Abstract
During sensory organ precursor (SOP) specification, a single cell is selected from a proneural cluster of cells. Here, we present evidence that Senseless (Sens), a zinc-finger transcription factor, plays an important role in this process. We show that Sens is directly activated by proneural proteins in the presumptive SOPs and a few cells surrounding the SOP in most tissues. In the cells that express low levels of Sens, it acts in a DNA-binding-dependent manner to repress transcription of proneural genes. In the presumptive SOPs that express high levels of Sens, it acts as a transcriptional activator and synergizes with proneural proteins. We therefore propose that Sens acts as a binary switch that is fundamental to SOP selection.
Keywords: Neuronal specification, proneural genes, Enhancer of split, Drosophila, peripheral nervous system development
Sensory organs serve as transducers that convert various physical stimuli into electrical signals. In Drosophila, a variety of internal and external sensory organs cover the body of larvae and adults (Hartenstein 1988; Hartenstein and Posakony 1989; Jan and Jan 1993). The early phase of sensory organ development is usually marked by low-level expression of one or more members of a group of basic helix-loop-helix (bHLH) genes, the proneural genes. These genes are expressed in a cluster of cells, the proneural cluster, and provide these cells with the competence to become neuronal precursors (Villares and Cabrera 1987; Cubas et al. 1991; Skeath and Carroll 1991; Jarman et al. 1993, 1994). The proneural competence is further refined to a smaller group of cells in each cluster, the proneural field, which accumulates relatively higher levels of proneural proteins (Cubas et al. 1991; Skeath and Carroll 1991). Ultimately, one cell of the proneural field will exhibit a yet higher level of proneural protein expression and become the SOP, whereas other cells will adopt an epidermal fate. The accumulation of large amounts of proneural proteins is thought to depend on the proneural gene activity via specific enhancers of proneural genes that show auto- and cross-regulatory characteristics (Van Doren et al. 1992; Modolell 1997; Culi and Modolell 1998).
Although all of the cells in a proneural field have the potential to become an SOP, only one (or a few) cell(s) will realize their potential. This is achieved by inhibitory cell-cell interactions mediated by members of the Notch (N) signaling pathway (Artavanis-Tsakonas et al. 1999). Binding of the ligand Delta to the N receptor of the neighboring cells initiates a series of proteolytic cleavages that ultimately release the intracellular domain of N (Nicd), which translocates into the nucleus. Together with another protein called Suppressor of Hairless [Su(H)], Nicd activates the transcription of the primary targets of N signaling, the genes of the Enhancer of split complex (E(spl)-C) (Delidakis and Artavanis-Tsakonas 1992; Knust et al. 1992; Schrons et al. 1992; Bailey and Posakony 1995; Lecourtois and Schweisguth 1995). Members of the E(spl)-C encode bHLH proteins that, together with the product of the neurogenic gene groucho (gro), suppress the expression of proneural genes and their targets (Paroush et al. 1994; Jimenez and Ish-Horowicz 1997; Culi and Modolell 1998; Giagtzoglou et al. 2003), thereby limiting the number of SOPs in each cluster to one or a few. It has been shown that proneural proteins are direct transcriptional activators of some of the key players of the lateral inhibition process, such as E(spl)m7 and E(spl)m8 (Kramatschek and Campos-Ortega 1994; Singson et al. 1994; Bailey and Posakony 1995; Cooper et al. 2000). Therefore, to accumulate proneural proteins in one cell and specify it as the SOP, this cell must somehow escape the inhibitory effect of the E(spl) proteins. This is, at least in part, mediated by lowering the transcription level of E(spl) in the SOP (Koelzer and Klein 2003).
sens encodes a zinc-finger transcription factor shown to be necessary for the formation of adult SOPs. We have shown that sens is genetically downstream of proneural genes and is, in turn, required for up-regulation of the proneural genes in the SOPs (Nolo et al. 2000). Loss- and gain-of-function phenotypes of sens indicate that it may help proneural proteins override the negative feedback they would promote by activating E(spl)-C transcription. sens clones lack sensory organs in adult flies, and overexpression of Sens is a potent inducer of extra sensory organ formation. Moreover, a recent study has shown a correlation between the proneural potency of various proneural proteins and their ability to induce Sens expression (Lai 2003). sens has two vertebrate homologs, Gfi-1 and Gfi-1B (Zweidler-Mckay et al. 1996; Tong et al. 1998). We have shown recently that Gfi-1 is required for inner ear hair cell differentiation and survival (Wallis et al. 2003).
The process of selecting one SOP from a group of equipotent cells seems to involve a dynamic interaction between proneural proteins, members of the N signaling pathway, and Sens. In this study, we have examined several aspects of these interactions at the molecular level. We find that both proneural and E(spl) proteins are involved in transcriptional regulation of sens. Sens, in turn, strongly activates the achaete (ac) promoter through a synergism with Ac and Daughterless (Da), and this activation is sensitive to the level of E(spl)m8 protein. Surprisingly, we observe that at low levels, Sens acts as a repressor of proneural gene expression, rather than an activator. These observations suggest a model in which Sens acts as a binary switch during neuronal precursor selection in the proneural field.
Results
Proneural proteins directly regulate sens transcription
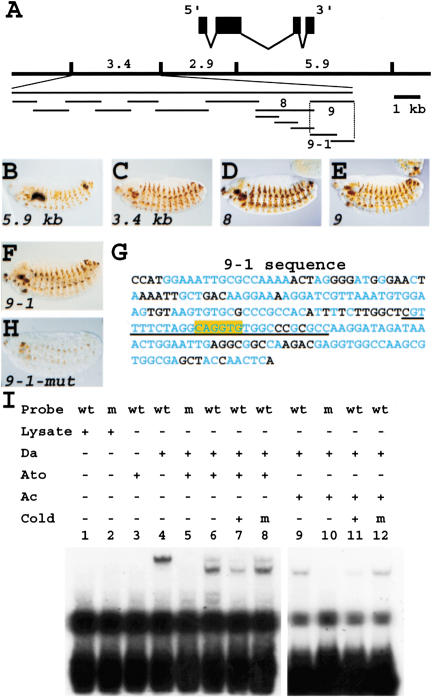
We have shown previously that proneural genes are required for sens expression (Nolo et al. 2000; Frankfort et al. 2001). To determine whether proneurals directly activate sens expression, we identified the putative enhancers of sens and scanned them for proneural protein-binding sites (E boxes). We have shown previously that an 11-kb genomic fragment containing the sens locus is able to rescue the sens mutant phenotype (Nolo et al. 2000). To identify the embryonic and imaginal disc enhancers, three genomic DNA fragments were used to create lacZ reporter transgenes (Fig. 1A). Both 5.9-kb and 3.4-kb fragments are sufficient to drive expression in the embryonic PNS in a pattern similar to endogenous sens (Fig. 1B,C). To refine sens enhancers, the 3.4-kb enhancer was divided into nine overlapping fragments (Fig. 1A). Fragments 8 and 9 induced lacZ expression in a pattern similar to the original 3.4-lacZ line, indicating that both contain regulatory elements sufficient for sens expression in the embryonic PNS (Fig. 1D,E). Fragments 8 and 9 were further divided into overlapping fragments. Only 9-1-lacZ expresses the reporter in a pattern similar to the 3.4-lacZ (Fig. 1F). Inspection of the 9-1 sequence showed that it contains a single E box (Fig. 1G, box). We also used the recently sequenced genome of Drosophila pseudoobscura, a species 25-30 myr divergent from Drosophila melanogaster (Russo et al. 1995) to align the genomic regions. The alignment showed that the E box, as well as several other elements in the 9-1 enhancer, is fully conserved (Fig. 1G, blue residues). Upon mutation of this E box from CAGGTG to CCGGTG, most of the PNS cells failed to express lacZ, and staining in other cells was much weaker than for the wild-type transgene (Fig. 1H). These data indicate that proneural genes directly regulate the transcription of sens.
Figure 1.
Proneural proteins directly regulate sens transcription. (A) Schematic of genomic fragments used to identify sens enhancers. (B-F) Patterns generated by various fragments identified in A in stage 14 or 15 embryos. Anterior is to the left, dorsal to the top. (G) Sequence of the 9-1 fragment showing a single E box (highlighted in yellow). Blue residues are identical to a similar fragment in the 5′-UTR of the sens homolog in D. pseudoobscura. (H) Expression of lacZ conferred by the E box-mutated 9-1 fragment in a stage 15 embryo. (I) EMSA using wild-type (wt) and mutant (m) probes that correspond to the sequence underlined in G. Note that Da, Da/Ato, and Da/Ac are all able to bind this site.
It is thought that the two core nucleotides of the E box as well as its flanking sequences are involved in the specificity of each E box for its cognate bHLH transcription factor (Singson et al. 1994; Jennings et al. 1999). We were intrigued by the observation that expression of the lacZ marker was almost abolished in chordotonal organs that are dependent on atonal (ato) (Jarman et al. 1993) as well as in external organs and multiple dendritic organs that are dependent on ac, sc, and amos (Cabrera et al. 1987; Goulding et al. 2000; Huang et al. 2000). Because the 9-1 fragment only contains a single E box, the data suggest that different proneural proteins can bind the same E box in vivo. We therefore performed EMSA to determine whether a variety of Da-proneural heterodimers can shift a wild-type or an E box-mutated probe taken from the 9-1 sequence. Whereas the lysate alone did not show a band-shift, Da homodimer, Ato/Da heterodimer, Ac/Da heterodimer (Fig. 2), and Sc/Da heterodimer (data not shown) were all able to bind to this E box. Mutation from A to C in the second position of the E box abolished binding for all protein combinations tested, suggesting that these interactions are sequence specific. We conclude that at least three proneural proteins (Ac, Sc, and Ato) directly regulate sens expression in the embryonic PNS, and that they may bind the same site in vivo.
Figure 2.
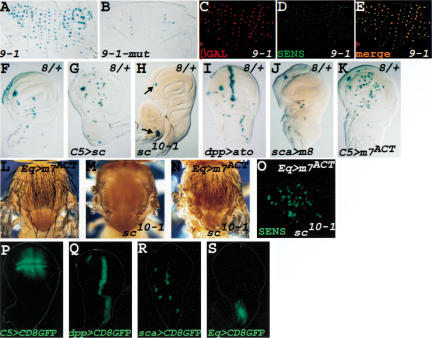
Proneural and E(spl) proteins regulate sens expression in wing imaginal discs and thorax. (A) Thorax of a 16-17-hour-old pupa that carries the 9-1-lacZ transgene. lacZ is expressed in most microchaetae precursors. (B) Same as in A, but the transgene carries a single nucleotide change in the E box (see Fig. 1G). (C-E) Double staining of a 16-17-hour-old 9-1-lacZ pupal notum with anti-βGAL (C) and anti-Sens (D) antibodies and the merged image (E). All of the lacZ-expressing cells correspond to SOPs. (F) lacZ expression pattern conferred by fragment 8 in Fig. 1A. (G) Same as in F, but the Sc protein is now expressed ectopically in the wing pouch. Note that many more cells express lacZ. (H) Same as in F, but derived from a larva that lacks expression of Sc and Ac. Note the absence of lacZ expression, except in clusters of cells that normally express Ato (arrows). (I) Same as in F, but Ato is expressed under control of the dpp-GAL4 driver. lacZ expression is induced at the A/P boundary, where Dpp is normally expressed. (J) E(spl)m8 protein is expressed under the control of sca-GAL4 in SOPs and the surrounding cells. Note the loss of lacZ expression in several SOPs. (K) E(spl)m7 fused to the VP16 transactivator domain is expressed in the wing pouch. This leads to ectopic expression of lacZ throughout the wing pouch. (L) Overexpression of E(spl)m7ACT in the anterior parts of the notum. Note that numerous extra bristles are formed. (M) Thorax of a sc10-1 fly. (N) Same as in L, but in a sc10-1 background. Note that several extra bristles are still formed (compare with M). (O) The anterior part of the presumptive notum in a third instar larva with the same genotype as N, stained with anti-Sens antibody. Note that several cells express Sens. (P-S) Pattern of GAL4 expression for C5-GAL4 (P), dpp-GAL4 (Q), sca-GAL4 (R), and Eq-GAL4 (S) revealed by CD8-GFP expression. For all wing imaginal discs, anterior is to the left, ventral to the top.
sens Expression in imaginal discs is controlled by proneural and E(spl) proteins
To examine whether sens regulation in the precursors of the adult PNS is also under direct proneural regulation, we compared the 9-1-lacZ and 9-1-mut-lacZ expression patterns in the SOPs of the thoracic microchaetae. Similar to what we observe in embryos, a single-nucleotide change in the 9-1 E box abolishes most of the lacZ expression in pupae of the same age (Fig. 2A,B), again suggesting direct regulation of sens by proneurals. All LacZ-positive cells are SOPs, as they coexpress Sens and βGAL (Fig. 2C-E).
We wished to assess the effects of loss- and gain-of-function of proneural genes on sens expression in the imaginal discs of third instar larvae. Because fragments 9 and 9-1 do not drive lacZ at this stage (data not shown), we used enhancer 8. The 8-lacZ transgene drives lacZ expression in several wing SOPs in late third instar larvae (Fig. 2F). To determine whether proneural genes are able to control 8-lacZ expression, we overexpressed Sc in the wing pouch using the C5-GAL4 driver (Fig. 2P; Yeh et al. 1995). As shown in Figure 2G, many more cells express lacZ in the wing pouch than in wild type (Fig. 2F), indicating that the Sc protein is able to induce lacZ expression ectopically. On the other hand, removal of the activity of both ac and sc genes results in loss of lacZ expression in all of the ac/sc-dependent SOPs (Fig. 2H). Note that the precursors of the ventral radius and the femoral chordotonal organs (arrows) still express lacZ, as these cells are dependent on Ato expression (Jarman et al. 1993). Moreover, upon Ato overexpression driven by dpp-GAL4 (Fig. 2Q), 8-lacZ is strongly induced at the A/P boundary (Fig. 2I). Together, these data indicate that proneural proteins regulate sens expression in the precursors of the adult PNS. Fragment 8 contains two E boxes, one of which is fully conserved between D. pseudoobscura and D. melanogaster. Band-shift experiments showed that the Ac/Da heterodimer can bind to a radioactive probe that contains the conserved E box of fragment 8 (data not shown), further suggesting that proneurals directly regulate sens expression.
E(spl) proteins are known to prevent SOP formation through transcriptional repression of proneural gene expression. We sought to examine whether they affect sens expression as well. We used scabrous (sca)-GAL4 (Fig. 2R; Abdelilah-Seyfried et al. 2000) to express E(spl)m8 in the SOPs and a few cells around the SOPs in third instar imaginal discs. Figure 2J shows that lacZ expression is abolished in most or all cells (Fig. 2, cf. F and J). Moreover, misexpression of an “activator” version of E(spl)m7 (m7ACT), in which the Gro-binding motif is replaced with the VP16 transactivator domain (Jimenez and Ish-Horowicz 1997), caused numerous extra lacZ-positive cells when driven in the wing pouch (Fig. 2K). These observations suggest that E(spl)m7 and E(spl)m8 proteins are also involved in the transcriptional regulation of sens and that proneural proteins and E(spl) proteins have an antagonistic relationship in transcriptional control of sens. E(spl) proteins are known to bind to proneural gene enhancers and m7ACT is able to activate ac and sc transcription (Jimenez and Ish-Horowicz 1997). Therefore, it is formally possible that m7ACT is indirectly activating the sens enhancer through its up-regulation of proneural proteins. On the other hand, it has been shown recently that even in the absence of endogenous ac and sc, overexpression of m7ACT causes extra bristle formation (Giagtzoglou et al. 2003), suggesting that the E(spl) proteins not only regulate proneural gene expression, but also regulate the expression of one or more of proneural target genes. Is m7ACT able to induce sens expression in the absence of ac and sc? To address this question, we confirmed that overexpression of m7ACT can produce several extra bristles in a sc10-1 background (Fig. 2, cf. M and N). Staining of the imaginal wing discs of these flies showed that there are many Sens-positive cells in the anterior part of the presumptive notum (Fig. 2O), where the Eq-GAL4 driver used in this experiment is expressed (Fig. 2S; Pi et al. 2001). Whereas we cannot exclude that a proneural protein other than ac or sc is mediating the activation of sens by m7ACT, our data suggest that sens is one of the targets of the E(spl) proteins. Altogether, sens enhancers seem to be able to integrate the positive and negative inputs from proneural and E(spl) proteins, respectively.
Sens physically interacts with a subset of the bHLH members of the E(spl) complex
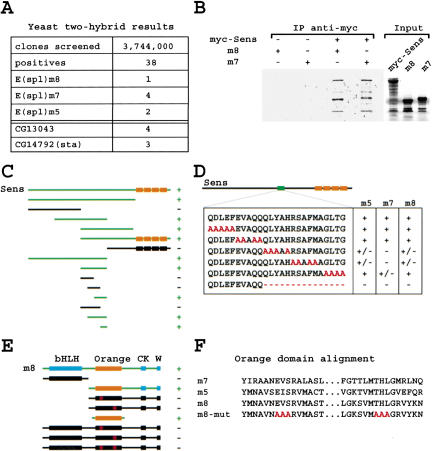
Protein-protein interactions play a significant role in determining how a transcription factor regulates its target genes. To identify proteins that bind Sens, we performed a yeast two-hybrid (YTH) screen. Of 38 positives sequenced from the screen, seven correspond to members of the E(spl) complex (Fig. 3A). To confirm the interactions identified in yeast, we performed coimmunoprecipitation (co-IP) assays using in vitro-translated E(spl) proteins and myc-tagged Sens. A monoclonal anti-myc antibody could precipitate E(spl)m7, E(spl)m8 (Fig. 3B), and E(spl)m5 (data not shown) only in the presence of myc-Sens. We then used the YTH assay to identify the interaction motif in each partner. Testing a series of Sens deletion constructs showed that a 25-amino acid fragment of Sens (amino acids 276-300) is necessary and sufficient for Sens/E(spl) interaction (Fig. 3C). To further delineate the interaction motif, we mutated the 25 amino acids to alanines five at a time and generated five mutant sens constructs (Fig. 3D). The YTH assays suggested that a 15-amino acid deletion (Sens-del) would abrogate the interaction for all three members of the E(spl) complex. This was indeed observed.
Figure 3.
Sens interacts physically with E(spl) proteins. (A) Three members of the E(spl) complex, as well as two other genes, were isolated in a two-hybrid screen in which Sens was used as bait. (B) Sens binds to E(spl)m7 and m8 in a co-IP assay. (C) A series of deletion constructs identify a 25-amino acid interaction domain of Sens that is necessary and sufficient to interact with full-length E(spl)m8 protein in a YTH assay. The boxes depict zinc fingers. (D) Site-directed mutagenesis of the 25-amino acid motif was used to refine the interaction motif. (E) The Orange domain of E(spl) is necessary and sufficient to interact with Sens in a YTH assay. (F) Alignment of the Orange domains of the three members of the E(spl) complex that interact physically with Sens. m8-mut shows the amino acids mutated in the Orange domain.
Proteins of the E(spl) complex have several conserved motifs, for example, a basic domain, a Helix Loop Helix (HLH) domain, an Orange domain, a caseine kinase-binding motif (CK), and a WRPW or Gro interaction domain (W). To find the interaction motif in the E(spl) proteins, we created deletion constructs of E(spl)m8 and tested their ability to bind Sens in yeast. As shown in Figure 3E, the Orange domain was necessary and sufficient for the Sens/E(spl)m8 interaction. The 25-amino acid motif of Sens in isolation interacts with the Orange domain of E(spl)m8 in isolation in the YTH assay (data not shown). The Orange domain is conserved in all members of the Hairy-E(spl) family of proteins and there is evidence that this domain is functionally important (Dawson et al. 1995; Giebel and Campos-Ortega 1997). Alignment of the Orange domains of E(spl)m5, E(spl)m7, and E(spl)m8 prompted us to mutate three amino acids in each of the two conserved motifs to alanine (Fig. 3F) and test the ability of mutant m8 proteins to interact with Sens. We find that replacement of EVS with AAA or THL with AAA is sufficient to abolish the interaction of E(spl)m8 with Sens in our yeast assay. In summary, our data indicate that Sens and E(spl) proteins interact in yeast and in vitro.
Sens exhibits a strong transcriptional synergism with Da and Ac proteins on the ac promoter
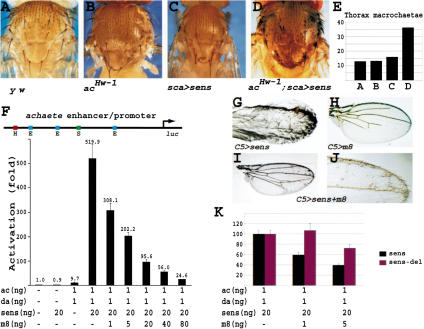
To further explore the mechanism by which Sens promotes SOP specification, we studied how Sens regulates proneural gene expression. We have shown previously that in sens mutant clones, proneural proteins fail to accumulate in the SOPs (Nolo et al. 2000). In the same study, a strong synergism was noted in the ability of Sens and Sc to promote extra bristle formation. Because in this study we have used Ac protein in our assays, we sought to establish whether there is also an in vivo synergy between Sens and Ac. Figure 4B shows that acHw-1 exhibits an occasional extra macrochaetae on the notum because of an increase in ac transcript level (Campuzano et al. 1986). Overexpression of Sens with sca-GAL4 also causes a number of extra micro- and macrochaetae on the notum (Fig. 4C). Comparison of Figure 4, C and D, indicates that overexpression of Sens in an acHw-1 background causes many more extra macrochaetae than the sum of the two genotypes alone, as quantified in Figure 4E. Therefore, there is a synergy between the bristle-promoting effects of the two proteins in vivo.
Figure 4.
Sens and Ac synergize in vivo and in vitro. (A) Thorax of a y w fly. (B) Thorax of an acHw-1 mutant. acHw-1 causes ectopic expression of Ac, leading to an occasional additional macrochaetae. (C) Thorax of a fly expressing Sens using a relatively weak UAS-sens transgene under the control of sca-GAL4 driver. A few extra bristles are observed in clusters. (D) Same as in C, but in an acHw-1 mutant background. Note the presence of numerous extra bristles, often in clusters. (E) Quantification of the number of macrochaetae in hemi-nota of flies shown in A-D. (F) Transcription assay in S2 cells using an enhancer/promoter fragment of the ac gene to drive luciferase (luc). H depicts Hairy/E(spl)-binding site. Note the 50-fold synergism between Sens and Ac/Da. (G-J) Wings of flies overexpressing Sens (G), E(spl)m8 (H), and both Sens and E(spl)m8 (I,J) in the wing pouch using C5-GAL4. (K) The transcriptional synergy obtained with Sens/Ac/Da (black) and Sens-del/Ac/Da (red) were normalized to 100%. Note that the ability of E(spl)m8 to antagonize the Sens-del synergy is significantly decreased compared with the wild-type Sens synergy.
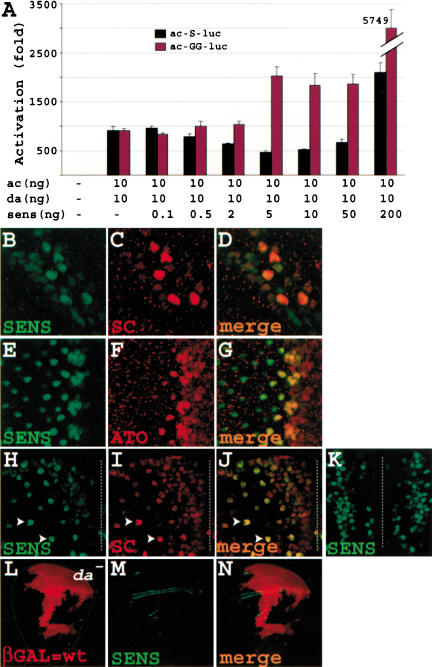
We next established an assay to determine whether Sens can affect ac gene transcription in Drosophila S2 cells. The reporter construct used in this assay was a 470-nucleotide fragment of the ac gene containing the ac promoter region fused to the firefly luciferase. This fragment contains the Hairy-E(spl)-binding site and the three E boxes that were shown to be involved in ac regulation by proneural and E(spl) proteins (Van Doren et al. 1992, 1994; Ohsako et al. 1994; Giagtzoglou et al. 2003). Moreover, it has been shown that during the course of microchaetae SOP specification, expression driven by the ac proximal enhancer/promoter refines from the proneural cluster to a single cell, supporting the notion that it can serve as an SOP-specific regulatory region (Modolell 1997; Pi et al. 2001). The constitutively active actin5 promoter was used to drive the expression of Da, Ac,
Sens, or E(spl)m8. As indicated in Figure 4F, Sens alone does not activate the ac470-luc construct. Cotransfection of minimal amounts of actin5-da and -ac activates the luciferase expression about 10-fold. However, adding an additional 20 ng of the actin5-sens leads to a dramatic activation of the ac promoter (>500-fold). We conclude that Sens can activate ac transcription through synergism with Ac/Da heterodimer in Drosophila S2 cells. These findings suggest that there is a parallel between the in vivo and transcriptional synergy observed between Sens and Ac.
Because E(spl)m8 strongly antagonizes SOP specification, we postulated that it may decrease the synergistic activation of ac by Sens. Cotransfection of 100 ng of actin5-E(spl)m8 with 1 ng actin5-da and -ac does not significantly repress the luciferase activity induced by these proneural proteins (data not shown). However, as shown in Figure 4F, cotransfection of actin5-E(spl)m8 together with sens, da, and ac constructs inhibits the synergy in a dose-dependent manner. In summary, Sens is able to strongly synergize with the proneural proteins in vivo and in vitro, and this synergism is antagonized by E(spl) proteins in a dose-dependent manner.
To determine whether the E(spl) antagonism of the Sens synergism operates in vivo, we first documented that overexpression of Sens at high levels in the wing pouch produces a vast excess of bristles in the wing (Fig. 4G). In addition, we also observe extra vein tissue and thickening of the wing veins. E(spl)m8 overexpression with the C5-GAL4 driver causes loss of wing vein tissue, as well as loss of some of the dorsal wing margin bristles (Fig. 4H; data not shown). When Sens and E(spl) are coexpressed, the two proteins suppress each other's phenotypes; the number of extra bristles is decreased significantly, and many wing veins are restored (Fig. 4I). A higher magnification (Fig. 4J) shows that there are still extra bristles as well as some aberrant vein tissue. Taken together, these data support the notion that Sens and E(spl)m8 have antagonistic effects at the level of proneural gene expression, in agreement with their in vivo effects on bristle formation. Finally, we sought to determine whether the physical interaction between Sens and E(spl) plays a role in their antagonism on ac enhancer in S2 cells. The Sens-del, which lacks the 15-amino acid E(spl)-interacting motif (Fig. 3D), can synergize with Ac and Da similar to wild-type Sens (data not shown). However, the ability of E(spl)-m8 to antagonize the synergy between Sens-del and proneural proteins is impaired when compared with its effect on the wild-type Sens/Ac/Da synergy (Fig. 4K), suggesting that the physical interaction between Sens and E(spl) plays a role in their antagonistic effect.
The Sens-binding site of the ac enhancer affects transcriptional activation by Sens, and bristle-promoting ability of an ac minigene
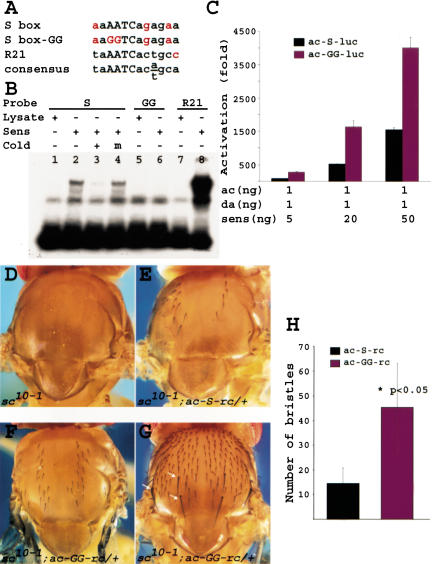
Sens can bind the consensus binding site of its vertebrate homolog Gfi-1 (Zweidler-Mckay et al. 1996; Nolo et al. 2000). Examination of the ac proximal enhancer showed that only one putative Sens-binding site is present between two of the E boxes in this enhancer (Fig. 4F, S box; Fig. 5A). Band-shift assays show that Sens can bind to S box in a sequence-specific manner (Fig. 5B). Mutating the core sequence from AATC to GGTC abolished Sens binding in this assay (Fig. 5B, lane 6). As a positive control, we used another oligo with 92% identity to the consensus (Fig. 5A, R21). Significantly more R21 probe was shifted by Sens than the endogenous oligo (Fig. 5B, lanes 7-8). These data indicate that there is a binding site for Sens in the ac promoter.
Figure 5.
Sens binds an AATC core in the ac enhancer and acts as a repressor in vivo. (A) Nucleotide consensus-binding element to which Sens binds in vitro using EMSA (B). (C) Mutational analysis of the ac enhancer shows that the ∼50-fold synergy documented in Fig. 4 becomes a ∼150-fold synergy under similar conditions when the Sens-binding site is mutated. (D-G) Thoraxes of adult flies that lack endogenous Sc and Ac proteins. (D) Note the complete absence of bristles in these flies. (E) In the presence of a wild-type ac minigene, some microchaetae are restored. (F) In the presence of a mutant ac minigene, we observe rescue of more microchaetae than with the wild-type minigene. (G) In one transgenic line, we even observe rescue of the ac-dependent macrochaetae (arrows). This was not observed with the wild-type minigene. (H) Quantification of the number of bristles rescued with the two transgenes: average number of bristles ± standard error of mean. The numbers are significantly different.
To determine whether S box mutations affect the synergy between Sens and Da/Ac, we repeated the transfections with an S box-mutant version of the ac-luc reporter. Quite unexpectedly, the mutant reporter construct consistently showed a three- to fourfold increase in synergism when compared with wild-type reporter (Fig. 5C). This suggests that DNA-binding has a negative regulatory role in the transcriptional activation of ac by Sens in S2 cells. To test the in vivo relevance of this observation, we examined whether the S box in the ac enhancer has a role in bristle formation in vivo. It is well established that sc10-1 mutant flies are devoid of thoracic bristles (Fig. 5D; Garcia-Bellido 1979). It has also been shown that two wild-type copies of a 2.2-kb ac minigene can restore some of the microchaetae on the notum of sc10-1 mutant flies (Van Doren et al. 1992). We therefore mutated the S box core from AATC to GGTC in a 2.2-kb ac genomic fragment and created transgenic animals. We obtained six wild-type and nine mutant transgenic strains. For each strain, we scored at least five flies containing the transgene in a sc10-1 background. Figure 5, E and F, show representative pictures of rescues conferred by wild-type or mutant ac minigenes, respectively. Comparison of the number of bristles restored by one copy of mutant versus wild-type transgene showed that, in agreement with our transcription assay results, the mutant ac minigene is more potent in promoting bristle formation than the wild-type transgene (Mann-Whitney U test: P <0.05; Fig. 5H). Interestingly, one of the mutant transgenes rescued almost all microchaetae on the notum, along with the three ac-dependent macrochaetae on each side (Fig. 5G, arrows; Simpson 1990; Gomez-Skarmeta et al. 1995). It is worth mentioning that none of the six wild-type transgenic lines show macrochaetae rescue. These data suggest that DNA binding is a negative modulator of the synergy between Sens and Ac/Da heterodimer in vivo.
Sens acts as a transcriptional repressor and activator
Because proneural gene expression precedes sens expression in most proneural clusters (see below), one can postulate that at least in a transitional period, Sens levels will be lower than proneural protein levels. Because we used a 1 proneural:20 sens ratio in previous experiments, we decided to reverse the ratio. Figure 6A (black bars) shows that Ac and Da can strongly induce luciferase gene expression, in agreement with a previous report (Van Doren et al. 1992). As the amount of sens construct is increased, we observe a gradual repression in luciferase activity, which reaches 50% of the Ac/Da activation. In agreement with our previous observations, when the ratio is 1 proneural:20 sens, we again observe synergism (∼2000-fold activation of baseline). We conclude that Sens can act both as a repressor and as an activator of ac transcription, depending on the ratio between Sens and Ac/Da.
Figure 6.
Sens acts as a dose-dependent transcriptional repressor and activator. (A) Transcription assays with higher proneural-to-Sens ratio show that Sens can act as a repressor. In addition, mutation of the Sens-binding site abolishes the repressive ability of Sens and causes synergy at levels that repress the wild-type enhancer. (B-K) Sens is not only expressed in SOPs but also in surrounding cells in the wing margin (B-D) and the eye imaginal disc (E-G) and pupal microchaetae field (H-K). Note that Sens is expressed at low levels in the nuclei of many cells in the wing margin of a Canton S third instar larva (B), and in 6-7 cells in each preommatidial cluster in the eye disc of a Canton S third instar larva (E). (H-J) Part of the hemi-notum of an 8-10-hour-old sensE2/+ pupa, which essentially has a wild-type bristle pattern. The future midline is depicted by the broken line. Note the field of cells closer to the midline that express low levels of Sens and Sc. Arrowheads show two examples of presumptive SOPs, which have up-regulated Sens and Sc. Note that the cells surrounding the SOPs show minimal Sens and Sc expression at this stage. (K) Part of the notum of a 7-9-hour-old Canton S pupa containing the medial aspects of both developing hemi-nota. Note columns of Sens-expressing cells on both sides of the midline. (L-N) Sens expression in the wing margin does not depend on da. Large mitotic clones for a null da allele are evident by the lack of βGAL staining (L). Note that in both the large clone covering the anterior margin and the small clone touching the posterior margin Sens expression is intact (N). Also note that in precursors of the ventral and dorsal radii, there seems to be a mutant cell at the border of the clone with Sens expression.
As our previous transfection and in vivo assays suggested a negative role for Sens DNA-binding in ac transcription and bristle promotion, we wished to determine whether the repressive role of low-level Sens is mediated via DNA binding. We therefore performed a similar transfection assay using the AATC to GGTC mutated ac enhancer as the reporter. Our results show that upon removal of the Sens-binding site, its ability to repress the luciferase level is lost (Fig. 6A, red bars). Moreover, the synergy between Sens and Ac/Da begins at a much lower sens:proneural ratio and reaches significantly higher levels. Therefore, the repressive effect of Sens seems to depend on its DNA binding.
The above findings prompted us to revisit Sens expression and its colocalization with proneural proteins. We find that Sens protein expression is not confined to the SOPs, in which it is abundantly expressed; it is also expressed at lower levels in cells surrounding the SOPs. This domain of expression is smaller than the proneural cluster and seems to be confined to the proneural field or even fewer cells. This is illustrated for the wing margin (Fig. 6B-D), the eye (Fig. 6E-G), and the microchaetae field of the pupal notum (Fig. 6H-K). In all of these extended proneural fields, low levels of Sens and proneural proteins are expressed in numerous cells that fail to become SOPs. However, in all of these tissues, cells that exhibit high levels of Sens also accumulate large amounts of proneural proteins. It is worth mentioning that we have not been able to detect similar low-level Sens expression in the typical single-SOP fields of notum macrochaetae, which could either be a technical issue or suggest that in these proneural fields, Sens expression is confined to SOPs.
In summary, our data suggest that low levels of Sens are present in cells that surround the presumptive SOPs of the notum microchaetae, wing margin, embryonic PNS (data not shown), as well as in cells that surround the presumptive R8 photoreceptors. Although all of the cells with low-level Sens expression also express low levels of proneural proteins, many of them will later lose proneural gene expression and adopt a non-neural fate. These observations are in agreement with the hypothesis that whereas high levels of Sens are required for proneural up-regulation in the SOP, low levels of Sens might repress proneural gene expression, and thus suppress neural potential.
Low levels of Sens expression do not seem to require proneural gene expression in the wing margin
Because Sens is expressed in the posterior wing margin, and as ac, sc, ato, and amos are not expressed in the posterior wing margin, we wished to determine whether expression of Sens in these cells is dependent on proneural gene expression by removing da. As shown in Figure 6, L-N (and data not shown), large clones of da do not cause a loss of early Sens expression at the anterior or posterior wing margin, suggesting that early Sens expression in these cells is under the control of other signaling pathways. This early expression of Sens is not affected in a sc10-1 animal either (data not shown). However, Sens expression is lost wherever the da clone encompasses an area other than the wing margin from which SOPs would normally arise. Finally, no Sens protein was detected in the developing notum of a 10-12-hour-old sc10-1 pupa (data not shown), suggesting that in the pupal microchaetae field, proneural proteins are the primary transcriptional activators of sens. Together, these data suggest that whereas the initiation and up-regulation of Sens in the majority of presumptive SOPs are under direct transcriptional control of proneural proteins, other proteins seem to be involved in the initiation of Sens expression in the wing margin.
Low levels of Sens are required for proneural protein down-regulation in vivo
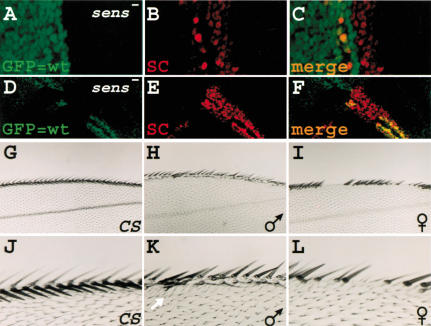
So far, we have provided evidence that low levels of Sens can act as a transcriptional repressor of ac in cell culture, and that in most proneural fields, low levels of Sens are present in the cells surrounding the presumptive SOP. To strengthen the hypothesis that Sens acts as a transcriptional repressor in the cells that express low levels of Sens, we generated sens clones in the wing imaginal discs of third instar larvae. If low levels of Sens repress proneural gene expression, and high levels promote SOP development, lack of Sens protein should lead to continued or slightly increased expression of proneural proteins in proneural fields, whereas causing a loss of up-regulation of proneural proteins in SOPs. We therefore expect to observe broad low levels of proneural proteins in sens mutant clones. We selected two different clones, one parallel to the dorso-ventral midline (Fig. 7A-C), and the other perpendicular to this midline (Fig. 7D-F). In both cases, the Sc expression fails to become restricted to single cells, as is observed in adjacent heterozygous or wild-type tissue. These observations provide further evidence that Sens is necessary to down-regulate proneural expression in the cells that will not adopt the SOP fate.
Figure 7.
Sens acts as a repressor in vivo. (A-F) Mitotic clones for the sensE2 allele are evidenced by the lack of GFP. Unlike the wild-type regions, the broad Sc expression in the clones fails to be down-regulated and become restricted to single cells. In A-C, the clone border almost coincided with the dorso-ventral boundary of the wing margin. (G-L) Depending on the expression level, Sens can preferentially result in gain or loss of bristles in the wing margin. Anterior is to the top, proximal to the left. (G,J) Canton S wing margins. (H,K) From a male progeny of the cross between UAS-sens; +; + and +; +; C96-GAL4. (I,L) From a female progeny of the same cross. The white arrow in K shows one of the extra bristles caused by Sens overexpression.
Finally, to provide additional evidence that Sens may act as a binary switch, we ectopically expressed varying levels of Sens to determine whether low levels of Sens expression prior to its normal onset of expression might cause bristle loss. We have shown previously that expression of Sens in the wing margin using the C96-GAL4 driver can result in wing-margin tissue loss, including bristles, similar to what is observed in Lyra mutants (Nolo et al. 2001). We crossed C96-GAL4 to our weakest UAS-sens transgene, which is inserted on the X chromosome, and compared females and males with one copy of the transgene in an otherwise identical genetic background and environment. Because males display dosage compensation, they should express more Sens protein than their sisters. As shown in Figure 7, H and K, most male progeny have a few extra bristles along the margin (Fig. 7, cf. G and J) when reared at 25°C. However, most female progeny display patches of wing margin bristle loss (Fig. 7I,L). These data suggest that lower amounts of exogenous Sens can preferentially lead to bristle loss.
Discussion
The selection of an SOP from a proneural cluster in the Drosophila PNS is one of the best-studied examples of neuroblast determination. Yet, an important question has remained unanswered. How precisely is one cell selected from surrounding cells, and what are the molecular mechanisms that underlie this selection? On the basis of our observations, we propose a model in which an intricate set of feedback loops between various transcription factors determines, through the action of Sens and E(spl), the selection of the adult SOP.
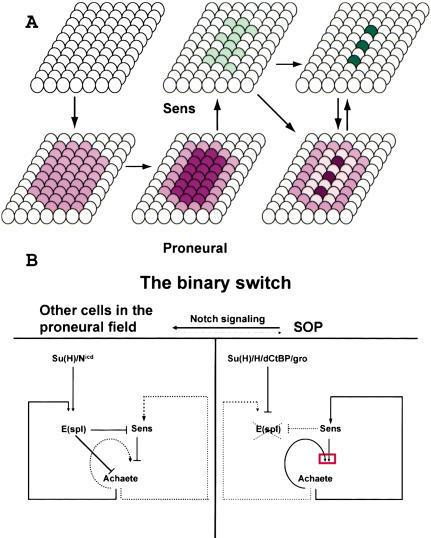
As shown in Figure 8A, most cells of a proneural cluster first express relatively low levels of proneural proteins. This leads to transcriptional activation of E(spl) genes in the cluster (Singson et al. 1994; Heitzler et al. 1996; Cooper et al. 2000). E(spl) proteins, together with the corepressor Gro, then prevent the up-regulation of proneural gene expression in the cluster (Paroush et al. 1994; Heitzler et al. 1996). It is thought that prepattern factors then lead to a higher level of proneural protein expression in a smaller group of cells of the proneural cluster, the proneural field (Modolell 1997). We propose that this higher level of proneural expression, probably together with the prepattern factors, induces low levels of Sens expression in the proneural field or an area that is even smaller. We observe consistent low levels of Sens staining in groups of cells in the pupal microchaetae field, embryos, wing, and eye discs. These domains that are part of the proneural cluster colabel with proneural proteins, and a single or a few cells are typically selected from these domains to induce higher levels of Sens (Fig. 6). We propose that Sens plays a critical role in the SOP through transcriptional synergy with proneural proteins. In addition, our data suggest that Sens plays a role in repressing proneural expression in non-SOP cells. Hence, we propose that Sens acts as a binary switch in the refinement of the proneural field that will lead to SOP selection.
Figure 8.
Model for Sens action. (A) Schematic of proneural and Sens expression in a spatial and temporal fashion. The darkness of the colors is meant to reflect levels of proteins. Arrows indicate temporal events. We have been unable to establish whether, in the SOP, the elevated Sens expression precedes elevated proneural expression, whether they occur simultaneously, or whether elevated proneural expression precedes elevated Sens expression. (B) Schematic of the mechanism by which the Sens binary switch operates. N signaling and low levels of Sens keep the levels of proneural proteins low in the proneural field. Upon N signaling, the Nicd together with Su(H) increase the expression of E(spl) proteins. Proneural proteins also increase the expression of E(spl). E(spl) in turn prevents proneural protein up-regulation by inhibiting expression and function of Sens and proneurals. Low levels of Sens also repress the expression of proneural genes. At a later stage, Sens levels in the presumptive SOP reach a point that is sufficient to initiate synergism (red box) between proneurals and Sens. Increased levels of proneurals in the presumptive SOP prevents this cell from receiving the N signal, and Su(H) together with Hairless and corepressors repress the expression of E(spl). In addition, high levels of Sens prevent the E(spl) proteins from repressing the proneurals.
Our data also suggest that sens transcription is mediated directly through proneural binding to E boxes in the sens enhancers (Figs. 1 and 2). In addition, sens enhancers integrate two opposing forces, the positive regulation by proneural and the negative regulation mediated by E(spl) proteins (Fig. 2), similar to SOP-specific enhancers of the proneural genes.
Because E(spl) prevents the up-regulation of the proneural gene and sens expression, this repressive effect must be overcome if some cells of the proneural field are to be selected as SOPs. In fact, it has been shown recently that by repressing E(spl)m8 and other repressors of sens, Su(H) plays a positive role in the SOP fate promotion (Koelzer and Klein 2003). It is also known that proneural proteins positively regulate E(spl) gene expression, which will prevent further up-regulation of proneural proteins (Singson et al. 1994; Cooper et al. 2000). This negative feedback has prompted the idea that to accumulate large amounts of proneural proteins in the SOP, the equilibrium between the proneural and E(spl) proteins should be displaced in favor of proneurals (Modolell 1997). We propose that the synergy between Sens and Da/Ac on the ac regulatory region is a key mechanism for the up-regulation of ac transcription. In this model, Sens accelerates proneural gene expression and proneural protein accumulation, overruling the negative feedback conferred by E(spl). This hypothesis is supported by the observation that the synergy between Sens and proneurals is highly sensitive to the levels of E(spl) protein in the transcription assay (Fig. 4F), as well as in vivo (Fig. 4G-J; data not shown). Ac up-regulation will lead to further Sens production and increased synergistic activation of ac transcription. In the absence of Sens, the presumptive SOPs fail to up-regulate proneural gene expression (Nolo et al. 2000; Frankfort et al. 2001). Hence, Sens will render the presumptive SOP less sensitive to N signaling. This is also supported by the observation that coexpression of Sens and proneurals is able to produce closely spaced bristles, indicating highly inefficient N signaling (Fig. 4D; Nolo et al. 2000; Lai 2003; data not shown). In summary, we propose that the balance between the levels of the Sens and E(spl) proteins determines the SOP selection.
The synergistic model of proneural gene activation predicts that low levels of Sens and proneural proteins may suffice to override the E(spl) inhibition. However, many cells that express sens and proneural genes fail to up-regulate proneural gene expression (Fig. 6B-J). We find that at low levels, Sens acts as a repressor of ac transcription (Fig. 6A), suggesting that in addition to the relative levels of E(spl), the relative levels of proneural proteins and Sens also play a critical role in SOP selection. In those areas of the proneural field in which Sens and proneural protein levels are low, not only is the transcriptional synergy absent, but there is also a weak repression of proneural gene expression. This should lead to a rapid loss of Sens expression and a failure to adopt the SOP fate. Analysis of the Sc expression pattern in sens clones that include the wing margin confirmed that in the absence of Sens function, the broad Sc expression in the wing margin persists, and at the same time, the presumptive SOPs fail to up-regulate Sc protein. This is further supported by the observation that overexpression of low levels of Sens causes bristle loss in the wing margin.
The mechanism by which Sens represses transcription of proneural genes is probably through DNA binding. As shown in Figure 6A, when the S box is mutated, Sens is unable to repress ac transcription. This finding is corroborated with our in vivo observations that the ac minigene with the mutated Sens-binding site is a more potent inducer of bristle formation than the wild-type minigene (Fig. 5D-H). We therefore conclude that the transcriptional repression of the ac promoter by Sens is mediated through DNA binding.
Altogether, our data support a model in which Sens promotes the SOP fate in one cell by activating ac transcription, whereas it prevents SOP fate in the neighboring cells by repressing ac transcription. The relative levels of Sens, proneural, and E(spl) proteins seem to be the major determinants of these fate decisions. Therefore, we propose that Sens acts as a binary switch in SOP determination by affecting a series of interconnected positive and negative regulatory loops to refine the potential for a specific fate from a group of cells to a single cell, the SOP.
Materials and methods
Plasmid constructs for lacZ reporters and rescue constructs
The 11-kb sens genomic fragment (Nolo et al. 2000) was digested with EcoRI, and the three resulting fragments were subcloned into the pCaSpeR-lacZ vector. Nine overlapping pieces of the 3.4-kb fragment, as well as 8-1, 8-2, 8-3, 9-1, and 9-2 were then generated by PCR and cloned into the pCaSpeR-lacZ vector. For E box mutagenesis, the 9-1 fragment was cloned into pBluescript vector (Stratagene) and the E box in the 9-1 fragment was mutated using QuikChange Site-Directed Mutagenesis kit (Stratagene). The 9-1 and 9-1-mut fragments were then inserted into pH-Pelican-lacZ vector (Barolo et al. 2000). The wild-type 2.2 kb ac genomic region (Van Doren et al. 1992) was subcloned in pBluescript and mutated at the S box. Both wild-type and mutant minigenes were subcloned into pCaSpeR-4vector. All constructs were integrated into the germ line (Rubin and Spradling 1982).
Fly strains
The following strains were used in this study: y w, Canton S, UAS-sens (C1), sensE2 FRT80B/TM6B (Nolo et al. 2000), UAS-sc, UAS-ato/TM3, Sb1 (Y.-N. Jan, University of California at San Francisco), Df(1)sc10-1, sc10-1/y1 acHw-1, y1 w*; P{w+mW.hs=GawBsca109-68/CyO (Bloomington, Stock Center), C5-GAL4 (Yeh et al. 1995), UAS-m8 (S. Bray, University of Cambridge), UAS-m7ACT (Jimenez and Ish-Horowicz 1997), UAS-CD8::GFP, y w hsFLP122; P{w+mC=ubi-GFP}61EF M(3)i(55) P{w+}70C FRT80B/TM6B, dpp-GAL4/TM6B, y w hsFLP122; M(2)24F FRT40A/CyO (G. Mardon, Baylor College of Medicine), Eq-GAL4/TM6B (Pi et al. 2001), C96-GAL4 (Nolo et al. 2001), da3 FRT40A/SM5-TM6 (K. Cadigan, University of Michigan). To generate sens and da mitotic clones, 1-h heat shock at 37° was used 24-48 h after egg laying to larvae of the following genotypes, respectively: y w hsFLP122; sensE2 FRT80B/P{w+mC=ubi-GFP}61EF M(3)i(55) P{w+}70C FRT80B and y w hsFLP122; da3 FRT40A/M(2)24F FRT40A.
EMSA
EMSAs were performed as described previously (Ou et al. 2000) using in vitro-translated proteins. Oligonucleotide sequences are available upon request.
YTH screen and interaction assays
Screen The MATCHMAKER Two-Hybrid System 3 was used for the YTH screen (Clontech). Full-length Sens was used as the bait. A Drosophila embryonic cDNA library cloned into the pACTII vector was used for the screen (S. Elledge, Baylor College of Medicine).
Sens-E(spl) interactions
Sens and E(spl) fragments were cloned in pGBKT7 and pACTII vectors, respectively, cotransformed into the AH109 yeast strain (Clontech), and plated on selective medium.
Biochemical interactions
Co-IPs were performed according to Clontech's MATCH-MAKER Co-IP Kit user manual, using 35S-myc-Sens and 35SE(spl) proteins generated from cDNA clones in pGBKT7 and pRSET (Invitrogen) vectors, respectively. The anti-myc antibody was from Oncogene.
Immunohistochemistry and X-Gal stainings
Primary antibodies were as follows: guinea pig anti-Sens (1:800) (Nolo et al. 2000), rabbit anti-βGAL (1:1000) (Cappel), mouse anti-βGAL (1:1000) (Promega), rabbit anti-Sc (1:100) (a gift from G. Boekhoff-Falk, University of Wisconsin at Madison), rabbit anti-Ato (1:2500) (Frankfort et al. 2001). Secondary antibodies were as follows: biotinylated anti-rabbit antibody (1:800) (Vector Laboratories), Alexa488-anti-guinea pig (1:500) (Molecular Probes) and Cy3-anti-rabbit (1:500), Cy3-anti-mouse (1:500) and Cy5-anti-guinea pig (1:500) (Jackson ImmunoResearch Laboratories). Confocal images were captured using a Zeiss microscope. X-Gal stainings were done according to Pi et al. (2001).
Plasmid constructs for the S2 cell assays
The 470-bp ac enhancer/promoter region was generated by PCR and cloned into the firefly reporter plasmid pGL3-Basic (Promega). The 470-bp ac enhancer/promoter region with the mutated S box was obtained by QuikChange Site-Directed Mutagenesis Kit in pBluescript vector and subcloned into pGL3-Basic reporter vector. Expression constructs were made by cloning the ORFs of ac, da, sens, E(spl)m8, and also sens-del into pAc5.1/V5-His-A expression vector (Invitrogen) for constitutive expression under the control of the actin5 promoter. As an internal control, we used pRL-CMV vector (Promega), which expresses the Renilla luciferase under the control of the CMV promoter.
S2 cell transfection and luciferase assays
A total of 0.5 mL of 5-7 × 105 cells/mL S2 cells was seeded in 24-well plates. Sixteen hours later, the cells were cotransfected with the reporter plasmid, expression plasmid(s), and the internal control plasmid pRL-CMV, using Cellfectin Reagent (Invitrogen) according to the manufacturer's protocol. The total amount of DNA used in each set of transfections was kept constant by adding the empty pAc5.1/V5-His-A vector. Cell lysis and luciferase assay were performed 48 h after transfection using the Dual Luciferase Assay System (Promega) and a TD-20/20 Luminometer with dual automatic injectors (Turner Designs). Each data point is the average of three to nine independent transfections.
Sequencing
All constructs were sequenced, and all primers used for sequencing are available upon request.
Acknowledgments
We thank S. Elledge, C.-T. Chien, S. Bray, D. Ish-Horowicz, Y.-N. Jan, J. Posakony, G. Boekhoff-Falk, G. Mardon, K. Cadigan, and the Bloomington Stock Center for reagents, G. Mardon, B. Frankfort, G. Halder, and V. Bayat for comments on the manuscript, and the members of the Bellen lab for their discussions and support. We also thank the reviewers who helped us improve this work significantly. This work was supported by a grant from the NASA. H. J.-N. is an associate and H.J.B. is an investigator of the HHMI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1122403.
References
- Abdelilah-Seyfried S., Chan, Y.M., Zeng, C., Justice, N.J., Younger-Shepherd, S., Sharp, L.E., Barbel, S., Meadows, S.A., Jan, L.Y., and Jan, Y.N. 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand, M.D., and Lake, R.J. 1999. Notch signaling: Cell fate control and signal integration in development. Science 284: 770-776. [DOI] [PubMed] [Google Scholar]
- Bailey A.M. and Posakony, J.W. 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 9: 2609-2622. [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver, L.A., and Posakony, J.W. 2000. GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29: 726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- Cabrera C.V., Martinez-Arias, A., and Bate, M. 1987. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell 50: 425-433. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Balcells, L., Villares, R., Carramolino, L., Garcia-Alonso, L., and Modolell, J. 1986. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell 44: 303-312. [DOI] [PubMed] [Google Scholar]
- Cooper M.T., Tyler, D.M., Furriols, M., Chalkiadaki, A., Delidakis, C., and Bray, S. 2000. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev. Biol. 221: 390-403. [DOI] [PubMed] [Google Scholar]
- Cubas P., de Celis, J.F., Campuzano, S., and Modolell, J. 1991. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes & Dev. 5: 996-1008. [DOI] [PubMed] [Google Scholar]
- Culi J. and Modolell, J. 1998. Proneural gene self-stimulation in neural precursors: An essential mechanism for sense organ development that is regulated by Notch signaling. Genes & Dev. 12: 2036-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S.R., Turner, D.L., Weintraub, H., and Parkhurst, S.M. 1995. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 15: 6923-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C. and Artavanis-Tsakonas, S. 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl. Acad. Sci. 89: 8731-8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu M.E., Zipursky, S.L., and Cagan, R.L. 1996. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122: 4139-4147. [DOI] [PubMed] [Google Scholar]
- Frankfort B.J., Nolo, R., Zhang, Z., Bellen, H., and Mardon, G. 2001. senseless repression of rough is required for R8 photo-receptor differentiation in the developing Drosophila eye. Neuron 32: 403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A. 1979. Genetic analysis of the achaete-scute system of Drosophila melanogaster. Genetics 91: 491-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagtzoglou N., Alifragis, P., Koumbanakis, K.A., and Delidakis, C. 2003. Two modes of recruitment of E(spl) repressors onto target genes. Development 130: 259-270. [DOI] [PubMed] [Google Scholar]
- Giebel B. and Campos-Ortega, J.A. 1997. Functional dissection of the Drosophila enhancer of split protein, a suppressor of neurogenesis. Proc. Natl. Acad. Sci. 94: 6250-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta J.L., Rodriguez, I., Martinez, C., Culi, J., Ferres-Marco, D., Beamonte, D., and Modolell, J. 1995. Cis-regulation of achaete and scute: Shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes & Dev. 9: 1869-1882. [DOI] [PubMed] [Google Scholar]
- Goulding S.E., zur Lage, P., and Jarman, A.P. 2000. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron 25: 69-78. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. 1988. Development of Drosophila larval sensory organs: Spatiotemporal pattern of sensory neurones, peripheral axonal pathways and sensilla differentiation. Development 102: 869-886. [Google Scholar]
- Hartenstein V. and Posakony, J.W. 1989. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107: 389-405. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Bourouis, M., Ruel, L., Carteret, C., and Simpson, P. 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122: 161-171. [DOI] [PubMed] [Google Scholar]
- Huang M.L., Hsu, C.H., and Chien, C.T. 2000. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron 25: 57-67. [DOI] [PubMed] [Google Scholar]
- Jan Y.N. and Jan, L.Y. 1993. The peripheral nervous system. In Development of Drosophila melanogaster (eds. M. Bates and A. Martinez-Arias), pp. 1207-1244. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jarman A.P., Grau, Y., Jan, L.Y., and Jan, Y.N. 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73: 1307-1321. [DOI] [PubMed] [Google Scholar]
- Jarman A.P., Grell, E.H., Ackerman, L., Jan, L.Y., and Jan, Y.N. 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398-400. [DOI] [PubMed] [Google Scholar]
- Jennings B.H., Tyler, D.M., and Bray, S.J. 1999. Target specificities of Drosophila enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 19: 4600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G. and Ish-Horowicz, D. 1997. A chimeric enhancer of-split transcriptional activator drives neural development and achaete-scute expression. Mol. Cell. Biol. 17: 4355-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Schrons, H., Grawe, F., and Campos-Ortega, J.A. 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132: 505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelzer S. and Klein, T. 2003. A Notch-independent function of Suppressor of Hairless during the development of the bristle sensory organ precursor cell of Drosophila. Development 130: 1973-1988. [DOI] [PubMed] [Google Scholar]
- Kramatschek B. and Campos-Ortega, J.A. 1994. Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development 120: 815-826. [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2003. Drosophila tufted is a gain-of-function allele of the proneural gene amos. Genetics 163: 1413-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M. and Schweisguth, F. 1995. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes & Dev. 9: 2598-608. [DOI] [PubMed] [Google Scholar]
- Modolell J. 1997. Patterning of the adult peripheral nervous system of Drosophila. Perspect Dev. Neurobiol. 4: 285-296. [PubMed] [Google Scholar]
- Nolo R., Abbott, L.A., and Bellen, H.J. 2000. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349-362. [DOI] [PubMed] [Google Scholar]
- ____. 2001. Drosophila Lyra mutations are gain-of-function mutations of senseless. Genetics 157: 307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako S., Hyer, J., Panganiban, G., Oliver, I., and Caudy, M. 1994. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes & Dev. 8: 2743-2755. [DOI] [PubMed] [Google Scholar]
- Ou X.M., Jafar-Nejad, H., Storring, J.M., Meng, J.H., Lemonde, S., and Albert, P.R. 2000. Novel dual repressor elements for neuronal cell-specific transcription of the rat 5-HT1A receptor gene. J. Biol. Chem. 275: 8161-8168. [DOI] [PubMed] [Google Scholar]
- Paroush Z., Finley Jr., R.L., Kidd, T., Wainwright, S.M., Ingham, P.W., Brent, R., and Ish-Horowicz, D. 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805-815. [DOI] [PubMed] [Google Scholar]
- Pi H., Wu, H.J., and Chien, C.T. 2001. A dual function of phyllopod in Drosophila external sensory organ development: Cell fate specification of sensory organ precursor and its progeny. Development 128: 2699-2710. [DOI] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling, A.C. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218: 348-353. [DOI] [PubMed] [Google Scholar]
- Russo C.A., Takezaki, N., and Nei, M. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12: 391-404. [DOI] [PubMed] [Google Scholar]
- Schrons H., Knust, E., and Campos-Ortega, J.A. 1992. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics 132: 481-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. 1990. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 109: 509-519. [DOI] [PubMed] [Google Scholar]
- Singson A., Leviten, M.W., Bang, A.G., Hua, X.H., and Posakony, J.W. 1994. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes & Dev. 8: 2058-2071. [DOI] [PubMed] [Google Scholar]
- Skeath J.B. and Carroll, S.B. 1991. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes & Dev. 5: 984-995. [DOI] [PubMed] [Google Scholar]
- Tong B., Grimes, H.L., Yang, T.Y., Bear, S.E., Qin, Z., Du, K., El-Deiry, W.S., and Tsichlis, P.N. 1998. The Gfi-1B protooncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol. Cell. Biol. 18: 2462-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Powell, P.A., Pasternak, D., Singson, A., and Posakony, J.W. 1992. Spatial regulation of proneural gene activity: Auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes & Dev. 6: 2592-2605. [DOI] [PubMed] [Google Scholar]
- Van Doren M., Bailey, A.M., Esnayra, J., Ede, K., and Posakony, J.W. 1994. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes & Dev. 8: 2729-2742. [DOI] [PubMed] [Google Scholar]
- Villares R. and Cabrera, C.V. 1987. The achaete-scute gene complex of D. melanogaster: Conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell 50: 415-424. [DOI] [PubMed] [Google Scholar]
- Wallis D., Hamblen, M., Zhou, Y., Venken, K.J., Schumacher, A., Grimes, H.L., Zoghbi, H.Y., Orkin, S.H., and Bellen, H.J. 2003. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130: 221-232. [DOI] [PubMed] [Google Scholar]
- Yeh E., Gustafson, K., and Boulianne, G.L. 1995. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc. Natl. Acad. Sci. 92: 7036-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-Mckay P.A., Grimes, H.L., Flubacher, M.M., and Tsichlis, P.N. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16: 4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]