This study reports alterations in the expression of microRNAs during replicative senescence in human trabecular meshwork cells. The results suggest that the upregulation of the anti-inflammatory miR-146a inhibits the excessive production of inflammatory mediators in senescent cells and limits their deleterious effects on the surrounding tissues.
Abstract
Purpose.
To investigate the alterations in microRNA (miRNA) expression during replicative senescence (RS) in human trabecular meshwork (HTM) cells.
Methods.
Two HTM cell lines were serially passaged until they reached RS. Changes in expression of 30 miRNAs were assessed by real-time quantitative (q)-PCR. The effects of miR-146a on gene expression were analyzed with gene arrays and the results confirmed by real-time q-PCR. Protein levels of IRAK1 and PAI-1 were analyzed by Western blot and those of IL6 and IL8 by ELISA. Senescence-associated markers were monitored by flow cytometry and cell proliferation by BrdU incorporation.
Results.
RS of HTM cells was associated with significant changes in expression of 18 miRNAs, including the upregulation of miR-146a. miR-146a downregulated multiple genes associated with inflammation, including IRAK1, IL6, IL8, and PAI-1, inhibited senescence-associated β-galactosidase (SA-β-gal) activity and production of intracellular reactive species (iROS), and increased cell proliferation. Overexpression of either IRAK1 or PAI-1 inhibited the effects of miR-146a on cell proliferation and iROS production in senescent cells.
Conclusions.
RS in HTM cells was associated with changes in miRNA expression that could influence the senescent phenotype. Upregulation of the anti-inflammatory miR-146a may serve to restrain excessive production of inflammatory mediators in senescent cells and limit their deleterious effects on the surrounding tissue. Among the different proteins repressed by miR-146a, the inhibition of PAI-1 may act to minimize the effects of senescence on the generation of iROS and growth arrest and prevent alterations of the extracellular proteolytic activity of the TM.
The trabecular meshwork (TM) from glaucoma donors is characterized by chronic activation of a stress response that leads to increased production of inflammatory markers.1–3 Chronic activation of a similar inflammatory response has been found during aging and certain age-related conditions in other tissues.4–12 One of the factors proposed to contribute to such a response is the increased presence of senescent cells.
Senescent cells have been shown to accumulate with age and in certain pathologic conditions in several tissues and organs,5,9,13 including the TM in glaucoma.14 The senescent response is associated with a series of phenotypic changes that have been proposed to disrupt the tissue microenvironment and contribute to pathologic alterations associated with aging.15,16 An important alteration observed in senescent cells that may contribute to tissue malfunction is the presence of a characteristic senescence-associated secretory phenotype (SASP).17–19 Such a secretory phenotype involves an increase in the release of inflammatory mediators and growth factors that can affect the function of adjacent cells and lead to a chronic activation of a stress response18 similar to that observed in the TM of glaucoma donors.3
The regulatory mechanisms that mediate the phenotypic changes in senescent cells and, in particular, those involved in the chronic activation of inflammatory mediators have not been completely elucidated.
MicroRNAs (miRNAs) are important regulators of gene expression and have been implicated in a variety of cellular functions, including differentiation, apoptosis, and cancer progression.20–22 miRNAs are transcribed as primary transcripts or pri-miRNAs that are converted in the nucleus into 70-nucleotide, stem-loop structures known as pre-miRNAs. These pre-miRNAs are then processed in the cytoplasm to mature miRNAs, 21 to 23 nucleotides in length, by the endonuclease Dicer, which also initiates the formation of the RNA-induced silencing complex (RISC).23 After integration into the active RISC, miRNAs bind to target sites in the 3′ untranslated region (UTR) of the specific mRNA transcripts and inhibit translation or induce mRNA degradation by argonaute proteins, the catalytically active members of the RISC.24 There is some experimental evidence suggesting that miRNAs play a role in cellular senescence. Several miRNAs such as miR-34 and -20a have been shown to induce senescent growth arrest.25,26 Ablation of Dicer in mouse embryonic fibroblasts also induces senescence by upregulating p53.27 Furthermore, we have recently shown that stress-induced premature senescence (SIPS) is associated with significant alteration in expression of several miRNAs in both human fibroblasts and TM cells.28 It has been hypothesized that miRNAs play both positive and negative roles in regulating the senescent response.29–31 Specifically, it has recently been reported that the anti-inflammatory miR-146 is upregulated in senescent fibroblasts in response to increased levels of inflammatory cytokines induced by the senescent response, generating a negative feedback loop that restrains excessive production of inflammatory mediators in senescent cells and limits the deleterious effects of the SASP on the surrounding tissues.31 However, there is still little information about the role that miRNAs play in modulating the senescent response.
We investigated changes in expression of miRNAs during the process of replicative senescence (RS) in human TM (HTM) cells. Since one of the miRNAs significantly upregulated during this process was miR-146a, which has been implicated in the modulation of the SASP in fibroblasts,31 we further investigated the effects of this miRNA on gene expression in HTM cells and the mechanisms by which miR-146a may contribute to the senescent response in the TM.
Methods
Cell Culture of Primary HTM Cells
Donor human eyes or cornea rings were obtained from the New York Eye Bank within 7 days after death, according to the tenets of the Declaration of Helsinki. HTM from a single individual was dissected from surrounding tissue and digested in 10 mg collagenase/20 mg bovine serum albumin (BSA)/5 mL phosphate-buffered saline (PBS). The cells were seeded on collagen I-coated 3-cm Petri dishes and maintained at 37°C in a humidified atmosphere of 5% CO2 in TM culture medium containing 20% fetal bovine serum (FBS). The TM culture medium was low-glucose Dulbecco's modified Eagle's medium (DMEM) with l-glutamine and 110 mg/L sodium pyruvate, supplemented with 100 μM nonessential amino acids, 100 U/mL penicillin, and 100 μg/mL streptomycin sulfate. All reagents were obtained from Invitrogen Corp. (Carlsbad, CA). The HTM636 and HTM1073 cell lines were generated from 22- and 26-year-old donors. These cells lines were used for experiments involving comparisons between late-passage (p15 or p11) and early-passage (p4–6) cells. The HTM682, HTM113, and HTM714 cell lines were generated from 47-, 49- and 42-year-old donors and were used at p4 to p7, to study the effects of miR-146a on gene and protein expression. None of the donors had a history of POAG or ocular diseases.
miRNA and Plasmid Transfection
Transfection of miRNAs or plasmids was performed with a nonviral transfection system (Nucleofector System; Amaxa Inc. Gaithersburg, MD), according to the manufacturer's instructions. miR-146a mimic (146aM) or negative miRNA control mimic (ConM, 120 pmol per 5 × 105 cells; Dharmacon, Inc. Chicago, IL), or PAI-1 plasmid (pPAI-1) (pCMV-SPORT6; Open Biosystems, Huntsville, AL), recombinant IRAK1 plasmid (pIRAK1; pENTR221, OHS4559-99869058; Open Biosystems), or psiCHECK2 vector (2 μg per 5 × 105 cells, pCon; Promega Corp., Madison, WI) were transfected into HTM cells (program T23; Amaxa). For experiments involving cotransfection of expression plasmids and miRNA mimics, 2 μg of plasmid (pPAI-1, pIRAK1, or pCon) and 120 pmol of miRNA mimic (either 146aM or ConM) were cotransfected into 5 × 105 HTM cells by using the same program. The culture medium was replaced with fresh DMEM 24 hours after transfection, and cell culture supernatant or cells were collected 72 hours after transfection.
PCR Quantification of Changes in Expression of miRNAs
Small RNA was isolated (mirVana miRNA Isolation Kit; Applied Biosystems, Inc. [ABI], Austin, TX), according to the manufacturer's instructions. cDNAs from 20 ng miRNAs were transcribed (TaqMan microRNA reverse transcription Kit; ABI) and amplified with specific primers (ABI). RNU6B (ABI) was used as the control for normalization.
Gene Microarray and Data Analysis
HTM cell cultures (HTM1073, p6) were transfected with miR-146a or control mimic for 3 days, total RNA was isolated and hybridized to a gene microarray (Human Genome U133 2.0 Array; Affymetrix, Santa Clara, CA) at the Duke University Microarray facility (Durham, NC). This array includes the Human Genome U133 Set plus 6500 additional genes for analysis of more than 47,000 transcripts. Raw data were normalized and analyzed (GeneSpring 10; Silicon Genetics, Wilmington, DE). The genes were filtered by intensity compared with the control channel. P ≤ 0.05, by paired student t-test, was considered significant. The list of genes that were significantly downregulated was compared to those on three databases that predict targets for miRNAs: Microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/; European Molecular Biology Laboratory, Heidelberg, Germany) TargetScan (http://www.targetscan.org/; Whitehead Institute for Biomedical Research, Cambridge, MA), and PicTar-Vert (http://pictar.mdc-berlin.de/; Max Delbruck Centrum, Berlin, Germany, and The Center for Comparative Functional Genomics, New York University, NY). To analyze the biological significance and regulatory pathways involved in the changes observed in the arrays, we performed network analysis of genes differentially expressed more than twofold (P ≤ 0.05; Metacore pathway analysis; GeneGo, St. Joseph, MI).
Quantitative PCR Analysis of Gene Expression
Total RNA was isolated (RNeasy kit; Qiagen Inc., Valencia, CA). RNA yields were measured with fluorescent dye (RiboGreen; Molecular Probes, Eugene, OR). First-strand cDNA was synthesized from total RNA (1 μg) by reverse transcription with oligodT and reverse transcriptase (Superscript II; Invitrogen Corp.). Quantitative (q)-PCR was performed in a 20-μL mixture that contained 1 μL of the cDNA preparation and 1× iQ SYBR Green mastermix (Supermix; Bio-Rad, Hercules, CA), with the following PCR parameters: 95°C for 5 minutes followed by 50 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 15 seconds. Each quantification was conducted in triplicate, and the experiments were performed in triplicate with three individual cell lines. The fluorescence threshold value (Ct) was calculated with the system software (iCycle; Bio-Rad). The absence of nonspecific products was confirmed by both the analysis of the melting curves to exclude primer-dimer and by electrophoresis in 3% agarose gels (Super AcrylAgarose; DNA Technologies, Inc., Gaithersburg, MD), to verify the correct product size. β-Actin was used as an internal standard of mRNA expression, to normalize the individual gene expression levels. The specific primer pairs used to amplify the genes are shown in Table 1.
Table 1.
Primer Pairs Used to Quantify Gene Expressions in HTM Cells
| Gene Name | Accession No.* | Primer Pairs |
|---|---|---|
| IRAK1 | NM_001025242 | For: 5′-ATT TAT GCT TGG GAG GTC GAG GCT |
| Rev: 5′-TCG CTT CTT GCT AGG ACT GAA CCA | ||
| IL11 | NM_000641 | For: 5′-AGA TAT CCT GAC ATT GGC CAG GCA |
| Rev: 5′-TTG GAC TTC AGT GAT CCA CTC GCT | ||
| PPP2R1B | NM_002716 | For: 5′-CTC GCA CAT CTG CAT GTG GTT TGT |
| Rev: 5′-TGT CAT CTG AGC ACA AGG AAC GGA | ||
| GALNT10 | NM_017540 | For: 5′-AGA GCC ACG AAT CTG CCT TTG AGA |
| Rev: 5′-TTC CTC GCC AGC TAC ACA TTC AGT | ||
| IL8 | NM_000584 | For: 5′-AGA AAC CAC CGG AAG GAA CCA TCT |
| Rev: 5′-CAC CTT CAC ACA GAG CTG CAG AAA | ||
| SLC10A3 | NM_019848 | For: 5′-TTC TGC ATC AAG GTC TCA CCT GCT |
| Rev: 5′-TCC ACT TTG CAC CCA AAC GAA CAC | ||
| HAS1 | NM_001523 | For: 5′-TTG AGG CCT GGT ACA ACC AGA AGT |
| Rev: 5′-ACC TGG AGG TGT ACT TGG TAG CAT | ||
| SERPINE1 | NM_000602 | For: 5′-AAT GTG TCA TTT CCG GCT GCT GTG |
| Rev: 5′-ACA TCC ATC TTT GTG CCC TAC CCT | ||
| CXCL3 | NM_002090 | For: 5′-ACC GAA GTC ATA GCC ACA CTC AAG |
| Rev: 5′-ACT TCT CTC CTG TCA GTT GGT GCT | ||
| CXCL6 | NM_002993 | For: 5′-AGA GCT GCG TTG CAC TTG TTT ACG |
| Rev: 5′-AAC TTG CTT CCC GTT CTT CAG GGA | ||
| CCL2 | NM_002982 | For: 5′-TCG CTC AGC CAG ATG CAA TCA ATG |
| Rev: 5′-TGG AAT CCT GAA CCC ACT TCT GCT | ||
| CCL20 | NM_004591 | For: 5′-AGT TTG CTC CTG GCT GCT TTG ATG |
| Rev: 5′-CTG CCG TGT GAA GCC CAC AAT AAA | ||
| PTGS1 | NM_080591 | For: 5′-GCA CCA ACC TCA TGT TTG CCT TCT |
| Rev: 5′-TGT CTC CAT AAA TGT GGC CGA GGT | ||
| IL6 | NM_000600 | For: 5′-AAA TTC GGT ACA TCC TCG ACG GCA |
| Rev: 5′-AGT GCC TCT TTG CTG CTT TCA CAC | ||
| β-Actin | NM_001101.3 | For: 5′-CCT CGC CTT TGC CGA TCC G |
| Rev: 5′-GCC GGA GCC GTT GTC GAC G |
GenBank, National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov/Genbank.
Assay of Intracellular Reactive Oxygen Species
The production of intracellular (i)ROS was measured by 2′-7′-dichlorofluorescein H oxidation.32 Briefly, 10 mM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; Invitrogen Corp.) was dissolved in methanol and diluted 500-fold in HBSS to give a 20-μM concentration of H2DCFDA. Cells grown in 12-well plates were exposed to H2DCFDA and collected after 15 minutes. The cells were then pelleted, washed once with PBS, and resuspended in 200 μL of PBS for flow cytometry (FACScalibur; BD Biosciences, San Jose, CA) with 488-nm excitation and emission filters appropriate for Alexa Fluor 488 dye.
Measurement of Mitochondrial Membrane Potential
Mitochondrial membrane potential (Δψm) was monitored in cells loaded with JC-1 dye (Invitrogen Corp.) as described previously, with minor modifications.1 Briefly, cells in 12-well plates were trypsinized and pelleted by centrifuge. The cells were then washed with PBS once, loaded with JC-1 in 1 mL of PBS to a final concentration of 2 μM, and incubated at 37°C, 5% CO2 for 15 minutes. The cells were pelleted and washed again with PBS, then resuspended in 200 μL of PBS. The cells were analyzed by flow cytometry with 488-nm excitation with emission filters appropriate for Alexa Fluor 488 dye and R-phycoerythrin.
Measurement of Endogenous Autofluorescence and SA-β-gal
Endogenous autofluorescence was quantified by flow cytometry. The fluorescence emitted by 10,000 cells in the FL-2 channel (563–607 nm) was recorded and analyzed (CELLQuest software; BD Biosciences). Activity of SA-β-gal was also measured by flow cytometry with the fluorogenic substrate 5-dodecanoylaminofluorescein di-β-D-galactopyranoside (C12FDG; Invitrogen Corp.) as previously described.33 Briefly, the cells were incubated with 300 μM chloroquine for 1 hour at 37°C to modulate the intracellular pH. The cells were then trypsinized and washed once with PBS, loaded with 2 mM C12FDG solution, and incubated 1 minute at 37°C. The cells were then diluted 10× in cold PBS, incubated on ice for 30 minutes, and analyzed by flow cytometer, using the FL-1 channel (Alexa Fluor 488 nm). The average number of cells analyzed for each experiment was 10,000.
Determination of Cell Proliferation
Cell growth was quantified with a BrdU cell proliferation assay (Calbiochem, San Diego, CA) according to the manufacturer's instructions. Briefly, 100 μL of cells (nontransfected p4 and p15, or transfected with miR-146a mimic [146aM] or control mimic [ConM]) at concentration of 4 × 104 cells/mL were seeded into a 96-well culture dish and incubated for 24 (p4 and p15) or 48 (146aM and ConM) hours. Culture medium was then replaced with 100 μL fresh DMEM containing 10% FBS and BrdU 1:10,000 dilution. After overnight incubation, the cells were fixed, and BrdU incorporation was measured by using anti-BrdU antibody and reconstituted peroxidase goat anti-mouse IgG HRP conjugate. The color was then developed by adding substrate solution to each well. After 15 minutes of incubation in the dark at room temperature (RT), blocking solution (Stop; Cell Signaling, Danvers, MA) was added to each well, and absorbance was measured with a spectrophotometric plate reader at dual wavelengths of 450 to 540 nm.
Protein Extraction and Immunoblot
Cells were washed twice in cold PBS. Total protein was extracted with RIPA buffer (150 mM NaCl, 10 mM Tris [pH 7.2], 0.1% SDS, 1.0% Triton X-100, 5 mM EDTA [pH 8.0]) containing 1× protease inhibitor cocktail (Roche, Inc., Indianapolis, IN). Protein concentration was determined with a protein assay (Micro BCA Protein Assay Kit; Pierce, Rockford, IL). Total protein extracts (40 μg) were separated by 8% SDS-PAGE and transferred to PVDF membrane (Bio-Rad). The membranes were blocked with 5% nonfat dry milk and incubated overnight with the primary antibodies anti-IRAK1 (Cell Signaling, Inc.) or anti-PAI-1/serpine-1 (Santa Cruz Biotechnology, Santa Cruz, CA). Then they were incubated with a secondary antibody conjugated to HRP for 1 hour at RT. Immunoreactive proteins were visualized by chemiluminescence (ECL Plus; GE Healthcare, Pittsburgh, PA). For detection of endogenous control, the membrane was stripped with stripping buffer (25 mM glycine ([pH 3.0] plus 1% sodium dodecyl sulfate [SDS]) and then incubated with anti-β-tubulin (SC-9935; Santa Cruz Biotechnology).
Quantification of IL6 and IL8
HTM cells were transfected with miR-146aM or ConM. Three days after transfection, cell culture supernatant was collected, and 25 μL culture medium was used to quantify protein levels of IL6 and IL8 (Human TH1/Th2 plex FlowCytomix kit; Render MedSystems, Inc., Burlingame, CA) according to the manufacturer's instructions. Briefly, 25 μL of samples or standards were mixed with 25 μL of bead mixture and 50 μL of biotin-conjugate mixture. After 2 hours' incubation at RT, the tubes were centrifuged at 200g for 5 minutes. The pellet was washed twice with assay buffer and then 50 μL of streptavidin-PE was loaded to each tube. After a 1-hour incubation, the pellet was spun down and washed twice with assay buffer. The samples were then analyzed on a flow cytometer in the FL3 channel.
Statistical Analysis
The data are presented as the mean ± SD. Statistical significance between groups was assessed by the Mann–Whitney U Test. P < 0.05 was considered statistically significant.
Results
Changes in miRNAs in Replicative Senescent HTM cells
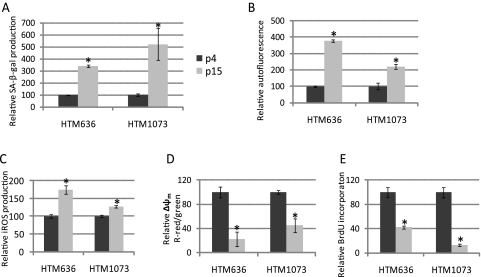
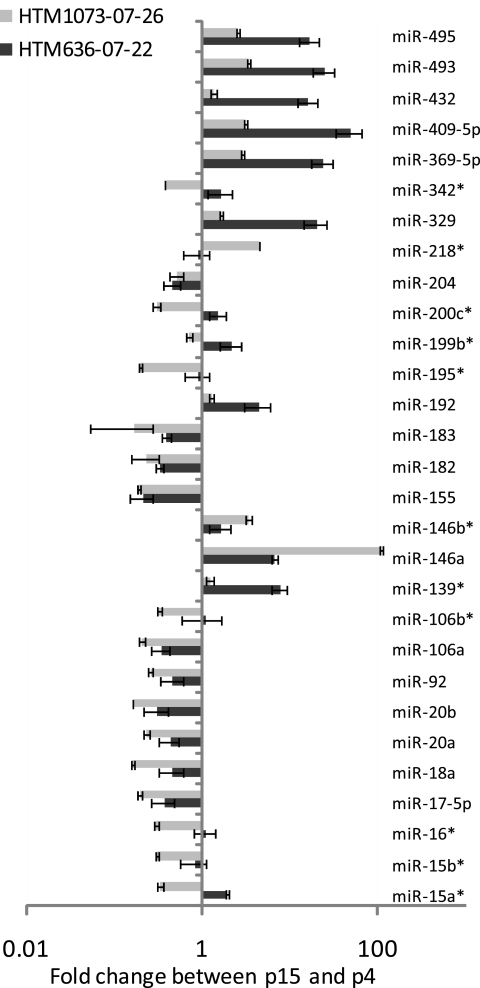
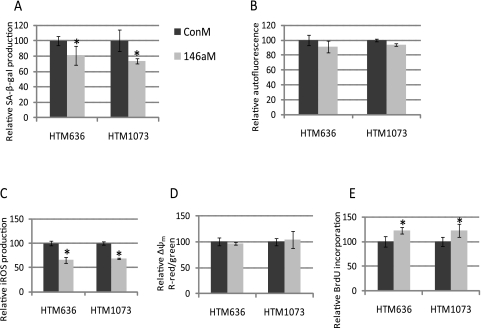
Primary cultures of two HTM (636-07-22 and 1073-07-26) cell lines were split from one to two cell culture plates when the cells reached 95% confluence. Both cell lines showed a significant increase in expression of cellular senescence markers34–36 at p15 (Fig. 1). miRNA expression was then analyzed by real-time q-PCR (TaqMan; ABI) at p15 versus the same HTM cell line at p4. Eighteen miRNAs (has-miR-17–5p, -18a, -20a, -20b, -92, -106a, -155, -146a, -182, -183, -192, -204, -329, -369–5p, -409–5p, -432, -493, and -495) showed significant up- or downregulation in both senescent HTM cell lines. MiR-146a showed particularly high levels of upregulation (111-fold) in the HTM1073 cell line and lower levels (6.8-fold) in the HTM636 cell line (Fig. 2).
Figure 1.
Evaluation of cellular senescence markers in two replicative senescent HTM cell lines. Two HTM (636-07-22 and 1073-07-26) cell lines showed significant expression of senescence markers at p15. Induction of SA-β-gal (A), autofluorescence (B), iROS (C), and Δψm (D) in p15 versus p4 cells was quantified by flow cytometry. The cell proliferation rate (E) was quantified by BrdU incorporation. Data represent the mean percentage change ± SD, n = 3. *P < 0.05, compared with p4 cultures; Mann–Whitney U test.
Figure 2.
Changes in miRNA expression in replicative senescent HTM cells. Small RNAs (20 ng) isolated from p4 and p15 of HTM (636-07-22 and 1073-07-26, respectively) cell lines were reverse transcribed and amplified with miRNA-specific primers and probes. Relative expression was calculated by the comparative Ct method, and miRNA abundance was normalized relative to human RNU6B miRNA. Results from p15 are expressed as the change of miRNA levels relative to p4 cultures. Data represent the mean changes ± SD, n = 3. P < 0.05, compared with p4 cultures; by Mann–Whitney U test. *No significant change in one cell line.
Effects of miR-146a on Gene Expression in HTM Cells
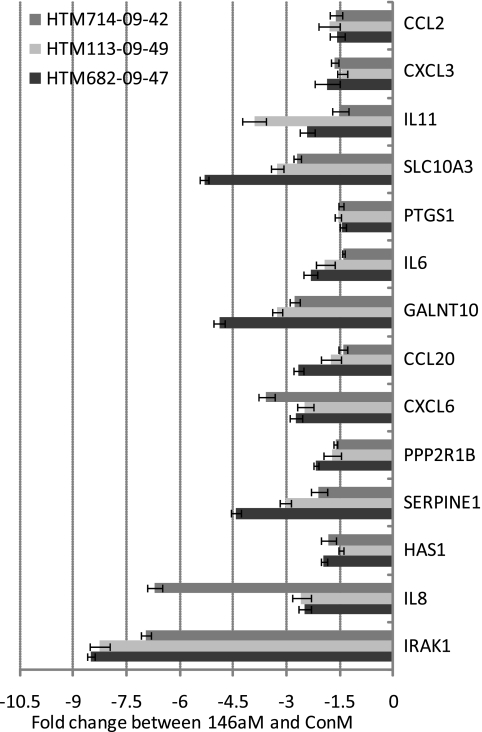
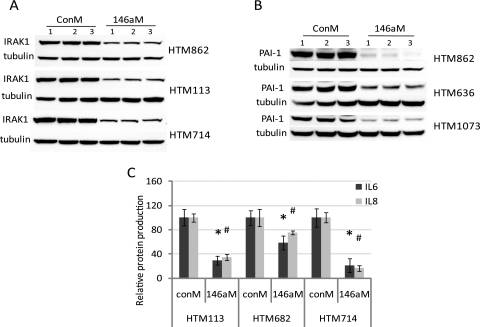
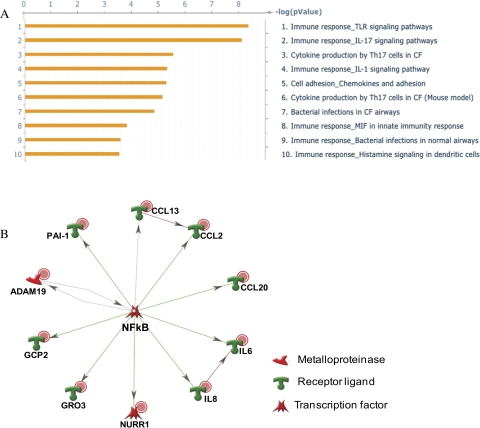
The effects of miR-146a in HTM cells were evaluated by gene array (Affymetrix) in one HTM cell line 3 days after miR-146a transfection. As shown in Table 2, 30 genes were significantly downregulated (by more than twofold), whereas only one gene (MAN1C1) showed significant upregulation (higher than twofold). Among the 30 downregulated genes, 11 (IRAK1, GALNT10, SLC10A3, HAS1, PHKB, SLC1A1, TIMM17B, HAS2, SLC2A3, SRPRB, and CARD10) showed potential targets of miR-146a in at least one of the three miRNA databases (Microcosm, TargetScan, or PicTar-Vert). Fourteen downregulated genes were further validated by real-time q-PCR in three additional HTM cell lines. All these genes showed significant downregulation by q-PCR and the levels of downregulation were higher than twofold, with the exception of CCL2, CXCL3, PTGS1, and HAS1, which showed significant levels of downregulation between 1.4- and 2-fold (Fig. 3). IRAK1, SERPINE1/PAI-1 (plasminogen activator inhibitor type 1), IL6 and IL8 protein levels were also significantly downregulated by miR-146a in all three individual HTM cell lines (Fig. 4). The pathways identified by pathway analysis (Metacore; GeneGo) as more likely to be involved with the changes induced by miR-146a were associated with immune response and cytokine production (Fig. 5A). The transcription regulation subnetwork with the highest score was that of NF-kB (Fig. 5B). Analysis of the pathway maps generated by the software also provided a direct relationship between the known target of miR-146a, IRAK1 and NF-kB through TRAF6-dependent phosphorylation of IKK-B and I-KB.37
Table 2.
Gene Microarray Showing the Effects of miR-146a on Gene Expressions in HTM Cells
| Gene Symbol | Change (x-fold) ([146a] vs. [ConM]) | Unigene* | P | Microcosm | PicTar-Vert | Targetscan |
|---|---|---|---|---|---|---|
| IRAK1 | −4.28 | Hs.522819 | 0.0011 | T | T | T |
| IL11 | −4.14 | Hs.467304 | 0.035 | |||
| PPP2R1B | −3.8 | Hs.584790 | 0.012 | |||
| GALNT10 | −3.49 | Hs.651323 | 0.0033 | T | ||
| IL8 | −3.32 | Hs.551925 | 6.89E-04 | |||
| SLC10A3 | −3.031 | Hs.522826 | 0.0038 | T | T | T |
| HAS1 | −2.74 | Hs.57697 | 0.049 | T | T | |
| SERPINE1/PAI-1 | −2.74 | Hs.414795 | 0.0022 | |||
| MANIC1 | +2.54 | Hs.197043 | 0.026 | |||
| PHKB | −2.54 | Hs.78060 | 0.0038 | T | T | |
| SLCIA1 | −2.52 | Hs.444915 | 0.0023 | T | T | |
| CXCL3 | −2.47 | Hs.89690 | 9.42E-04 | |||
| CXCL6 | −2.39 | Hs.164021 | 0.0048 | |||
| HYOU1 | −2.33 | Hs.277704 | 0.0062 | |||
| CCL2 | −2.31 | Hs.303649 | 0.014 | |||
| TIMMI7B | −2.3 | Hs.30570 | 8.57E-04 | T | ||
| HAS2 | −2.21 | Hs.159226 | T | T | ||
| ADAM19 | −2.16 | Hs.483944 | 0.011 | |||
| WSB2 | −2.12 | Hs.506985 | 0.006 | |||
| SLC2A3 | −2.11 | Hs.419240 | 0.022 | T | ||
| RCAN1 | −2.10 | Hs.282326 | 0.0024 | |||
| SRPRB | −2.10 | Hs.584950 | 0.0038 | T | T | |
| CCL20 | −2.09 | Hs.75498 | 0.027 | |||
| PTGS1 | −2.09 | Hs.201978 | 0.0049 | |||
| CARD10 | −2.06 | Hs.57973 | 0.041 | T | T | |
| NR4A2 | −2.06 | Hs.563344 | ||||
| LIF | −2.06 | Hs.2250 | 0.044 | |||
| IL6 | −2.05 | Hs.654458 | ||||
| XYLT1 | −2.03 | Hs.22907 | 0.011 | |||
| PTHLH | −2.02 | Hs.591159 | ||||
| RY1 | −2.00 | Hs.54649 | 0.0018 |
UniGene, National Center for Biotechnology Information, Bethesda, MD; http://www.ncbi.nlm.nih.gov/UniGene.
Figure 3.
Genes downregulated by miR-146a in three independent HTM cell lines. Three additional individual HTM cell lines (714-09-42, 113-09-49, and 682-09-47) were transfected with 146aM and ConM. Three days later, total RNAs were isolated, and q-PCR was performed with SYBR green master mix with specific primers (Table 1). The results were normalized with β-actin, and the gene expressions in 146aM-transfected cells were expressed as the change in levels of specific genes relative to that in ConM-transfected cells. Data represent the mean change ± SD, n = 3. P < 0.05, compared with ConM transfected cells; Mann–Whitney U test.
Figure 4.
Effects of miR-146a on production of IL6, IL8, IRAK1, and PAI-1 in HTM cells. Three individual HTM cell lines were transfected with 146aM or ConM. Three days later, cell culture supernatant and total proteins were collected. IL6 and IL8 levels in cultured supernatant were measured by flow cytometry. Production of IRAK1 and PAI-1 from total protein were determined by Western blot and normalized by β-tubulin (A, B, n = 3). The percentage changes in IL6 and IL8 in 146aM transfected cells compared with their individual ConM-transfected cells. Data are expressed as the mean percentage of change ± SD, n = 3. (C) *#P < 0.05 compared with ConM transfected cells; Mann–Whitney U test.
Figure 5.
Pathway analysis of the genes showing changes in expression higher than twofold with P < 0.05 in the gene microarrays. (A) The pathways identified as the more significantly affected by miR-146a at a threshold of 0.001 and P = 0.05. (B) The NF-kB transcription regulation subnetwork. NF-kB was identified as the transcription factor with the highest ranking in terms of P-value and gene ontology interpretation with 12 nodes, 11 root nodes; P-value of 7.10e-36, z-score of 129.23, and g-score of 129.23. Red dots: downregulation in the gene array analysis; green symbols: induction; red symbols: inhibition; gray arrows: unspecified interaction.
Components of the SASP in HTM Cells Regulated by miR-146a
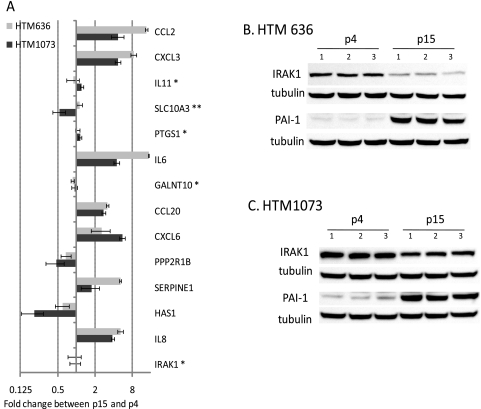
To study whether the 14 genes significantly downregulated by miR-146a potentially involved in the SASP were also differentially expressed in replicative senescent HTM cells, realtime q-PCR was conducted in two HTM cell lines (HTM1073-07-26 and HTM636-07-22; p15 versus 4). As shown in Figure 6A, seven genes (IL8, PAI-1, CXCL6, CCL20, IL6, CXCL3, and CCL2) were significantly upregulated and two genes (HAS1 and PPP2R1B) were significantly downregulated in both senescent HTM cell lines. Consistently, PAI-1 protein production was also clearly increased in both senescent HTM cell lines. However, production of IRAK1 decreased in the protein level, but not in the message level (Fig. 6B).
Figure 6.
Identification of components of the SASP in HTM cells regulated by miR-146a. Total RNAs (500 ng) isolated from p4 and p15 of HTM cell lines (636-07-22 and 1073-07-26) were reverse transcribed and amplified with specific primers (Table 1) and SYBR green master mix. Relative expression was calculated by the comparative cycle threshold method and normalized relative to human β-actin. Results from p15 are expressed as the change in RNA levels relative to p4 cultures. Data represent the mean changes ± SD, n = 3. P < 0.05 compared with p4 cultures; Mann–Whitney U test. (A) **No significant change in one cell line; *no significant changes in any of the cell lines. Total proteins were isolated from p4 and p15 of HTM cells lines (B) 636-07-22 and (C) 1073-07-26), n = 3. Western blot analysis was conducted with 8% SDS PAGE, and the membrane was stained with IRAK1- and PAI-1-specific antibodies. The same membrane was stripped and restained with β-tubulin (B, C).
Effects of miR-146a on the Expression of Cellular Senescence Markers in HTM Cells
To investigate whether miR-146a can also affect cellular senescence markers in HTM cells, two HTM cell lines (HTM1073-07-26 and HTM636-07-22) in p11 were transfected with 146M or ConM. Since senescent cultures at p15 proved to be technically difficult to transfect at high efficiency, the experiments were conducted in presenescent cells at p11 that were already showing a significant increase in expression of senescent markers. As shown in Figure 7, production of SA-β-gal and iROS was significantly decreased, and BrdU incorporation was significantly increased by miR-146a in both HTM cell lines. However, miR-146a did not alter the production of either lipofuscin accumulation or mitochondria membrane potential.
Figure 7.
Effects of miR-146a on the expression of cellular senescence markers in HTM cells. Two HTM (636-07-22 and 1073-07-26) cell lines p11 transfected with 146aM or ConM. Induction of SA-β-gal (A), autofluorescence (B), iROS (C), Δψm (D) in 146aM-transfected cells (146aM) versus ConM-transfected cells (ConM) were quantified by flow cytometry. The proliferation rate (E) was quantified by BrdU incorporation. Data show the percentage of increase or decrease compared with their individual ConM-transfected cells and represent the mean percentage change ± SD, n = 3. *P < 0.05, compared with ConM-transfected cells; Mann–Whitney U test.
Role of PAI-1 and IRAK1 in the Effects of miR-146a on the Expression of Cellular Senescence Markers in HTM Cells
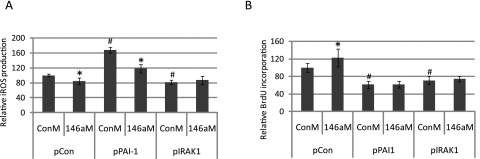
Given the central role played by IRAK1 on the effects mediated by miR-146a30 and the reported role of PAI-1 in cellular senescence as an important component of the SASP,38,39 we investigated whether the observed effects of miR-146a on SA-β-gal activity, iROS, and cell proliferation in senescent HTM cells were mediated by the downregulation of either the direct target IRAK1 or the secondary target PAI-1. The overexpression of IRAK1 or PAI-1 did not result in significant changes in SA-β-gal (data not shown). Although overexpression of PAI-1 led to an increase of approximately twofold in the production of iROS compared with cells transfected with a control vector, miR-146a still generated a significant decrease in iROS in cells overexpressing PAI-1. Overexpression of IRAK1 lacking the 3′ UTR that contains the target site for miR-146a completely prevented the effects of miR-146a on iROS production (Fig. 8A). Overexpression of both PAI-1 and IRAK1, lacking the miR-146a target site, inhibited the effects of miR-146a on cell proliferation (Fig. 8B).
Figure 8.
Role of PAI-1 and IRAK1 on the effects of miR-146a on the production of iROS and BrdU incorporation. Presenescent HTM cells at p11 were cotransfected with 2 μg of either a negative control plasmid (pCon), a plasmid expressing PAI-1 (pPAI-1), or a plasmid expressing IRAK1 (pIRAK1, which lacked the 3′UTR that contains the target site for miR-146a) and 120 pmol of either a miR-146a mimic (146aM) or a scrambled miRNA used as a negative control (ConM). Three days after transfection, production of iROS was determined by DCFH oxidation, and cell proliferation was measured by BrdU incorporation. (A) Comparison cells cotransfected with pPAI-1/ConM with those cotransfected with pCon/ConM showed that overexpression of PAI-1 resulted in an increase of almost twofold in the generation of iROS. However, a comparison between cells cotransfected with pPAI-1/146aM or pPAI-1/ConM showed that miR-146 decreased the production of iROS in cells overexpressing PAI-1. In contrast, expression of IRAK1 lacking the 3′UTR prevented completely the decrease in iROS production mediated by miR-146a. (B) Overexpression of either PAI-1 or IRAK1 lacking the 3′UTR prevented the increase in cell proliferation induced by miR-146a. Data represent the mean percentage changes ± SD, n = 3–4. *#P < 0.05, compared with ConM+pCon by Mann–Whitney U test.
Discussion
We observed significant changes in miRNA expression in replicative scenescent HTM cells. There were several differences between the two cell lines analyzed, suggesting some level of variability in the alterations of miRNA expression induced by RS. However, 18 microRNAs of the 30 analyzed were consistently up- or downregulated in both cell lines.
Similar to what was previously observed in SIPS, RS of TM cells was associated with the downregulation of several members of the miR-106b family (miR-17–5p, -18a, -20a, -20b, -106a, and -106b) that are located in the oncogenic miRNA polycistronic clusters 17–5p-92a1, 106b-25, and 106a-363. These clusters are frequently upregulated in cancer, and their oncogenic effects are mediated at least in part by members of the miR-106b family that are known to promote cell cycle progression.40–42 The consistent downregulation of these miRNAs observed in both stress-induced and RS appears to be a common feature of senescent cells that could contribute to their permanent growth arrest.
A downregulation of members of the miR-15 family, similar to that previously observed in SIPS, was found in one of the cell lines (HTM1073-07-26), whereas only miR-15a was significantly downregulated in HTM636-07-22. Given the proapoptotic role of miR-15a, the downregulation of this miRNA in senescent cells could contribute to the increased resistance to the apoptosis characteristic of senescent cells.
RS was also associated with changes in miRNAs that have not been observed in SIPS. These changes included a particularly notable upregulation of several miRNAs: miR-146a, -329, -369, -409–5p, -432, -493, and -495.
One of the miRNAs more clearly upregulated in senescent HTM cells, miR-146a, has also been found to be upregulated in senescent human fibroblasts and is believed to be implicated in modulating the inflammatory response29 and in particular the senescence-associated inflammatory mediators IL6 and IL8.43 miR-146a is known to target IRAK1, a key activator of the innate immune system signaling cascade that leads to the induction of inflammatory target gene expression.43 Upregulation of miR-146a/b in senescent fibroblasts has been hypothesize to serve as a mechanism aimed at preventing excessive production of inflammatory mediators by senescent cells, thus limiting the impact of the SASP on adjacent cells.31
Although in senescent fibroblasts it has been reported that both miR-146a and -146b were upregulated during RS,31 HTM cells showed a notable upregulation only of miR-146a, whereas they showed a relatively low upregulation of miR-146b (3.4-fold in HTM1073 cells and 1.67-fold in HTM636 cells). These results suggest cell-type–specific differences in the changes in expression of these miRNAs during RS.
Analysis of the changes in gene expression induced by miR-146a in HTM cells was consistent with the anti-inflammatory role proposed for this miRNA. In addition to downregulation of the well-characterized target IRAK1 and the concomitant downregulation of IL6 and IL8 observed in senescent fibroblasts, miR-146a also decreased the expression of multiple genes involved in the inflammatory response including: IL11, IL8, CXCL3 (GRO3), CXCL6 (GCP2), CCL2, CCL20, PTGS1, and IL6. None of these genes contains any predicted target sequences or miR-146a and should be considered secondary targets of this miRNA.
Pathway analysis indicated that several these changes in gene expression could result from direct targeting of IRAK1 by miR-146a through activation of NF-êB.44
Several of the genes downregulated by miR-146a were found to be significantly upregulated in senescent TM cells compared with their expression in the low-passage control cells. This upregulation could contribute to the SASP and the activation of an inflammatory response in senescent HTM cells. These genes included IL6, IL8, PAI-1, CXCL3, CXCL6, and CCL2, which have been reported to be part of the SASP in other cell types,18,45 as well as the additional member of the chemokine signaling pathway CCL20.
Of interest, the overall levels of upregulation of these genes in senescent cells were lower in HTM1073 cells, where miR-146a was upregulated 111-fold, compared with expression in HTM636 cells, in which this miRNA was upregulated sevenfold. This observation is consistent with the concept that the increase in expression of miR-146a observed in senescent cells antagonizes with the increased expression of inflammatory mediators associated with the senescence response.
The observed increased expression of PAI-1 could contribute to the pathogenesis of primary open-angle glaucoma. PAI-1 is the principal inhibitor of tissue plasminogen activator (tPA) and urokinase (uPA), the activators of plasminogen. Therefore, an increase in PAI-1 may lead to decreased fibrinolysis and extracellular proteolysis in aqueous humor and the trabecular meshwork, which could result in a decrease in aqueous humor outflow facility by increasing protein deposition and obstruction.46,47 In addition, increased expression of TGF-B2, which leads to increased outflow resistance and elevated intraocular pressure, has been reported to result in increased PAI-1 production in cell culture and organ culture models.48,49 Similarly, corticosteroid treatment, which also results in increased outflow resistance, has been shown to induce a decrease in activity of tissue plasminogen activator.50 Therefore, the inhibitory effect of miR-146a on the upregulation of PAI-1 observed in senescent cells could contribute to preventing a decrease in the activity of the plasminogen system and a concomitant increase in outflow resistance.
Some of the components of the SASP, such as PAI-1, are believed to contribute to the reinforcement of the senescence phenotype by autocrine and paracrine mechanisms.38,39 Our results showed that transfection of presenescent cells with miR-146a mimic had significant inhibitory effects on SA-β-gal activity, production of ROS, and cell proliferation. The effects of miR-146a on ROS generation and cell proliferation were prevented by overexpression of IRAK1, suggesting that the changes in expression of multiple genes involved in the SASP induced by the inhibition of IRAK1 contribute to increased ROS production and growth arrest in senescent cells. Since overexpression of PAI-1 was enough to prevent the effects of miR-146a on cell proliferation, our results also point to PAI-1 as an important component of the SASP of HTM cells that is negatively modulated by miR-146a.
In conclusion, RS in HTM cells was associated with significant changes in miRNA expression that may contribute to the modulation of the senescent response. Specifically, the upregulation of miR-146a antagonized the induction of inflammatory mediators: the increased production of ROS and the decrease in proliferative capacity characteristic of the SASP. The observed inhibition of PAI-1 by miR-146a may limit the alterations in the extracellular proteolytic activity of the TM during aging and minimize the autocrine and paracrine effects of PAI-1, including increased generation of iROS and decreased cell proliferation. These results are consistent with the concept that upregulation of miR-146a in senescent HTM cells may serve to prevent an excessive increase in the production of inflammatory mediators and limit some of the potentially deleterious effects of the SASP on the physiology of the TM.
Acknowledgments
The authors thank the Flow Cytometry Core Facility at the Duke Cancer Center.
Footnotes
Supported by National Eye Institute Grants EY01894, EY016228, and EY05722, and Research to Prevent Blindness.
Disclosure: G. Li, None; C. Luna, None; J. Qiu, None; D.L. Epstein, None; P. Gonzalez, None
References
- 1.Li G, Luna C, Liton PB, Navarro I, Epstein DL, Gonzalez P. Sustained stress response after oxidative stress in trabecular meshwork cells. Mol Vis 2007;13:2282–2288 [PMC free article] [PubMed] [Google Scholar]
- 2.Liton PB, Luna C, Challa P, Epstein DL, Gonzalez P. Genome-wide expression profile of human trabecular meshwork cultured cells, nonglaucomatous and primary open angle glaucoma tissue. Mol Vis 2006;12:774–790 [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N, Chintala SK, Fini ME, Schuman JS. Activation of a tissue-specific stress response in the aqueous outflow pathway of the eye defines the glaucoma disease phenotype. Nat Med 2001;7:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Chen M, Forrester JV. Para-inflammation in the aging retina. Prog Retin Eye Res 2009;28:348–368 [DOI] [PubMed] [Google Scholar]
- 5.Aikata H, Takaishi H, Kawakami Y, et al. Telomere reduction in human liver tissues with age and chronic inflammation. Exp Cell Res 2000;256:578–582 [DOI] [PubMed] [Google Scholar]
- 6.Castro P, Giri D, Lamb D, Ittmann M. Cellular senescence in the pathogenesis of benign prostatic hyperplasia. Prostate 2003;55:30–38 [DOI] [PubMed] [Google Scholar]
- 7.Castro P, Xia C, Gomez L, Lamb DJ, Ittmann M. Interleukin-8 expression is increased in senescent prostatic epithelial cells and promotes the development of benign prostatic hyperplasia. Prostate 2004;60:153–159 [DOI] [PubMed] [Google Scholar]
- 8.Flanary BE, Sammons NW, Nguyen C, Walker D, Streit WJ. Evidence that aging and amyloid promote microglial cell senescence. Rejuvenation Res 2007;10:61–74 [DOI] [PubMed] [Google Scholar]
- 9.Kitada T, Seki S, Kawakita N, Kuroki T, Monna T. Telomere shortening in chronic liver diseases. Biochem Biophys Res Commun 1995;211:33–39 [DOI] [PubMed] [Google Scholar]
- 10.Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I. Endothelial cell senescence in human atherosclerosis: role of telomere in endothelial dysfunction. Circulation 2002;105:1541–1544 [DOI] [PubMed] [Google Scholar]
- 11.Muller M. Premature cellular senescence induced by pyocyanin, a redox-active Pseudomonas aeruginosa toxin. Free Radic Biol Med 2006;41:1670–1677 [DOI] [PubMed] [Google Scholar]
- 12.Vasile E, Tomita Y, Brown LF, Kocher O, Dvorak HF. Differential expression of thymosin beta-10 by early passage and senescent vascular endothelium is modulated by VPF/VEGF: evidence for senescent endothelial cells in vivo at sites of atherosclerosis. FASEB J 2001;15:458–466 [DOI] [PubMed] [Google Scholar]
- 13.Manestar-Blazic T, Volf M. The dynamic of senescent cells accumulation can explain the age-specific incidence of autoimmune diseases. Med Hypotheses 2009;73:667–669 [DOI] [PubMed] [Google Scholar]
- 14.Liton PB, Challa P, Stinnett S, Luna C, Epstein DL, Gonzalez P. Cellular senescence in the glaucomatous outflow pathway. Exp Gerontol 2005;40:745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YH, Govinda B, Kim JC, et al. Oxidative stress resistance through blocking Hsp60 translocation followed by SAPK/JNK inhibition in aged human diploid fibroblasts. Cell Biochem Funct 2009;27:35–39 [DOI] [PubMed] [Google Scholar]
- 16.Hasenmaile S, Pawelec G, Wagner W. A lack of telomeric non-reciprocal recombination (TENOR) may account for the premature proliferation blockade of Werner's syndrome fibroblasts. Biogerontology 2003;4:253–273 [DOI] [PubMed] [Google Scholar]
- 17.Fumagalli M, d'Adda di Fagagna F. SASPense and DDRama in cancer and ageing. Nat Cell Biol 2009;11:921–923 [DOI] [PubMed] [Google Scholar]
- 18.Coppe JP, Patil CK, Rodier F, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 2008;6:2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann BD, Paine MS, Brooks AM, et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Res 2008;68:7864–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc 2009;84:55–71 [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Jima DD, Jacobs C, et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 2009;113:4586–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaddar T, Rouault JP, Chien WW, et al. Two new miR-16 targets: caprin-1 and HMGA1, proteins implicated in cell proliferation. Biol Cell 2009;101:511–524 [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001;409:363–366 [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 25.Christoffersen NR, Shalgi R, Frankel LB, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 2010;17(2):236–245 [DOI] [PubMed] [Google Scholar]
- 26.Scherr M, Venturini L, Battmer K, et al. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res 2007;35:e149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mudhasani R, Zhu Z, Hutvagner G, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol 2008;181:1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev 2009;130(11–12):731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett 2009;583(20):3349–3355 [DOI] [PubMed] [Google Scholar]
- 30.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006;103:12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaumik D, Scott GK, Schokrpur S, et al. MicroRNAs miR-146a/b negatively modulate the senescence associated inflammatory mediators IL-6 and IL-8. Aging 2009;1:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Cui G, Tzeng NS, et al. Femtomolar concentrations of dextromethorphan protect mesencephalic dopaminergic neurons from inflammatory damage. FASEB J 2005;19:489–496 [DOI] [PubMed] [Google Scholar]
- 33.Fiering SN, Roederer M, Nolan GP, Micklem DR, Parks DR, Herzenberg LA. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry 1991;12:291–301 [DOI] [PubMed] [Google Scholar]
- 34.Lu T, Finkel T. Free radicals and senescence. Exp Cell Res 2008;314:1918–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 1995;92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Zglinicki T, Nilsson E, Docke WD, Brunk UT. Lipofuscin accumulation and ageing of fibroblasts. Gerontology 1995;41(suppl 2):95–108 [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Dziarski R, Kirschning CJ, Muzio M, Gupta D. Micrococci and peptidoglycan activate TLR2→MyD88→IRAK→TRAF→NIK→IKK→NF-kappaB signal transduction pathway that induces transcription of interleukin-8. Infect Immun 2001;69:2270–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acosta JC, O'Loghlen A, Banito A, Raguz S, Gil J. Control of senescence by CXCR2 and its ligands. Cell Cycle 2008;7:2956–2959 [DOI] [PubMed] [Google Scholar]
- 39.Peiretti F, Bernot D, Lopez S, et al. Modulation of PAI-1 and proMMP-9 syntheses by soluble TNFalpha and its receptors during differentiation of the human monocytic HL-60 cell line. J Cell Physiol 2003;196:346–353 [DOI] [PubMed] [Google Scholar]
- 40.Ivanovska I, Cleary MA. Combinatorial microRNAs: working together to make a difference. Cell Cycle 2008;7:3137–3142 [DOI] [PubMed] [Google Scholar]
- 41.Fontana L, Fiori ME, Albini S, et al. Antagomir-17–5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE 2008;3:e2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim TE, Kim BH, Kim JR, et al. Effect of food on the pharmacokinetics of the oral phosphodiesterase 5 inhibitor udenafil for the treatment of erectile dysfunction. Br J Clin Pharmacol 2009;68:43–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell Signal 2008;20:269–276 [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Zhu YY, Zhang XJ, et al. TSPAN1 protein expression: a significant prognostic indicator for patients with colorectal adenocarcinoma. World J Gastroenterol 2009;15:2270–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer 2009;9:81–94 [DOI] [PubMed] [Google Scholar]
- 46.Epstein DL, Hashimoto JM, Grant WM. Serum obstruction of aqueous outflow in enucleated eyes. Am J Ophthalmol 1978;86:101–105 [DOI] [PubMed] [Google Scholar]
- 47.Meyer MW, von Depka M, Wilhelm C, Schroder A, Erb C. Plasminogen activator inhibitor-1 mRNA expression in cultured pigmented ciliary epithelial cells of the porcine eye. Graefes Arch Clin Exp Ophthalmol 2002;240:679–686 [DOI] [PubMed] [Google Scholar]
- 48.Fuchshofer R, Welge-Lussen U, Lutjen-Drecoll E. The effect of TGF-beta2 on human trabecular meshwork extracellular proteolytic system. Exp Eye Res 2003;77:757–765 [DOI] [PubMed] [Google Scholar]
- 49.Fuchshofer R, Yu AH, Welge-Lussen U, Tamm ER. Bone morphogenetic protein-7 is an antagonist of transforming growth factor-beta2 in human trabecular meshwork cells. Invest Ophthalmol Vis Sci 2007;48:715–726 [DOI] [PubMed] [Google Scholar]
- 50.Snyder RW, Stamer WD, Kramer TR, Seftor RE. Corticosteroid treatment and trabecular meshwork proteases in cell and organ culture supernatants. Exp Eye Res 1993;57:461–468 [DOI] [PubMed] [Google Scholar]