Abstract
In a recent study, a single nucleotide polymorphism (SNP) of the Toll-like receptor 4 (TLR4) gene (c.1196C>T [rs4986791, p.T399I]) emerged as conferring protection from fibrosis progression compared to a major, wild-type (WT) CC allele (p.T399). The present study examined the functional linkage of this SNP, along with another common, highly cosegregated TLR4 SNP (c.896A>G [rs4986790, p.D299G]), to hepatic stellate cell (HSC) responses. Both HSCs from TLR4−/− mice and a human HSC line (LX-2) reconstituted with either TLR4 D299G and/or T399I complementary DNAs were hyporesponsive to lipopolysaccharide (LPS) stimulation compared to those expressing WT TLR4, as assessed by the expression and secretion of LPS-induced inflammatory and chemotactic cytokines (i.e., monocyte chemoattractant protein-1, interleukin-6), down-regulation of bone morphogenic protein and the activin membrane-bound inhibitor expression (an inhibitory transforming growth factor β pseudoreceptor), and activation of a nuclear factor κB (NF-κ)–responsive luciferase reporter. In addition, spontaneous apoptosis, as well as apoptosis induced by pathway inhibitors of NF-κB, extracellular signal-regulated kinase (ERK), and phosphatidylinositol 3-kinase were greatly increased in HSCs from either TLR4−/− or myeloid differentiation factor 88−/− (a TLR adaptor protein) mice, as well as in murine HSCs expressing D299G and/or T399I SNPs; increased apoptosis in these lines was accompanied by decreased phospho-ERK and Bcl-2.
Conclusion
TLR4 D299G and T399I SNPs that are associated with protection from hepatic fibrosis reduce TLR4-mediated inflammatory and fibrogenic signaling and lower the apoptotic threshold of activated HSCs. These findings provide a mechanistic link that explains how specific TLR4 SNPs may regulate the risk of fibrosis progression.
The natural history and progression of fibrosis are highly variable.1 While a number of host factors including age, sex, body mass index, and alcohol use contribute to this variable risk (reviewed in Asselah et al.2), a significant component is attributable to genetic determinants that have not been fully identified. Several reports have described genetic variants that are associated with fibrosis progression. We recently conducted a gene-centric functional genome scan in patients with chronic hepatitis C virus, which yielded a Cirrhosis Risk Score signature consisting of seven single nucleotide polymorphisms (SNPs) that may predict the risk of developing cirrhosis.3 Among these, a major CC allele of Toll-like receptor 4 (TLR4) encoding a threonine at amino acid 399 (p.T399) was the second most predictive SNP among the seven, indicating a protective role in fibrosis progression of its c.1196C>T (rs4986791) variant at this location (p.T399I), along with another highly cosegregated c.896A>G (rs4986790) SNP located at coding position 299 (p.D299G). These SNPs have previously been related to a blunted response to lipopolysaccharide (LPS)4 and to susceptibility to infectious diseases and sepsis.5,6
TLR4 is a transmembrane pattern recognition receptor that plays a key role in innate immunity by provoking inflammatory responses to its main ligand, LPS.7 TLR4 signals through adaptor proteins, including myeloid differentiation factor 88 (MyD88),8 in activating downstream effectors that include nuclear factor κB (NF-κB),9 mitogen-activated protein kinase, and phosphatidylinositol 3-kinase (PI3K).10 Collectively, these pathways regulate the expression of proinflammatory cytokines and genes that control cell survival and apoptosis.11 In liver, TLR4 signaling contributes to hepatic inflammation and injury of many etiologies.12,13 While the function of TLR4 in LPS-stimulated proinflammatory responses of Kupffer cells (resident hepatic macrophages) has been well studied,14,15 more recent findings underscore the importance of TLR4 to fibrogenic signaling of hepatic stellate cells (HSCs). TLR4 mediates LPS-triggered inflammatory phenotype of culture-activated stellate cells9 and enhances transforming growth factor β (TGF-β) responsiveness in HSCs by down-regulation of bone morphogenic protein and the activin membrane-bound inhibitor (BAMBI), an inhibitory TGF-β pseudo-receptor protein, thus stimulating hepatic fibrosis.16
Hepatic stellate cells are the major fibrogenic cell type in injured liver, and their activation is a well-characterized phenotypic response.17 Moreover, activated stellate cells become resistant to proapoptotic stimuli.18 Induction of stellate cell apoptosis has been proposed as a strategy to treat liver fibrosis (reviewed in Elsharkawy et al.19). An antiapoptotic effect of TLR4 signaling has been reported in macrophages20 and cancer cells.21 In hepatic stellate cells, however, while the role of TLR4 in the cell fibrogenesis has been scrutinized, its capacity to regulate survival has not been explored.
In this study, we explored the functional mechanisms underlying the emerging genetic association between TLR4 polymorphisms and fibrosis risk. Specifically, we have examined the impact of both TLR4 D299G and T399I SNPs on stellate cell responsiveness and clarified their potential mechanistic links to the inflammatory response, regulation of fibrogenesis, cell growth, and apoptotic sensitivity.
Materials and Methods
Vector Construction
TLR4 D299G, T399I, and dual D299G/T399I SNPs were generated by site-directed mutagenesis (see primer sequences in Supplementary Table 1) and a Quikchange®II-E site-directed mutagenesis kit (Strategene, La Jolla, CA) on an original pcDNA3 construct containing a full length hu-TLR4 complementary DNA (cDNA) with the TLR4 p.T399 and p.D299 major allele (wild-type [WT]). A FLAG epitope coding sequence was added at the C-terminus of the TLR4 WT and SNP cDNA with polymerase chain reaction (PCR) amplification using Pfx50™ DNA Polymerase (Invitrogen Corporation, Carlsbad, CA). The products were then TOPO-cloned into pCR®8/GW/TOPO® entrez vector (Invitrogen) and further transferred into destination vectors via LR recombination reactions. The destination vectors selected were pcDNA-DEST40 Gateway™ vector (Invitrogen) for the transfection of LX-2 cells, an immortalized human stellate cell line,22 and Plenti4/TO/V5-DEST Gateway™ vector (Invitrogen) for lentivirus-mediated transduction of hu-TLR4 cDNAs into mouse HSC lines, described as below. The vector sequences were validated by commercial sequencing (GENEWIZ, Inc., South Plainfield, NJ).
Assessment of LPS Responsiveness in LX-2 Cells
LX-2 cells were transfected with a mixture of three independent small interfering RNA (siRNA) sequences targeted against human TLR4 (Invitrogen™ Life Technologies), or a validated negative control siRNA using Lipofectamine™ RNAiMAX. They were then treated with 100 ng/mL LPS (Escherichia coli serotype 0111:B4, Sigma-Aldrich, Poole, U.K.) in 0.2% bovine serum albumin (BSA) or 0.2% BSA alone for 12 hours prior collected for messenger RNA (mRNA) and protein analysis. For NF-κB responsive reporter assays, a NF-κB responsive firefly luciferase reporter plasmid23 and a Renilla luciferase expression construct were transiently cotransfected with TLR4 siRNA or control siRNA into LX-2 cells. The cells were treated with LPS for 12 hours and collected for NF-κB reporter activity determination as described below.
For assessment of the LPS responsiveness in LX-2 cells expressing TLR4 SNPs, the cells were transfected with pcDNA-DEST40-TLR4-Flag WT and SNP cDNAs, as well as a pcDNA-DEST40 LacZ control vector using Fu-GENE HD reagent (Roche). The transfected cells were treated with 100 ng/mL LPS or vehicle for 12 hours as described above, and then collected for RNA and protein analyses. For NF-κB responsive reporter assays, LX-2 cells were transiently cotransfected with pcDNA-DEST40-TLR4-Flag WT or SNP cDNA, or LacZ control constructs along with the NF-κB responsive reporter plasmid and the Renilla control construct (in a ratio of 1:1:0.005) before LPS stimulation.
Isolation and Immortalization of Mouse Stellate Cells
Mouse hepatic stellate cells (mHSCs) were isolated from C57/Bl6 WT, TLR4−/−, and MyD88−/− mice by enzymatic digestion and Percoll density gradient centrifugation,24 with modifications. Following two to three passages, 50% subconfluent mHSCs were transfected with pCMV Simian Virus 40 Large T antigen22 for 48 hours followed by selection with 100 mg/mL hygromycin B (Invitrogen, San Diego, CA) and grown at 33°C. Immortalized WT, TLR4−/−, and MyD88−/− mHSC lines were generated by single cell clonal expansion and were genotyped as described for the TLR4 knockout mice.25,26
Reconstitution of hu-TLR4 SNPs Expression in mHSC Lines
Replication-incompetent lentiviral vectors were prepared by cotransfection by lipofectamine™ 2000 of a ViraPower™ Packaging Mix (Invitrogen), along with Flag-tagged TLR4 WT or SNP-expressing pLenti4/TO/V5-GW constructs, or a pLenti4/TO/V5-GW/LacZ control plasmid, into 293FT cells. The virus-containing supernatants were cleared and applied in a 10-fold dilution to mouse WT, TLR4−/−, and MyD88−/− HSCs at 30% to 50% confluence. The infected cells were selected and maintained with 500 μg/mL Zeocin™ (Invitrogen).
Assessment of LPS Responsiveness in mHSC Lines Reconstituted with hu-TLR4 WT SNP cDNAs
mHSC lines stably expressing human TLR4 WT or SNP(s) were plated and treated with LPS or vehicle and analyzed for RNA analysis or western blot. For the assessment of NF-κB responses, the mHSC lines were transiently transfected with the NF-κB responsive luciferase reporter plasmid described above, along with a Renilla luciferase expression contruct (in a ratio of 1:0.005) for 12 hours, using lipofectamine 2000 reagent (Invitrogen). The transfected cells were treated with LPS or vehicle and collected at 12 hours thereafter.
NF-κB Luciferase Reporter Assay
Cell lysates were prepared using a dual-luciferase reporter assay system (Promega). Changes in firefly luciferase activity were normalized with Renilla luciferase activity, and fold changes of NF-κB activity with or without LPS stimuli of each HSC line were compared.
Reverse-Transcription and Real-Time Quantitative PCR
RNA was extracted from the cells and reverse-transcribed into cDNA using an RNeasy® kit (Qiagen, Valencia, CA) and Omniscript RT Kit (Qiagen), respectively, and analyzed by quantitative PCR using SYBR green qPCR Master Mix (Roche) on the lightCycler®480 System (Roche). Data are represented as the fold induction of inflammatory cytokines (MCP-1, IL-6, IL-1β) by LPS as well as BAMBI and other fibrogenic-related genes (TGF-β, TGF-β receptor II, α-smooth muscle actin, collagen type I, and connective tissue growth factor) relative to cells not exposed to LPS. The primers used are listed in Supplementary Table 1.
Cytokine Assays
The culture supernatants were collected from cells in 12-well plates used for mRNA analysis at 12 hours post LPS stimulation. MCP-1 and IL-6 secretion was measured using enzyme-linked immunosorbent assay (ELISA) kits (human: R&D Inc., Minneapolis, MN; mouse: Biosource, Invitrogen) according to the manufacturer’s instructions.
Assessment of HSC Proliferation
mHSC lines reconstituted with different TLR4 SNP(s) were plated in 24-well plates at 50% confluence. They were treated with 10% fetal bovine serum/Dulbecco’s modified Eagle’s medium containing Isolution™ NF-κB activation inhibitor (a NF-κB inhibitor), PD98059 (an ERK inhibitor), or LY294002 (a PI3K inhibitor) that were purchased from Calbiochem (CN Biosciences, Beeston, U.K.) in 5-μM, 10-μM, and 20-μM concentrations, respectively. Incorporation of 3[H]-thymidine (1 μCi per well in 500 μL medium) was used for assaying DNA synthesis as a measurement of cell proliferation.
Assessment of HSC Apoptosis
mHSC lines were serum-starved in 0.2% BSA for 24 hours or incubated with PI3 kinase, ERK, or NF-κB inhibitors. Apoptosis was assessed by flow cytometry using a fluroescence-activated cell sorting (FACS) instrument (Facscan; Becton Dickinson, Franklin Lakes, NJ) and an annexin-fluorescein iso-thiocyanate (FITC) apoptosis detection kit (BD Biosciences). The percentages of cells in early apoptosis (single-stained by Annexin V-FITC), and late apoptosis (double-stained with Annexin V-FITC and propidium iodide), were analyzed by CellQuest Pro software. Data were expressed as the induction rate of apoptosis (which includes both early and late apoptosis) in each of the cell line.
Western Blot
Western blots of cell extracts were prepared by pelleting the cells with lysis buffer (Roche Diagnostics, Indianapolis, IN) complemented with protease inhibitor (Roche) and protein phosphatase inhibitor cocktails (Upstate, Temecula, CA). Protein concentration was determined with a Bio-Rad DC kit (Bio-Rad). Antibodies for phospho-ERK (p-ERK) (Thr202/Tyr204), total ERK, and β-actin were purchased from Sigma. Antibodies for phospho-Akt (p-Akt) (Ser473), total Akt, Bcl-2, and Bax were from Cell Signaling (Danvers, MA).
For the determination of cleaved poly (ADP-ribose) polymerase (PARP) as a marker of apoptosis, the same number of cells reconstituted with different TLR4 sequences were plated in 6-well plates using 10% fetal bovine serum/Dulbecco’s modified Eagle’s medium. After 24 hours, cells were lysed and pelleted. The supernatants were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot with an anti-PARP antibody (Cell Signaling).
Statistics
Results are expressed as the mean ± standard error of the mean (SEM). P values (Student two-tailed, unpaired t test) of at least three independent determinations were calculated with Microsoft Excel software. Data were considered to be statistically significant at P < 0.05.
Results
LX-2 Cells Have Intact TLR4 Signaling
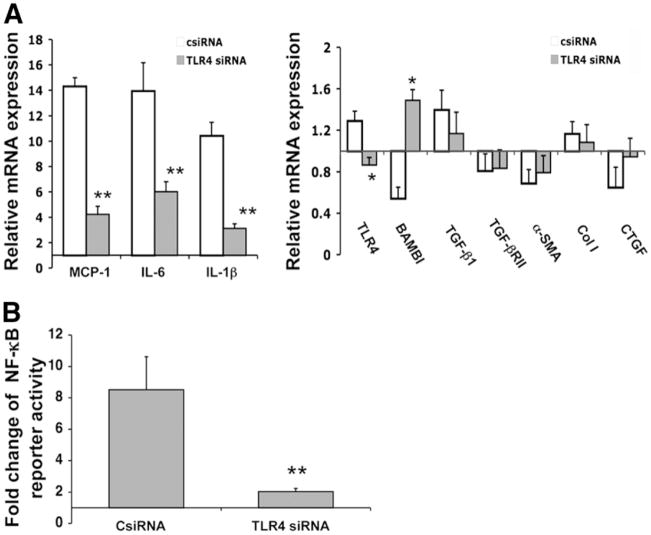
To confirm that LX-2 cells respond to LPS through TLR4, we first examined the impact of TLR4-specific siRNAs on NF-κB activation and NF-κB targets in response to LPS. As shown in Fig. 1, LPS stimulation induced marked up-regulation of MCP-1, IL-6, and IL-1β and down-regulation of BAMBI compared to control siRNA-transfected cells (Fig. 1A). The LPS responsiveness was associated with marked activation of NF-κB activity (Fig. 1B). In contrast, siRNA to TLR4 abrogated LPS responsiveness of these same target genes (Fig. 1A), indicating that their regulation by LPS is TLR4-dependent.
Fig. 1.
TLR4 siRNA abrogates LPS responsiveness in LX-2 cells. Following transfection with TLR4 siRNAs alone for mRNA analysis by real-time PCR or cotransfection with an NF-κB responsive reporter plasmid for the assessment of NF-κB activation, LX-2 cells were stimulated with LPS (100 ng/mL) for 12 hours. (A) Relative mRNA expression of inflammatory cytokines and fibrogenic genes in LX-2 cells transfected with TLR4 siRNAs (TLR4 siRNA) or control siRNA (csiRNA) in response to LPS. Data are depicted relative to expression in cells without LPS or siRNA, which are assigned a value of 1. (B) Fold change of NF-κB responsive reporter activity in LX-2 cells transfected with TLR4 siRNAs or control siRNA in response to LPS. Data are depicted relative to expression in cells without LPS or siRNA, which are assigned a value of 1. TLR4 siRNA abrogated LPS-induced up-regulation of MCP-1 and IL-6, and down-regulation of BAMBI, coincident with a reduced responsiveness of NF-κB activity. Each column represents the mean ± SEM of n = 6 per group in three independent experiments. *P < 0.05; **P < 0.01 when compared with the control siRNA transfected cells.
TLR4 SNPs Confer Decreased NF-κB Activation and Attenuate MCP-1, IL-6, and BAMBI Responses
Flag-tagged human WT or SNP TLR4 proteins were amply expressed in LX-2 cells or mouse TLR4−/− stellate cells following reconstitution of their cDNAs based on Western blot and immunofluoresence; the latter was suggestive of cell surface expression (Supplementary Fig. 1). Expression of TLR4 polymorphisms in transfected mHSC lines was also confirmed by restriction fragment length polymorphism analysis.27 Of note, LX-2 cells express WT TLR4 (data not shown).
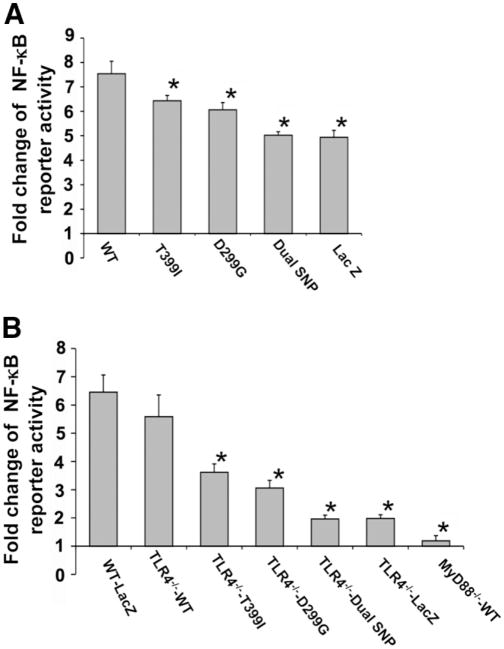
As determined by transfection of an NF-κB responsive luciferase reporter, LX-2 cells or mHSC lines expressing either the single or dual SNPs displayed less NF-κB activation (Fig. 2A,B). The impact of TLR4 SNP(s) on down-regulation of NF-κB responses was even more striking in the TLR4−/− HSCs reconstituted with hu-TLR4 cDNAs. In addition, as shown in Fig. 2B, TLR4−/− and MyD88−/− cells displayed almost no NF-κB activation.
Fig. 2.
TLR4 SNPs confer decreased NF-κB activation to LPS stimulation in HSCs. An NF-κB responsive reporter plasmid was cotransfected with TLR4 cDNAs into LX-2 cells or transfected into mHSCs stably reconstituted with human TLR4 cDNAs. The cells were stimulated with LPS (100 ng/mL) for 12 hours. The activation of NF-κB responsive reporter was determined with a dual-luciferase reporter assay system. (A) Fold change of NF-κB reporter activity in LX-2 cells transfected with TLR4 WT or SNP sequences, or a LacZ control vector. (B) Fold change of NF-κB reporter activity in TLR4−/− mouse HSCs transfected with TLR4 WT or SNP sequences, or a LacZ control vector, and in MyD88−/− HSCs reconstituted with WT human TLR4 sequence. Cells expressing either the single or dual SNPs had less NF-κB activation. Each column represents the mean ± SEM of n = 6 per group in more than three independent experiments. *P < 0.05 when compared with the WT TLR4 cDNA-transfected stellate cells.
We also assessed the impact of TLR4 polymorphisms on the LPS responsiveness of NF-κB target genes in LX-2 cells and TLR4−/− mHSCs reconstituted with human TLR4 cDNAs. As shown in Fig. 3A,B, mRNA expression of MCP-1, IL-6, and BAMBI was proportionately reduced in TLR4 WT, T399I, D299G, and dual D299G/T399I SNP cells, respectively. These data for mRNA expression of MCP-1 and IL-6 corresponded to parallel changes in protein secretion, as assessed by specific ELISAs (Fig. 3C,D).
Fig. 3.
TLR4 SNPs attenuate the response of NF-κB target genes in HSCs following LPS stimulation. LX-2 cells or TLR4−/− mHSC lines that were transiently or stably reconstituted with human TLR4 cDNAs were stimulated with 100 ng/mL LPS for 12 hours. The mRNA expression of inflammatory cytokines MCP-1 and IL-6, and BAMBI in LX-2 cells (A) and mHSCs (B) was determined by real-time quantitative PCR. The parallel changes in protein secretion of MCP-1 and IL-6 in the cell culture supernatants of LX-2 cells (C) and mouse HSCs (D) were assessed by ELISA. The fold changes of MCP-1 and IL-6 mRNAs after LPS exposure were reduced in the presence of single or dual SNPs, whereas the extent of BAMBI down-regulation was attenuated. Changes of α-smooth muscle actin and collagen type I mRNAs were not statistically different following reconstitution with TLR4 SNPs (not shown). Each column represents the mean ± SEM of n = 6 per group in three independent experiments. *P < 0.05 when compared with the WT TLR4 cDNA-transfected stellate cells.
Unlike these NF-κB target genes downstream of TLR4, the expression of α-smooth muscle actin and type I collagen did not change significantly in response to LPS stimulation (data not shown) or TLR4 knockdown (Fig. 1), suggesting that these markers of stellate cell activation were not directly regulated by TLR4 signaling.
TLR4 SNPs Reduce mHSC Growth and Enhance Cell Growth Inhibition by Pathway Inhibitors
As shown in Fig. 4A, cells expressing the T399I, D299G, or combined T399I/D299G SNPs had significantly reduced growth compared to cells expressing WT TLR4, as assessed by tritiated thymidine incorporation. Moreover, as shown in Fig. 4B, cells with either single or dual TLR4 SNPs were more growth-inhibited following incubation with pathway(s) inhibitors than WT TLR4 cells.
Fig. 4.
TLR4 SNPs reduce mHSC growth and enhance growth inhibition by pathways inhibitors. TLR4−/− mHSC lines that were stably reconstituted with human TLR4 cDNAs were treated with PI3K inhibitor (LY 294002), ERK inhibitor (PD 98059), or NF-κB inhibitor (20 μM, 10 μM, and 5 μM, respectively). DNA synthesis was assessed by 3[H]-incorporation. (A) DNA synthesis in TLR4−/− mHSCs transfected with human TLR4 WT or SNP cDNAs and MyD88−/− HSCs reconstituted with human TLR4 WT. (B) Growth reduction rate following addition of a PI3K inhibitor, ERK inhibitor, or NF-κB inhibitor. Cells with either single or dual SNPs had slower cell growth and more growth reduction following pathway inhibition than WT TLR4 cells. Each column represents the mean ± SEM of n = 6 per group in three independent experiments. *P < 0.05 when compared with the WT TLR4 cDNA-transfected HSCs.
TLR4 SNPs Increase Spontaneous Apoptosis and Pathway Inhibitor-Induced mHSC Apoptosis
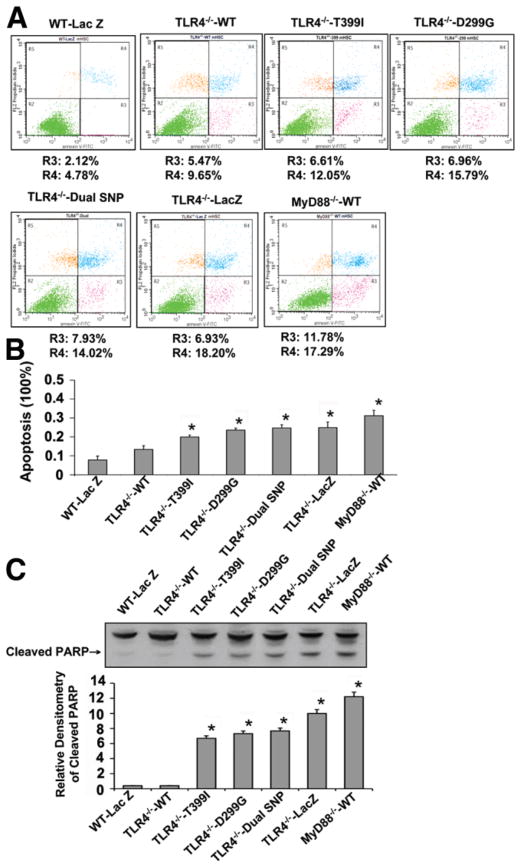
Using FACS and PARP cleavage as markers of apoptosis, we examined the impact of TLR4 SNPs on apoptotic responses of cultured HSCs. As shown in Fig. 5, the expression of single T399I, D299G, or dual T399I/D299G TLR4 SNPs conferred a greatly increased rate of spontaneous apoptosis in mHSCs (Fig. 5A,B) compared with WT TLR4-expressing mHSCs. These findings were associated with increased PARP cleavage (Fig. 5C).
Fig. 5.
TLR4 SNPs increase the spontaneous apoptosis of mHSCs. Spontaneous cell apoptosis was assessed in TLR4−/− mHSC lines that were stably reconstituted with human TLR4 cDNAs by FACS and by western blot analysis of cleaved PARP. (A) FACS analysis of spontaneous apoptosis of TLR4−/− mouse HSCs transfected with human TLR4 cDNAs and MyD88−/− HSCs reconstituted with WT TLR4. (B) Apoptosis rate (including both the early apoptotic rate [R3] and late apoptotic rate [R4]) was determined by FACS analysis of each cell line. (C) Western blot analysis of cleaved PARP in each cell line (representative western blotted membrane and relative densitometric quantitation are shown). Each column represents the mean ± SEM of n = 3 per group in three independent experiments. *P < 0.01 when compared with WT TLR4 cDNA-transfected stellate cells. Cells expressing single or dual SNPs showed higher spontaneous apoptosis than WT TLR4-transfected cells.
The impact of these SNPs in response to either serum starvation or pathway inhibition by either the PI3K, ERK, or NF-κB inhibitors is shown in Fig. 6. Cells expressing either single or dual TLR4 SNPs were less tolerant to either serum starvation or pathway inhibition than WT TLR4-expressing cells. In other words, TLR4 SNPs lowered the apoptotic threshold, the impact of which in vivo would be increased clearance of activated HSCs.
Fig. 6.
TLR4 SNPs lower the apoptotic threshold of activated HSCs. TLR4−/− mHSC lines that were stably reconstituted with TLR4 cDNAs were either serum-starved for 24 hours or incubated with pathway inhibitors including PI3K inhibitor (LY 294002), ERK inhibitor (PD 98059), or NF-κB inhibitor for 12 hours (20 μM, 10 μM, and 5 μM, respectively). The apoptosis rate was analyzed by FACS. Cells expressing either single or dual TLR4 SNPs have higher apoptotic induction rates under serum starvation or pathway inhibition than WT TLR4-expressing cells. Each column represents the mean ± SEM of n = 3 per group in three independent experiments. *P < 0.01 when compared with WT TLR4 cDNA-transfected HSCs.
The Expression of Apoptosis-Related Proteins Is Altered in mHSCs Expressing TLR4 SNPs
Finally, we examined several key downstream effectors regulating apoptosis, including p-ERK, p-Akt, Bcl-2, and Bax. As assessed by Western blot (Fig. 7), the basal levels of Bcl-2 and p-ERK were lower in TLR4−/−, MyD88−/−, and TLR4 T399I, D299G, or dual T399I/D299G SNPs expressing TLR4−/− mHSCs when compared with WT TLR4-expressing mHSCs, while Bax expression in all these cells was similar. p-ERK was inducible in response to LPS in the WT and TLR4−/− mHSCs reconstituted with WT hu-TLR4 cDNA, whereas this modest but significant responsiveness was abrogated in the cells that were TLR4−/− or MyD88−/−, or expressing TLR4 single or dual SNPs. Although the p-Akt level was low in TLR4−/− and MyD88−/− mHSCs and was not responsive to LPS stimulation, its expression in SNP-expressing TLR4−/− stellate cells was similar to WT mHSCs or TLR4−/− cells reconstituted with TLR4 WT.
Fig. 7.
TLR4 SNPs alter the level of apoptosis-related proteins in mHSCs. TLR4−/− mHSC lines that were stably reconstituted with human TLR4 cDNAs were stimulated with 100 ng/mL LPS for 12 hours. Western blot of apoptosis-regulating proteins was performed, including total ERK (t-ERK), p-ERK, total Akt (t-Akt), p-Akt, Bcl-2, and Bax (representative western blotted membrane and relative densitometric quantitation to β-actin are shown). Two key downstream effectors regulating apoptosis (p-ERK and Bcl-2) were reduced in TLR4 knockout and T399I, D299G, dual D299G/T399I SNP-expressing stellate cells. The figure is a representative of three independent experiments. *P < 0.01 compared with WT TLR4 cDNA-transfected HSC. #P < 0.05 compared with the cells without LPS treatment.
Discussion
Linking SNPs associated with disease risk to functional pathways underlying these same diseases remains a major goal of studies in the emerging era of “personalized medicine.”28 The strong association of specific TLR4 SNPs with fibrosis risk led us to explore the most direct pathway(s) through which these SNP sequences might exert their protective activity. Specifically, we examined whether TLR4 SNPs altered the biology and response of the key fibrogenic cell type in liver, the activated HSCs, while recognizing that the SNPs also likely affect fibrogenesis through responses in other cell types as well.
Here we show either the absence of TLR4 or expression of TLR4 T399I and/or D299G SNPs conferred reduced LPS responsiveness in cultured human or mouse HSCs. The SNPs reduced NF-κB activation and proinflammatory cytokine expression, and attenuted down-regulation of TGF-β pseudoreceptor BAMBI in HSCs after LPS stimulation. These SNPs also reduced cell growth and lowered the apoptotic threshold in mHSCs following apoptotic stress. Both the TLR4 T399I and D299G SNPs attenuated receptor signaling, with the TLR4 D299G SNP exhibiting a greater inhibitory effect, and the dual D299G/T399I cosegregating SNPs exhibiting the greatest attenuation, in line with a previous study.29
The TLR4 T399I and D299G are two common, highly cosegregated nonsynonymous polymorphisms within the extracellular domain of TLR4 protein, which may affect the strength of interactions with either agonists and/or coreceptors, leading to a decreased recognition of ligands in an agonist-independent manner.29 The genetic variation in the TLR4 gene, especially the D299G SNP, is associated with susceptibility to infectious diseases and severe malaria,30 as well as diseases as disparate as inflammatory bowel disease,31 Helicobacter pylori infection, and gastric cancer.32 In contrast to these diseases, however, the same SNPs conferred delayed hepatic fibrosis progression.33 Thus, whereas specific SNPs confer LPS hyporesponsiveness and increased infection susceptibility, they reduce the likelihood of end-organ damage due to progressive scarring. Hepatic stellate cells play a key role in the fibrogenesis of injured liver. Apart from the important fibrogenic activity of HSCs, the cells have emerged as key effectors of the liver’s inflammatory response, rather than simply targets of inflammation. In this context, the critical activity of TLR4 signaling in triggering the inflammatory phenotype of HSCs and sensitizing HSCs to TGF-β signaling via BAMBI regulation is increasingly relevant.9,16 Although in two studies there was no association between TLR4 polymorphisms and the outcome of liver transplantation34 or in circulating cytokine levels in response to sub-maximal exposure to LPS,35 these analyses did not assess the polymorphisms of the allografts. Moreover, whereas MCP-1 was the most responsive cytokine in our study, this was not assessed in these two earlier reports. Additionally, the study assessing response to LPS used limited time points of sample collection and very low LPS dosage.
Our most striking finding is the importance of TLR4 signaling in maintaining HSC survival, and the ability of protective TLR4 SNPs to lower the apoptotic threshold. Both TLR4 and MyD88 appear to preserve the basal levels of NF-κB activity and p-ERK and p-Akt in mHSCs. However, the involvement of Akt may be less important than NF-κB and ERK, since cells reconstituted with TLR4 SNPs showed an intermediate rate of cell growth and apoptosis, which is consistent with the reduced basal as well as LPS-stimulated up-regulation of NF-κB activity and p-ERK, while their levels of p-Akt were near normal. The difference might be due to SNPs that alter the magnitude of the TLR4 signal; alternatively, there may be self-regulation of Akt activation in HSCs, which may serve as a complementary pathway for apoptosis regulation by NF-κB and ERK. Of note, the effect of NF-κB pathway inhibition was much greater than inhibiting either PI3K or ERK. This finding may reflect the fact that NF-κB is a common downstream pathway linked to both PI3K and ERK signaling,10 which may account for the greater effect compared to inhibition of either pathway alone.
In the present study, the TLR4 and MyD88 knockout cells and HSCs reconstituted with protective TLR4 SNPs had reduced NF-κB activity. They expressed less Bcl-2, an antiapoptotic protein that regulates mitochondrial pathways of apoptosis,36 while the Bax levels were not changed compared to WT HSCs. Reduced levels of Bcl-2 in TLR4−/− or MyD88−/− cells, as well as cells expressing TLR4 SNPs, may be at least one mechanism underlying their lowered apoptotic threshold to both physiological and exogenous stress.
Spontaneous apoptosis in HSCs is increased in LPS-free culture when the cells are TLR4−/− or express TLR4 SNPs. Other TLR4 ligands may exist in the medium to induce TLR4 signaling and render an anti-apoptotic effect in WT TLR4-expressing cells. Potential endogenous substrates include low molecular weight hyaluronic acid, saturated fatty acid, fibrinogen, fibronectin, heat shock protein 60 and 70, and high mobility group box-1 (reviewed in Guo and Friedman37). In vivo, damage signals and extracellular matrix degradation can activate TLR4. It is likely that TLR4 signaling becomes more critical to fibrogenesis once liver injury is initiated.38
In conclusion, TLR4 –MyD88 –NF-κB signaling mediates an LPS-stimulated inflammatory phenotype of activated HSCs that contributes to cell survival. While its impact on other cell types in liver (e.g, hepatic macrophages) remains to be explored, TLR4 signaling in HSCs is affected by SNPs that are associated with protection from hepatic fibrosis in large, well-characterized clinical cohorts with hepatitis C virus. These findings confirm the critical role of TLR4 signaling in regulating HSC activation and validate the power of unbiased genetic studies to identify novel biologic pathways affecting the risk of hepatic fibrosis progression.
Supplementary Material
Acknowledgments
Supported by National Institutes of Health Grant RO1DK56631 and by Celera Diagnostics.
We thank Prof. Ruslan M. Medzhitov of Yale University Medical School for providing human the TLR4 cDNA construct and Dr. Maria T. Abreu of the Inflammatory Bowel Disease Center of Mount Sinai School of Medicine for providing mouse colonies.
Abbreviations
- BAMBI
bone morphogenic protein and activin membrane-bound inhibitor
- BSA
bovine serum albumin
- cDNA
complementary DNA
- ELISA
enzyme-linked immunosorbent assay
- ERK
extracellular signal-regulated kinase
- FACS
fluorescence-activated cell sorting
- FITC
fluorescein isothiocyanate
- HSC
hepatic stellate cell
- IL-1β
interleukin-1β
- IL-6
interleukin-6
- LPS
lipopolysaccharide
- MCP-1
monocyte chemoattractant protein-1
- mHSC
mouse hepatic stellate cell
- mRNA
messenger RNA
- MyD88
myeloid differentiation factor 88
- NF-κB
nuclear factor κB
- p-Akt
phospho-Akt
- PARP
poly (ADP-ribose) polymerase
- PCR
polymerase chain reaction
- p-ERK
phospho-ERK
- PI3K
phosphatidylinositol 3-kinase
- SEM
standard error of the mean
- siRNA
small interfering RNA
- SNP
single nucleotide polymorphism
- TGF-β
transforming growth factor β
- TLR4
Toll-like receptor 4
- WT
wild-type
Footnotes
Presented in abstract form as an oral presentation at the 2007 American Association for the Study of Liver Diseases Annual meeting and published in abstract form.
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825–832. doi: 10.1016/s0140-6736(96)07642-8. [DOI] [PubMed] [Google Scholar]
- 2.Asselah T, Bieche I, Paradis V, Bedossa P, Vidaud M, Marcellin P. Genetics, genomics, and proteomics: implications for the diagnosis and the treatment of chronic hepatitis C. Semin Liver Dis. 2007;27:13–27. doi: 10.1055/s-2006-960168. [DOI] [PubMed] [Google Scholar]
- 3.Huang H, Shiffman ML, Friedman S, Venkatesh R, Bzowej N, Abar OT, et al. A 7 gene signature identifies the risk of developing cirrhosis in patients with chronic hepatitis C. Hepatology. 2007;46:297–306. doi: 10.1002/hep.21695. [DOI] [PubMed] [Google Scholar]
- 4.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 5.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 6.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 7.Beutler B. Inferences, questions and possibilities in Toll-like receptor signaling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 8.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 9.Paik YH, Schwabe RF, Bataller R, Russo MP, Jobin C, Brenner DA. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 2003;37:1043–1055. doi: 10.1053/jhep.2003.50182. [DOI] [PubMed] [Google Scholar]
- 10.Lee JY, Ye J, Gao Z, Youn HS, Lee WH, Zhao L, et al. Reciprocal modulation of Toll-like receptor-4 signaling pathways involving MyD88 and phosphatidylinositol 3-kinase/AKT by saturated and polyunsaturated fatty acids. J Biol Chem. 2003;278:37041–37051. doi: 10.1074/jbc.M305213200. [DOI] [PubMed] [Google Scholar]
- 11.Hsu LC, Park JM, Zhang K, Luo JL, Maeda S, Kaufman RJ, et al. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428:341–345. doi: 10.1038/nature02405. [DOI] [PubMed] [Google Scholar]
- 12.Yohe HC, O’Hara KA, Hunt JA, Kitzmiller TJ, Wood SG, Bement JL, et al. Involvement of Toll-like receptor 4 in acetaminophen hepatotoxicity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1269–G1279. doi: 10.1152/ajpgi.00239.2005. [DOI] [PubMed] [Google Scholar]
- 13.Takayashiki T, Yoshidome H, Kimura F, Ohtsuka M, Shimizu Y, Kato A, et al. Increased expression of toll-like receptor 4 enhances endotoxin-induced hepatic failure in partially hepatectomized mice. J Hepatol. 2004;41:621–628. doi: 10.1016/j.jhep.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Uesugi T, Froh M, Arteel GE, Bradford BU, Thurman RG. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology. 2001;34:101–108. doi: 10.1053/jhep.2001.25350. [DOI] [PubMed] [Google Scholar]
- 15.Su GL, Klein RD, Aminlari A, Zhang HY, Steinstraesser L, Alarcon WH, et al. Kupffer cell activation by lipopolysaccharide in rats: role for lipopolysaccharide binding protein and toll-like receptor 4. Hepatology. 2000;31:932–936. doi: 10.1053/he.2000.5634. [DOI] [PubMed] [Google Scholar]
- 16.Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 17.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novo E, Marra F, Zamara E, Valfre di Bonzo L, Monitillo L, Cannito S, et al. Overexpression of Bcl-2 by activated human hepatic stellate cells: resistance to apoptosis as a mechanism of progressive hepatic fibrogenesis in humans. Gut. 2006;55:1174–1182. doi: 10.1136/gut.2005.082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elsharkawy AM, Oakley F, Mann DA. The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis. 2005;10:927–939. doi: 10.1007/s10495-005-1055-4. [DOI] [PubMed] [Google Scholar]
- 20.Lombardo E, Alvarez-Barrientos A, Maroto B, Bosca L, Knaus UG. TLR4-mediated survival of macrophages is MyD88 dependent and requires TNF-alpha autocrine signaling. J Immunol. 2007;178:3731–3739. doi: 10.4049/jimmunol.178.6.3731. [DOI] [PubMed] [Google Scholar]
- 21.He W, Liu Q, Wang L, Chen W, Li N, Cao X. TLR4 signaling promotes immune escape of human lung cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Mol Immunol. 2007;44:2850–2859. doi: 10.1016/j.molimm.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, Arditi M. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609–1616. doi: 10.4049/jimmunol.167.3.1609. [DOI] [PubMed] [Google Scholar]
- 24.Blomhoff R, Berg T. Isolation and cultivation of rat liver stellate cells. Methods Enzymol. 1990;190:58–71. doi: 10.1016/0076-6879(90)90009-p. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 26.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 27.Torok HP, Glas J, Tonenchi L, Mussack T, Folwaczny C. Polymorphisms of the lipopolysaccharide-signaling complex in inflammatory bowel disease: association of a mutation in the Toll-like receptor 4 gene with ulcerative colitis. Clin Immunol. 2004;112:85–91. doi: 10.1016/j.clim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Jain KK. Personalized medicine. Curr Opin Mol Ther. 2002;4:548–558. [PubMed] [Google Scholar]
- 29.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, et al. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 30.Mockenhaupt FP, Cramer JP, Hamann L, Stegemann MS, Eckert J, Oh NR, et al. Toll-like receptor (TLR) polymorphisms in African children: common TLR-4 variants predispose to severe malaria. Proc Natl Acad Sci U S A. 2006;103:177–182. doi: 10.1073/pnas.0506803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning BL, Huebner C, Petermann I, Gearry RB, Barclay ML, Shelling AN, et al. Has toll-like receptor 4 been prematurely dismissed as an inflammatory bowel disease gene? Association study combined with meta-analysis shows strong evidence for association. Am J Gastroenterol. 2007;102:2504–2512. doi: 10.1111/j.1572-0241.2007.01463.x. [DOI] [PubMed] [Google Scholar]
- 32.Hold GL, Rabkin CS, Chow WH, Smith MG, Gammon MD, Risch HA, et al. A functional polymorphism of toll-like receptor 4 gene increases risk of gastric carcinoma and its precursors. Gastroenterology. 2007;132:905–912. doi: 10.1053/j.gastro.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 33.Marra F, Romanelli RG, Giannini C, Failli P, Pastacaldi S, Arrighi MC, et al. Monocyte chemotactic protein-1 as a chemoattractant for human hepatic stellate cells. Hepatology. 1999;29:140–148. doi: 10.1002/hep.510290107. [DOI] [PubMed] [Google Scholar]
- 34.Kovalovich K, Li W, DeAngelis R, Greenbaum LE, Ciliberto G, Taub R. Interleukin-6 protects against Fas-mediated death by establishing a critical level of anti-apoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J Biol Chem. 2001;276:26605–26613. doi: 10.1074/jbc.M100740200. [DOI] [PubMed] [Google Scholar]
- 35.Taudorf S, Krabbe KS, Berg RM, Møller K, Pedersen BK, Bruunsgaard H. Common studied polymorphisms do not affect plasma cytokine levels upon endotoxin exposure in humans. Clin Exp Immunol. 2008;152:147–152. doi: 10.1111/j.1365-2249.2008.03612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams JM, Cory S. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem Sci. 2001;26:61–66. doi: 10.1016/s0968-0004(00)01740-0. [DOI] [PubMed] [Google Scholar]
- 37.Guo J, Friedman S. Hepatic fibrogenesis. Semin Liver Dis. 2007;27:413–426. doi: 10.1055/s-2007-991517. [DOI] [PubMed] [Google Scholar]
- 38.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Conditional signaling by Toll-like receptor 4. FASEB J. 2005;19:872–874. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.