Abstract
Kinetochores are multiprotein complexes that assemble on centromeric DNA and attach chromosomes to spindle microtubules. Over the past six years, the number of proteins known to localize to the Saccharomyces cerevisiae kinetochore has increased from around 10 to over 60. However, relatively little is known about the protein-protein interactions that mediate kinetochore assembly or about the overall structure of microtubule-attachment sites. Here we used biophysical techniques, affinity purification, mass spectrometry, and in vivo assays to examine the state of association of 31 centromere-binding proteins, including six proteins newly identified as kinetochore subunits. We found that yeast kinetochores resemble transcriptional enhancers in being composed of at least 17 discrete subcomplexes that assemble on DNA to form a very large structure with a mass in excess of 5 MD. Critical to kinetochore assembly are proteins that bridge subunits in direct contact with DNA and subunits bound to microtubules. We show that two newly identified kinetochore complexes, COMA (Ctf19p-Okp1p-Mcm21p-Ame1p) and MIND (Mtw1p including Nnf1p-Nsl1p-Dsn1p) function as bridges. COMA, MIND, and the previously described Ndc80 complex constitute three independent and essential platforms onto which outer kinetochore proteins assemble. In addition, we propose that the three complexes have different functions with respect to force generation and MT attachment.
Keywords: Kinetochore, chromosome segregation, mitosis, centromere, proteomics, hydrodynamics
Kinetochores perform three related and essential functions in the mechanical and error-checking processes necessary for accurate chromosome segregation. First, they form bridges between centromeric DNA (CEN DNA) and the plus-ends of spindle microtubules (MTs). Second, they maintain plus-end attachment during cycles of MT shrinkage and growth, thereby generating forces required for chromosome movement. Third, they act as sensors of MT attachment and regulate the onset of the metaphase-to-anaphase transition via the spindle-assembly checkpoint. In carrying out these functions, kinetochores must bind in a tight but flexible manner to polymers that are among the largest assemblies in the nucleus: MTs have a diameter of 250Å, and growing and shrinking MTs differ in helical pitch along the last 500Å of the polymer. Although kinetochores were discovered over 50 years ago (Luykx 1965 and references therein), we know remarkably little about their overall organization. An important goal of kinetochore research is to develop an accurate picture of MT attachment from a structural and functional perspective, and thereby uncover the mechanisms of force generation and checkpoint signaling.
The simplest known kinetochores are those found in budding yeast. However, even these kinetochores consist of at least 65 different protein subunits (McAinsh et al. 2003). The assembly of Saccharomyces cerevisiae kinetochores begins with the binding of the CBF3 complex onto the CDEIII element of CEN DNA. CBF3 is composed of four subunits: Ndc10p, Ctf13p, Cep3p, and Skp1p (Ng et al. 1986; Lechner and Carbon 1991; Goh and Kilmartin 1993; Strunnikov et al. 1995; Connelly and Hieter 1996), and is required for the recruitment of all known kinetochore proteins to CEN DNA in vivo. Mutations in CBF3 proteins result in a complete loss of chromosome-MT attachment in vivo and in vitro (He et al. 2001; Sorger et al. 1994; Goh and Kilmartin 1993). Initially, it was thought that CBF3 and an associated kinesin-related protein (KRP) might be sufficient for kinetochore-MT attachment (Hyman et al. 1992). However, recent studies have revealed that at least seven kinetochore proteins are likely to contact MTs, including Dam1p (a subunit of the DASH complex; Cheeseman et al. 2001; Janke et al. 2002; Li et al. 2002), Stu2p, Bik1p, Bim1p, and the KRPs Cin8p, Kip1p, and Kip3p (Enquist-Newman et al. 2001; He et al. 2001; Jones et al. 2001; McAinsh et al. 2003; J. Tytell and P. Sorger, unpubl.). As none of these MT-binding components seem to bind directly to CBF3, it is thought that other kinetochore subunits act as linkers, bridging CBF3 and MT-binding components (He et al. 2001; Cheeseman et al. 2002a; McAinsh et al. 2003). The current, albeit limited, understanding of how kinetochore proteins interact is based on co-immune precipitation experiments, high-throughput two-hybrid screens, and genetic interaction analysis (Ching et al. 2003; Ito et al. 2001; Uetz et al. 2000; for review, see Cheeseman et al. 2002a). The best-characterized linker component is the conserved Ndc80 complex, which is required for MT-kinetochore attachment and checkpoint signaling (He et al. 2001; Janke et al. 2001; McCleland et al. 2003; Wigge and Kilmartin 2001). The Dam1p-containing DASH complex, Stu2p, Cin8p, and Kip1p require the Ndc80 complex, which, in turn, requires CBF3 for association with the centromere (He et al. 2001; Janke et al. 2001, 2002; Wigge and Kilmartin 2001; J. Tytell and P. Sorger, unpubl.). One can therefore imagine the kinetochore as comprising three layers that associate with CEN DNA in a hierarchical fashion: a DNA-binding layer, a linker layer, and an MT-binding layer. The assignment of proteins to the “linker” remains imprecise, and we cannot exclude the possibility that so-called linkers actually bind to DNA or MTs in collaboration with other kinetochore proteins.
To understand the overall organization of the S. cerevisiae kinetochore, the protein-protein and protein-DNA interactions necessary for assembling an MT-attachment site must be identified. As a first step, we need a list of key kinetochore proteins and knowledge of which ones bind to each other in vitro and in vivo. We can imagine three possibilities: (1) that kinetochores can, like ribosomes, be isolated more or less intact as very large complexes, (2) that kinetochore proteins, like components of transcriptional enhancers, are present in subcomplexes substantially smaller than intact kinetochores but typically containing more than one subunit, (3) that the majority of kinetochore proteins are monomeric prior to their assembly on CEN DNA. The available data are ambiguous with respect to these three possibilities. On the one hand, it has been shown that Ndc80p is a component of a kinetochore subcomplex containing only four proteins (Janke et al. 2001; Wigge and Kilmartin 2001), but it has also been suggested that Ctf19p is part of a very large complex that might comprise much of the kinetochore (Cheeseman et al. 2002b). In the present study we analyzed 19 known kinetochore proteins using biophysical methods, and we established that they lie in 14 discrete multiprotein complexes. Several of these complexes have unknown subunits, which we identified by affinity purification and mass spectrometry. Four of the newly identified kinetochore proteins are components of two essential complexes: the COMA complex (Ctf19p-Okp1p-Mcm21p-Ame1p) and the MIND complex (Mtw1p including Nnf1p-Nsl1p-Dsn1p). The COMA and MIND complexes, and the previously characterized Ndc80 complex, assemble independently of each other onto CEN DNA. They appear to form a linker layer that recruits multiple MT-binding components.
Results
Hydrodynamic analysis of kinetochore proteins
To determine the extent to which a set of 19 known S. cerevisiae kinetochore subunits are associated with each other and with unidentified proteins, endogenous genes were replaced with HA, FLAG, or protein-A (ProA) C-terminal fusions (Supplemental Table 1). Tagged proteins were checked for function and extracted from cells under conditions previously shown to dissociate CBF3 from CEN DNA (Sorger et al. 1994). Whole-cell extracts were then analyzed, using quantitative Western blotting, on calibrated velocity gradients and size-exclusion chromatography columns (Fig. 1). For each protein, the combined hydrodynamic analysis yielded the diffusion constant and Svedberg sedimentation coefficient, which were converted into an estimate of native molecular mass of the complex, Stokes radius, frictional coefficient, and axial ratio (the degree of elongation). Among the 19 proteins analyzed, only Mif2p had a native molecular weight consistent with a monomer (Fig. 1A, B; Table 1, line 1). The other tagged proteins were present in species with molecular weights ranging from 89 kD to 440 kD. Because most of these species were elongated, estimates of molecular mass derived from a single technique are inaccurate, emphasizing the importance of combining analysis on gradients and columns (Table 1). In a few cases, data already in the literature suggest a likely composition for these subcomplexes, whereas in other cases, the patterns of co-elution and measured molecular mass suggest the presence of unidentified subunits (Fig. 1; Table 1).
Figure 1.
Hydrodynamic analysis of kinetochore proteins. Whole-cell extracts were prepared from strains expressing either Slk19p-TAP or Stu2p-TAP (A,B), Ndc80p-6HA, Nuf2p-6HA, Spc24p-3HA or Spc25p-3HA, Dam1p-3Myc and Spc19p-ProA, Dam1p-3Myc and Spc34p-ProA (C,D), Mcm21p-TAP, Ctf19p-TAP, Okp1p-TAP, or Cse4p-TAP (E,F), and Mtw1p-ProA (G,H). Equal amounts of each extract were analyzed on a HiPrep 16/60 Sephacryl S-500 HR size-exclusion chromatography column and 5%-40% glycerol-density gradients. Fractions were immunoblotted with antisera that recognize the HA, Myc, TAP, or ProA epitope tags. Mif2p (A,B) and Ndc10p and Cep3p (E,F) were detected with polyclonal antibodies raised against recombinant protein.
Table 1.
Hydrodynamic properties of some S. cerevisiae kinetochore subcomplexes
| Analyzed protein | Subcomplex | Diffusion coefficient (10−7 cm2 s−1) | Svedberg coefficient (10−13 s) | Measured mass (kD) | Presumed composition1 and mass (kD)2 of the subcomplex | Stokes radius (Å) | Friction coefficient f/fo | Axial ratio (a:b) | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Mif2p | Mif2p | 10.2 ± 2.4 | 3.2 ± 0.2 | 36 ± 16 | Mif2p monomer (62 kD) | 24 ± 5 | 1.1 ± 0.5 | 1.6 ± 0.5 |
| 2 | Ndc80p-6HA | Ndc80 complex | 2.4 ± 0.2 | 5.6 ± 0.4 | 196 ± 11 | Ndc80p-6HA, Nuf2p, Spc24p, and Spc25 (188 kD) | 87 ± 3 | 2.2 ± 0.1 | 19 ± 1 |
| 3 | Dam1p-ProA | DASH complex | 3.0 ± 0.2 | 7.7 ± 0.5 | 216 ± 13 | Dam1p-ProA, Duo1p, Spc19p, Spc34p, Dad1p, Dad2p, Dad3p, Dad4p, Hsk3p, and Ask1p (218 kD) | 70 ± 4 | 1.75 ± 0.5 | 10 ± 1 |
| 4 | Slk19p-TAP | Slk19p | 1.8 ± 0.1 | 9.3 ± 0.8 | 434 ± 21 | Slk19p-TAP tetramer (460 kD) | 108 ± 5 | 1.85 ± 0.3 | 16 ± 1 |
| 5 | Stu2p-TAP | Stu2p | 2.4 ± 0.2 | 6.0 ± 0.4 | 210 ± 12 | Stu2p-TAP dimer (230 kD) | 85 ± 4 | 2.1 ± 0.1 | 17 ± 1 |
| 6 | Amelp-TAP | COMA complex | C1) 3.5 ± 0.3 | C1) 7.3 ± 0.3 | C1) 183 ± 18 | Ctf19p, Amelp-TAP, Mcm21p, and Okplp (185 kD) | C1) 62 ± 4 | C1) 1.6 ± 0.1 | C1) 9 ± 0.1 |
| C2) 4.1 ± 0.5 | C2) 4.2 ± 0.5 | C2) 94 ± 8 | C2) 44 ± 4 | C2) 1.4 ± 0.2 | C2) 5 ± 1.5 | ||||
| 7 | Cep3p | CBF3 complex | 6.1 ± 0.4 | 8.4 ± 0.5 | 124 ± 22 | Cep3p dimer (126 kD) | 37 ± 3 | 1.1 ± 0.5 | 1.5 ± 0.5 |
| Ndc10p | 2.6 ± 0.2 | 7.5 ± 0.4 | 232 ± 16 | Ndc10p dier (224 kD) | 82 ± 3 | 2 ± 0.5 | 14 ± 1 | ||
| 8 | Mtw1p-ProA | MIND complex | M1) 2.8 ± 0.2 | M1) 7.3 ± 0.5 | M1) 215 ± 10 | Mtw1p-ProA, Nnf1p, Nsl1p, Dsn1p (162 kD) | M1) 88 ± 4 | M1) 2.1 ± 0.2 | M1) 18 ± 1 |
| M2) 3.2 ± 0.2 | M2) 5.6 ± 0.4 | M2) 153 ± 18 | M2) 66 ± 4 | M2) 1.6 ± 0.2 | M2) 12 ± 1 | ||||
| M3) 4.5 ± 0.5 | M3) 3.7 ± 0.3 | M3) 89 ± 7 | M3) 49 ± 5 | M3) 1.7 ± 0.1 | M3) 8 ± 2 | ||||
| 9 | Nkplp-ProA | Nkp1 complex | 1) 4.1 ± 0.4 | N.D.3 | N.D. | Nkp1p-ProA and proteins to be determined (N.D.) | 1) 54 ± 5 | N.D. | N.D. |
| 2) 5.8 ± 0.9 | 2) 39 ± 5 | ||||||||
| 10 | Mcm22p-ProA | Mcm22 complex | 1) 2.2 ± 0.1 | N.D. | N.D. | Mcm22p-ProA, Ctf3p, and and Mcm16p (N.D.) | 1) 92 ± 3 | ||
| 2) 3.0 ± 0.2 | 2) 70 ± 4 | ||||||||
| 3) 3.8 ± 0.4 | 3) 58 ± 4 | ||||||||
| 11 | Bovine catalase | Bovine catalase4 | 5.1 ± 0.5 | 10.4 ± 0.5 | 225 ± 15 | Catalase (4 × 58 KD) | 55 ± 5 | 1.2 ± 0.2 | 2.1 ± 0.3 |
For references, see text.
Sum of the masses of the known subcomplex components.
N.D.: not determined.
Included as a control (Horiike et al. 1983).
Ndc80 complex Ndc80p, Nuf2p, Spc24p, and Spc25p, which have been proposed to associate in solution (Janke et al. 2001; Wigge and Kilmartin 2001) comigrated as a single peak on gradients and gel filtration columns (Fig. 1C, D). The measured molecular mass of this peak, 196 kD (Table 1, line 2) was consistent with the predicted heterotetrameric complex containing one copy of each of the Ndc80p, Nuf2p, Spc24p, and Spc25p proteins (188 kD; Fig. 1C, D; Table 1, line 2).
DASH complex Dam1p, Spc19p, and Spc34p are three proteins whose co-elution and native molecular mass (∼216 kD) are consistent with previously published data (Cheeseman et al. 2001, 2002b; Janke et al. 2002; Li et al. 2002). These three proteins are subunits of a 10-protein DASH complex, which, in our hands, eluted as a single peak that was distinct from the peak of the Ndc80 complex. Thus the Ndc80 and DASH complexes exist as distinct multiprotein assemblies (Fig. 1C, D; Table 1, lines 2,3).
Slk19p Slk19p had a native mass consistent with that of a homotetramer, and affinity purification followed by SDS-PAGE failed to reveal any tightly associated proteins (Fig. 1A, B; Table 1, line 4; data not shown).
Stu2p Stu2p-TAP (121 kD) had a native mass (210 kD) consistent with that of a homodimer (Fig. 1A, B; Table 1, line 5), supporting the recent finding by van Breugel et al. (2003) using recombinant protein.
Ctf19p-Okp1p-Mcm21p complex Ctf19p was present in two peaks with native masses of ∼94 kD and ∼183kD (Fig. 1E; Table 1, line 6). Both of these species were larger than expected for monomeric Ctf19p, consistent with previous data that Ctf19p associates, by two-hybrid and immune precipitation assays, with kinetochore proteins Okp1p and Mcm21p, as well as with the centromeric histone Cse4p, and subunits of the CBF3 complex (Ortiz et al. 1999). This finding is interesting, as no other kinetochore proteins have been shown to bind to CBF3, even though the assembly of the kinetochore requires the binding of CBF3 to CEN DNA. When Okp1p and Mcm21p were tagged, both comigrated with Ctf19p on gradients and on columns. However, none of these three proteins comigrated with Ndc10p or Cep3p-Ctf13p-Skp1p, the primary subassemblies of CBF3 in the absence of CEN DNA (Fig. 1E, F; Table 1, line 7), or with Cse4p (Fig. 1E, F). Thus, although Ctf19p is clearly part of one or more multiprotein complexes, it is stably associated with only a subset of the binding partners identified by two-hybrid and co-immune precipitation studies. Moreover, the predicted mass of a Ctf19p-TAP-Okp1p-Mcm21p heterotrimer (147 kD) is not similar to the measured masses for either of the peaks that contain all three proteins. Thus, additional unidentified proteins are likely to be associated with Ctf19p.
Mtw1p complex Mtw1p was present in multiple peaks of 89, 153, and 215 kD that did not co-elute on either gradients or columns with any of the other 19 proteins in our study (Fig. 1G, H; Table 1, line 8).
Purification of proteins associated with Ctf19p
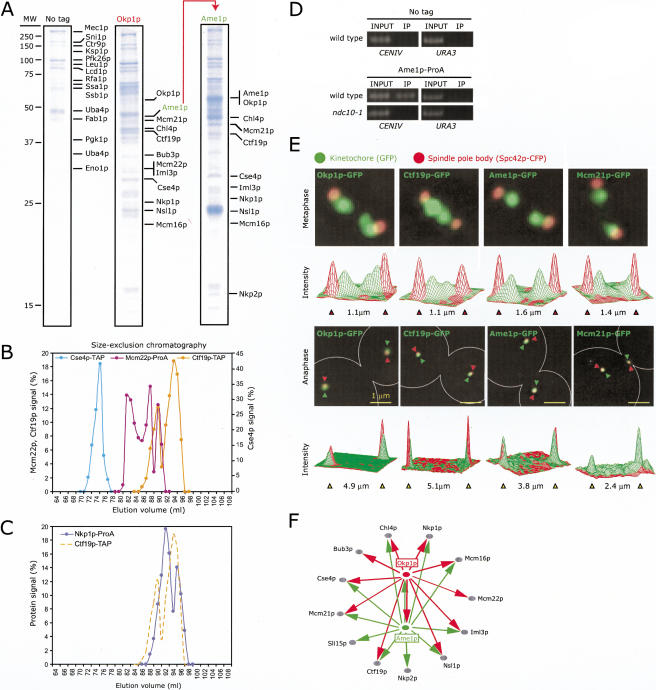
Of the 19 proteins we examined, the Ctf19p-Okp1p-Mcm21p and Mtw1p complexes stood out as poorly understood in terms of structure and function. We therefore sought to define them better using affinity purification and mass spectrometry, methods that have proven effective in the analysis of proteins in the DNA damage signaling and repair pathways (Ho et al. 2002). In our initial experiments, we tagged Okp1p because it is easier to be sure that an essential protein is functional after tagging. Whole-cell extracts were prepared from asynchronous cells carrying Okp1p-3FLAG and complexes isolated by immune precipitation using anti-FLAG beads followed by elution with FLAG peptide. Eluates were analyzed on SDS-PAGE and sequenced using microcapillary liquid chromatography-tandem mass spectrometry (LC-MS/MS). The untagged parent strain was used as a specificity control for immune purification. A total of 29 proteins were represented by multiple peptides in the mass spectrometry of Okp1p-3FLAG immune purifications, had sequence coverage above 10%, and were strongly enriched relative to control purifications (Fig. 2A, F; Supplemental Table 2). The protein that was most enriched was Ame1p, an essential protein of unknown function reported to localize to MTs and SPBs (http://www.yeastgenome.org/). Eleven other copurified proteins were known or suspected kinetochore subunits (Fig. 2A, F; Supplemental Table 2), and the remainder were proteins frequently found as contaminants in single-step affinity purifications from yeast (Ho et al. 2002; Supplemental Materials and Methods). Of the co-enriched kinetochore proteins, Chl4p, Mcm22p, Mcm16p, and Iml3p are thought to be components of a conserved but nonessential complex involved in the assembly or modification of centromeric heterochromatin (Takahashi et al. 2000; Measday et al. 2002; Pot et al. 2003), Cse4p is a specialized histone H3 protein localized to kinetochores (Meluh et al. 1998), Bub3p is a spindle assembly checkpoint protein, and Nkp1p is an uncharacterized kinetochore protein (Cheeseman et al. 2002b). However, neither Mcm22p, Cse4p, nor Nkp1p co-eluted with Ctf19p on gradients and columns (Figs. 1E, 2B, C), indicating that none are stably associated with Ctf19p. Our attention therefore shifted to the possibility that Ame1p might be a kinetochore protein bound to Okp1p.
Figure 2.
Purification and identification of kinetochore proteins associated with Okp1p and Ame1p. (A) Left lane shows a colloidal blue-stained SDS PAGE gel of a mock immune purification from a wild-type strain (no tag). The labeled proteins are commonly found as contaminants in single-step affinity purifications from yeast. The center and right lanes are purifications from the Okp1p-3FLAG and Ame1p-3FLAG strains. Proteins that were highly enriched and absent in the control are labeled. The positions of some proteins differ from the expected molecular weights (Suppl. Table 2) as a consequence of protein degradation. (B) Hydrodynamic analysis of Ctf19p-TAP on size-exclusion chromatography compared to Cse4p-TAP, Mcm22p-ProA, and (C) Nkp1p-ProA. (D) Crosslinking of Ame1p-ProA to CENIV DNA in wild-type or ndc10-1 cells at 37°C using ChIP. DNA in immune complexes (IP) was amplified with primers to CENIV, or as negative controls, TELVI-R and URA3, and compared to DNA in whole-cell extracts (labeled “1% INPUT”). (E) Images of metaphase (upper panels) and anaphase (lower panels) cells carrying Spc42p-CFP (red) and Okp1p-GFP, Ctf19p-GFP, Ame1p-GFP, or Mcm21p-GFP (green). Images represent projections of 3D image stacks containing 10 to 20 0.2-μm sections and collected on a DeltaVision deconvolution microscope. The graphs show the intensity of the CFP (red) and GFP (green) signals along the axis of the spindle in arbitrary units. (F) Network summarizing interactions found among proteins present in Okp1p and Ame1p immune purifications (see Suppl. Table 2 for further information).
To determine whether Ame1p associates with CEN DNA in vivo, we compared the extent of crosslinking to CENIV and control telomere-proximal and URA3 sequences using chromatin immunoprecipitation (ChIP). We found that Ame1p-ProA was crosslinked specifically to centromeres in an Ndc10p-dependent manner (Fig. 2D; data not shown). As an independent assay for kinetochore association, we localized Ame1p by tagging it with GFP in a strain that carries a fusion between Spc42p and the CFP (cyan fluorescent protein) to mark the locations of SPBs. In metaphase cells, Ame1p localized to two lobes that lay along the spindle axis but were internal to the SPBs (Fig. 2E, upper panel). This bilobed pattern is typical of kinetochore proteins during metaphase and arises from a superposition of 16 pairs of sister kinetochores attached to spindle MTs (He et al. 2000). Kinetochore proteins have previously been found on other MT-based structures, such as spindle MTs, SPBs, astral MTs, and cortical tips (He et al. 2000). However, in our images, Ame1p-GFP was not present at a detectable level at any of these locations, or at any other obvious nuclear structure (Fig. 1E, lower panel; data not shown). This suggests that Ame1p is associated primarily, if not exclusively, with kinetochores. This also appears to be true of Okp1p-GFP, Ctf19p-GFP, and Mcm21p-GFP (Fig. 2E).
Having established that Ame1p is a kinetochore protein, we attempted to identify its binding partners by immune purification followed by LC-MS/MS. The set of 12 proteins associated with Ame1p-3FLAG was highly enriched in known or suspected kinetochore subunits (Fig.2 A,F; Supplemental Table 2). Ctf19p, Okp1p, and Mcm21p were all present, consistent with the idea that all four proteins might be components of a common complex. However, the total mass of the 12 Ame1p-associated proteins (∼500 kD) was much larger than the measured native mass of even the largest Okp1p- and Ame1p-containing complexes (∼183kD). In repeat purifications, some proteins were always present, whereas others came and went (data not shown). Thus, we conclude that immune purification and LC-MS/MS of Okp1p and Ame1p are effective at identifying sets of interacting proteins but not at revealing which proteins are actually bound to each other in stable kinetochore subcomplexes (Fig. 2F).
Ctf19p, Okp1p, Mcm21p, and Ame1p are components of the COMA complex
To define the composition of Okp1p- and Ame1p-containing complexes, we turned to hydrodynamic analysis of wild-type and mutant extracts. When Ctf19p, Okp1p, Mcm21p, and Ame1p were analyzed in parallel, all four proteins co-eluted on gradients and columns as two distinct species: C1 of ∼183kD, and a C2 species of ∼94 kD (Fig. 3A,C; Table 1, line 6). The measured native mass of the C1 species was similar to the predicted mass of a Ctf19p-Okp1p-Mcm21p-Ame1p heterotetramer containing one copy of each protein (Fig. 3B). The composition of the lighter C2 complex was compatible with almost any homo- or heterodimer containing Ctf19p, Okp1p, Mcm21, or Ame1p, as all four proteins have molecular weights in the 40-50-kD range (Fig. 3A). When extracts were prepared from Ame1p-TAP okp1-5 cells grown at 24°C, both the C1 and C2 complexes were present on glycerol gradients. However, C1 and C2 were absent in extracts from cells grown at 37°C for 4 h, which arrests cells with a large budded morphology (Fig. 3C; Ortiz et al. 1999). Moreover, Western blotting showed that Ame1p-TAP was present in okp1-5 cells grown at 24°C but was undetectable in 37°C extracts. In contrast, α-tubulin and Nkp1p (a kinetochore protein that does not co-elute with Ame1p; Fig. 2C), were present at similar levels in both 24°C and 37°C extracts (Fig. 3D). Thus, the inactivation of Okp1p results in the loss of detectable Ame1p protein from cell extracts (Fig. 3D). However, as few as one copy of a protein per kinetochore (32 per cell) is potentially sufficient for function, and we have observed that reductions in protein levels seen in Western blots do not necessarily result in significant changes in the amount of centromere-bound protein (Table 2). We therefore examined Ame1p-GFP in okp1-5 cells at 24°C and 37°C by imaging and found that Ame1p-GFP was not detectable at kinetochores at 37°C (Fig. 3E). We therefore conclude that Ame1p is not expressed, or is unstable, in cells in which Okp1p has been inactivated. The same was true for TAP-tagged Mcm21p and Ctf19p, both of which were undetectable on Western blots of extracts prepared from okp1-5 cells shifted to 37°C (Fig. 3D). As a specificity control, we showed that the C1 and C2 complexes were present in extracts from mtw1-1 cells grown at 24°C or 37°C (Fig. 3F). Mtw1p is an essential kinetochore protein that is not associated with Ame1p by hydrodynamic analysis (Figs. 1G,H, 3A; Table 1, lines 6,8). Thus, Ame1p, Mcm21p, and Ctf19p are destabilized when their binding partner, Okp1p, is inactivated, but are largely unaffected by the inactivation of unrelated kinetochore proteins. This finding is consistent with data from previous studies showing that subunits of multiprotein complexes are more stable and soluble when assembled into complexes than when present on their own (Sterner et al. 1999; Bell and Dutta 2002).
Figure 3.
Ctf19p, Okp1p, Mcm21p, and Ame1p are components of the COMA complex. (A) Co-elution of Ctf19p-TAP, Okp1p-TAP, Mcm21p-TAP, and Ame1p-TAP by high-resolution size-exclusion chromatography. (B) Model for the predicted organization of Ame1p, Ctf19p, Mcm21p, and Okp1p subunits in the C1 and C2 COMA complexes based on the MW of the proteins: Ctf19p, 42 kD; Okp1p, 47 kD; Mcm21p, 37 kD; Ame1p, 37 kD; TAP tag, 20 kD. (C) Glycerol-density gradient analysis of Ame1p-TAP in okp1-5 cells at the permissive (24°C) and restrictive (37°C) temperatures. (D) Immunoblots using anti-ProA antibodies to detect the Ctf19p-ProA, Mcm21p-ProA, and Ame1p-TAP proteins in okp1-5 cells grown at 24°C or 37°C. The same extracts were probed with an anti-tubulin antibody as a loading control. The Nkp1p-ProA is included as a negative control. (E) Localization of Ame1p-GFP (green) in okp1-5 cells at 24°C and 37°C. (F) Glycerol-density gradient analysis of Ame1p-TAP in mtw1-1 cells grown at 24°C or 37°C and of Ame1p-TAP in (G) wild-type, ctf19Δ, mcm21Δ, and (H) mcm21Δctf19Δ cells. (I) Glycerol-density gradient analysis of Ame1p-TAP in cycling and α-factor arrested cells. (J) Localization of Ame1p-GFP (green) in wild-type, ctf19Δ, mcm21Δ, and mcm21Δctf19Δ cells.
Table 2.
Summary of kinetochore subcomplex interdependencies
| Protein | Genetic background | ChIP | Imaging | Western blot | Glycerol-gradient ultracentrifugation |
|---|---|---|---|---|---|
| Ame1p | wild-type | +++ | +++ | +++ | C1 and C2 |
| COMA complexes | |||||
| ndc10-1 | — | N.D.1 | + | N.D. | |
| ndc80-1 | +++ | ++ | + | N.D. | |
| mtw1-1 | +++ | ++ | + | C1 and C2 | |
| COMA complexes | |||||
| okp1-5 | — | — | — | — | |
| Mtw1p2 | wild-type | +++ | +++ | +++ | M1, M2, and M3 |
| ndc80-1 | +++ | ++ | N.D. | N.D. | |
| okp1-5 | +++ | + | ++/+++ | M1, M2, and M3 | |
| MIND complexes | |||||
| nnf1-17 | — | N.D. | — | — | |
| spc25-7 | +++ | N.D. | ++ | N.D. | |
| Ndc80p | wild-type | +++ | +++ | +++ | Ndc80 complex |
| mtw1-1 | +++ | ++ | ++ | N.D. | |
| okp1-5 | +++ | ++ | N.D. | N.D. |
N.D.: not determined.
Mtw1p-Myc protein levels are unaffected in ndc10-2 cells grown at 37°C (Goshima and Yanagida 2000).
To further define the composition of the C1 and C2 complexes, we deleted the nonessential genes CTF19 and MCM21, individually or in combination, in Ame1p-TAP cells. Neither deletion altered the overall level of Ame1p, but both caused the larger C1 complex to shift to a smaller size without significantly changing the size of the C2 complex (Fig. 3G). When both MCM21 and CTF19 were deleted, only a single complex in the location of C1 was observed (Fig. 3H). Taken together, these data indicate that C1 is a heterotetramer containing Ame1p, Okp1p, Mcm21p, and Ctf19p in one copy each. We will refer henceforth to the heterotetrameric Ctf19p-Okp1p-Mcm21p-Ame1p assembly as the COMA complex. We conclude that the other proteins found in Okp1p and Ame1p immune complexes are not core COMA components but instead are weakly or transiently bound. Because Ame1p is lost from the C2 complex in okp1-5 cells (no Ame1p is present in these cells when shifted to 37°C), but the mass of C2 does not change upon CTF19 or MCM21 deletion, we believe that the C2 peak corresponds to two different species: a heterodimer of Okp1p-Ame1p and a heterodimer of Mcm21p-Ctf19p (both with masses of ∼100-105 kD).
What is the functional relationship between the C1 and C2 COMA complexes? Only the C2 complex was seen when Ame1p-TAP was analyzed in extracts from cells arrested at START using α-factor (Fig. 3I). In addition, Western blots show that Ame1p-TAP and Okp1p-TAP are expressed at similar relatively high levels, whereas Mcm21p-TAP and Ctf19p-TAP are expressed at much lower levels (data not shown). Thus Ctf19p and Mcm21p may be auxiliary subunits of the COMA complex that play a nonessential but cell cycle-regulated role in COMA assembly. Neither Ctf19p nor Mcm21p appeared to be necessary for the binding of COMA to kinetochores, because the levels of Ame1p-GFP on kinetochores were similar in wild-type and MCM21Δ CTF19Δ cells (Fig. 3J).
Purification of proteins associated with Mtw1p
Next, we investigated the proteins associated with Mtw1p. Identifying Mtw1p interactors is of considerable interest, because Mtw1p is essential for chromosome segregation in S. cerevisiae, S. pombe, and human cells (Goshima et al. 1999, 2003; Goshima and Yanagida 2000). Mtw1p was tagged at its C-terminus with a 3FLAG epitope, isolated from extracts by immune precipitation, and associated proteins were identified by LCMS/MS. Fifteen kinetochore proteins were associated with Mtw1p, including components of the Ndc80 and COMA complexes (Fig. 4A; solid arrows in Fig. 4D; Supplemental Table 2). In repeat purifications, the spectrum of associated proteins was somewhat different, and also included components of the DASH complex (Spc19p), the CBF3 complex (Cep3p, Ctf13p), and Sli15 complex (Ipl1p, Sli15p; dashed arrows in Fig. 4D; Supplemental Table 2). However, four proteins were consistently present that had not previously been shown to bind to kinetochores: Nnf1p, Dsn1p, Nsl1p, and Spc105p. All four proteins could be crosslinked specifically to CEN DNA, but not to telomere or URA3 DNA by ChIP (Fig. 4B; data not shown). When tagged with GFP, all proteins exhibited a bilobed localization typical of kinetochores (Fig. 4C). Like Spc24p, Spc25p, Spc19p, and Spc34p, Spc105p was originally thought to be an SPB component (Wigge et al. 1998), but can now be assigned to kinetochores. Nnf1p, Dsn1p, and Nsl1p were previously shown to interact with each other and with Mtw1p in synthetic lethal, dosage suppression, and two-hybrid assays (Euskirchen 2002). Other than this, however, little is known about their function.
Figure 4.
Purification and identification of kinetochore proteins associated with Mtw1p. (A) Purification of Mtw1p-3FLAG, Nnf1p-3FLAG, and Mcm21p-3FLAG was carried out as described in the Figure 2A legend. (B) ChIP analysis and (C) localization of Nnf1p-GFP, Dsn1p-GFP, Nsl1p-GFP, Spc105p-GFP, and Cnn1p-GFP were as carried out as described in the Figure 2D,E legend. The expression levels of Cnn1p were very low, so we could not use Spc42p-CFP due to bleed-through of the CFP signal into the GFP channel. (D) Networks summarizing the interactions found among Mtw1p, Nnf1p, Mcm21p, and other kinetochore components. The solid lines depict the interactions identified by the proteins purifications shown in panel (A). The dashed lines show additional interactions identified in repeat purifications (see Suppl. Table 2 for further information).
To probe the network of protein-protein interactions involving Mtw1p, we sequenced proteins that copurified with Nnf1p. Four proteins were present at high levels in Nnf1p-3FLAG complexes: Nnf1p itself, Mtw1p, Nsl1p, and Dsn1p (Fig. 4A). Other proteins were present at lower levels, including the known kinetochore proteins Nuf2p, Slk19p, Bik1p, Bir1p, Cep3p, Stu2p, and Cse4p (Fig. 4A). Several proteins encoded by open reading frames of unknown function were also found. Of these we have examined only YFR046C, which we have named Cnn1p (Copurified with Nnf1p; Fig. 4A; Supplemental Table 2). Cnn1p is a kinetochore protein by ChIP and imaging (Fig. 4B,C), but is nonessential and present at low levels. As a second interactor, we purified Mcm21p from cells. It was associated with a large number of kinetochore proteins, including Dsn1p, Nnf1p, Mtw1p, Ndc80p, Nuf2p, Mif2p, and Spc105p (Fig. 4A; solid arrows in Fig. 4D; Supplemental Table 2). Thus, Mtw1p-3FLAG, Nnf1p-3FLAG, and Mcm21p-3FLAG purifications yielded a protein-interaction network that was highly enriched in kinetochore subunits.
Hydrodynamic analysis argues against the stable association of Mtw1p with previously described kinetochore proteins, leaving only Nnf1p, Dsn1p, Nsl1p, Spc105p, and Cnn1p as potential binding partners. On gradients and columns, Mtw1p was found in three distinct complexes: M1 (∼215 kD), M2 (∼153 kD), and M3(∼89 kD; Fig. 5A). Nnf1p-ProA and Nsl1p-ProA co-elute with Mtw1p-ProA in all three complexes, but Dsn1p-ProA was found only in the larger M1 and M2 complexes. The molecular masses of the complexes were consistent with the possibility that the M3complex represents an Mtw1p-Nnf1p-Nsl1p heterotrimer (predicted mass, 97 kD), and that M2 represents a heterotetramer of Dsn1p-Mtw1p-Nnf1p-Nsl1p (predicted mass, 162 kD; Fig. 5B; Table 1). M1 is ∼50 kD heavier than M2, suggesting that it contains an additional copy of Dsn1p, Nsl1p, Nnf1p, or Mtw1p or an unidentified protein. It seems reasonable to propose that M2 and M3are intermediates in the assembly of the larger M1 complex.
Figure 5.
The MIND complex contains Mtw1p, Nnf1p, Nsl1p, and Dsn1p. (A) Mtw1p-ProA, Dsn1p-ProA, Nsl1p-ProA, and Nnf1p-ProA co-elute on high-resolution size-exclusion chromatography. (B) Model for the predicted organization of Mtw1p, Dsn1p, Nsl1p, and Nnf1p in the M1, M2, and M3subcomplexes based on the molecular weight of the individual proteins: Mtw1p, 33 kD; Dsn1p, 65 kD; Nsl1p, 25 kD; Nnf1p, 24 kD; ProA tag, 15 kD. (C) Anti-ProA immunoblots of nnf1-17 cells carrying Nsl1p-ProA, Dsn1p-ProA, or Mtw1p-ProA grown at 24°C or 37°C. (D) Crosslinking of Nsl1p-ProA and Dsn1p-ProA to CEN DNA in nnf1-17 cells grown at 24°C or 37°C. (E) Anti-ProA immunoblots of whole-cell extracts, and (F) crosslinking of Nsl1p-ProA, Dsn1p-ProA, and Nnf1p-ProA to CEN DNA in mtw1-1 cells grown at 24°C or 37°C. (G) As a control, extracts were prepared from Mtw1p-ProA okp1-5 cells grown at 24°C or 37°C and analyzed by glycerol-density gradient ultracentrifugation.
Conditional mutations are available for NNF1 and MTW1 (Shan et al 1997; Goshima and Yanagida 2000), and we therefore asked what effect these mutations would have on the assembly of Mtw1p-containing complexes. Western blots revealed that the levels of Nsl1p, Dsn1p, and Mtw1p were dramatically reduced in extracts from nnf1-17 cells shifted to 37°C for 3 h, relative to cells maintained at 24°C. CEN-association in vivo was also disrupted, as determined by ChIP (Fig. 5C,D). Similarly, in mtw1-1 cells grown at 37°C, Nnf1p, Nsl1p, and Dsn1p were virtually absent (Fig. 5E,F), but the inactivation of the essential COMA protein Okp1p had no effect on M1, M2, or M3(Fig. 5G). Thus, the proteins associated with Mtw1p behave in a manner similar to COMA subunits in that a mutation in one member of a set of interacting proteins results in the instability of the other members. In contrast, a mutation in a protein that is a component of a distinct kinetochore complex has little or no effect (summarized in Table 2). When considered together, these data strongly suggest that Mtw1p, Dsn1p, Nnf1p, and Nsl1p are components of a kinetochore complex that associates with CEN DNA in vivo and is essential for cell viability. We will refer to it as the MIND complex (Mtw1p including Nnf1p/Nsl1p and Dsn1p), leaving open the possibility that one or more additional subunits remain to be identified.
Assembly of the COMA and MIND complexes at kinetochores
Having established which kinetochore proteins are tightly bound to each other in multiprotein subcomplexes, the difficult task of examining dependencies among dozens of proteins could be reduced to the simpler task of examining how COMA, MIND, and the Ndc80 complexes interact. When the crosslinking of Ame1p to CEN DNA was examined by ChIP, we found that COMA does not bind to kinetochores in ndc10-1 or okp1-5 mutants under restrictive conditions (37°C; Figs. 2D, 6A; Table 2). However, COMA was kinetochore-bound in mtw1-1 and ndc80-1 cells at 37°C (Fig. 6A; Table 2). As expected, Ame1p-GFP was undetectable at kinetochores in an okp1-5 mutant, but it still localized to kinetochores in mtw1-1 and ndc80-1 mutants at 37°C (Fig. 6B; Table 2). The intensity of Ame1p-GFP signals at kinetochores appeared to be reduced in mtw1-1 and ndc80-1 mutants relative to wild-type cells, but this may partly reflect a reduction in the extent of kinetochore clustering (Table 2; see below for further details). Unfortunately, it is not possible to accurately compare the integrated intensities of diffuse and punctate signals. Kinetochore association by Mtw1p required Ndc10p and to a lesser extent, NNF1, but not Okp1p or Spc25p (Fig. 6C; Table 2; Goshima and Yanagida 2000). This result is consistent with functional data demonstrating that the Ndc80 complex, but not MIND, is required for the spindle checkpoint (Goshima and Yanagida 2000; Janke et al. 2001; McCleland et al. 2003). Finally, Ndc80p was crosslinked to CEN DNA in okp1-5 and mtw1-1 cells at 37°C (Fig. 6D; Table 2), but not in ndc10-1 cells (He et al. 2001; data not shown). Overall, these experiments suggest that the MIND, COMA, and Ndc80 complexes associate independently with CEN DNA. One possibility is that they may represent three separate sites for the recruitment of outer kinetochore proteins, including those that bind to MTs (such as Cin8p; Fig. 6D).
Figure 6.
Assembly of the COMA, MIND, and Ndc80 complexes at kinetochores. (A) Crosslinking of Ame1p-GFP to CEN DNA in wild-type, okp1-5, ndc80-1, and mtw1-1 cells grown at 37°C. (B) Localization of Ame1p-GFP (green) in wild-type, okp1-5, ndc80-1, and mtw1-1 cells grown at 37°C. (C) Crosslinking of Mtw1p-ProA to CEN DNA in spc25-7 cells grown at 24°C or 37°C and in wild-type, okp1-5, and nnf1-17 cells grown at 37°C. Ame1p-ProA acts as the positive control. (D) Crosslinking of Ndc80p-GFP to CEN DNA in wild-type, okp1-5, and mtw1-1 cells grown at 37°C. Crosslinking of Cin8p-GFP to CEN DNA in wild-type and ndc80-1 cells grown at 37°C (negative control).
Function of the COMA and MIND complexes in chromosome segregation
If the COMA, MIND, and Ndc80 complexes have distinct functions at kinetochores, we might expect their inactivation to give rise to distinct defects in chromosome segregation. To investigate this possibility, we first measured the extent of transient sister-chromatid separation. A tag was introduced into chromosome IV (chrIV) by integrating a tandem array of 225 TetO sequences 2 kb away from the centromere (CENIV) in cells that express TetR-GFP (He et al. 2000). The binding of TetR-GFP to the TetO array creates a very bright spot that is readily visible by fluorescence microscopy. In the absence of tension at kinetochores, centromere-proximal tags on sister chromatids are unresolvable, and give rise to one bright dot. When kinetochores come under tension, as a result of pulling forces exerted by MTs, tags separate and two dots are visible (He et al. 2000). To quantitate transient sister separation, wild-type, mtw1-1, nnf1-17, okp1-5, and ndc80-1 cells were synchronized in α-factor, shifted to 37°C, and released into fresh medium. The fraction of cells with separated sisters was then counted at various times after release. In wild-type cells, the degree of transient separation reached a maximum of about 50% during late metaphase, 75 min after α-factor release. In mtw1-1, nnf1-17, okp1-5, and ndc80-1 cells, transient separation was five- to tenfold lower than in wild-type cells, showing that the amount of force exerted on sister chromatids was dramatically reduced (Fig. 7A).
Figure 7.
Functional analysis of the COMA and MIND complexes in chromosome segregation. (A) Strains carrying a centromere-proximal chrIV tag and Spc42p-GFP (He et al. 2000) were synchronized with α-factor and released into prewarmed media at 37°C. Cells were fixed after 75 min, and the extent of transient sister separation was determined in metaphase cells (as determined by nuclear morphology and spindle length). Data for ndc80-1 are provided for comparison (He et al. 2001). (B) okp1-5 and mtw1-1 cells containing Ndc80p-GFP (green) and ndc80-1 cells containing Mtw1p-GFP (green) were shifted to 37°C for 3 h, fixed, and imaged as described in the Figure 2E legend. All strains contained Spc42p-CFP (red) to mark the position of the SPBs, which are indicated by the red arrows. (C) Schematic of the chrIV tag and Spc42p-GFP SPB tag showing the spindle-centered cylindrical reference system and three key parameters used to characterize chromosome motion: d1, the distance from the reference spindle pole to one chromatid tag; d2, the distance from the reference spindle pole to the sister chromatid; and d3, the distance between the SPBs. (D-O, left column) Scatter plots of the positions of chromosome tags over 300 sec in a time-lapse movie in wild-type, ndc80-1, okp1-5, and mtw1-1 cells at 37°C. The length of the spindle is shown in red; in general, spindles are longer in cells with kinetochore mutants than in wild-type cells, as a consequence of reduced inward directed spindle forces (Goshima and Yanagida 2000). Separate green and black lines are seen only when chromatids are transiently separated. The positions of chromosomes are viewed along the spindle axis. (D-O, middle column). Scatter plots with the spindle axis rotated -45° and elevated 20°. (D-O, right column) Plots of spindle length, d3 in part C (red), the distance between a reference SPB and the first chromosome tag, d1 (black) and between the reference SPB and the second chromosome tag, d2 (green). Distinct black and green lines are seen only when sister chromatids are undergoing transient separation.
In a second assay, kinetochores were tagged in wild-type and mutant cells using a GFP fusion to a protein that remains CEN-bound under nonpermissive conditions (Ndc80p-GFP for okp1-5 and mtw1-1 mutants, and Mtw1p-GFP for ndc80-1 mutants; refer to ChIP data in Fig. 6). This makes it possible to determine the overall distribution of kinetochores (Fig. 7B). In ndc80-1 cells, kinetochores were visible as 7-15 foci scattered throughout the nucleus, each of which presumably represented one or more pairs of sister kinetochores (Fig. 7B). The scattering of kinetochores is a consequence of the complete detachment of chromosomes from MTs (He et al. 2001; Janke et al. 2001; Wigge and Kilmartin 2001). Kinetochores in okp1-5 and mtw1-1 cells, in contrast, were found in clusters along the spindle axis. Although the normal bilobed distribution of kinetochores was disrupted (Fig. 7B), the maintenance of close association between spindles and chromosomes in okp1-5 and mtw1-1 cells shows that neither mutation completely abolishes chromosome-MT attachment.
In a final assay, TetO/TetR-GFP tagged chromosomes were tracked in living cells to determine patterns of chromosome movement and to visualize force-generating processes (Michaelis et al. 1997; Goshima and Yanagida 2000; He et al. 2000). Wild-type cells exhibited the expected pattern of transient sister-chromatid separation and the oscillatory movement of sister chromatids along the spindle axis (Fig. 7D-F; He et al. 2000, 2001). ndc80-1 mutants, in contrast, exhibited a pattern of undirected chromosome motion that took place well away from the spindle and resembled a random walk, as described previously (Fig. 7G-I; He et al. 2001). In 16 okp1-5 cells analyzed in detail, chrIV remained closely associated with the spindle axis relative to wild-type cells (Fig. 7J-L), even following prolonged arrest. This pattern of movement was similar to that observed in stu2-277 mutants and is consistent with bipolar attachment and reduced force generation (He et al. 2001). In mtw1-1 cells, chromosomes also remained associated with the spindle (Fig. 7M-O). In some metaphase cells, chromosomes appeared to detach from microtubules, although the range of motion was less than that seen in ndc80-1 (data not shown). After prolonged periods of metaphase arrest (>45 min) mtw1-1 cells progress into anaphase, but this is associated with a reduction in cell viability (Goshima and Yanagida 2000), and we have not analyzed late phenotypes in great detail. We do not yet have the computational tools to interpret relatively subtle differences in the phenotypes of mtw1-1 and okp1-5 cells from a mechanistic perspective, but it is our distinct impression that the patterns of motion differ from each other and from patterns in ndc80-1 cells. Overall, we can conclude from these data that the loss of the Ndc80 complex, COMA, or MIND disrupts force generation at kinetochores. In the case of ndc80-1 mutants, this reflects a complete dissociation of chromosomes from MTs. In okp1-5 and mtw1-1 mutants, chromosomes probably remain associated with the spindle, in a state of bipolar attachment, but force generation is disrupted. Thus, both functional and assembly data support the hypothesis that the Ndc80, COMA, and MIND complexes have distinct roles at the kinetochore.
Discussion
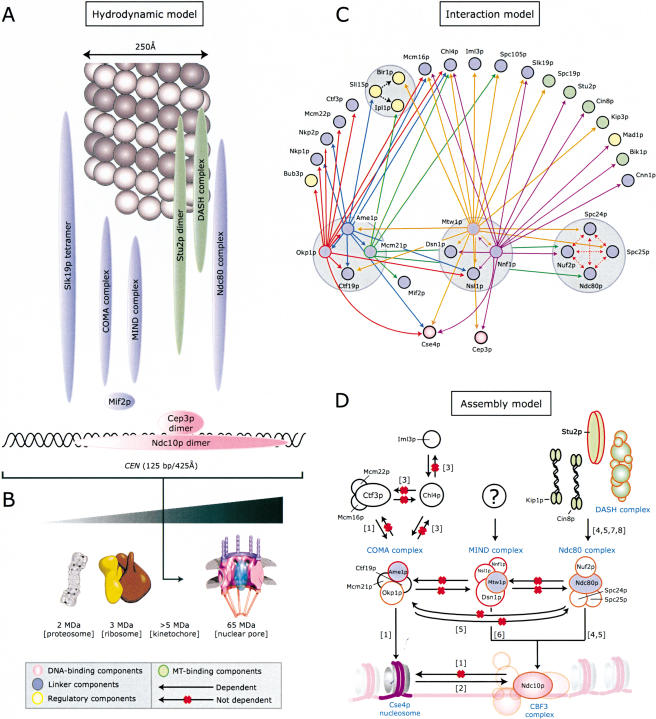
Here we report the analysis, using biophysical methods, chromatin immunoprecipitation, and live-cell imaging, of 19 known S. cerevisiae kinetochore proteins, with the aim of identifying key protein-protein interactions and building a model of kinetochore architecture (Fig. 8A-D). Using mass spectrometry, we also identified one nonessential (Cnn1p) and five essential proteins (Ame1p, Nnf1p, Nsl1p, Dsn1p, and Spc105p) of unknown function as kinetochore subunits. In addition, we assigned four of the new and 26 known kinetochore proteins to one of 14 multiprotein complexes found in cell extracts. The kinetochore subcomplexes have an average of four subunits and molecular weight of 200 kD. Assuming that only one copy of each complex is present per kinetochore, the lower limit on kinetochore mass is approximately 5 MD, twice the size of the budding yeast proteasome or ribosome (Fig. 8B). However, we observed that GFP-tagged kinetochore proteins differ in fluorescent intensities when bound to CEN DNA. Although we cannot exclude the possibility that different GFP fusions have different fluorescent yields, it seems likely that some proteins are present in several copies per kinetochore. Thus, the possibility exists that even the simple kinetochores of budding yeast have a mass considerably greater than 5 MD.
Figure 8.
Models of the structure, organization, and assembly of budding yeast kinetochores. (A) Hydrodynamic data from Table 1 were used to draw 3D ellipsoids for each kinetochore complex such that the volume was proportional to the molecular weight, and the axial ratio is represented as the degree of elongation of the ellipse. The single MT and CEN DNA are drawn approximately to scale. (B) Predicted size of the yeast kinetochore assuming that each known subunit is present in a single copy unless the subunit is known to be a homomultimer. (C) Protein-interaction model summarizing interactions among Okp1p (solid red arrows), Ame1p (solid blue arrows), Mcm21p (solid green arrows), Mtw1p (solid orange arrows), and Nnf1p (solid purple arrows). Interactions among Ndc80p complex subunits (dotted red arrows) are described by Wigge and Kilmartin (2001) and Janke et al. (2001). Interactions found with Sli15p (dotted black arrows) are described by Cheeseman et al. (2002b). (D) Assembly model for the COMA, MIND, and Ndc80 complexes at kinetochores. Black arrows represent dependencies (CEN binding by Mtw1p requires NDC10, and the arrow therefore connects the MIND and CBF3 complexes); a red “X” represents a lack of dependency. (For references see: [1] Measday et al. 2002, [2] Ortiz et al. 1999, [3] Pot et al. 2003, [4] He et al. 2001, [5] Janke et al. 2001, [6] Goshima and Yanagida 2000, [7] Janke et al. 2002, [8] J. Tytell and P. Sorger, unpubl.)
General implications for protein-interaction maps and protein complexes
An as-yet unresolved question is the relationship between interaction maps and protein structure derived from more conventional biochemical approaches. In the past few years, high-throughput methods, including directed and random two-hybrid analysis, systematic synthetic lethality screens, and rapid mass spectrometry have been developed to study protein-protein interactions. These methods yield interaction maps that embed proteins in a network of contacts (for review, see Bader et al. 2003). The affinity-based purifications and LC-MS/MS described here are essentially identical to the methods used successfully in high-throughput analysis of the yeast proteome (Ho et al. 2002). By applying standard thresholds for specificity and yield, we generated a rich network of protein-protein interactions involving kinetochore subunits (Fig. 8C). However, the network does not distinguish clearly between proteins that are present in stable subcomplexes and those that are more loosely—or even adventitiously—associated. Moreover, the precise spectrum of proteins associated with a target protein varied to some extent from purification to purification, whereas hydrodynamic analysis was consistent. We therefore conclude that, in the case of kinetochores, standard biophysical methods and protein-interaction maps do not yield the same information. The utility of subcomplexes as defined conventionally is clear for reconstitution and high-resolution structural analysis: we have found that the co-expression of all of the members of kinetochore subcomplexes in insect cells and Escherichia coli yields stable and soluble entities, whereas the absence of even one component leads to failure (C. Espelin and J. Miranda, unpubl.). The utility of mass spectrometry in the identification of novel components of protein complexes is also clear. However, the value of interaction maps in guiding biochemical analysis remains to be determined. Our findings suggest that caution is warranted as we turn increasingly to high-throughput data for mechanistic insight.
Constructing a structural model for kinetochores
Developing an accurate three-dimensional model of kinetochore-mediated MT attachment must start with answers to some basic questions: Do kinetochore proteins form discrete subcomplexes in the absence of CEN DNA, and do these subcomplexes have distinct functions? Can a hierarchy be established for the association of proteins with CEN DNA that tells us something about their position in the overall structure and their order of assembly? Can mutant phenotypes be mapped onto the architecture to give an initial sense that structure and function are meaningfully related? Does the structure of yeast kinetochores hint at the existence of features that have been conserved through evolution?
We have shown that the majority of kinetochore proteins exist in cell extracts as subcomplexes with molecular weights larger than the proteins themselves, yet smaller than intact kinetochores (Fig. 8A). A subcomplex in this context refers to a set of proteins that bind to each other in solution to form a discrete high-affinity homo- or hetero-oligomeric assembly, which then associates in a stepwise fashion with other subcomplexes to form a complete kinetochore. It is useful to think in terms of subcomplexes—rather than individual subunits—if most subunits are members of one, or at most a small set, of subcomplexes with well defined composition. In contrast, if the number of subcomplexes is large relative to the number of subunits, or if most proteins are components of several different subcomplexes, it is likely that random fragments of a structure that has fallen apart in an arbitrary fashion are being examined. Importantly, some large multiprotein structures, such as ribosomes, can be isolated from cells as a single species, whereas other large multiprotein assemblies, such as transcriptional enhancers, can only be isolated as subcomplexes that do not appreciably interact with each other in the absence of DNA (for review, see Merika and Thanos 2001; Struhl 2001). In the case of kinetochores, our data strongly suggest the existence of discrete and well defined subcomplexes and an enhancer-like model of assembly in which proteins associate soon after synthesis into stable species that then assemble on centromeric DNA in a stepwise manner.
Evidence described in this paper for the stepwise assembly of kinetochores derives primarily from studies of protein hydrodynamics, stability, and centromere binding. By comparing the native molecular weights of proteins in cells carrying different mutations we can infer, with considerable reliability, which proteins associate with one another in solution. Hydrodynamic measurements of native molecular mass have also proven reliable, with coefficients of variation of about ±10%-15%. One complicating factor is the existence of multiple COMA and MIND complexes. We believe that these represent assembly intermediates whose abundance is cell cycle-regulated, but we have not yet examined this possibility in great detail.
A powerful source of information on subcomplex composition is the examination of protein stability by ChIP, in vivo imaging, and Western blotting (Table 2). We have found that a mutation in one subunit of a multiprotein kinetochore complex frequently results in the disappearance of other subunits, whereas a mutation in an unrelated protein has little or no effect. The selective loss of kinetochore proteins within a single complex is consistent with data from other studies showing that subunits of multiprotein structures are significantly more stable when assembled (see, for example, the 1.8-MD SAGA transcriptional repressor complex, Sterner et al. 1999). We have not determined whether changes in kinetochore protein levels are caused by lower expression, reduced solubility, or increased degradation. The latter seems most likely, but whatever the mechanism, the analysis of interdependencies of protein expression is an effective means to identify proteins that interact in high-affinity subcomplexes.
Hierarchical assembly
Data from several sources indicate that kinetochores assemble from subcomplexes in a hierarchical fashion (see the introduction of this article). Only three protein species have been shown to bind to centromeric DNA in a sequence-specific manner: CBF3, CBF1, and Ndc10p (Cai and Davis 1990; Lechner and Carbon 1991, Espelin et al. 2003). Nucleosomes containing the kinetochore-localized Cse4p histone are also likely to be DNA-bound (Meluh et al. 1998), although it is not known whether binding occurs to CEN DNA itself or to flanking DNA. Other kinetochore proteins are thought to bind directly to MTs, based either on homology to MAPs or on in vitro binding assays. These proteins include Cin8p, Kip3p, and Kip1p KRPs and the Stu2p, Bik1p, and the DASH complex MAPs (Cheeseman et al. 2001; He et al. 2001; J. Tytell and P. Sorger, unpubl.). However, many other essential kinetochore proteins have been assigned to a loosely defined linker layer that lies between MTs and DNA-binding proteins. The best characterized linker is the Ndc80 complex. As expected from a hierarchical model for kinetochore assembly, in vivo ChIP and imaging show that centromere association by the Ndc80 complex requires functional CBF3, but that CBF3 binds DNA in the absence of functional Ndc80p (He et al. 2001; Janke et al. 2001). The Ndc80 complex is required for centromere association by the MT-binding Stu2p, Cin8p, and Kip1p proteins and the DASH complex (Figs. 6D, 8D; He et al. 2001; Janke et al. 2001; J. Tytell and P. Sorger, unpubl.). As expected, mutations in these MT-binding proteins do not affect the association of Ndc80p with kinetochores; J. Tytell and P. Sorger, unpubl.). Our data suggest that the COMA and MIND complexes comprise two additional linkers that bind to centromeres and function independently of each other and of the Ndc80 complex, as judged by ChIP and imaging in mutant cells. Both COMA and MIND also require CBF3. This model is consistent with functional data demonstrating that Ndc80 complex mutants (such as spc25-7) have a defective spindle checkpoint, whereas mtw1-1, nnf1-17, and okp1-5 mutants have a functional checkpoint (Goshima and Yanagida 2000; Janke et al 2001; data not shown). COMA appears to be involved in the recruitment of Ctf3p, Chl4p, and associated proteins to kinetochores (Fig. 8D; Measday et al. 2002; Pot et al. 2003), but we have not yet identified proteins that require MIND for centromere association. The relative complexity of kinetochore linkers is consistent with data that kinetochore-MT attachment involves multiple proteins with discrete functions that cooperate in the formation of a mature attachment site.
Although the COMA, MIND, and Ndc80 complexes assemble independently, several lines of evidence suggest complicated patterns of interaction. For example, the overall level of the COMA subunit Ame1p is reduced in ndc10-1 and ndc80-1 relative to wild-type cells, even though ChIP suggests no significant change in the levels of CEN-bound Ame1p (Table 2). These and other findings hint at poorly understood differences between bound and unbound protein populations. Many kinetochore proteins are found at levels well above the 32 copies required for one molecule to be present on each centromere, but it is not known whether the unbound populations have special functions. In future studies it will be important to differentiate between chromatin-bound and soluble protein populations and to determine whether the existence of multiple COMA and MIND complexes is a reflection of regulated steps in kinetochore assembly.
Mapping function onto structure
We showed previously that mutations in different kinetochore proteins give rise to distinct phenotypes with respect to chromosome-MT attachment (He et al. 2000). Complete inactivation of kinetochores, in ndc10-1 cells, causes chromosomes to dissociate from MTs and float free in the nucleus. In contrast, ipl1 and dam1 mutations give rise to monopolar attachment in which kinetochores remain associated primarily with one SPB (He et al. 2001; Janke et al. 2002; Tanaka et al. 2002). Perhaps the most interesting type of defect, previously seen only in stu2 mutants, is bipolar binding without the generation of sufficient pulling force to cause centromere stretching and transient sister-chromatid separation (He et al. 2001). Both the MIND and COMA complexes appear to create this type of defect, although patterns of chromosome movement differ between the two mutants in ways we do not yet understand. We can conclude that the processes of MT attachment and force generation not only involve different MAPs and motors but also different components of a tridentate linker layer. Moreover, two of the three yeast linker complexes analyzed here contain at least one subunit that is homologous to a known kinetochore protein in animal cells. An additional similarity between simple and complex kinetochores is that both appear to contain a large number of highly elongated, fibrous components. Our hydrodynamic data suggest that the MIND, COMA, DASH, and Ndc80 complexes have axial ratios of 10-20 or more. This may allow them to form a socket-like structure in which an MT can become deeply embedded.
Materials and methods
See Supplementary Materials and Methods for details.
Yeast strains and manipulations
All strains used in this study are congenic with W303 (Supplemental Table 1). Genetic manipulations and growth conditions were as described (Guthrie and Fink 1991). For temperature shift experiments, cells were grown at 24°C until mid-log phase, spun down, resuspended in prewarmed (37°C) medium, and grown for another 3-4 h.
Biochemical methods
Anti-FLAG affinity purifications were performed as described (Ho et al. 2002). Proteins in the column eluate were precipitated with trichloroacetic acid (TCA), rinsed with acetone, resuspended in SDS-loading buffer, and separated by 12.5% SDS PAGE. Proteins were stained with Colloidal Blue (Invitrogen), and identified by microcapillary liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
Chromatin immunoprecipitations (ChIP) of GFP, ProA, and TAP-tagged proteins were performed based on the protocol described by He et al. (2001). A 200-bp fragment of the centromere of chromosome IV (He et al. 2001), a 200-bp region of the URA3 gene of chromosome V (negative control; He et al. 2001), and a 400-bp region located 600 bp from the TG repeat of telomere VI-R (negative control; Martin et al. 1999) were analyzed by PCR.
Whole-cell extracts expressing an epitope-tagged protein were prepared as described (Sorger and Pelham 1987), and size-exclusion chromatography and glycerol-density gradient analysis were done as described (Russell et al. 1999). The molecular weight, size (Stokes radius), and shape (axial ratio) of protein complexes were calculated from measured diffusion coefficients and Svedberg sedimentation constants using a series of well established equations (Harding and Colfen 1995; Schuyler and Pellman 2002).
Microscopy and image analysis
Microscopy and image analyses were carried out on a DeltaVision deconvolution microscope as described (He et al. 2001; Rines et al. 2002; Thomann et al. 2002; Rines et al. 2003).
Acknowledgments
We thank B. Aldrige, A. Amon, D. D'Amours, B. Hughes, J. Lechner, D. Rines, E. Schiebel, and M. Yanagida for reagents and assistance, and members of the Sorger and Amon labs for comments on the manuscript. P.D.W. is supported by the Charles A.
King Trust Medical Foundation, and A.D.M. by the Jane Coffin Childs Fund for Medical Research. Research in this paper was supported by NIH grants GM51464 and GM64524.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1144403.
References
- Bader G.D., Heilbut, A., Andrews, B., Tyers, M., Hughes, T., and Boone, C. 2003. Functional genomics and proteomics: Charting a multidimensional map of the yeast cell. Trends Cell. Biol. 13: 344-356. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Brew, C., Wolyniak, M., Desai, A., Anderson, S., Muster, N., Yates, J.R., Huffaker, T.C., Drubin, D.G., and Barnes, G. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155: 1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Drubin, D.G., and Barnes, G. 2002a. Simple centromere, complex kinetochore: Linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157: 199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates 3rd, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. 2002b. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163-172. [DOI] [PubMed] [Google Scholar]
- Ching S., Hazbun, T.R., Cheeseman, I.M., Aranda, J., Fields, S., Drubin, D.G., and Barnes, G. 2003. Kinetochore protein interactions and their regulation by the aurora kinase Ipl1p. Mol. Biol. Cell 14: 3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C. and Hieter, P. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman M., Cheeseman, I.M., Van Goor, D., Drubin, D.G., Meluh, P.B., and Barnes, G. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12: 2601-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin C.W., Simons, K.T., Harrison, S.C., and Sorger, P.K. 2003. Binding of the essential S. cervisiae Kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell 14: 4557-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen G.M. 2002. Nnf1p, Dsn1p, Mtw1p, and Nsl1p: A new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot. Cell 1: 229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P.Y. and Kilmartin, J.V. 1993. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121: 503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G. and Yanagida, M. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100: 619-633. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh, S., and Yanagida, M. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 13: 1664-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu, T., Yoda, K., and Yanagida, M. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160: 25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. and Fink, G.R. 1991. Guide to yeast genetics and molecular biology, Vol. 194. Academic Press, San Diego, CA.
- Harding S.E. and Colfen, H. 1995. Inversion formulae for ellipsoid of revolution macromolecular shape functions. Anal. Biochem. 228: 131-142. [DOI] [PubMed] [Google Scholar]
- He X., Asthana, S., and Sorger, P.K. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101: 763-775. [DOI] [PubMed] [Google Scholar]
- He X., Rines, D.R., Espelin, C.W., and Sorger, P.K. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195-206. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180-183. [DOI] [PubMed] [Google Scholar]
- Horiike K., Tojo, H., Yamano, T., and Nozaki, M. 1983. Interpretation of the stokes radius of macromolecules determined by gel filtration chromatography. J. Biochem. 93: 99-106. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Middleton, K., Centola, M., Mitchison, T.J., and Carbon, J. 1992. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature 359: 533-536. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M.M., Schramm, C., and Schiebel, E. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20: 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz, J., Tanaka, T.U., Lechner, J., and Schiebel, E. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21: 181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.H., He, X., Giddings, T.H., and Winey, M. 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. 98: 13675 -13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J. and Carbon, J. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64: 717-725. [DOI] [PubMed] [Google Scholar]
- Li Y., Bachant, J., Alcasabas, A.A., Wang, Y., Qin, J., and Elledge, S.J. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes & Dev. 16: 183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luykx P. 1965. The structure of the kinetochore in meiosis and mitosis in Urechis eggs. Exp. Cell Res. 39: 643-657. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche, T., Suka, N., Grunstein, M., and Gasser, S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 91: 621-633. [DOI] [PubMed] [Google Scholar]
- McAinsh A.D., Tytell, J.D., and Sorger, P.K. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19: 519-539. [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Gardner, R.D., Kallio, M.J., Daum, J.R., Gorbsky, G.J., Burke, D.J., and Stukenberg, P.T. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes & Dev. 17: 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Hailey, D.W., Pot, I., Givan, S.A., Hyland, K.M., Cagney, G., Fields, S., Davis, T.N., and Hieter, P. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes & Dev. 16: 101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607-613. [DOI] [PubMed] [Google Scholar]
- Merika M. and Thanos, D. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11: 205-208. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk, R., and Nasmyth, K. 1997. Cohesins: Chromosomal proteins that prevent mature separation of sister chromatids. Cell 91: 34-45. [DOI] [PubMed] [Google Scholar]
- Ng R., Ness, J., and Carbon, J. 1986. Structural studies on centromeres in the yeast Saccharomyces cerevisiae. Basic Life Sci. 40: 479-492. [DOI] [PubMed] [Google Scholar]
- Ortiz J., Stemmann, O., Rank, S., and Lechner, J. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes & Dev. 13: 1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot I., Measday, V., Snydsman, B., Cagney, G., Fields, S., Davis, T.N., Muller, E.G., and Hieter, P. 2003. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell 14: 460-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rines D.R., He, X., and Sorger, P.K. 2002. Quantitative microscopy of green fluorescent protein-labeled yeast. In Guide to yeast genetics and molecular biology, pp. 16-34. Academic Press, San Diego, CA. [DOI] [PubMed]
- Rines D.R., Thomann, D., Dorn, J.F., Goodwin, P., and Sorger, P.K. 2003. Live cell imaging of yeast. In Live cell imaging: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (in press). [DOI] [PubMed]
- Russell I.D., Grancell, A.S., and Sorger, P.K. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145: 933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C. and Pellman, D. 2002. Analysis of the size and shape of protein complexes from yeast. Methods Enzymol. 351: 150-168. [DOI] [PubMed] [Google Scholar]
- Shan X., Xue, Z., Euskirchen, G., and Melese, T. 1997. NNF1 is an essential yeast gene required for proper spindle orientation, nucleolar and nuclear envelope structure and mRNA export. J. Cell Sci. 110: 1615-1624. [DOI] [PubMed] [Google Scholar]
- Sorger P.K. and Pelham, H.R. 1987. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 6: 3035-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P.K., Severin, F.F., and Hyman, A.A. 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127: 995-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant, P.A., Roberts, S.M., Duggan, L.J., Belotserkovskaya, R., Pacella, L.A., Winston, F., Workman, J.L., and Berger, S.L. 1999. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19: 86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. 2001. Gene regulation. A paradigm for precision. Science 293: 1054-1055. [DOI] [PubMed] [Google Scholar]
- Strunnikov A.V., Kingsbury, J., and Koshland, D. 1995. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128: 749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen, E.S., and Yanagida, M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215-2219. [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317-329. [DOI] [PubMed] [Google Scholar]
- Thomann D., Rines, D.R., Sorger, P.K., and Danuser, G. 2002. Automatic fluorescent tag detection in 3D with super-resolution: Application to the analysis of chromosome movement. J. Microsc. 208: 49-64. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Drechsel, D., and Hyman, A. 2003. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 161: 359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A. and Kilmartin, J.V. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152: 349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen, O.N., Holmes, S., Soues, S., Mann, M., and Kilmartin, J.V. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141: 967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]