas ligand system and other modulators of apoptosis in the cornea. Invest Ophthalmol Vis Sci 1996;37:1582–1592 [PubMed] [Google Scholar]
- 47.Pei Y, Reins RY, McDermott AM. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp Eye Res 2006;83:1063–1073 [DOI] [PubMed] [Google Scholar]
- 48.Estey T, Cantore M, Weston PA, Carpenter JF, Petrash JM, Vasiliou V. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem 2007;282:4382–4392 [DOI] [PubMed] [Google Scholar]
- 49.Stagos D, Chen Y, Cantore M, Jester JV, Vasiliou V. Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res Bull 2010;15;81:211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niakan KK, McCabe ER. DAX1 origin, function, and novel role. Mol Genet Metab 2005;86:70–83 [DOI] [PubMed] [Google Scholar]
- 51.Zieske JD, Takahashi H, Hutcheon AE, Dalbone AC. Activation of epidermal growth factor receptor during corneal epithelial migration. Invest Ophthalmol Vis Sci 2000;41:1346–1355 [PubMed] [Google Scholar]
- 52.Block ER, Matela AR, SundarRaj N, Iszkula ER, Klarlund JK. Wounding induces motility in sheets of corneal epithelial cells through loss of spatial constraints: role of heparin-binding epidermal growth factor-like growth factor signaling. J Biol Chem 2004;279:24307–24312 [DOI] [PubMed] [Google Scholar]
- 53.Bek EL, McMillen MA. Endothelins are angiogenic. J Cardiovasc Pharmacol 2000;36:S135–S139 [DOI] [PubMed] [Google Scholar]
- 54.Osada M, Park HL, Park MJ, et al. A p53-type response element in the GDF15 promoter confers high specificity for p53 activation. Biochem Biophys Res Commun 2007;354:913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kempf T, Eden M, Strelau J, et al. The transforming growth factor-beta superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ Res 2006;98:351–360 [DOI] [PubMed] [Google Scholar]
- 56.Badea T, Niculescu F, Soane L, et al. RGC-32 increases p34CDC2 kinase activity and entry of aortic smooth muscle cells into S-phase. J Biol Chem 2002;277:502–508 [DOI] [PubMed] [Google Scholar]
- 57.Fosbrink M, Cudrici C, Niculescu F, et al. Overexpression of RGC-32 in colon cancer and other tumors. Exp Mol Pathol 2005;78:116–122 [DOI] [PubMed] [Google Scholar]
- 58.Saigusa K, Imoto I, Tanikawa C, et al. RGC32, a novel p53-inducible gene, is located on centrosomes during mitosis and results in G2/M arrest. Oncogene 2007;26:1110–1121 [DOI] [PubMed] [Google Scholar]
- 59.Manda R, Kohno T, Matsuno Y, Takenoshita S, Kuwano H, Yokota J. Identification of genes (SPON2 and C20orf2) differentially expressed between cancerous and noncancerous lung cells by mRNA differential display. Genomics 1999;61:5–14 [DOI] [PubMed] [Google Scholar]
- 60.Matusiak D, Glover S, Nathaniel R, Matkowskyj K, Yang J, Benya RV. Neuromedin B and its receptor are mitogens in both normal and malignant epithelial cells lining the colon. Am J Physiol Gastrointest Liver Physiol 2005;288:G718–G728 [DOI] [PubMed] [Google Scholar]
- 61.Siegfried JM, Krishnamachary N, Gaither Davis A, Gubish C, Hunt JD, Shriver SP. Evidence for autocrine actions of neuromedin B and gastrin-releasing peptide in non-small cell lung cancer. Pulm Pharmacol Ther 1999;12:291–302 [DOI] [PubMed] [Google Scholar]
- 62.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell Signal 2008;20:269–276 [DOI] [PubMed] [Google Scholar]