Abstract
We determined the crystal structure of a bifunctional group I intron splicing factor and homing endonuclease, termed the I-AniI maturase, in complex with its DNA target at 2.6 Å resolution. The structure demonstrates the remarkable structural conservation of the β-sheet DNA-binding motif between highly divergent enzyme subfamilies. DNA recognition by I-AniI was further studied using nucleoside deletion and DMS modification interference analyses. Correlation of these results with the crystal structure provides information on the relative importance of individual nucleotide contacts for DNA recognition. Alignment and modeling of two homologous maturases reveals conserved basic surface residues, distant from the DNA-binding surface, that might be involved in RNA binding. A point mutation that introduces a single negative charge in this region uncouples the maturase and endonuclease functions of the protein, inhibiting RNA binding and splicing while maintaining DNA binding and cleavage.
Keywords: Homing endonuclease, maturase, group I intron, crystal structure, DNA binding
Multifunctional nucleic acid binding proteins are a small but growing class of cellular factors. These include a number of well known transcription factors that not only bind DNA but also RNA (Cassiday and Maher 2002). Here we present the first structure of a bifunctional protein that assists RNA splicing of a Group I intron and also cleaves a DNA target site to initiate mobilization of the same intron. A mutational and biochemical analysis, based on the structure, demonstrates the uncoupling of endonuclease and maturase activities of the protein and the use of different protein surfaces for DNA and RNA binding.
Homing is the lateral transfer of an intervening sequence (either an intron or intein) to a homologous allele that lacks the sequence (Dujon 1989;Belfort and Perlman 1995;Jurica and Stoddard 1999;Lambowitz et al. 1999;Chevalier and Stoddard 2001). DNA transfer is catalyzed by a homing endonuclease (encoded within the mobile intervening sequence) that recognizes and cleaves a DNA target site, generates a double-strand break, and induces cellular mechanisms to repair the break. If the intron- or intein-containing allele is used as a template for repair, the endonuclease open reading frame (ORF) is duplicated into the target site (Chevalier and Stoddard 2001). Homing endonucleases are highly specific and have evolved to recognize long target sequences (14-40 bp;Chevalier and Stoddard 2001). They tolerate some individual base variations at their homing site, ensuring their mobility despite sequence drift of their target site (Argast et al. 1998;Chevalier et al. 2003).
Many group I intron-encoded proteins also function as maturases, by facilitating the folding and efficient splicing of their cognate introns (Ho et al. 1997;Lambowitz et al. 1999). These proteins do not cleave the RNA directly; instead they aid in folding the RNA into a conformation that favors self-splicing. While many intron-encoded proteins function solely as endonucleases or maturases, some are bifunctional. The precise relationship of DNA- and RNA-binding activities and surfaces used by these proteins has not been determined.
The majority of known group I homing endonucleases and all known maturases are members of the LAGLIDADG family, which contains dozens of functionally characterized proteins and several times that number of sequenced ORFs (Belfort and Roberts 1997;Chevalier and Stoddard 2001). Residues within the conserved motif participate in dimer or domain packing and in coordination of catalytic metal ions. These enzymes are usually pseudosymmetric monomers that contain two copies of their namesake sequence motif, but can also be homodimers with one copy of the motif per subunit (Chevalier and Stoddard 2001). Monomeric homing endonucleases are also found associated with inteins (Pietrokovski 1998). Homodimeric enzymes recognize palindromic target sites and pseudopalindromic variants, whereas enzyme monomers recognize asymmetric DNA target sites. Despite their relatively small size, all LAGLIDADG endonucleases recognize long DNA target sites (average length ∼20 bp), cleaving across the minor groove within the site to generate cohesive four-base, 3′ overhangs (Chevalier and Stoddard 2001).
The I-AniI protein is a monomeric 250-residue LAGLIDADG protein encoded within a group I intron in the Aspergillus nidulans (A.n.) mitochondrial apocytochrome b gene (Ho et al. 1997). I-AniI recognizes and cleaves a 19-bp DNA target site (Fig. 1A), generating overhangs of sequence 5′-GTTT-3′ on the upper strand and its complement on the lower strand (Ho et al. 1997). The enzyme binds its DNA substrate with ∼1 nM affinity, and cleaves the noncoding upper strand more rapidly than the coding lower strand (Geese and Waring 2001; Chatterjee et al. 2003).
Figure 1.
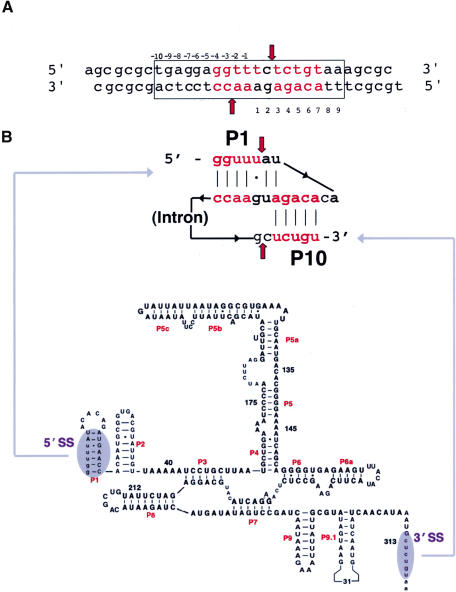
(A) Sequence of the DNA target site for I-AniI and the design of the oligonucleotide construct used for the structure determination. Cleavage sites are indicated by arrows; red bases indicate those similar to their aligned counterparts in the RNA splice junctions (panel B). (B) Schematic representation of A.n.COBme pre-RNA secondary structure around the 5′ and 3′ splice sites. The P1/P10 pseudoknot is highlighted. Arrows point to the splice sites. The sequence and predicted secondary structure of the A.n.COB intron is shown below for reference.
The cognate intron for I-AniI (A.n.COB;Fig. 1B) does not exhibit self-splicing activity under physiological conditions. RNase protection assays indicate that the intron is incapable of forming a stable, fully reactive folded conformation in the absence of a protein cofactor. This is consistent with the lack of a full complement of RNA sequence signatures indicative of stabilizing tertiary interactions between the intron's P4-P6 domain and its catalytic core (Ho and Waring 1999). I-AniI binds the A.n.COB RNA with a picomolar dissociation constant, reducing dependence on Mg2+ for splicing and imparting a 30-fold rate enhancement to splicing relative to optimal self-splicing conditions (Ho and Waring 1999;Solem et al. 2002).
Results
Structural analysis
The complex of I-AniI and its corresponding target DNA duplex crystallized in space group P21. A single protein-DNA complex is present in the crystallographic asymmetric unit. The data and refinement statistics are presented in Table 1.
Table 1.
X-ray data and refinement statistics
| Crystallographic data | |
|---|---|
| Space group | P21 |
| Cell parameters | a = 60.46 b = 72.84 c = 65.23 β = 108.21 |
| Number of complexes per AU | 1 |
| Resolution (Å) | 2.6 |
| Number of reflections measured | 58,621 |
| Number of unique reflections | 16,749 |
| Redundancy | 12.7 |
| Completeness (2.69-2.6 Å) | 90.4 (66.2) |
| Average I/σ(I) (2.69-2.6 Å) | 9.5 (3.2) |
| Rsym (2.69-2.6 Å) | 4.0 (21.0) |
| Refinement | |
|---|---|
| Resolution range (Å) | 0-2.6 |
| R factor (%) | 23.8 |
| R-free (%) | 26.2 |
| Number of protein atoms | 1973 |
| Number of nucleic acid atoms | 1176 |
| Ramachandran plot (percent core, allowed, generous, disallowed) | 81.6, 18.4, 0, 0 |
| rms on atoms (Å) | 0.014 |
| rms on angles (°) | 1.933 |
| Average B (Å2) (protein, DNA) | 65.01, 79.32 |
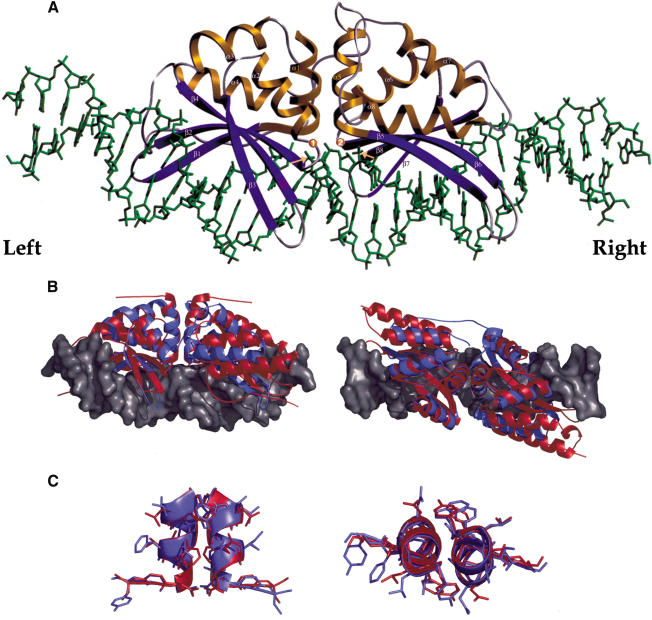
The refined I-AniI/DNA complex structure (Fig. 2A) consists of a full-length enzyme monomer and a 31-bp DNA duplex. The DNA construct contains the asymmetric 19-bp sequence of the native homing site in the Aspergillus mitochondrial COB intron (Figs. 1A, 3A). A pair of bound magnesium ions are present in the structure, and the DNA is cleaved on both strands at positions corresponding to the known cleavage pattern in the recognition site. The entire complex is ∼80 Å long, burying ∼3400 Å2 of surface area each from the endonuclease and DNA in the interface (Fig. 2B). The enzyme monomer of I-AniI is a single polypeptide chain with two pseudosymmetric domains of mixed α/β topology, similar to the fold and topology of other members of the LAGLIDADG enzyme family. The core endonuclease fold (α-ββ-α-ββ-α) is structurally conserved with previously determined structures of LAGLIDADG enzymes (Duan et al. 1997;Heath et al. 1997;Jurica et al. 1998;Silva et al. 1999;Ichiyanagi et al. 2000; Chevalier et al. 2002, 2003; Moure et al. 2002), and the β-sheets involved in DNA contacts from each domain/subunit display similar twist and curvature as they follow the major groove of their respective target halfsites (Fig. 2B). The C-terminal end of the I-CreI subunits are longer than the corresponding ends of the I-AniI domains and contain an additional α-helix.
Figure 2.
(A) Ribbon diagram of I-AniI maturase bound to its target DNA site. The left and right halfsites are noted here and in Figure 3 as described in the text. The bound magnesium ions are shown here as spheres labeled “1” and “2” (also in Figs. 3, 4). Adjacent scissile phosphates are indicated by orange arrows. (B) Backbone trace of I-AniI (blue ribbons) bound to its DNA target (gray space-filled model) superimposed against backbone ribbon of I-CreI (red ribbons). (C) Superposition of LAGLIDADG helices of I-AniI (blue) and I-CreI (red). Panel A and Figure 3A were made with the program SETOR (Evans 1993); panels B and C and Figures 5 and 7 were made with the program PYMOL (DeLano 2002).
Figure 3.
DNA recognition and contacts by I-AniI. (A) Ribbon diagram of DNA-binding β-strands and their side chains in each DNA target halfsite. Cleavage sites are indicated by red spheres. (B) Schematic representation of direct protein interactions to nucleotide base pairs and the phosphate backbone in the DNA interface. Bound metal ions are shown as green spheres as in Figures 2 and 4. Colors represent contacts to phosphate backbone (blue), to nucleotide bases (red), and to divalent metal ions (green).
The two conserved LAGLIDADG motifs are found at the core of the structure, with residues from these sequences forming two parallel α-helices that pack against one another at the domain interface (Fig. 2C). The helices are the most structurally conserved elements between I-CreI and I-AniI, with a superimposed rmsd of 1.5 Å. The linker peptide that connects the N- and C-terminal enzyme domains is exceptionally long, consisting of an extended chain spanning residues 110-127. This linker curves around the exterior of the N-terminal domain over a distance of ∼35 Å and leads into the second LAGLIDADG helix at the domain interface. The structure of this linker is strikingly different from that observed for the monomeric I-DmoI homing endonuclease (Silva et al. 1999). For that enzyme, the linker is quite short (three residues) and directly links the C-terminal helix of domain 1 and the N-terminal helix of domain 2.
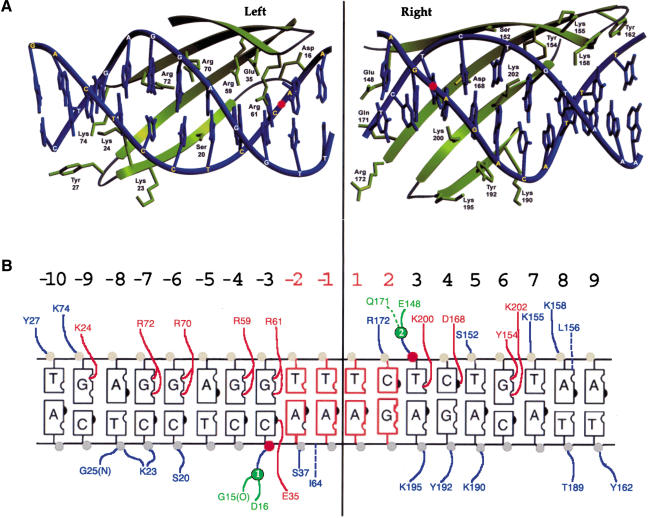
In each domain of I-AniI, four α-helices form a hydrophobic core packed against one side of an extended, four-stranded antiparallel β-sheet that represents the DNA binding surface of the enzyme (Fig. 3A). The β-sheet from each domain contacts nucleotide bases and phosphate backbone atoms corresponding to a single DNA target halfsite, such that recognition of the halfsites is accomplished in a modular manner. All contacts to nucleotide bases are made in the major groove. Side chains from each β-sheet contact nucleotide atoms starting just outside the scissile phosphates (positions ±3) and continuing to the most distal ends of the target site (Fig. 3B). Seventeen of the 38 phosphate groups are in direct contact with the protein. A total of 14 direct contacts, with a distance of 3.2 Å or less, are made between potential hydrogen-bond partners on the protein and corresponding hydrogen-bond donors and acceptors on DNA bases in the target site. This corresponds to roughly 25% of the potential hydrogen-bond sites across the DNA target's major groove. The direct contacts are distributed asymmetrically across the target, with 10 contacts made to bases in the left halfsite, and four to bases in the right halfsite.
Virtually all of the direct contacts to nucleotide bases are made to the top (noncoding) strand of the target DNA (Fig. 3B). A significant fraction of the direct contacts to nucleotide bases are between arginine residues and a run of purine bases in the left halfsite. These contacts correspond to the DNA target sequence 5′-GGAGG-3′ from base pair -7 to -3, and protein residues Arg 72, Arg 70, Arg 59, and Arg 61. These interactions are all identical, corresponding to a bifurcated pair of contacts between the terminal guanidino nitrogens of the arginine side chains and the O6/N7 H-bond acceptors of each guanine base.
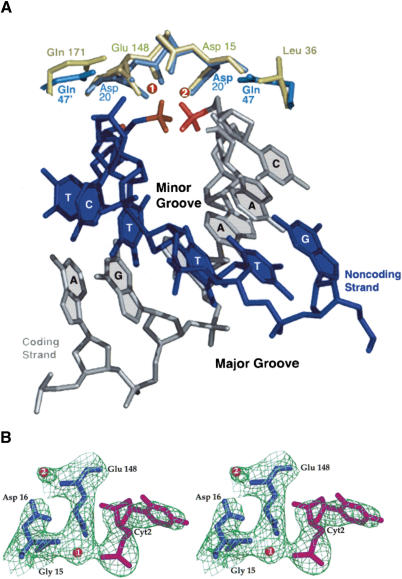
The target DNA is gradually bent around the endonuclease binding surface, giving an overall curvature across the entire length of the site of ∼45° that is similar to the bend observed for bound I-CreI in either its un-cleaved substrate or cleaved product complex (Jurica et al. 1998;Chevalier et al. 2001b). These bends are induced by I-AniI and I-CreI through dissimilar patterns of individual base pair deformations across their two respective target sites. The DNA target in the I-AniI complex is bent asymmetrically, with the major bend observed across base pairs +1 to +3 of the right halfsite. In contrast, bending in the I-CreI/DNA complex is symmetric, resulting from the combination of two identical kinks in the DNA halfsites (Jurica et al. 1998). The central four base pairs display negative roll values for both of these structures, as is also observed for bound DNA in complex with the PI-SceI intein (Moure et al. 2002). For all observed LADLIDADG endonuclease-DNA complexes, bending of the DNA narrows the minor groove across the cleavage site. As a result, the scissile phosphates in the I-AniI complex are positioned ∼5 Å apart and are located near the conserved acidic residues and bound magnesium ions in the active sites (Fig. 4).
Figure 4.
Structure of I-AniI active sites. (A) Superposition of active-site residues of I-AniI (gold) and I-CreI (light blue). Bound metals from the I-AniI complex are shown as green spheres; coding and noncoding DNA strands are shown in gray and dark blue, respectively. The four central bases at the cleavage site and a single flanking base to each side are shown for each DNA strand. The view is looking straight through the minor groove, which is significantly narrowed due to the negative roll angles for these bases. The scissile phosphates, which are ∼5 Å apart, are shown in red. (B) Experimental SAD electron density. Both bound metal ions and the two conserved LAGLIDADG acidic residues (Asp 16 and Glu 148) are shown, as well as the 3′ base of one strand with a cleaved phosphate (in contact with magnesium 1). The third oxygen of the free phosphate is pointing away from the viewer and is not visible.
Each active site of I-AniI contains a single conserved acidic residue from an LAGLIDADG motif (Asp16 and Glu148, respectively) in complex with a single bound divalent magnesium ion that itself is in contact with a single scissile phosphate group. This pattern of metal binding was also observed in the DNA-bound complex of PI-SceI (Moure et al. 2002). We do not observe an additional metal bridging the scissile phosphates and active sites as observed for I-CreI (Chevalier et al. 2001a). The differences between these studies may indicate a real divergence between the catalytic mechanisms of the homodimeric and monomeric endonuclease subfamilies, or may reflect a lower metal occupancy for that site. Beyond the minimal conserved elements described above, the two active sites in I-AniI are structurally diverged from one another. One bound metal and phosphate is in contact with a glutamine residue (Gln 171), whereas the opposing active site contains a leucine residue (Leu 36) at the equivalent position. These two residues in I-AniI occupy the same position as a catalytically essential glutamine residue and its symmetry mate (Gln 47 and 47′) in I-CreI (Fig. 4).
Biochemical analyses of DNA recognition
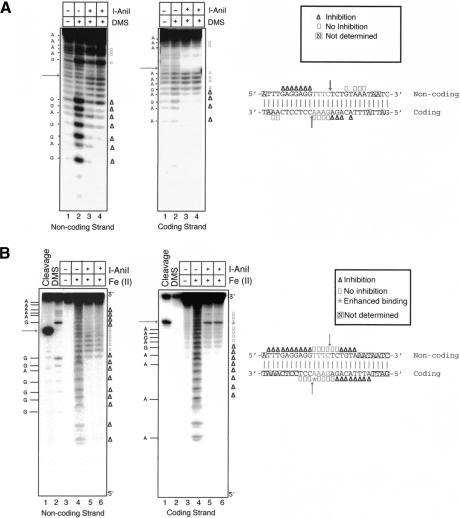
Base alkylation and hydroxyl radical cleavage analyses were used to define nucleotide base contacts that are required for I-AniI binding (Fig. 5). Modified DNA was incubated with protein, and stable complexes were recovered by filtration through nitrocellulose membranes. Denaturing gel electrophoresis was used to visualize gaps in the resulting DNA cleavage ladder that were caused by modification of specific nucleotides. Both analyses indicate that bases immediately outside and adjacent to the four-base cleavage center are most important for recognition and binding. See Figure 6 for the precise results described below.
Figure 5.
(A) DMS modification interference analysis. Oligonucleotides of either the coding or noncoding strand were 5′-end-labeled and hybridized to the corresponding unlabeled DNA strand, modified with DMS, and incubated with limiting concentrations of I-AniI. Bound DNAs were recovered by nitrocellulose filter binding. Modifications that inhibit binding are indicated by gaps in the cleavage patterns relative to the minus I-AniI lane. The red arrow demarcates the I-AniI cleavage product. The data are summarized schematically to the right. (Lane 1) Untreated target-site DNA. (Lane 2) DMS-treated target-site DNA. (Lane 3) Target-site DNA (1 μM) bound to 0.75 μM I-AniI. (Lane 4) Target-site DNA (1 μM) bound to 0.375 μM I-AniI. (B) Hydroxyl radical nucleoside deletion analysis. Oligonucleotides of either the coding or noncoding strand were 5′-end-labeled and hybridized to the corresponding unlabeled DNA strand, cleaved by hydroxyl radicals, and incubated with limiting concentrations of I-AniI; bound DNAs were recovered by nitrocellulose filter binding. Modifications that inhibit binding are indicated by gaps in the cleavage patterns relative to the minus I-AniI lane. The red arrow demarcates the I-AniI cleavage product. The data are summarized schematically to the right. (Lane 1) I-AniI cleavage of target-site DNA. (Lane 2) Sequencing markers derived from DMS-treated DNA. (Lane 3) Untreated target-site DNA. (Lane 4) Hydroxyl radical-treated target-site DNA. (Lane 5) Target-site DNA (405 nM) bound to 500 nM I-AniI. (Lane 6) Target-site DNA (405 nM) bound to 250 nM I-AniI.
Figure 6.
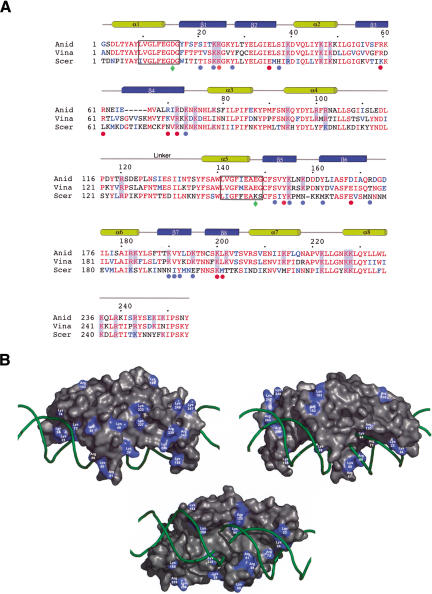
(A) Sequence alignment of proteins encoded by the third intron of the apocytochrome b gene in S. cerevisiae (Sccobi3; Scer; Lazowska et al. 1989) and the fifth intron of the same gene in V. inaequalis (Vicobi5; Vina;Zheng and Koller 1997) with the I-AniI maturase. The secondary structure of I-AniI is indicated above the aligned sequences; conserved basic residues in the aligned sequences are highlighted. The LAGLIDADG signatures are outlined with boxes; the residue from each signature involved in binding a catalytic metal ion is indicated with a green diamond. Residues that contact DNA nucleotide bases are indicated with red dots, and residues that contact DNA phosphate backbone are indicated with blue dots. (B) Solvent-accessible surface of I-AniI colored to indicate positions of basic surface residues conserved between I-AniI and Vicobi5/Sccobi3 maturases (alignment shown in Fig. 1B). The protein surface is shown from both sides (top and middle panels) and from the bottom (the DNA-binding surface, bottom panel). Note that one side and end of the protein surface is populated with conserved basic residues that are not involved in DNA contacts, including Lys 46, Lys 183, Lys 222, Lys 227, Lys 236, Lys 247, Lys 249, Arg 239, and Arg 243.
Dimethyl sulfate (DMS) alkylates the N-7 and N-3 positions of guanine and adenines, respectively (Siebenlist and Gilbert 1980). On the noncoding strand, methylation of a contiguous run of purines from Gua -9 to Gua -3 in the left halfsite (immediately 5′ to the cleavage site) inhibited binding (Fig. 5A). In contrast, methylation of Gua +6, Ade +8, Ade +9, and Ade +10 on the same strand (bases that are nearer to the end of the DNA target and distal to the cleavage site) had less significant effects on binding. A qualitatively similar result was observed across the coding strand, where modification of several bases directly 5′ to the site of cleavage caused inhibition of binding.
Hydroxyl radical cleavage of double-stranded DNA results in the release of nucleosides (a base and its ribose), creating “gaps” in the DNA molecule (Hayes and Tullius 1989). On the noncoding strand, elimination of any nucleosides in the left halfsite 5′ to the site of cleavage inhibited I-AniI binding (Fig. 5B). Four sequential bases on the same strand within the right halfsite are also important for binding; however, the intervening nucleosides between and near the cleavage sites are less crucial. On the coding strand, elimination of most bases in the right halfsite 5′ to the cleavage site inhibited binding, whereas deleting bases within and immediately 3′ to the cleavage overhang region was less deleterious.
Alignment and homology modeling of maturase homologs
An unresolved question for I-AniI and related endonuclease/maturase proteins is whether the DNA- and RNA-binding properties and associated biochemical activities utilize a common protein surface and nucleic acid recognition mechanism. The crystal structure of I-AniI reveals several clusters of solvent-exposed, basic residues outside of the DNA-binding surface that might participate in a separate RNA-binding surface. To address this question, we compared the I-AniI structure with homologous proteins encoded by the third intron (Sccobi3) of the apocytochrome b gene in Saccharomyces cerevisiae (Lazowska et al. 1989) and the fifth intron (Vicobi5) of the same gene in Venturia inaequalis (Fig. 6; Zheng and Koller 1997). Both introns are inserted at an equivalent location relative to the A.n.COB intron, and both encode maturases (Lazowska et al. 1989;Bassi et al. 2002; Y.K. Kwon and R.B. Waring, unpubl.). These two proteins are the only known close homologs of I-AniI, displaying 49% and 65% sequence identity, respectively (Fig. 6A). The Vina maturase is capable of facilitating the splicing of the A.n.COB intron, implying that the structural features of the proteins that are involved in RNA recognition and binding are similar (Y.K. Kwon and R.B. Waring, unpubl.).
Homology models of the yeast and Venturia maturase proteins were constructed and energy-minimized using the program SWISS-MODEL (Guex and Peitsch 1997). The ORFs display very high identity across their entire length and no heterogeneous insertions or deletions in their sequences (Fig. 6A), resulting in highly reliable models for comparative analyses. The three proteins harbor a large number of conserved, surface-exposed basic residues, as one would expect for proteins capable of binding multiple nucleic acid substrates. Many of these residues are involved in contacts to DNA bases and their corresponding phosphate backbone groups in I-AniI, as described above. In addition, two additional clusters of conserved basic residues are found at residues 39, 46, and 48 of the N-terminal domain of I-AniI, and residues 222, 227, 228, 236, and 239 of the C-terminal domain. These residues are distributed asymmetrically across the surface of I-AniI (Fig. 6B). The majority of the residues are clustered on one side and end of the protein. The surface area populated by these residues is -3000-4000 Å2, and is convex in shape. This surface is not obviously similar to RNA-binding surfaces of other proteins, but is large enough to form extensive interactions to a complex folded RNA structure. An additional three conserved basic surface residues (K183, R102, and 182) are found ∼20 Å distant. This region is on the opposite end of the protein surface, distal from the dense cluster noted above, and also far away from the DNA-binding surface.
Mutational uncoupling of DNA endonuclease and RNA maturase activities
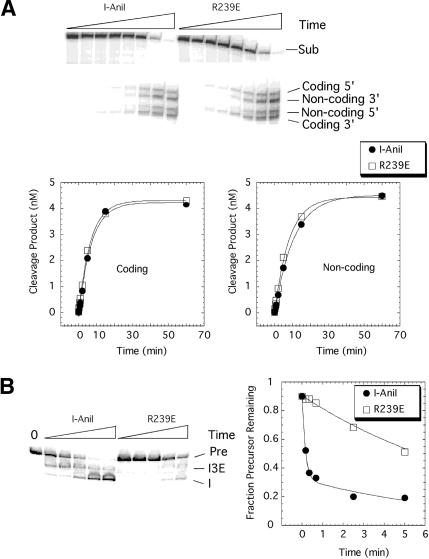
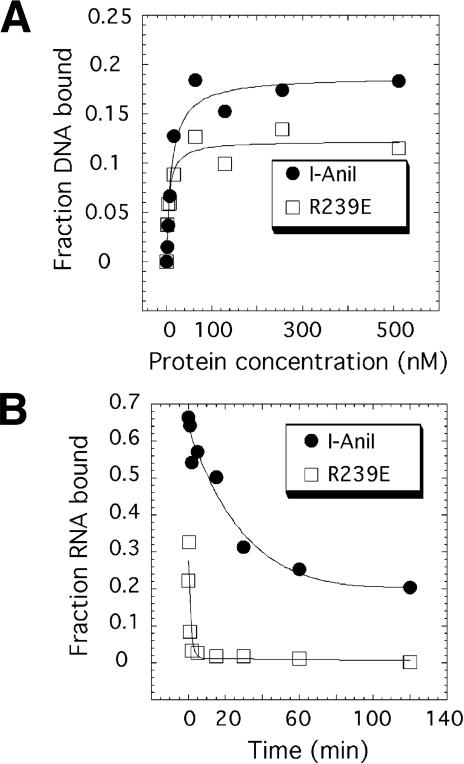
The middle residue of three closely packed, conserved basic residues mentioned above (K236, R239, and R243; Fig. 6B) in the C-terminal domain was chosen for point mutagenesis, to further test the possibility that the two nucleic-acid binding activities of the protein reside on different surfaces and might be uncoupled. An arginine to glutamate substitution was made at residue 239; the DNA endonuclease and RNA splicing and binding properties of the mutant were then evaluated. Under conditions where I-AniI is saturating, the R239E mutant cleaved the DNA target site at the same rate as the wild-type protein (Fig. 7A). Equilibrium binding experiments show that mutant protein binds the target site DNA with the same affinity as the wild-type protein (Fig. 8A). In contrast, under similar conditions, the rate of R239E-facilitated RNA splicing was 10-fold slower than for the wild-type maturase (Fig. 7B). The primary defect in RNA splicing by R239E appears to be caused by compromised binding affinity (Fig. 8B). Wild-type I-AniI binds the A. n.COB pre-RNA with picomolar affinity due to an extremely slow off-rate (∼0.02 min-1;Solem et al. 2002). The off-rate for the mutant protein is 35-fold faster than I-AniI, indicating that the R239E/A.n.COB pre-RNA complex is destabilized. Thus, a single amino acid substitution on a surface distinct from the DNA-binding face of I-AniI predominantly affects the intron RNA-binding and maturase function of the protein.
Figure 7.
Endonuclease and splicing activity of wild-type vs. R239E I-AniI. (A) Endonuclease assay. Double-stranded cleavage of an internally radiolabeled 200-bp DNA (3 nM) containing the I-AniI binding site was initiated by the addition of wild-type or mutant protein (200 nM), and aliquots of the reactions were stopped by organic extraction; the products were separated by gel electrophoresis (Chatterjee et al. 2003). Plots of the coding and noncoding strand cleavage are shown below. Two independent experiments gave average rates for coding strand cleavage of 0.13 min-1 (range 0.14-0.11 min-1) and 0.13 (range 0.15-0.11 min-1) for the wild-type and R239E, respectively. Likewise, average rates for noncoding strand cleavage were 0.09 (range 0.09-0.09 min-1) and 0.1 (range 0.12-0.08 min-1) for the wild-type and R239E. (Sub) Substrate. (B) Splicing assay. Splicing of radiolabeled COBme-pre-RNA (0.07 nM) was initiated by the addition of wild-type or mutant protein (500 nM), and aliquots of the reactions were stopped by organic extraction; the products were separated by gel electrophoresis (Solem et al. 2002). A plot of the data is shown to the right. Two independent experiments gave average splicing rates of 3 min-1 (range 2-4 min-1) and 0.15 (range 0.2-0.1 min-1) for the wild-type and R239E, respectively. (Pre) Precursor;(I3E) intron-3′exon intermediate;(I) free intron; (0) zero time point.
Figure 8.
DNA and RNA intron target-site binding of R239E. (A) Equilibrium binding for DNA target site. Trace amounts of radiolabeled DNA were incubated with a range of protein concentrations and filtered through nitrocellulose. Two independent experiments gave average equilibrium constants (Kd) of 11 nM (range 9-13 nM) and 7 nM (range 6-8 nM) for the wild-type and R239E, respectively. (B) Dissociation of protein/COBme-pre-RNA complexes. The rate of dissociation was measured by diluting the protein/radiolabeled RNA complexes into buffer containing an 800-molar excess of unlabeled COBme-pre-RNA, and aliquots were withdrawn and filtered through nitrocellulose at the times indicated. Two independent experiments gave average dissociation rates of 0.02 min-1 (range 0.01-0.02 min-1) and 0.7 min-1 (range 0.7-0.7 min-1) for the wild-type and R239E, respectively.
Discussion
DNA recognition and binding
LAGLIDADG homing endonucleases can be subdivided into several distinct, highly divergent subfamilies (Pietrokovski 1994;Belfort and Roberts 1997;Dalgaard et al. 1997;Turmel et al. 1997;Lucas et al. 2001). Structures of two members of a homodimeric subfamily (I-CreI and I-MsoI) have been determined in complex to a DNA target (Chevalier et al. 2003), as has the structure of a single domain from a monomeric enzyme (I-DmoI) in the context of an engineered artificial enzyme chimera (Chevalier et al. 2002), and a DNA-bound structure of an intein-associated endonuclease (PI-SceI;Moure et al. 2002). In all of these structures, four-stranded antiparallel β-sheets are used to form the interface and make contacts to each DNA target halfsite.
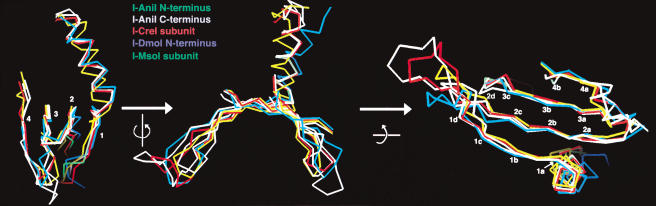
The use of the β-sheet binding motif by homing endonucleases appears to be more uniform and predictable than motifs and contacts formed by restriction enzymes in complex with DNA. A structural alignment of five individual domains and subunits from free-standing endonucleases in their DNA-bound conformations (single subunits from I-CreI and I-MsoI, the N- and C-terminal domains from I-AniI, and the I-DmoI N-terminal domain) indicates that the conformation of the central core of the β-sheets is well conserved (Fig. 9). At least 12 Cα positions within these β-sheets are in close juxtaposition. These residues span all four β-strands within the sheets and have a Cα rmsd of ∼0.7 Å. These positions correspond to residues that make contacts to base pairs ±1 to 6 in each DNA halfsite, which are typically recognized with higher specificity than bases at the more distant flanks of the DNA target. The conformations of the more distant ends of the β-strands and their connecting turns are much less well conserved, displaying rmsd values of >3 Å for DNA-contacting residues. Additional alignments of intein-associated endonuclease domains from PI-SceI indicate a less conserved structure of the β-sheet motifs, reflecting the divergence of intron- and intein-encoded homing endonucleases.
Figure 9.
Superposition of β-sheet DNA binding motifs for five independent LAGLIDADG domains and subunits, from their DNA-bound structures: subunits from I-CreI and I-MsoI, the N-terminal domain from I-DmoI, and the N- and C-terminal domains from I-AniI. Cα positions corresponding to residues side chains that interact with the DNA target, and that superimpose closely (less than 1 Å rmsd each) are labeled.
Approximately 25% of potential major-groove H-bond donor and acceptor atoms in the DNA target are in direct contact with protein side chains. This result was also observed in an initial low-resolution structure of the I-CreI/DNA complex. Subsequent high-resolution (1.9 Å) structures of the same complex and that of an isoschizomer (I-MsoI) demonstrated that an equivalent number of water-mediated contacts are also formed by those enzymes (Chevalier et al. 2003). These contacts appear to be nearly as important for sequence-specific DNA recognition as direct contacts. Assuming that I-AniI behaves in a similar manner, approximately one-half of the potential sequence-specific contact atoms across the target-site major groove are engaged by the protein, either directly or through water-mediated contacts. In addition, one would predict that base pairs -7 to -3 in the left halfsite would be most specifically defined across the targets that can be cleaved by I-AniI, followed closely by base pairs +3, +4, and +7 in the right halfsite.
Taken together, the DMS and hydroxyl radical cleavage patterns indicate that I-AniI does not contact nucleosides or bases within the 4-bp “window” defined by the two cleavage sites, but does contact the immediately flanking sequences. This is confirmed in the I-AniI/DNA cocrystal structure, and in cocrystal structures of related homing endonucleases. The DMS modification analysis (Fig. 5A) correlates well with the series of arginine-guanine contacts observed on the noncoding strand in the left halfsite (Fig. 3). The counterparts to those bases in the right halfsite on the coding strand also display sensitivity to DMS modification but are not in direct contact with protein side chains. One possible explanation for this result is that these bases make water-mediated contacts to the protein; these contacts are not explicitly modeled in our structure (because water assignments are not reliable at 2.6 Å resolution), but the closest residues are capable of participating in such contacts.
A recent study compared I-CreI/DNA contacts with the polymorphisms of alternate target-site variants recognized and cleaved by I-CreI (Chevalier et al. 2003). That study indicated that “information content” (a measure of specificity derived at each base pair;Schneider et al. 1986) is distributed nonuniformly across the site, and is correlated with total contacts made by the enzyme to each base pair. Conservatively, I-CreI recognizes 1 in 109 random sequences of length 22 bp, making it specific enough to act in a gene-specific manner. A comparison of the length and number of contacts in that structure to the I-AniI DNA complex indicates that the latter protein may be less specific (perhaps by an order of magnitude).
RNA recognition and binding
Group I introns catalyze their own posttranscriptional excision from precursor RNA. A variety of in vitro studies have demonstrated that many group I introns fold inefficiently (Treiber and Williamson 2001), and only self-splice under nonphysiological conditions such as elevated temperatures or very high concentrations of salt or divalent cations (Coetzee et al. 1994;Ho et al. 1997). Many if not all of these introns are associated with tightly bound proteins that function to promote a stable and active RNA conformation (Lambowitz and Perlman 1990;Lambowitz et al. 1999). The mechanisms by which these proteins recognize their cognate introns and induce a well defined RNA conformation are not well understood.
Group I intron splicing cofactors include various nuclear-encoded factors that often have an additional RNA-related activity, and maturase proteins such as I-AniI that are encoded within the introns themselves. Two nuclear-encoded group I intron splicing factors have been extensively characterized. The yeast CBP2 protein specifically promotes splicing of the fifth intron (bi5) in the mitochondrial cytochrome b (COB) gene (McGraw and Tzagoloff 1983). In contrast, the Neurospora mitochondrial tyrosyl tRNA synthetase (termed CYT-18) promotes splicing of a wide variety of group I introns (Mannella et al. 1979;Walweber et al. 1997). These two unrelated proteins use quite different mechanisms to bind their RNA targets and facilitate splicing. CBP2 promotes splicing by a “tertiary capture” mechanism, where an unstable but significantly folded RNA structure is captured and stabilized (McGraw and Tzagoloff 1983; Weeks and Cech 1995a,b, 1996). CYT-18 uses a different mechanism, where the protein binds to an early intermediate in the intron folding pathway (via a stable P4-P6 RNA subdomain) and then acts as a scaffold for the nucleation and assembly of the complete folded intron structure (Saldanha et al. 1995, 1996; Caprara et al. 1996a,1996b, 2001).
I-AniI binds its cognate intron with a 1:1 stoichiometry and forms a very stable ribonucleoprotein (RNP) complex facilitating correct folding of the intron catalytic core (Ho and Waring 1999;Solem et al. 2002). Association kinetics of protein/RNA complex formation and splicing revealed that I-AniI binding is a multistep process that includes an intermediate “encounter” complex that is relatively slowly resolved into a native, splicing competent complex (Solem et al. 2002). Deletion of sequences and secondary structure near the 5′ and 3′ splice sites have minimal effects on binding (Fig. 1B; Solem et al. 2002). These results suggest that I-AniI binds to regions of the A.n.COB pre-RNA that are far apart in primary sequence but brought together in close proximity in the folded core (Geese and Waring 2001; Solem et al. 2002).
The similarity between respective DNA binding sites and the RNA structure near the 5′ and 3′ splice sites (Fig. 1) suggests that the same protein surface, using analogous sets of interactions, is used to perform both functions. However, competition experiments indicate that I-AniI can bind either the wild-type A.n.COB intron RNA or a low-affinity mutant intron in the presence of saturating concentrations of its DNA target site substrate, suggesting that the protein might use different binding sites for DNA and RNA (Chatterjee et al. 2003; Geese et al. 2003). In addition, two different mutants of I-AniI (in which the conserved LAGLIDADG acidic residues were individually changed to alanine, inhibiting DNA binding and cleavage) have little effect on the protein's RNA maturation activity (Chatterjee et al. 2003). As RNA splicing is likely a secondary adaptation of the protein (and may play a role in “cementing” the ORF into the intron), it has been proposed that this and other homing endonucleases may have acquired maturase function by utilizing a previously “nonfunctional” protein surface (Chatterjee et al. 2003;Geese et al. 2003). Identifying mutations that affect only the RNA binding and splicing activity of a bifunctional maturase, such as R239E in I-AniI as described above, constitutes strong evidence for independent functional surfaces for these two activities.
Introns that require embedded maturase genes for splicing do not share obvious common signature sequences or strictly conserved features of unusual secondary structure, suggesting that individual maturases, similar to nuclear-encoded splicing factors, recognize a variety of diverse features within their RNA targets. In addition, at least one intron was recently shown to require both a nuclear-encoded protein and an intron-encoded, LAGLIDADG maturase to be spliced in vitro (Bassi et al. 2002). The extent to which individual maturase-intron pairs have evolved unique, widely diverse mechanisms of molecular recognition and RNA splicing enhancement is unknown, and awaits analyses of multiple model systems, particularly for introns with significantly different structural elaborations relative to A.n- COB.
In conclusion, the structure of the I-AniI/DNA complex and the associated studies described here provide insight into both its endonuclease and maturase functions, and a testable hypothesis regarding the identity of the protein surfaces and corresponding residues involved in binding the A.n.COB intron. The structure also provides details on the association, conformation, and contacts formed by an asymmetric homing endonuclease to its DNA target; these data are crucial for understanding the specificity and information content of homing endonuclease-DNA complexes and for future engineering of homing endonucleases. A future structural study of the RNP complex formed between I-AniI and its cognate intron should contribute greatly to our understanding of the evolution of RNA chaperone and folding cofactor activities from preexisting functional protein scaffolds, such as tRNA synthetases and homing endonucleases.
Materials and Methods
Crystallographic structure determination
The I-AniI reading frame, starting six residues upstream of the first LAGLIDADG motif, was subcloned as an N-terminally His-tagged fusion protein into a derivative pET vector, and this plasmid (pDN2) was used to transform Escherichia coli HMS 174(DE3) for wild-type protein expression. Five-milliliter overnight cultures were grown at 37°C from single colonies in SOC media in the presence of kanomycin. The following morning, liter flasks were inoculated with overnight cultures, and growth was allowed to continue to an O.D.600 of 0.7-0.9. Isopropyl-thio-β-D-galactosidase (IPTG) was added to 0.5 mM concentration, the temperature was reduced to 30°C, and growth continued for 18-20 h. Cells were harvested by centrifugation and frozen at -80°C for later use. For expression of selenomethionine-derivatized protein, I-AniI was expressed in M9 minimal media in the E. coli strain BL21(DE3) adapted for growth with methionine pathway inhibition (Doublie 1997). Cells were grown in minimal media at 37°C to an O.D.600 of 0.8, and the following amino acids were added to inhibit the methionine biosynthetic pathway: 100 mg/L lysine, threonine, and phenylalanine;75 mg/L selenomethionine;50 mg/L leucine, isoleucine, and valine. Following a 15-min incubation at 37°C, 0.5 mM IPTG was added to induce expression, and the cultures were grown at 30°C for 10-14 h.
For purification, the cells were lysed by sonication in 100 mM NaCl, 20 mM Tris at pH 7.9, 5 mM imidazole, and 100 μM PMSF. After clarification by centrifugation, the supernatant was mixed with nickel NTA resin (QIAGEN) at 4°C for 1 h. The protein-resin slurry was transferred to the column and washed with 30 mM imidazole, 500 mM KCl, 20 mM Tris at pH 7.9. The protein was then eluted with 100 mM imidazole, 50 mM KCl, 20 mM Tris at pH 7.9, pooled, dialyzed against a solution of 100 mM NaCl, 50 mM Tris at pH 7.9, 50 mM MgCl2, 50% glycerol, and concentrated to ∼10 mg/mL using an Amicon pressure cell filtration unit under nitrogen and a centricon spin filter unit for the final reduction in volume. The protein was exchanged into a final storage buffer of 100 mM NaCl, 50 mM Tris at pH 7.9, 50 mM MgCl2, 5% glycerol, and stored in aliquots at -80°C. The His-tag was not removed from the protein and is present in the crystallization construct; the residues of the tag are not visible in the final structure.
DNA oligonucleotides were purchased from Oligos Etc. (1 μmole synthesis scale, HPLC-purified research grade), suspended in 5 mM TE buffer. Complementary DNA strands were annealed by incubating for 5 min at 90°C and then allowed to cool to room temperature. The protein was diluted to 5 mg/mL concentration and mixed with a 1.2 molar excess of DNA duplex at room temperature and used directly for cocrystallization. Crystals were grown using two separate oligonucleotide duplex constructs; the construct used for the structure determination is shown in Figure 1. The crystals were grown at 22°C by vapor diffusion. A 500-μL reservoir containing 100 mM KCl, 20 mM MgCl2, 50 mM sodium citrate at pH 5.0 and 16%-22% PEG 3350 was equilibrated against a 2-μL drop containing a 1:1 mixture of the protein/DNA complex and the reservoir solution. Crystals grew as plates within 2 wks. The crystals are space group P21 and diffract to ∼2.6 Å resolution. Data and phasing statistics are provided in Table 1.
The structure of the I-AniI/DNA complex was solved by the single anomalous dispersion (SAD) phasing method using selenomethionyl protein. This phasing method proved superior to the MAD method, due to crystal decay during data collection. There are two methionine residues in the protein reading frame, in addition to the N-terminal residue which is disordered. Data were collected at the ALS synchrotron beamline 5.0.2 (Lawrence Berkeley National Laboratory) using a four-panel ADSC CCD area detector and a wavelength of 0.9794 Å, tuned to the absorption and fluorescence edge of the Se atoms. Data were collected to 2.6 Å resolution using an inverse phiflipping strategy with 30° data wedges; a full 360° of data was collected to maximize completeness and redundancy. Data were processed and scaled using the program HKL2000 (Otwinowski and Minor 1997). Subsequent data analysis was performed using the CNS package (Brunger et al. 1998). Anomalous difference Patterson maps contained exceptionally strong peaks and cross-peaks (exceeding 14 σ contour height) corresponding to the expected selenium positions. An interpretable electron density map was obtained after density modification with solvent flattening using a solvent content of 60%, and an initial model was built using the program XtalView (McRee 1999). Ten percent of the data was excluded from the refinement, beginning at the initial stages of model building, for the calculation of the cross-validating free R-factor (Brunger 1993). The model was refined with program CNS (Brunger et al. 1998), and the model geometry was checked with program PROCHECK (Laskowski et al. 1993). Refined model statistics are proved in Table 1. The structure has been deposited into the protein data bank (RCSB) with the accession code 1P8K.
The alignment of I-AniI with the Vina and Scer maturases (Fig. 6) was performed using CLUSTAL W (Thompson et al. 1994) and visualized using BOXSHADE, version 3.21 (K. Hofmann and M. Baron, public domain).
Nucleoside deletion analysis
To cleave the DNA backbone with hydroxyl radicals, hybridized DNAs were treated with 2 mM Fe(II), 2.5 mM EDTA at pH 8, 0.07% H2O2, and 6 mM Na-Ascorbate at pH 7 for 20 min at 37°C. The reaction was quenched by the addition of 10 mM thiourea. Target-site DNA (405 nM) was incubated with either 500 or 250 nM I-AniI in 50 mM Tris-HCl, 50 mM NaCl, and 10 mM CaCl2 for 10 min at 37°C. The mixture was filtered through nitrocellulose, and the bound DNA was recovered by organic extraction and ethanol precipitation. The cleaved products were separated by electrophoresis through a denaturing 15% polyacrylamide gel. The modification patterns were quantified with a PhosphorImager (Molecular Dynamics) and the data normalized. Modifications that reduced binding by 2.5-fold or more were considered inhibitory.
DMS modification/interference analysis
5′ 32P-labeled oligonucleotide (either the coding or noncoding strand) was hybridized with a twofold excess of unlabeled opposite-strand oligonucleotide by incubating the 100-μL reaction at 90°C for 2 min and slow cooling to 37°C over a 20-min period. The samples were treated with 10 μL of a 1/80 dilution of DMS in ethanol for 5 min at 37°C. The reactions were quenched by adding 2 mL of β-mercaptoethanol. Target-site DNA (1 μM) was incubated with either 0.75 or 0.375 μM I-AniI in 50 mM Tris-HCl, 50 mM NaCl, and 10 mM CaCl2 for 10 min at 37°C. The mixture was filtered through nitrocellulose, and the bound DNA was recovered by organic extraction and ethanol precipitation. To cleave at DMS-modified adenines and guanines, the DNA was dissolved in 10 mM phosphate buffer at pH 7.5 and incubated at 90°C for 15 min. NaOH was added to a final concentration of 100 mM, and the reaction was incubated at 90°C for 30 min, after which the reaction was neutralized by the addition of HCl to 100 mM. The reactions were ethanol-precipitated twice, and the products were separated by electrophoresis through a denaturing 15% polyacrylamide gel. The modification patterns were quantified with a phosphorimager (Molecular Dynamics) and the data normalized. Modifications that reduced binding by fivefold or more were considered inhibitory.
RNA splicing, DNA cleavage, and nucleic acid binding
The preparation of radiolabeled A.n.COB pre RNA and 200-bp target-site DNA as well as the kinetic and equilibrium analyses and data fits were carried out as described (Solem et al. 2002; Chatterjee et al. 2003).
Site-directed mutagenesis
The I-AniI R239E mutation was created using the Stratagene QuikChange® kit following the standard protocols supplied by the vendor. The low GC content of this region necessitated using long (49-base) oligonucleotide primers. The primer sequences used were 5′-CAATATTTATTATGGTTAAAACAGCTGGAGAAAA TATCTAGATATTCAG-3′ and its complement.
Acknowledgments
We thank Django Sussman for assistance with data collection and processing; Betty Shen for advice during structure determination and modeling; the staff of ALS beamline 5.0.2 for technical assistance; Yong Kwon for assistance with sequence alignments; and Meg Holmes, Roland Strong, and Adrian Ferre-D'Amare for advice and assistance. Funding for this project is from the NIH GM49857 (B.L.S. and J.B.), NIH GM62853 (M.G.C.) and MCB-0130991 (R.W.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1109003.
References
- Argast G.M., Stephens, K.M., Emond, M.J., and Monnat, R.J. 1998. I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 280: 345-353. [DOI] [PubMed] [Google Scholar]
- Bassi G.S., Oliveira, D.M.D., White, M.F., and Weeks, K.M. 2002. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc. Natl. Acad. Sci. 99: 128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M. and Perlman, P.S. 1995. Mechanisms of intron mobility. J. Biol. Chem. 270: 30237-30240. [DOI] [PubMed] [Google Scholar]
- Belfort M. and Roberts, R.J. 1997. Homing endonucleases—Keeping the house in order. Nucleic Acids Res. 25: 3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. 1993. Assessment of phase accuracy by cross validation: The free R value. Methods and Applications. Acta Cryst. D49: 24-36. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905-921. [DOI] [PubMed] [Google Scholar]
- Caprara M.G., Lehnert, V., Lambowitz, A., and Westhof, E. 1996a. A tyrosyl-tRNA synthetase recognizes a conserved tRNA-like structural motif in the group I intron catalytic core. Cell 87: 1135-1145. [DOI] [PubMed] [Google Scholar]
- Caprara M.G., G. Mohr, and A. Lambowitz. 1996b. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol. 257: 512-531. [DOI] [PubMed] [Google Scholar]
- Caprara M.G., Myers, C.A., and Lambowitz, A. 2001. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4-P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 308: 165-190. [DOI] [PubMed] [Google Scholar]
- Cassiday L.A. and Maher III, L.J. 2002. Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 30: 4118-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P., Brady, K.L., Solem, A., Ho, Y., and Caprara, M.G. 2003. Functionally distinct nucleic acid binding sites for a group I intron-encoded RNA maturase/DNA homing endonuclease. J. Mol. Biol. 329: 239-251. [DOI] [PubMed] [Google Scholar]
- Chevalier B.S. and Stoddard, B.L. 2001. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nuc. Acids. Res. 29: 3757-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B., Monnatt, R.J., and Stoddard, B.L. 2001a. The LA GLIDADG homing endonuclease I-CreI shares three divalent cations between two active sites. Nat. Struct. Biol. 8: 312-316. [DOI] [PubMed] [Google Scholar]
- Chevalier B.S., Monnat Jr., R.J., and Stoddard, B.L. 2001b. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 8: 312-316. [DOI] [PubMed] [Google Scholar]
- Chevalier B.S., Kortemme, T., Chadsey, M.S., Baker, D., Monnat, J.R.J., and Stoddard, B.L. 2002. Design, activity and structure of a highly specific artificial endonuclease. Mol. Cell 10: 895-905. [DOI] [PubMed] [Google Scholar]
- Chevalier B., Turmel, M., Lemieux, C., Monnat, R.J., and Stoddard, B.L. 2003. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 329: 253-269. [DOI] [PubMed] [Google Scholar]
- Coetzee T., Herschlag, D., and Belfort, M. 1994. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes & Dev. 8: 1575-1588. [DOI] [PubMed] [Google Scholar]
- Dalgaard J.Z., Klar, A.J., Moser, M.J., Holley, W.R., Chatterjee, A., and Mian, I.S. 1997. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 25: 4626-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. 2002. The PYMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- Doublie S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276: 523-530. [PubMed] [Google Scholar]
- Duan X., Gimble, F.S., and Quiocho, F.A. 1997. Crystal structure of PI-SceI, a homing endonuclease with protein splicing activity. Cell 89: 555-564. [DOI] [PubMed] [Google Scholar]
- Dujon B. 1989. Group I introns as mobile genetic elements: Facts and mechanistic speculations—A review. Gene 82: 91-114. [DOI] [PubMed] [Google Scholar]
- Evans S. 1993. SETOR: Hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11: 134-138. [DOI] [PubMed] [Google Scholar]
- Geese W.J. and Waring, R.B. 2001. A comprehensive characterization of a group IB intron and its encoded maturase reveals that protein-assisted splicing requires an almost intact intron RNA. J. Mol. Biol. 308: 609-622. [DOI] [PubMed] [Google Scholar]
- Geese W.J., Kwon, Y.K., Wen, X., and Waring, R.B. 2003. In vitro analysis of the relationship between endonuclease and maturase activities in the bifunctional group I intron-encoded protein, I-AniI. Eur. J. Biochem. 270: 1543-1554. [DOI] [PubMed] [Google Scholar]
- Guex N. and Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modelling. Electrophoresis 18: 2714-2723. [DOI] [PubMed] [Google Scholar]
- Hayes J.J. and Tullius, T.D. 1989. The missing nucleoside experiment: A new technique to study recognition of DNA by protein. Biochemistry 28: 9521-9527. [DOI] [PubMed] [Google Scholar]
- Heath P.J., Stephens, K.M., Monnat, R.J., and Stoddard, B.L. 1997. The structure of I-CreI, a group I intron-encoded homing endonuclease. Nat. Struct. Biol. 4: 468-476. [DOI] [PubMed] [Google Scholar]
- Ho Y. and Waring, R.B. 1999. The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J. Mol. Biol. 292: 987-1001. [DOI] [PubMed] [Google Scholar]
- Ho Y., Kim, S.J., and Waring, R.B. 1997. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease Proc. Natl. Acad. Sci. 94: 8994-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K., Ishino, Y., Ariyoshi, M., Komori, K., and Morikawa, K. 2000. Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 300: 889-901. [DOI] [PubMed] [Google Scholar]
- Jurica M.S. and Stoddard, B.L. 1999. Homing endonucleases: Structure, function and evolution. Cell Mol. Life Sci. 55: 1304-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Monnat Jr., R.J., and Stoddard, B.L. 1998. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell 2: 469-476. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M. and Perlman, P.S. 1990. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci. 15: 367-382. [DOI] [PubMed] [Google Scholar]
- Lambowitz A., Caprara, M.G., Zimmerly, S., and Perlman, P.S. 1999. Group I and group II ribozymes as RNPs: Clues to the past and guides to the future. In The RNA world II (eds. R.F. Gesteland, J.F. Atkins, and T.R. Cech), pp. 451-485. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Laskowski R.J., Macarthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereo-chemical quality of protein structures. J. Appl. Crystall. 26: 283-291. [Google Scholar]
- Lazowska J., Claisse, M., Gargouri, A., Kotylak, Z., Spyridakis, A., and Slonimski, P.P. 1989. Protein encoded by the third intron of cytochrome b gene in Saccharomyces cerevisiae is an mRNA maturase. Analysis of mitochondrial mutants, RNA transcripts proteins and evolutionary relationships. J. Mol. Biol. 205: 275-289. [DOI] [PubMed] [Google Scholar]
- Lucas P., Otis, C., Mercier, J.P., Turmel, M., and Lemieux, C. 2001. Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res 29: 960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C.A., Collins, R.A., Green, M.R., and Lambowitz, A.M. 1979. Defective splicing of mitochondrial rRNA in cytochrome-deficient nuclear mutants of Neurospora crassa. Proc. Natl. Acad. Sci. 76: 2635-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P. and Tzagoloff, A. 1983. Assembly of the mitochondrial membrane system. Characterization of a yest nuclear gene involved in the processing of the cytochrome b premRNA. J. Biol. Chem. 258: 9459-9468. [PubMed] [Google Scholar]
- McRee D.E. 1999. A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156-165. [DOI] [PubMed] [Google Scholar]
- Moure C.M., Gimble, F.S., and Quiocho, F.A. 2002. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol. 9: 764-770. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307-326. [DOI] [PubMed] [Google Scholar]
- Pietrokovski S. 1994. Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 3: 2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1998. Modular organization of inteins and C-terminal autocatalytic domains. Protein Sci. 63: 64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha R.J., Patel, S.S., Surendran, R., Lee, J.C., and Lambowitz, A.M. 1995. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry 34: 1275-1287. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Analysis of the CYT-18 protein binding site at the junction of stacked helices in a group I intron RNA by quantitative binding assays and in vitro selection. J. Mol. Biol. 261: 23-42. [DOI] [PubMed] [Google Scholar]
- Schneider T.D., Stormo, G.D., Gold, L., and Ehrenfeucht, A. 1986. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188: 415-431. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. and Gilbert, W. 1980. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl. Acad. Sci. 77: 122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva G.H., Dalgaard, J.Z., Belfort, M., and Roey, P.V. 1999. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 286: 1123-1136. [DOI] [PubMed] [Google Scholar]
- Solem A., Chatterjee, P., and Caprara, M.G. 2002. A novel mechanism for protein-assisted group I intron splicing. RNA 8: 412-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber D.K. and Williamson, J.R. 2001. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 11: 309-314. [DOI] [PubMed] [Google Scholar]
- Turmel M., Otis, C., Cote, V., and Lemieux, C. 1997. Evolutionarily conserved and functionally important residues in the I-CeuI homing endonuclease. Nucleic Acids Res. 25: 2610-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walweber G.J., Mohr, S., Rennard, R., Caprara, M.G., and Lambowitz, A.M. 1997. Characterization of neurospora mitochondrial group I introns reveals different CYT-18 dependent and independent splicing strategies and an alternative 3′ splicing site for an intron ORF. RNA 2: 114-131. [PMC free article] [PubMed] [Google Scholar]
- Weeks K.M. and Cech, T.R. 1995a. Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry 34: 7728-7738. [DOI] [PubMed] [Google Scholar]
- ____. 1995b. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell 82: 221-230. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science 271: 345-348. [DOI] [PubMed] [Google Scholar]
- Zheng D. and Koller, W. 1997. Characterization of the mitochondrial cytochrome b gene from Venturia inaequalis. Curr. Genet. 32: 361-366. [DOI] [PubMed] [Google Scholar]