Abstract
A novel peak alignment algorithm using a distance and spectrum correlation optimization (DISCO) method has been developed for two-dimensional gas chromatography time-of-flight mass spectrometry (GC×GC/TOF-MS) based metabolomics. This algorithm uses the output of the instrument control software, ChromaTOF, as its input data. It detects and merges multiple peak entries of the same metabolite into one peak entry in each input peak list. After a z-score transformation of metabolite retention times, DISCO selects landmark peaks from all samples based on both two-dimensional retention times and mass spectrum similarity of fragment ions measured by Pearson’s correlation coefficient. A local linear fitting method is employed in the original two-dimensional retention time space to correct retention time shifts. A progressive retention time map searching method is used to align metabolite peaks in all samples together based on optimization of the Euclidean distance and mass spectrum similarity. The effectiveness of the DISCO algorithm is demonstrated using data sets acquired under different experiment conditions and a spiked-in experiment.
Keywords: GC×GC/TOF-MS, metabolomics, retention time alignment, DISCO
INTRODUCTION
Metabolomics is the systematic analysis of the metabolome, which is a complete set of small-molecule metabolites in a biological system. Such analyses include the study of metabolites’ dynamics, composition, interactions, and responses to interventions or to changes in the biological environment. Even though significant progress in metabolomics has been made in the last decade due to advances in the high-throughput enabling technologies such as mass spectrometry, the number of types of metabolites present in a metabolome and their relative concentrations still remain largely unknown.1 A comprehensive and individual characterization of all the metabolites is a very challenging task.2 The metabolites present in a metabolome must be separated first to simplify the complexity of the metabolite composition. Currently, gas chromatography (GC), high performance liquid chromatography (HPLC), and capillary electrophoresis (CE) are widely used as separation methods. Due to the large number and low concentration of many intracellular metabolites and the changes in their concentrations with environment and cell history, it is impossible to analyze the intracellular metabolite profile in one run using a single chromatographic or electrophoretic technique.3 Multidimensional separation techniques therefore are a more practical choice for metabolomics analysis in order to separate as many metabolites as possible.4,5 An emerging technology, comprehensive two-dimensional gas chromatography coupled with time-of-flight mass spectrometry (GC×GC/TOF-MS), brings much increased signal-to-noise ratio, dynamic range, separation capacity, chemical selectivity, and sensitivity to metabolomics analyses.6, 7
GC×GC/TOF-MS has recently gained considerable attention in metabolomics analysis, in particular for many complex samples, such as metabolites in plants,8–11 microorganisms,12–16 mammals,17–22 and the other metabolite samples.23–25 It is very common that multiple samples are analyzed in a metabolomics study, such as for metabolite biomarker discovery experiments. After analyzing these samples using GC×GC/TOF-MS, it is necessary to recognize molecular features of the same metabolite occurring in different samples from each of the raw instrument data. Ideally, the same metabolite should have the identical retention times in the two-dimensional GC if the instrument configuration is the same. However, this is not true due to experimental variations. Metabolite retention times may shift in both GC dimensions as a result of several, sometimes uncontrollable factors such as temperature and pressure fluctuations, matrix effects on samples, and stationary phase degradation. Retention time shifts introduce difficulty in the data processing for metabolomics analyses. Therefore, the retention time variation must be adjusted, i.e., aligned, before applying statistical data mining methods.
Metabolite peak alignment can be achieved using two approaches. One is to use raw instrument data as input material to adjust the retention times of each raw data to a common retention time scale. The other approach is that the peak lists of all biological samples are used as input material for alignment. To enable the second approach, the raw instrument data are first subjected for spectrum deconvolution to generate a list of metabolite peaks for each sample, of which each metabolite peak is characterized by multiple molecular features including retention times in the two-dimensional GC, peak area, fragment spectrum, and other associated features. The choice of the alignment approach depends on the strategy of the downstream data analysis. Alignment methods using the first approach require comparing the aligned raw signals to find molecular expression differences between samples, while methods using the second approach lead to statistical multivariate data analysis.
Currently, few studies addressed alignment issue for the two-dimensional GC separations using the raw instrument data as input material. Fraga et al. developed a rank-based algorithm using the generalized rank annihilation method (GRAM) to correct retention time variations in the two- dimensional GC.26, 27 Mispelaar et al. developed a correlation-optimized shifting-based algorithm to align a local region of a GC×GC chromatogram.28 These two methods can only be used to align small regions of interest in the two-dimensional GC data set. To correct the entire chromatogram in both GC dimensions, Pierce et al. proposed an indexing scheme together with a piecewise retention time alignment algorithm.29 Zhang et al. developed a two dimensional correlation optimized warping (2-D COW) method by extending the correlation optimized warping method from 1-D to 2-D.30, 31 However, these methods align the GC×GC/TOF-MS data based on two-dimensional retention times alone, even through the signature feature of a metabolite, i.e., mass spectrum of fragment ions, is readily available in the raw instrument data. Aligning metabolite peaks solely based on the two dimensional retention times may introduce a high rate of false alignment because some metabolites with similar chemical functional groups have similar retention times in both GC dimensions. For this reason, we developed MSort software which employs two dimensional retention times and the mass spectrum of metabolite fragment ions for metabolite alignment.32 This method greatly reduced the rate of false alignment compared to existing alignment approaches. However, MSort software uses a user-defined retention time window with a fixed size in the two retention time dimensions. The size of the retention time window affects the reliability and efficiency of the software and the algorithm is not able to handle non-linear retention time distortion. Another limitation of the MSort algorithm is that it uses a lot of computer memory, which severely limits its application to large scale metabolomics study. Meanwhile, Msort software aligns different samples based on a fixed retention time variation window, which makes it impossible to align experimental data acquired under different experimental conditions, such as different temperature gradients.
To overcome the limitations of current alignment algorithms, this paper reports a novel distance and spectrum correlation optimization (DISCO) alignment algorithm for GC×GC/TOF-MS based metabolomics. The DISCO algorithm is based on the following assumption: There is a set of metabolite peaks, termed as landmark peaks, present in each biological sample. The retention time shifts of these landmark peaks in each sample should closely reflect the retention time shifts of all other metabolite peaks in the same sample. DISCO first selects landmark peaks using the Euclidean distances of two-dimensional retention times and mass spectrum correlation of two corresponding metabolite peaks. Then, it corrects retention time shifts using a local partial linear fitting method to handle non-linear retention time distortion. Finally, DISCO aligns the metabolite peaks of all samples using a progressive retention time map searching method. The performance of DISCO is demonstrated using experimental data. This proposed algorithm has been implemented into a software package using MATLAB R2008b.
EXPERIMENTAL SECTION
Mixture of Standard Compounds
A mixture of 76 compounds (8270 MegaMix, Restek Corp., Bellefonte, PA) and C7-C40 saturated n-alkanes (Sigma-Aldrich Corp., St. Louis, MO) were spiked with a deuterated six component semi-volatiles internal standard (ISTD) mixture (Restek Corp., Bellefonte, PA) at a concentration of 2.5 μg/mL prior to GC×GC/TOF-MS analysis. Table 1 lists all of the mixed standard components.
Table 1.
The list of mixed standard compounds.
| 76 standard compounds (STs) | ||||
|---|---|---|---|---|
| Aniline | Dimethyl phthalate | Phenol, 2,4-dichloro- | Benzene,1,4-dinitro- | N-Nitrosodimethylamine |
| Azobenzene | Ethane,hexachloro- | Phenol, 2,4,6-trichloro- | Benzene,1,3-dinitro- | Phenol,4-chloro-3-methyl- |
| Anthracene | Acenaphthene | Benzo[ghi]perylene | Benzene,1,2-dinitro- | Propane, 2,2′-oxybis[1-chloro- |
| Fluorene | Naphthalene,2-chloro- | Di-n-octyl phthalate | Benzene,1,3-dichloro- | 1-Propanamine,N-nitroso-N-propyl- |
| Pyridine | Naphthalene,1-methyl- | Phenol,2-nitro- | Benzene,1,4-dichloro- | 2-Cyclohexen-1-one,3,5,5-trimethyl- |
| Pyrene | Naphthalene,2-methyl- | Phenol,3-methyl- | Benzene,1,2-dichloro- | Methane, bis(2-chloroethoxy)- |
| Chrysene | Benz[a]anthracene | Phenol,4-methyl- | Benzene, hexachloro- | 1,3-Butadiene,1,1,2,3,4,4-hexachloro- |
| p-Nitroaniline | Benzene, nitro- | Phenol,2,4-dimethyl- | Benzyl butyl phthalate | 1,3-Cyclopentadiene,1,2,3,4,5,5-hexachloro- |
| Dibenzofuran | Dibutyl phthalate | Phenol,2-methyl- | Benzenamine,N-phenyl- | Hexanedioic acid, bis(2-ethylhexyl) ester |
| Carbazole | m-Nitroaniline | Phenol,2-chloro- | Benzo(b)fluoranthene | Bis(2-ethylhexyl)phthalate(dioctyl |
| Fluoranthene | Diethyl Phthalate | Phenol, pentachloro- | Benzo[k]fluoranthene | Benzene, 2-methyl-1,3-dinitro- |
| Phenol | Benzo(a)pyrene | Phenol,2,4-dinitro- | Phenol,2,3,4,6-tetrachloro- | Benzene, 1-methyl-2,4-dinitro- |
| Phenanthrene | Acenaphthylene | Phenol,4-nitro- | Phenol,2,3,5,6-tetrachloro- | Benzene,1-chloro-4-phenoxy- |
| Naphthalene | Benzyl Alcohol | 1,2,4-Trichlorobenzene | Dibenz(a,h)anthracene | Benzene,1-bromo-4-phenoxy- |
| o-Nitroaniline | Bis(2-chloroethyl) ether | 2,4,5-trichlorophenol | Indeno[1,2,3-cd]pyrene | Phenol,2-methyl-4,6-dinitro- |
| p-Chloroaniline | ||||
| C7-C40 saturated alkanes | ||||||
|---|---|---|---|---|---|---|
| Heptane | Dodecane | Heptadecane | Docosane | Hexacosane | Dotriacontane | Heptatriacontane |
| Octane | Tridecane | Octadecane | Tricosane | Octacosane | Tritriacontane | Octatriacontane |
| Nonane | Tetradecane | Nonadecane | Tetracosane | Nonacosane | Tetratriacontane | Nonatriacontane |
| Decane | Pentadecane | Eicosane | Pentacosane | Triacontane | Pentatriacontane | Tetracontane |
| Undecane | Hexadecane | Heneicosane | Heptacosane | Hentriacontane | Hexatriacontane | |
| Dueterated six compenont semivolatiles | |||||
|---|---|---|---|---|---|
| 1,4-Dichlorobenzene-d4 | Naphthalene-d8 | Acenaphthene-d10 | Phenathrene-d10 | Chrysene-d12 | Perylene-d12 |
Spiked-in Sample
A 100 uL rat plasma sample was mixed with 900 μL of organic solvent mixture (methanol:water = 8:1, v/v) and vortexed for 15 s. After sitting at −20°C for 30 min, the mixture was centrifuged with 16000 g at 4°C for 15 min. Supernatants from the mixture were collected and evaporated to dryness with a SpeedVac and then redissolved in 100 μL of pyridine. 50 μL of the metabolite extract was treated with 100 μL of 50 mg/mL ethoxyamine hydrochloride pyridine solution for 30 min at 60°C. Subsequently, the extracts were derivatized with 100 μL of MTBSTFA for 1 h at 60°C. The derivatized sample was spiked with ISTD mixture at a concentration of 2.5 μg/mL right before the GC×GC/TOF-MS analysis.
GC×GC/TOF-MS Analysis
All GC×GC/TOF-MS analyses were performed on a LECO Pegasus® 4D time-of-flight mass spectrometer (TOF-MS) with a Gerstel MPS2 auto-sampler. The Pegasus 4D GC×GC/TOF-MS instrument was equipped with an Agilent 7890 gas chromatograph featuring a LECO two stage cryogenic modulator and secondary oven. A 30 m × 0.25 mm id. × 0.25 μm film thickness, Rxi-5ms GC capillary column (Restek Corp., Bellefonte, PA) was used as the primary column for the GC×GC/TOF-MS analysis. A second GC column of 1.2 m × 0.10 mm id. × 0.10 μm film thickness, BPX-50 (SGE Incorporated, Austin, TX) was placed inside the secondary GC oven after the thermal modulator. The helium carrier gas flow rate was set to 1.0 mL/min at a corrected constant flow via pressure ramps. A 1 μL liquid sample was injected into the liner using the splitless mode with the injection port temperature set at 260°C. The primary column temperature was programmed with an initial temperature of 60°C for 0.5 min and then ramped at a variable temperature gradient to 315°C. The secondary column temperature program was set to an initial temperature of 65°C for 0.5 min and then also ramped at the same temperature gradient employed in the first column to 320°C accordingly. The thermal modulator was set to +20°C relative to the primary oven and a modulation time of 5 s was used. The MS mass range was 10–750 m/z with an acquisition rate of 150 spectra per second. The ion source chamber was set at 230°C with the MS transfer line temperature set to 260°C and the detector voltage was 1800 V with an electron energy of 70 eV.
The mixture of standard compounds was analyzed using three different temperature gradients, i.e., 5°C/min, 7°C/min, and 10°C/min. The spiked-in experiment was operated using a temperature gradient of 7°C/min.
Raw Data Reduction
The LECO ChromaTOF software version 3.41 equipped with the National Institute of Standards and Technology (NIST) MS database (NIST MS Search 2.0, NIST/EPA/NIH Mass Spectral Library; NIST 2002) was used for instrument control, spectrum deconvolution and metabolite identification. We used the manufacturer recommended parameters for ChromaTOF to reduce the raw instrument data to a metabolite peak list. These parameters are: baseline offset = 0.5; smoothing = Auto; peak width in first dimension = 6 s; peak width in the second dimension = 0.1 s; signal-to-noise ratio = 100.0; match required to combine peaks = 500; R.T. shift = 0.08 s; minimum similarity match = 600. The peak true spectrum was also exported as part of the information for each peak in absolute format of intensity values.
THEORETICAL BASIS
DISCO uses the peak list generated by the LECO ChromaTOF software as its input data. It employs a sequential two-stage alignment framework: primary alignment and on-the-fly alignment. The methodology under these two stages is the same, i.e., aligning the samples based on a landmark peak list. In this work, the landmark peaks are a list of metabolite peaks that are present in every sample processed during the primary alignment. The only difference is that the primary alignment selects a certain number of samples from the sample pool and constructs the so-called landmark peak table and aligns all of these selected samples, while the on-the-fly alignment aligns the rest samples in the sample pool one by one based on the landmark peak list. The purpose of having the two-stage alignment design is to avoid the limitation of computer memory. During the primary alignment, the data of all selected samples are uploaded into the computer memory for landmark peak detection and alignment. After the primary alignment, only the information of landmark peaks and the aligned peak table of the selected samples are kept in the computer memory while all other intermediate information is deleted. Then the on-the-fly alignment process starts if there are unprocessed data files in the sample pool. During the on-the-fly alignment, DISCO only uploads one sample data at a time into computer memory and aligns it to the existing alignment table generated during the process of primary alignment. The alignment table is updated and the sample data of the current sample is discarded. Then, DISCO uploads the next unprocessed data file. This process is repeated until all sample files in the sample pool are processed.
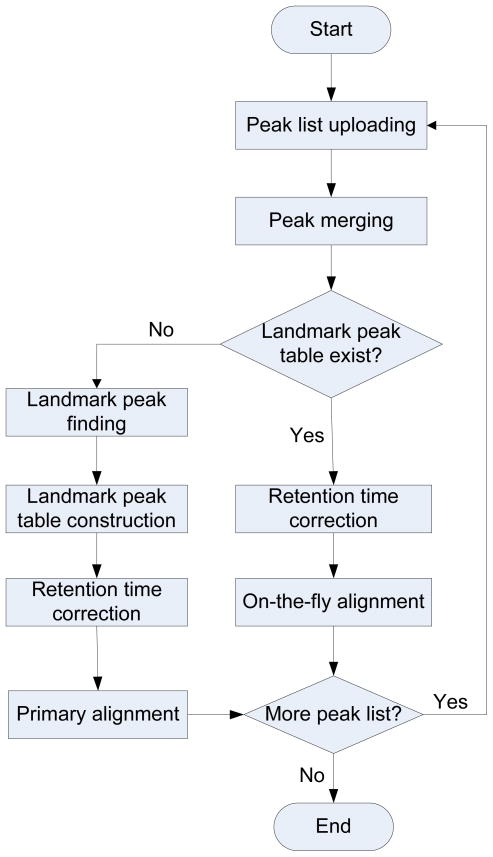
It is possible that the primary alignment can fulfill the peak alignment task if the size of data files and the intermediate files does not reach the hardware limitation. That is to say, if the size of the samples to be aligned is not very large (determined based on the limitation of the computer system, i.e., most 32-bit computers limit the virtual memory available to 2 GB, while for 64-bit computers the limitation is based on the amount of memory available on the computer), only primary alignment is employed. Otherwise, DISCO will first randomly select a certain number of samples or use the samples selected by user for the primary alignment and then process the remaining samples using on-the-fly alignment. Figure 1 shows the workflow of the DISCO software.
Figure 1.
The workflow of DISCO alignment algorithm.
Peak Entries Merging
Ideally, all instrument signals generated by one type of metabolite should be reported as a single peak in the output file of ChromaTOF software, i.e., one peak entry in the metabolite peak list. However, multiple peak entries of the same metabolite can be reported due to the abnormal metabolite peak shape and/or the high sensitivity of the peak detection algorithm. In order to minimize data variations to be introduced into the downstream statistical analysis, it is necessary to detect all metabolites with multiple peak entries and merge all peak entries of same metabolite as one entry in each peak list.
Multiple entries of the same metabolite can be recognized using combined information based on the mass spectrum similarity and the metabolite’s retention time in the first dimension GC. For a given GC×GC system, the metabolites from the first GC column are trapped for a fixed modulation time (Δ) before they are released into the second GC column. Therefore, the retention time of the metabolite peaks in the first dimension GC increases in a fixed interval Δ. The DISCO algorithm considers that all peak entries with the mass spectrum similarity ≥ R0 and retention time difference ≤ Δ are originated from the same type of metabolite, where R0 is a predefined threshold value of the correlation coefficient between two mass spectra of two metabolites. These multiple peak entries are then merged into one representative peak entry. The fragment ion mass spectrum of the representative peak is calculated by the sum of all the mass spectra of the member peaks, i.e., the multiple peak entries, according to the m/z value of each fragment ion. It is well known that peak tailing in GC can be caused by column contamination, too low of a split ratio, solvent-phase polarity mismatch, and so on. For this reason, the values of the first dimension retention time and the second dimension retention time of the representative peak entry are determined by the peak-area-weighted average values that are calculated as follows:
| (1) |
| (2) |
where Apn denotes peak area of the representative peak, Api is peak area of the ith multiple entry to be merged, k is the index number of multiple peak entries to be merged, RTpn denotes retention time of the representative peak, RT pi is the retention time of the ith multiple entry to be merged. Here, the retention time is either the first dimension retention time or the second dimension retention time.
ChromaTOF uses the peak area of the quantitation (quant) mass to represent the abundance of a metabolite. Quant mass refers to a fragment ion that was generated solely by one type of metabolite of multiple co-eluting metabolites. Due to the complexity of the metabolome, it is possible that two or more metabolites are not well separated and the overlapped portion of these metabolites will enter the mass spectrometer together for electron ionization and subsequent fragmentation. In this case, ChromaTOF may select different fragment ions as the quant mass for each of the multiple peak entries even though all of these entries actually originated from the same type of metabolite. If this happens, DISCO selects the quant mass of a peak entry with the maximum peak area as the quant mass of the representative peak. The peak areas of other quant masses used in the remaining peak entries are converted accordingly based on the relative abundances of these fragment ions in the mass spectrum.
Landmark Peak Discovery
DISCO selects landmark peaks using Euclidean distances of the two-dimensional retention times and mass spectrum correlation of two corresponding metabolite peaks. It should be noted that the value of the retention time in the first dimension GC is typically much larger than the retention time in the second dimension GC because of the instrument setup. This imbalance between the two-dimensional retention times weights the contribution of the first dimension retention time much more than the second dimension retention time in the Euclidean distance. To balance the contribution of the two dimensional retention times, the retention time values in both the first and the second dimension GC are transformed into z-scores as following:
| (3) |
where RT1Z is the z-score value after transformation, RT1 is the original value of the first dimension retention time, RT1μ is the mean value of the original first dimension retention times within a peak list, RT1σ is the standard deviation of the original first dimension retention times. Accordingly, the symbols in the second dimension retention time have similar meanings.
After standardizing the two dimensional retention times into z-score values, DISCO aligns GC×GC/TOF-MS data based on a list of landmark peaks found in all peak lists during the primary alignment. The retention times of the landmark peaks are used as reference markers of the retention times in the two GC dimensions to enable effective and accurate alignment. Landmark peak discovery is accomplished by an optimization process which searches peaks using two criteria simultaneously: one is the Euclidean distance of two metabolites in the two-dimensional retention time space after z-score transformation; the other is fragment ion mass spectrum similarity.
For a set of samples S (s1, s2,⋯, sN), DISCO randomly selects one of them as reference sample sR and aligns the remaining samples S̄ = {si} (i = 1,⋯, N − 1) to the level of sR based on a set of landmark peaks. The method of selecting the reference sample will not affect the results of identifying landmark peaks because of the definition of landmark peaks. The following section describes the optimization procedure of the landmark peaks discovery:
Assigning all peaks PR (pr1, pr2,⋯, prk) in the reference sample sR as potential landmark peaks PL (pl1, pl2,⋯, plk), where k is the number of landmark peaks. R0′ is a predefined threshold value of the correlation coefficient between two fragment ion mass spectra of two metabolites from different samples. A variable t used to trace the analyzed samples is initialized as t = 2;
Selecting a sample randomly as the target sample sT from the set of remaining samples S̄, then S̄ = {si} (i = 1,⋯, N − t);
Selecting the first peak from the landmark peaks PL as the working landmark peak pl;
Calculating the Euclidean distances between peak pl and all peaks in the target sample sT to get a set of distances D = {dij} (j = 1,⋯J), where J is the number of metabolite peaks in sT. Sort D in an ascending order to obtain a sorted distance set D′;
-
Selecting the smallest distance in the sorted distance set D′ as the current distance dc, the corresponding metabolite peak in the target sample sT associated with dc is considered as the current peak pc. Hereafter, the correlation coefficient R(pc, pl) between fragment ion mass spectra of peak pc and peak pl is calculated using Pearson’s correlation method, which assumes that a linear function best describes the relationship of fragment ion mass spectra between two metabolite peaks pc and pl.
(4) where Ic and Il are the fragment ion peak intensities of the same m/z value in the two fragment ion spectra, n is the number of data points in the spectra, i.e., the number of the m/z values.
If R (pc, pl) ≥ R0′, peak pc is considered as a corresponding landmark peak of the target sample sT for the landmark peak pl of the reference sample sR and the entire sorted distance set D′ is deleted. Otherwise, the current distance value dc in the sorted distance set D′ is deleted, and DISCO goes to step 5) until a corresponding landmark peak is identified. In case of no match for peak pl in target sample sT, the working landmark peak pl will be removed from the landmark peak set PL;
Going to step 3) to select the next unprocessed peak in the landmark peaks PL as pl and run steps 4)-6) until all the peaks in PL are selected;
Going to step 2), t = t + 1 and run steps 3)-7) until the remaining samples S̄ is empty or half of the computer memory is occupied;
After this analysis, all peaks in the landmark peak list PL have corresponding landmark peaks in all samples involved in this process.
Removal of Potential False-Positives from the Landmark Peak List
The accuracy of determining landmark peaks is critical to the success of the DISCO algorithm. It is likely that the initial landmark peak list PL may contain false-positive landmark peaks. In order to detect and remove these false-positives, we expect that the same two-dimensional GC column configurations were used during the experiments. The same two-dimensional GC column configurations ensure that the elution order of landmark peaks in different samples is the same, even though the elution order of two metabolites with similar retention times may vary under different experiment conditions. The DISCO algorithm employs the metabolite elution order to detect and remove potential false-positive landmark peaks by requiring that all landmark peaks should have the same elution order in both the first and the second GC columns across all samples, respectively.
Each target sample sT and the reference sample sR can form a sample pair (sR, sT). In each sample pair (sR, sT), a landmark peak plr in the reference sample sR has a corresponding landmark peak plt in the target sample sT. These two landmark peaks can form a landmark peak pair (plr, plt). Therefore, k landmark peaks pairs can be formed between the target sample sT and the reference sample sR, where k is the number of landmark peaks.
Based on the value of the retention time of each landmark peak, DISCO first ranks the elution order of all landmark peaks in the reference sample sR and the target sample sT, respectively. It then calculates the absolute value of rank-order difference of each landmark peak pair. The landmark peaks in a landmark peak pair with the maximum rank-order difference are considered as potential false-positive landmark peaks and are removed from the two landmark peak lists. If there are more than one and the landmark peak pairs have the same maximum rank-order difference, the DISCO algorithm removes the one with the maximum retention time difference. If their retention time differences are also the same, all of them are removed. This process is repeated until the maximum rank-order difference is zero. All the corresponding landmark peaks in the rest of sample are also removed if a landmark peak is removed from the reference sample sR.
An example of removing false-positive landmark peaks from a sample pair with 10 original potential landmark peak pairs is showed in Figure 2. After calculating the rank-order of all landmark peaks in the reference sample sR and the target sample sT, the landmark peak pair highlighted in blue has the largest rank-order difference of 5. The two landmark peaks in this landmark peak pair are removed (Step 1). The rank-order of the remaining landmark peaks in the reference sample and the target sample are evaluated again, respectively. The two landmark peak pairs with a rank-order difference of 2 have the largest retention time difference. The two peaks that formed the blue landmark peak pair are then removed from the landmark peak lists (Step 2). This process is repeated in Step 3 to remove the two landmark peaks whose landmark peak pair has a rank-order difference of 1. After this process, the maximum rank-order difference of all landmark peak pairs is zero and we consider that these remaining landmark peaks are the true landmark peaks.
Figure 2.
A sample process of detecting and removing potential false positive landmark peaks from the initial landmark peak list. DISCO first ranks the elution order of all landmark peaks in the reference sample and the target sample, respectively. It then calculates the absolute value of rank-order difference of each landmark peak pair. The landmark peaks in a landmark peak pair with the maximum rank-order difference are considered as potential false-positive landmark peaks and removed from the two landmark peak lists. This process is repeated until all landmark peak pairs have zero rank-order difference.
It is possible that some true positive landmark peaks may be removed during this elution order filtering process, especially in the second dimension GC retention time domain. However, the accuracy of landmark peaks is much more important because the retention times of all metabolites in each sample will be adjusted based on the retention times of the landmark peaks. In metabolomics studies, there are many metabolites present in each of the biological samples even though the relative abundance of these metabolites may vary from sample to sample because of biological variations and changes in biological environment. The number of landmark peaks is usually quite large, which is actually computational expensive for the downstream analysis. Removing some of the true positive landmark peaks will not affect the accuracy of alignment.
Retention Time Correction
Based on the retention times of the landmark peaks, the DISCO algorithm corrects the two dimensional retention time shifts of all metabolite peaks present in each peak list using a two-step approach. It first assigns the values of the retention time of a landmark peak in the reference sample to the retention time of all corresponding landmark peaks in the remaining samples. It then uses a local partial linear fitting function to interpolate the retention time of non-landmark peaks located between two landmark peaks in each retention time dimension, respectively. Because multiple landmark peaks can be detected in a set of experimental data, adjusting retention time shifts using two adjacent landmark peaks is capable of correcting non-linear retention time shifts. This process works as follows:
For a sample pair (sR, sT), where sR is the reference sample and sT is the target sample, its landmark peak pair list is (PR, PT), where PR = (pR1, pR2,⋯, pRk), PT = (pT1, pT2,⋯, pTk), and k is the number of landmark peaks. We denote retention times of all peaks, i.e., landmark peaks and non-landmark peaks, in the reference sample as RTR = (rtR1, rtR2,⋯, rtRA), while retention times of all peaks in the target sample as RTT = (rtT1, rtT2,⋯, rtTB). The DISCO algorithm first partitions the RTT and RTR into k + 1 segments based on the landmark peaks list, i.e.,
| (5) |
| (6) |
where rtPR0 = rtR1, rtPRk+1 = rtRA, rtpT0 = rtT1, rtpTk+1 = rtTB
Then DISCO stretches or compresses each retention time segment rtTSi in RTT into the section of its corresponding segment rtRSi in RTR based on the corresponding landmark peaks in sR and sT, and corrects the value of two dimensional retention times of all peaks in the segment using a linear fitting method:
| (7) |
The DISCO algorithm first assigns the values of the retention times of the landmark peaks in the reference sample to the retention time of all corresponding landmark peaks in the target sample. A local partial linear fitting function is then employed to interpolate the retention time of non-landmark peaks located between two landmark peaks, as shown in Figure 3. For non-landmark peaks prior to the first landmark and after the last landmark peak, DISCO applies a linear fitting method to adjust their retention time based on the closest landmark peak.
Figure 3.
Schematic of retention time correction. DISCO first assigns the values of the retention time of a landmark peak in the reference sample to the retention time of all corresponding landmark peaks in the target sample. It then uses a local partial linear fitting function to interpolate the retention time of non-landmark peaks located between two landmark peaks in each retention time dimension, respectively.
Peak Alignment
After correcting the retention time shifts of all peaks in every sample, DISCO aligns metabolite peaks in all samples into a single peak table. This table is constructed based on the landmark peaks using a progressive retention time map searching methodology described as follows:
Selecting a reference sample sR from sample set S(s1, s2,⋯,sN) and denoting the remaining samples as S̄= {si} (i = 1,⋯, N − 1).
The corresponding peak list of the reference sample is defined as the reference table (RefTbl), and the corresponding peak lists of the remaining samples are designated as search tables (SchTbl) numbered from SchTbl1 to SchTbln−1. A variable m is initialized m = 1;
Selecting the mth search table SchTblm from the search tables and matching each landmark peak in SchTblm to the landmark peaks in RefTbl.
-
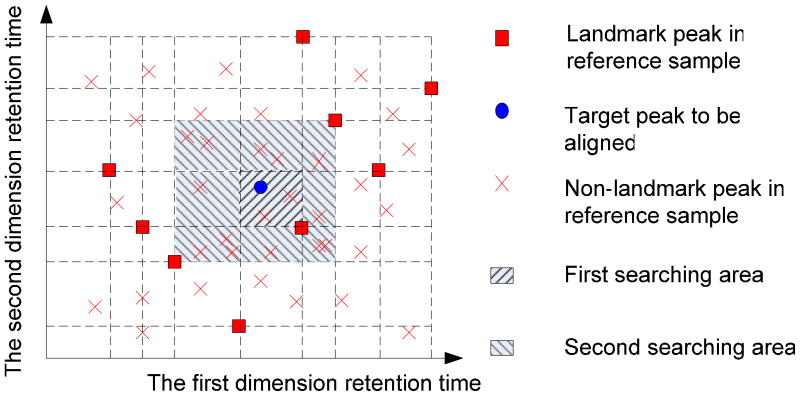
To align the non-landmark peaks in the two-dimensional retention time space, DISCO partitions SchTblm and RefTbl into patches in the two-dimensional retention time space based on their landmark peaks as Figure 4;
For each non-landmark peak in SchTblm, DISCO designates the patch where the non-landmark peak is located as the first searching area and the contiguous patches as the secondary searching areas; DISCO searches the matching peak from all peaks of RefTbl in the first searching area by optimizing the balance between the Euclidean distance and the fragment ion (EI) mass spectrum correlation described in the landmark peak discovery process. If a matching peak is not found, the algorithm searches the matching peak in the secondary searching area. If there is no matching peak in the secondary searching area, the non-landmark peak will be added into RefTbl as a new reference peak; otherwise, DISCO aligns this non-landmark peak to the matching peak in RefTbl, and the reference values in RefTbl are updated as follows for searching the next peak table SchTblm+1:(8) (9) (10) where AR is the peak area of matching peak in RefTbl, AS is the peak area of the non-landmark peak in SchTblk, RTR1 and RTR2 are the first and second dimension retention time of the matching peak in RefTbl, respectively, RTS1 and RTS2 are the first and second dimension retention time of the non-landmark peak in SchTblm, respectively, and are peak area weighted first and second dimension retention time of the aligned peak, respectively. Accordingly, SR, SS, are fragment ion mass spectra of the matching peak in RefTbl, the non-landmark peak in SchTblm and the aligned new reference peak, respectively.
Updating m = m +1 and going to step 3). The peak alignment is completed when m = N − 1.
Figure 4.
Schematic of progressive retention time map searching to align a non-landmark peak in a target sample to another non-landmark peaks in the reference sample. DISCO searches the matching peak from all peaks of reference sample in the first searching area by optimizing the balance between the Euclidean distance and the mass spectrum correlation. If a matching peak is not found, the algorithm searches the matching peak in the secondary searching area. If there is no matching peak in the secondary searching area, the non-landmark peak will be considered as a new reference peak; otherwise, this non-landmark peak is aligned to the matching peak in reference sample.
RESULTS AND DISCUSSION
In large scale metabolomics, tens or even hundreds of biological samples are analyzed in one study to obtain more statistically sound conclusions. A high-throughput instrument like GC×GC/TOF-MS can generate a significant amount of experimental data in a relatively short period of time. The raw instrumental data of a GC×GC/TOF-MS experiment can easily reach 1 GB for a 40 min analysis of a biological sample, such as human plasma extract. Using raw instrument data for alignment is a time consuming process and causes a significant computer memory challenge. For this reason, we used the instrument control software ChromaTOF to process each raw data file for metabolite identification and peak picking. The output of the ChromaTOF software is a list of metabolite peaks detected in each sample, in which each metabolite peak is characterized by a series of features including retention times in the two-dimensional GC, peak area, and the mass spectrum containing peaks (m/z values and ion intensities).
We tested the performance of DISCO by aligning three datasets, which are replicate analyses of the same sample using an identical two dimensional GC configuration with different column temperature gradients, i.e., 5°C/min, 7°C/min, and 10°C/min, respectively. To evaluate the performance of our proposed algorithm more objectively, the experiments ramped at different temperature gradients have been repeated different times, i.e., 10 replicate analyses for 5°C/min, 3 replicate analyses for 7°C/min, and 4 replicate analyses for 10°C/min. We denote each experimental result as Sab, where a refers to the temperature gradient, and b is the experimental run. For example, S51 denotes the first run in 5°C/min temperature gradient.
Peak Entry Merging
As a preprocessing task to peak alignment, peak entry merging is very important for its influence on the quality of the downstream multivariate analysis. Multiple peak entries of the same metabolite can be recognized using the combined information based on the spectrum similarity between metabolites and the metabolite retention time in the first dimension GC. The DISCO algorithm automatically detects the value of the modulation time Δ in each peak list using the minimum non-zero retention time gap among all peaks in the peak list. It then considers all peak entries with the spectrum similarity ≥R0 (a predefined threshold value of the spectrum correlation coefficient) and retention time difference ≤Δ were originated from the same type of metabolite, and merges these multiple entries into one representative peak entry.
During the process of peak merging and landmark peak discovery, the values of threshold R0 are important to the decision whether the metabolite peaks under consideration are generated by the same type of metabolite. In order to find an optimal threshold value R0, we manually picked all mass spectra of the standard compounds from all 17 experiments and calculated Pearson’s correlation coefficients between every mass spectrum pair. In this process, correlation values of 770,995 mass spectrum pairs were generated and then the relation between true positive rate (TPR) and false positive rate (FPR) were studied at different correlation threshold. Here, the ‘positive’ means that a pair of mass spectra is from the same compound and the ‘negative’ denotes a pair of mass spectra is from two different standard compounds. During TPR and FPR calculation, a mass spectrum pair is classified as positive if their correlation value is higher than the threshold; otherwise it is classified as negative. Let TP be the number of true positives, i.e., spectrum pairs classified to be positive pairs that actually are from the same standard compound, and let FP be the number of false positives, i.e., spectrum pairs classified to be positive pairs are in fact from different standard compounds. In addition, let TN be the number of true negatives, and FN the number of false negatives. Then TPR and FPR for a given threshold can be computed as follows:
| (11) |
Because Pearson’s coefficient has the interval of [−1, 1], we changed the threshold value from −1 to 1 and calculated the values of TPR and FPR. The receiver operating characteristic (ROC) curve can be obtained as shown in Figure 5 by varying the threshold of the correlation coefficient. Every point on the ROC curve has a corresponding threshold value. Ideally, the fragment ions of the same type of standard compound should have the identical mass spectrum, and therefore the Pearson’s correlation coefficient should be 1.0. If a higher threshold R0 is selected, higher matching accuracy of merging peaks and landmark peaks can be guaranteed. However, this is not true because of various experimental variations contributing to the resulting mass spectrum, which makes that mass spectra originated from the same type of compound not identical anymore. An extremely high value of R0 can cause significant false-negatives during peak merging and landmark peaks discovery, and therefore, this induces less number of landmark peaks and potentially poor accuracy in retention time correction. In this work, we set the threshold of correlation value R0 as 0.90, which results in TPR = 0.959 and FPR = 0.038.
Figure 5.
Receiver operation characteristic (ROC) curve. TPR is true positive rate, and FPR is false positive rate. Each point on the ROC curve has a corresponding threshold value. Therefore, the threshold of spectrum similarity can be decided by the expected TPR and FPR.
Figure 6(a) shows the performance of peak entry merging in sample S51. There were 224 metabolite peak entries in the original peak list generated by the ChromaTOF software. After peak entry merging, the number of peaks was reduced to 195. During this peak entry merging process, DISCO first found Δ = 4.99 s from the peak list. The modulation time defined during experiments was 5 s. In order to take this time difference into account, DISCO increased the detected value of Δ to a factor of 1.1 times so that Δ = 5.49 sec. Figure 6(b) is a subset of Figure 6(a) highlighted by a circle. There are four peaks, i.e., peak 1, 2, 3 and 4, in the original peak list highlighted in blue. The retention times of these four peaks in the two dimensional GC are (329.75, 0.937), (409.69, 0.950), (414.69, 0.891) and (414.69, 0.96) (all values in s), respectively. Due to the fact that the difference of the first dimension retention time between peak 1 and 2 is larger than Δ, these two peaks were not merged. However, the difference of first dimension retention time between peak 2 and 3 is smaller than Δ and the spectrum correlation coefficient is larger than 0.90, so peak 2 and 3 were treated as multiple peak entries generated from the same metabolite. A similar decision was made by DISCO for peak 3 and 4. After identifying all peak entries generated by the same metabolite, these peak entries were merged and a representative peak 2′ colored in red was generated and inserted into the peak list as a substitute peak of the identified multiple peak entries, i.e., peak 2, 3, and 4. Table 2 shows the number of molecules in each sample after peak entry merging, from which we can find about 10% of peaks in the peak lists were merged. It should be noted that we did not identify all molecules in our sample due to experimental variations, sample concentration, and limited accuracy of metabolite identification software.
Figure 6.
Results of peak entry merging in sample S51. (a) displays the original peaks (colored in blue stars) reported by ChromaTOF software and the merged peaks (colored in red circles). (b) is a subset of (a) highlighted by a circle. Peak 1 was not merged with other peaks while peaks 2, 3, and 4 are merged and a representative peak 2′ is generated as substitute of these three peak entries. A peak-area-weighted method was used to determine the retention times of the representative peak 2′ in the two-dimensional retention time space.
Table 2.
The number of compounds in each sample after peak entry merging.
| Sample | Number of peaks | Number of peaks assigned | |||
|---|---|---|---|---|---|
| Original | After merging | STa | Alkaneb | ISTDc | |
| S51 | 224 | 195 | 56 | 28 | 6 |
| S52 | 214 | 194 | 59 | 28 | 6 |
| S53 | 202 | 180 | 56 | 28 | 6 |
| S54 | 190 | 174 | 55 | 28 | 6 |
| S55 | 182 | 165 | 56 | 27 | 6 |
| S56 | 177 | 159 | 54 | 26 | 6 |
| S57 | 209 | 187 | 56 | 27 | 6 |
| S58 | 198 | 172 | 58 | 28 | 6 |
| S59 | 204 | 175 | 58 | 27 | 6 |
| S510 | 202 | 176 | 55 | 28 | 6 |
| S71 | 156 | 142 | 56 | 27 | 6 |
| S72 | 201 | 178 | 55 | 27 | 6 |
| S73 | 154 | 141 | 56 | 27 | 6 |
| S101 | 187 | 171 | 59 | 26 | 6 |
| S102 | 168 | 157 | 56 | 25 | 6 |
| S103 | 145 | 129 | 57 | 26 | 6 |
| S104 | 157 | 146 | 57 | 25 | 6 |
The mixture of 76 compounds
C7-C4c saturated alkanes
Dueterated six component semivolatiles internal standard (1STD) mixture
Performance of Z-score Transformation
As described before, the original values of the retention times should be converted into a z-score transformation space to balance the contribution of the two dimensional retention times to the Euclidean distance. As a data normalization method, z-score transformation can also reduce inter-experiment variations induced by factors such as differences of the instrumental parameter settings and experiment-to-experiment variations.
To demonstrate the performance of the DISCO algorithm in analyzing experimental data acquired under different experiment conditions, we intentionally varied the temperature gradient from 5°C/min, 7°C/min, to 10°C/min. Figure 7(a) shows the distribution of metabolite peaks in the two dimensional retention time space. We set the maximum column temperature in the first and the second GC column to 315°C and 320°C, respectively. Therefore, the different temperature gradients result in a different retention time scale for the elution of all metabolites from the two dimensional GC columns. A large temperature gradient induces a small retention time scale. For the datasets acquired under the temperature gradient of 5°C/min, all metabolites eluted from the first dimension GC column in 3000 s. For the temperature gradient of 7°C/min and 10°C/min, the same set of metabolites eluted from the first dimension GC column in 2500 and 2000 s, respectively. There is no significant difference among the second dimension retention time within the three datasets acquired under the three different temperature gradients. After a z-score transformation, the scales of the two-dimensional retention times are similar between the three temperature gradients (Figure 7(b)), i.e., −1.8 to + 2.5 in the first dimension and −2.1 to + 5.0 in the second dimension. Therefore, using the z-score transformed retention time values for the calculation of Euclidean distance guarantees the contribution of the first and the second dimension retention times are equally weighted.
Figure 7.
Retention time distributions of standard compounds, analyzed under different temperature gradients, in the two-dimensional retention time space before and after z-score transformation. (a) is the distribution of metabolite peaks in the original two-dimensional retention time space before z-score transformation and (b) is the distribution of metabolites in the z-score transformation space. Blue stars (*) are metabolites detected during the temperature gradient of 5°C/min. Red circles (○) are metabolites detected during 7°C/min and black triangles (Δ) are metabolites detected during 10°C/min.
Landmark Peak Discovery
Landmark peak discovery is a key step in the DISCO algorithm because retention time shifts of every peak are corrected based on the retention times of all landmark peaks. After landmark peak selection, the two-dimensional retention times of all peaks in all samples are corrected to the same scale so that they can be compared directly to each other for the downstream statistical analysis such as pattern recognition or statistical significance tests. To enable such an analysis process, a reference sample should be chosen as a standard to which the remaining samples can be aligned.
The DISCO algorithm randomly selects a reference sample from the sample pool. By the definition of landmark peaks, the method of selecting a reference sample should not affect the final results of landmark peak detection. In practice, it is required that a quality assessment (QA) process such as the Kolmgorov-Smirnov test33 should be applied to the peak lists of all samples to detect the outlier samples that may have few peaks detected or have a peak distribution significantly different from that of the samples acquired from the same biological cohort. This QA process will make sure that the samples subjected to alignment contain limited technical or biological variations. Therefore, the peak list of each sample contains metabolites that are also detected, if not all, in the other samples. Any of these samples can be selected as a reference sample. In this work, sample S51 was selected as the reference sample and the rest of the samples therefore are considered as target samples.
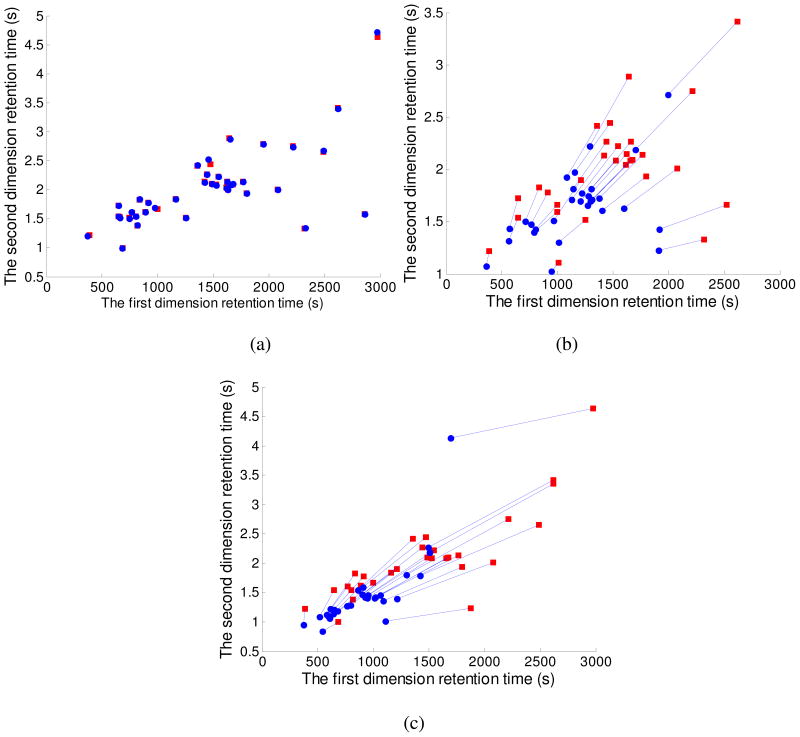
For the landmark peak detection and alignment, the user can set another threshold of spectrum similarity R0′ to determine whether the two mass spectra acquired in different samples were generated by the same type of metabolites. The value of R0′ is usually set to a value smaller than R0′ because the spectrum variation across experiments is larger than that within an experiment. In this work, we set R0′ to 0.90 for the analysis of mixture of standard compounds. S51 and S 2 are two replicate analysis of the same sample using GC×GC/TOF-MS. Figure 8(a) displays the coordinates of the landmark peaks between S51 and S52 in the original two-dimensional retention time space. It can be seen that the landmark peaks found by the DISCO algorithm are correct because their spatial positions are superposed on top of each other. Figures 8(b) and (c) display the coordinates of the landmark peaks between S51 and S71, as well as S51 and S101. Figure 8 demonstrates the performance of the DISCO algorithm in discovering landmark peaks from experimental data acquired under different experimental conditions. The retention times of the corresponding landmark peaks in different samples shift in the same direction, and the shift is roughly proportional to the value of the retention time. Even though the retention time shifts caused by different experimental conditions are very large, i.e., over 1000 s in the first GC dimension and over 1 s in the second GC dimension retention time; the corresponding peaks generated by the same metabolite can still be detected by the DISCO algorithm. This result, in return, proves our assumption that the same group of molecules has the same elution orders in an identical two dimension GC column configuration.
Figure 8.
Corresponding landmark peaks discovered between two samples. (a) between S51 and S52; (b) between S51 and S71; (c) between S51 and S101. In the figure, blue circles are landmark peaks in sample S51, and red squares are corresponding landmark peaks in sample S52 in (a), S71 in (b) and S101 in (c), respectively.
Fifty landmark peaks were detected from the 17 peak lists, among them 6 were from alkanes, 42 from the 76 standard compounds and 2 from ISTD. After removing false-positives, 34 landmark peaks were used for the retention time correction. These landmark peaks cover 98.3% of the retention time space in the first dimension and 93.5% in the second dimension. The large percentage of retention time space coverage demonstrates the rationality of landmark peaks selection, and insures the reliability for retention time correction and peak alignment.
Retention Time Correction
Based on the landmark peaks, the DISCO algorithm corrects retention time shifts based on the distribution of landmark peaks in the reference sample and the target samples. If a peak is in the landmark peak list, its two-dimensional retention times in the target samples are corrected to the same values of the corresponding landmark peak in the reference sample. Otherwise, its retention times are corrected along with stretching or compression, also known as warping, in the two retention time dimensions using a local linear fitting algorithm.
To enable retention time correction of non-landmark peaks, DISCO segments the entire two-dimensional retention time space into many small adjoining sections in each sample based on the distribution of landmark peaks. For each section in a target sample, DISCO corrects the retention time of all peaks located in this section using a linear interpolation function in the two retention time dimensions based on the retention times of the two adjacent landmark peaks, respectively. The distributions of retention times of each sample before and after correction are showed in Figure 9. It can be seen from Figure 9(a) and 9 (c), that the distributions of retention time of the three datasets acquired under different temperature gradients are different in the original first dimension retention time space and irregular distributed in the second-dimension. After retention time correction, the retention time distributions are very similar (Figure 9(b) and 9 (d)). This result demonstrates that the retention times of all peaks in each sample have been corrected to the same level of retention times of corresponding metabolite peaks in the reference sample S51. A similar result can be obtained by choosing a different reference sample (data not shown here).
Figure 9.
Distribution of retention times before and after retention time correction. Probability density function (PDF) of the retention time in each sample is computed using the normal distribution. (a) is the first dimension retention time before correction. (b) is the first dimension time after correction. (c) is the second dimension retention time before correction. (d) is the second dimension retention time after correction.
Analysis of Spiked-in Experiment Data
Furthermore, we evaluated the performance of DISCO by analyzing real biological samples in a spiked-in experiment, i.e., a MTBSTFA derivatized metabolites extracted from rat plasma with spiked-in ISTD compounds was analyzed five times on GC×GC/TOF-MS platform. After processing the instrumental data using ChromaTOF, five peak lists were generated. Each of them consisted of 759, 733, 695,727 and 661 peak entries, respectively.
One of the ISTD spiked-in compounds, 1,4-dichlorobenzene-D4, was found in only one experimental data after ChromaTOF analysis, while the other five compounds were identified in all of the five experiments. We evaluated the performance of DISCO based on the aligned results of the remaining five compounds presented in all of the five peak lists generated by ChromaTOF, where the parameter setting of ChromaTOF is the same as the analysis of the mixture of the standard compounds.
The five peak lists were first aligned using the default spectrum similarity setting for peak merging, i.e., R0′ = 0.90 One of the five compounds, acenaphthene-D10, was aligned into two groups. One group consisted of spiked-in samples 1, 2 and 3 and the other group consists of spiked-in samples 4 and 5. The other four compounds were perfectly aligned in all samples. When the value of R0′ was set to 0.85, all of the five compounds were correctly aligned together in all of the five samples. This means that the threshold of mass spectrum similarity R0′ = 0.90 was too large to align all corresponding peaks acquired in different experiments because the variations of mass spectra between experiments are larger than the mass spectrum variations within an experiment. We manually compared the mass spectra of the compound acenaphthene-D10 from the five samples and its spectrum recorded in the NIST database (Figure 10). The spectrum similarity between the NIST standard spectrum and spectra obtained in our spiked-in experiments fluctuated from 0.916 to 0.965. However, the spectrum pairs among our five experiments have poor similarity, as shown in Table 3. The spectrum similarity between the injection 1 and the injection 5 is only 0.865. This explains why our DISCO correctly aligned Acenaphthene-D10 compound in all of the five spiked-in experiments when the threshold of spectrum similarity was set to R0′ = 0.85, but failed when the value of R0′ was set to 0.90.
Figure 10.
Comparison of the mass spectra of compound acenaphthene-D10 acquired in the five replicate spiked-in experiments and the standard spectrum in NIST database. The value of CC is the Pearson’s correlation coefficient between the spiked-in spectrum and the standard spectrum.
Table 3.
The spectrum similarity across different spiked-in experiments for compound acenaphthene-d10.
| Spiked-in1 | Spiked-in2 | Spiked-in3 | Spiked-in4 | Spiked-in5 | |
|---|---|---|---|---|---|
| Spiked-in1 | 1.00 | ||||
| Spiked-in2 | 0.997 | 1.00 | |||
| Spiked-in3 | 0.996 | 0.999 | 1.00 | ||
| Spiked-in4 | 0.893 | 0.903 | 0.899 | 1.00 | |
| Spiked-in5 | 0.865 | 0.886 | 0.885 | 0.975 | 1.00 |
DISCO set the default threshold R0 = 0.90 for peak merging based on the ROC calculation of the 17 experiments of 117 standard compounds. However, this default correlation threshold is higher for R0′, which is the threshold used for peak alignment across different experiments, especially for the analyses of biological samples. With the increase of sample complexity in the biological samples, the chance of peak overlapping is increased and therefore, more variability is introduced in the metabolite mass spectrum during spectrum deconvolution. In this sense, the threshold of spectrum correlation R0′ should be set to a lower value based on the sample complexity. However, significantly lowering the threshold of spectrum correlation diminishes the contribution of mass spectrum similarity to the alignment and increases the chance of false alignment. We tested the alignment performance of DISCO in analysis of the spiked-in experimental data under different values of R0 and R0′, and found that DISCO can correctly align all of the five ISTD compounds when R0 was set to 0.90 and R0′ was ranged from 0.50 to 0.85. Meanwhile, the compound 1,4-dichlorobenzene-D4 was not aligned to any other metabolites in the samples. This, on the other hand, indicates the high specificity of DISCO. However, false negative alignment appears in the ISTD compounds when the value of R0′ is smaller than 0.50. For example, phenathrene-D10 can not be aligned in all of the five samples when R0′ = 0.45. This observation indicates that using the two dimensional retention times as the only input information for metabolite alignment can cause high rate of false alignment for GC×GC/TOF-MS platform based metabolomics. The combination of the metabolite two dimensional retention times and mass spectrum similarity can provide high accuracy of alignment and it is not necessary to set a high threshold of mass spectrum similarity. For this reason, we suggest the threshold of mass spectrum similarity should be adjusted to 0.80 for biological samples. In our DISCO software, this threshold can be changed by the user.
CONCLUSIONS
A new peak alignment algorithm named DISCO is proposed to align the GC×GC/TOF-MS data for metabolomics. The DISCO algorithm uses the output of the instrument control software, ChromaTOF, as its input data. It first detects and merges multiple peak entries of the same metabolite into one peak. After z-score transformation of the metabolite retention times, DISCO selects landmark peaks from a certain number of samples based on both two-dimensional retention times and the mass spectrum similarity measured by Pearson’s correlation coefficient. Then the original two-dimensional retention time space is split into many sections using the landmark peaks for the correction of retention time shifts using a local linear fitting function in each retention time dimension, respectively. A progressive retention time map searching method is used to align metabolite peaks in all samples together based on the optimization of Euclidean distance and mass spectrum similarity.
DISCO reduces the rate of false-positive alignment by employing the two dimensional GC retention times and fragment ion spectrum correlation. Another advantage of DISCO compared to all other warping algorithms is that it partitions each retention time section automatically based on the landmark peaks. The section parameters like section length and maximum warping are not required. This can avoid the problems caused by improper parameter selection and inconsistency among samples. Compared to our previous MSort algorithm which uses spectrum similarity and a fixed retention time windows to align corresponding peaks in different samples, the Euclidean distance in z-score transformation space is used in DISCO for landmark peaks searching which can handle the retention time shift cased by different experimental conditions. Meanwhile, DISCO employs a local linear fitting method for retention time correction and peak alignment based on the landmark peaks, which enables DISCO to deal with the nonlinear distortion of retention time shift caused by experimental errors. DISCO also enables on-the-fly alignment to avoid the problem of consuming large computer memory and therefore, it is able to process data generated from large scale experiments.
We tested the performance of the DISCO algorithm by aligning metabolite peak lists of samples analyzed under different experiment conditions. A higher percentage of the landmark peaks coverage in the two-dimensional retention time space demonstrates the effectiveness of landmark peaks selection in DISCO, and guarantees the reliability of peak retention time shift correction and peak alignment. Further, DISCO was evaluated by a spiked-in experiment. The results show that our algorithm can work well in alignment of metabolites from real biological samples. The excellent performance of the DISCO algorithm makes it possible to perform inter-laboratory studies or to re-analyze historical data as long as the experimental data were acquired using identical two dimensional GC configurations. With the rapid development of high-throughput instruments and the large number of metabolites to be discovered from biological samples, hundreds of biological samples can be analyzed in one experimental project with an increased volume of experimental data. As a critical step of data pre-processing, the alignment results achieved by DISCO can be used effectively for further analysis such as pattern recognition and statistical significance testing in metabolomics study.
Acknowledgments
This work was supported by NIH grant 1RO1GM087735-01. The authors also thank University of Louisville for partial financial support by the Competitive Enhancement Grant.
References
- 1.Dettmer K, Aronov PA, Hammock BD. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imasaka T, Nakamura N, Sakoda Y, Yamaguchi S, Watanabe-Ezoe Y, Uchimura T. Analyst. 2009;134:712–718. doi: 10.1039/b815630a. [DOI] [PubMed] [Google Scholar]
- 3.Jia L, Liu BF, Terabe S, Nishioka T. Anal Chem. 2004;76:1419–1428. doi: 10.1021/ac035039b. [DOI] [PubMed] [Google Scholar]
- 4.Pierce KM, Hoggard JC, Mohler RE, Synovec RE. J Chromatogr A. 2008;1184:341–352. doi: 10.1016/j.chroma.2007.07.059. [DOI] [PubMed] [Google Scholar]
- 5.Bedair M, Sumner LW. Trac-Trends in Analytical Chemistry. 2008;27:238–250. [Google Scholar]
- 6.Ong RC, Marriott PJ. J Chromatogr Sci. 2002;40:276–291. doi: 10.1093/chromsci/40.5.276. [DOI] [PubMed] [Google Scholar]
- 7.Shellie R, Marriott PJ. Anal Chem. 2002;74:5426–5430. doi: 10.1021/ac025803e. [DOI] [PubMed] [Google Scholar]
- 8.Kusano M, Fukushima A, Kobayashi M, Hayashi N, Jonsson P, Moritz T, Ebana K, Saito K. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:71–79. doi: 10.1016/j.jchromb.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Perera RMM, Marriott PJ, Galbally IE. Analyst. 2002;127:1601–1607. doi: 10.1039/b208577a. [DOI] [PubMed] [Google Scholar]
- 10.Hope JL, Prazen BJ, Nilsson EJ, Lidstrom ME, Synovec RE. Talanta. 2005;65:380–388. doi: 10.1016/j.talanta.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Pierce KM, Hope JL, Hoggard JC, Synovec RE. Talanta. 2006;70:797–804. doi: 10.1016/j.talanta.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 12.Mohler RE, Dombek KM, Hoggard JC, Pierce KM, Young ET, Synovec RE. Analyst. 2007;132:756–767. doi: 10.1039/b700061h. [DOI] [PubMed] [Google Scholar]
- 13.Mohler RE, Dombek KM, Hoggard JC, Young ET, Synovec RE. Anal Chem. 2006;78:2700–2709. doi: 10.1021/ac052106o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohler RE, Tu BP, Dombek KM, Hoggard JC, Young ET, Synovec RE. J Chromatogr A. 2008;1186:401–411. doi: 10.1016/j.chroma.2007.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo X, Lidstrom ME. Biotechnol Bioeng. 2008;99:929–940. doi: 10.1002/bit.21652. [DOI] [PubMed] [Google Scholar]
- 16.David F, Tienpont B, Sandra P. J Sep Sci. 2008;31:3395–3403. doi: 10.1002/jssc.200800215. [DOI] [PubMed] [Google Scholar]
- 17.Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmermann R, Fiehn O. Metabolomics. 2005;1:65–73. doi: 10.1016/j.chroma.2005.05.088. [DOI] [PubMed] [Google Scholar]
- 18.Shellie RA, Welthagen W, Zrostlikova J, Spranger J, Ristow M, Fiehn O, Zimmermann R. J Chromatogr A. 2005;1086:83–90. doi: 10.1016/j.chroma.2005.05.088. [DOI] [PubMed] [Google Scholar]
- 19.Sinha AE, Hope JL, Prazen BJ, Nilsson EJ, Jack RM, Synovec RE. J Chromatogr A. 2004;1058:209–215. [PubMed] [Google Scholar]
- 20.O’Hagan S, Dunn WB, Knowles JD, Broadhurst D, Williams R, Ashworth JJ, Cameron M, Kell DB. Anal Chem. 2007;79:464–476. doi: 10.1021/ac061443+. [DOI] [PubMed] [Google Scholar]
- 21.Tranchida PQ, Costa R, Donato P, Sciarrone D, Ragonese C, Dugo P, Dugo G, Mondello L. J Sep Sci. 2008;31:3347–3351. doi: 10.1002/jssc.200800289. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Xu ZL, Lu X, Yang XH, Yin PY, Kong HW, Yu Y, Xu GW. Analytica Chimica Acta. 2009;633:257–262. doi: 10.1016/j.aca.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 23.Koek MM, Muilwijk B, van Stee LL, Hankemeier T. J Chromatogr A. 2008;1186:420–429. doi: 10.1016/j.chroma.2007.11.107. [DOI] [PubMed] [Google Scholar]
- 24.Lacorte S, Ikonomou MG, Fischer M. Journal of Chromatography A. 2010;1217:337–347. doi: 10.1016/j.chroma.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Huang XD, Regnier FE. Analytical Chemistry. 2008;80:107–114. doi: 10.1021/ac071263f. [DOI] [PubMed] [Google Scholar]
- 26.Fraga CG, Prazen BJ, Synovec RE. Anal Chem. 2001;73:5833–5840. doi: 10.1021/ac010656q. [DOI] [PubMed] [Google Scholar]
- 27.Prazen BJ, Synovec RE, Kowalski BR. Analitical Chemistry. 1998;70:205–429. [Google Scholar]
- 28.van Mispelaar VG, Tas AC, Smilde AK, Schoenmakers PJ, van Asten AC. J Chromatogr A. 2003;1019:15–29. doi: 10.1016/j.chroma.2003.08.101. [DOI] [PubMed] [Google Scholar]
- 29.Pierce KM, Wood LF, Wright BW, Synovec RE. Anal Chem. 2005;77:7735–7743. doi: 10.1021/ac0511142. [DOI] [PubMed] [Google Scholar]
- 30.Zhang D, Huang X, Regnier FE, Zhang M. Anal Chem. 2008;80:2664–2671. doi: 10.1021/ac7024317. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen NPV, Carstensen JM, Smedsgaard J. Journal of Chromatography A. 1998;805:17–35. [Google Scholar]
- 32.Oh C, Huang X, Regnier FE, Buck C, Zhang X. J Chromatogr A. 2008;1179:205–215. doi: 10.1016/j.chroma.2007.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Press WH. Numerical recipes in C++: the art of scientific computing. 2. Cambridge University Press; Cambridge, UK; New York: 2002. [Google Scholar]