Abstract
Methylation of lysine residues, catalyzed by histone methyltransferase (HMT) enzymes, is one of many modifications of core histone proteins that regulate transcription and chromatin structure. G9a is the predominant HMT in mammalian euchromatin and recent data suggest that it is required to perpetuate a malignant phenotype in cancer cells and is implicated in metastasis, supporting this HMT as a therapeutic target for cancer and other diseases associated with epigenetic regulation. Of the methods currently used to measure methyltransferase activity, many involve a separation step or utilize coupling enzymes complicating implementation and data interpretation. Here we describe a homogeneous assay to measure G9a HMT activity using the chemiluminescence-based AlphaScreen immunoassay technology. Methylation of biotinylated-histone peptide is measured through specific antibody-based detection, in conjunction with streptavidin-coated donor and secondary antibody-coated acceptor beads. The method is particularly well suited for detection of inhibitors acting by the desired histone peptide competitive mechanism and is applicable to testing other HMTs, demonstrated here with the G9a homolog EHMT1, also known as GLP.
INTRODUCTION
The N-terminal tails of histone proteins are subject to a variety of covalent post-translational modifications, including acetylation, methylation, and phosphorylation, that alter chromatin structure to effect changes in gene transcription. Methylated lysine residues serve as epigenetic markers for recruitment of effector or adaptor proteins that modify local chromatin structure to elicit their functional consequences. Members of the SET domain-containing superfamily of histone lysine methyltransferases (HMTs) catalyze the transfer of methyl groups from S-adenosylmethionine (SAM) cofactor to the ε-amine of lysine via their C-terminal catalytic domain and thus regulate the methylation states of histone residues. G9a and other members of the SUV39 family of SET-domain HMTs have been shown to specifically methylate Lys9 of histone H3 (H3K9) 1. G9a catalyzes the mono- and di-methylation of H3K9 in mammalian euchromatic regions 2, 3, where the resulting H3K9me2 is indicative of transcriptional repression 4.
G9a has been recognized as a potential drug target for the treatment of several human diseases, including cancer where it is anticipated that inhibition of G9a, the predominant H3K9 methyltransferase in mammalian euchromatin 5, will result in transcriptional activation and work synergistically with DNA methyltransferase and histone deacetylase inhibitors to kill cancer cells 6, 7. Most inhibitors of HMTs identified to date, including methylthioadenosine, sinefungin, and S-adenosylhomocysteine (SAH), are competitive with the SAM cofactor and thus nonselective in their inhibition of methyltransferases 8. The diazepin-quinazolinamine derivative BIX-01294 was identified as an inhibitor of G9a HMT activity in a screen of 125,000 compounds and did not compete with SAM cofactor 7. Furthermore, treatment of mouse embryonic stem cells and mouse embryonic fibroblasts with BIX-01294 led to a decrease of H3K9me2 at promoter regions of G9a target genes.
Several methods are currently used to measure methyltransferase activity. Many of these involve a separation step which largely precludes execution of high-throughput screening (HTS) of target enzymes. Homogeneous assays for methyltransferases have been described and involve the coupling enzymes SAH hydrolase or SAH nucleosidase to metabolize SAH product to homocysteine or S-ribosylhomocysteine, respectively 9–12. Homocysteine is in turn conjugated nonenzymatically to thiol-reactive fluorescent reagents, such as ThioGlo 12, or reaction intermediates are further metabolized enzymatically to generate analyte 9. In addition to the reduced limit of detection of enzyme-coupled assays compared to direct measurement of activity 13, the coupled enzymes complicate assay development and high-throughput screening as each must be optimized, stable performance of multiple enzyme reagents must be ensured, and hit compounds validated in separate assays to eliminate false positives. A competitive fluorescence polarization assay was developed that measures formation of SAH by the use of an antibody, and the assay was both homogeneous and sensitive 13. However, the antibody cross-reactivity with SAM limits the cofactor concentration to 3 µM, conditions that can limit the testable range for enzyme activity of HMTs possessing Km values of 1 – 25 µM for SAM 14–17. In addition, high concentrations of cofactor are preferred for HTS of SAM-dependent methyltransferases to restrain nonspecific hits that are competitive with SAM. Another homogeneous assay was developed that measures the transfer of radiolabeled methyl groups to Flash Plate-bound peptides 18. While the assay is sensitive and transferrable to other methyltransferases, its use of radioactivity is highly undesirable in high-throughput settings. ELISA and DELFIA assays utilizing antibody detection of methylated product or SAH to measure methyltransferase activity has been accompanied by an increase in sensitivity and throughput 7, 19, 20, but are non-homogeneous assays and require washing steps, making them unsuitable for rapid HTS.
Here we present a simple, homogeneous assay utilizing AlphaScreen chemiluminescence technology to measure histone methyltransferase activity in 1,536-well microplate format. The assay is sensitive to competitive inhibitors of the peptide substrate and is applicable to a multitude of histone-modifying enzymes. Identification of such compounds will aid in the understanding of the biological roles of individual histone-modifying enzymes as well as provide a starting point for the development of therapeutic agents for the treatment of epigenetic diseases.
MATERIALS AND METHODS
Proteins and Reagents
G9a, euchromatic histone-lysine N-methyltransferase 2, was recombinantly expressed in E.coli (residues 913–1,193) to include the catalytic SET domain and purified. Residues 951–1,235 of euchromatic histone-lysine N-methyltransferase 1 (EHMT1, also known as GLP), were similarly recombinantly expressed and purified. S-adenosylmethionine (SAM), 2-(Hexahydro-4-methyl-1H-1,4-diazepin-1-yl)-6,7-dimethoxy-N-[1-(phenylmethyl)-4-piperidinyl]4-quinazolinamine (BIX-01294), and Reactive Blue 2 were obtained from Sigma-Aldrich Co. (St. Louis, MO). Histone H3 11-mer peptides (non-, mono-, and di-methylated at K9) with an N-terminal biotin tag (ARTKQTARKST) were synthesized, HPLC-purified to >90% purity, and mass-analyzed by the Tufts University Department of Physiology Core Facility (Boston, MA). Peptides corresponding to amino acids 1–21 of human histone H3 (ARTKQTARKSTGGKAPRKQLA) with a C-terminal GG-linker and biotinylated Lys were purchased from Millipore (Billerica, MA). Purity of peptides was >90% by HPLC. Polyclonal rabbit IgG antibody used for detection of methylated H3K9 was raised against a synthetic human H3 histone peptide monomethylated at Lys9 and conjugated to KLH (Abcam Inc., Cambridge, MA). White solid-bottom 384- and 1,536-well plates were obtained from Greiner Bio-One (Monroe, NC). Assays were performed in PBS buffer, pH 7.4, containing 0.01% Tween-20.
Antibody Specificity
Specificity of the polyclonal anti-monomethyl histone H3K9 antibody toward the methylation reaction products was measured with histone H3 peptides mono- or di-methylated at Lys9, referred to as b-H3(1–11)K9me1 and b-H3(1–21)K9me2, respectively. Methylated peptides were titrated in 384-well plates (0.1 nM – 1 µM), alongside their unmethylated versions, in the presence of 0.5 µg/mL antibody and 20 µg/mL streptavidin-coated donor and anti-rIgG acceptor AlphaScreen beads (PerkinElmer, Waltham, MA). Plates were read in an EnVision multilabel plate reader (PerkinElmer) following a 30 min incubation at room temperature, with the 384 plate HTS AlphaScreen aperture (excitation time 35 ms, measurement time 100 ms). Antibody was titrated (0.1 – 2.5 µg/mL) in 384-well plates in incubations containing 1.0 nM b-H3(1–11)K9me1 and 20 µg/mL streptavidin-coated donor and anti-rIgG acceptor AlphaScreen beads. Reactions were incubated and plates were read as described above.
Miniaturized Assay
To each well, 2 µL of HMT enzyme (final 20 nM) was added using a BioRAPTR (Beckman Coulter, Fullerton, CA) flying reagent dispenser (FRD). The G9a- and GLP-inhibitor BIX-01294 was used as an intraplate control, with a 16-point titration in duplicate. A Kalypsys pin-tool was employed to transfer 23 nL of BIX-01294 solution in DMSO to each well, yielding a final concentration range of 0.35 nM – 12 µM. Following a 15 min incubation of enzyme with inhibitor at room temperature, 1 µL mixture of b-H3K9 peptide substrate (final 500 nM) and SAM cofactor (final 20 µM) was added. Reactions were allowed to proceed at room temperature for 2 hours. Methylated b-H3K9 peptide product was detected with 0.5 µg/mL rabbit polyclonal anti-monomethyl histone H3K9 antibody and 20 µg/mL each streptavidin-coated donor and anti-rabbit IgG acceptor AlphaScreen beads. Antibody and beads were added in a 1 µL FRD dispense, for a final volume of 4 µL, and plates were incubated protected from the light for 10 min at room temperature. Microplates were read on an EnVision multilabel plate reader using the 1,536 plate HTS AlphaScreen aperture (excitation time 80 ms, measurement time 240 ms). Data were normalized to the mean of the no-enzyme and uninhibited controls, and IC50 values were determined by nonlinear regression fits using GraphPad Prism4 software.
Counterscreen
Biotinylated rabbit-IgG (b-rIgG) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was added to 1,536-well plates in a 3 µL FRD dispense. Compound was added in a 23 nL pin-transfer step, and plates were incubated at room temperature for 15 min. Streptavidin-coated donor and anti-rIgG acceptor AlphaScreen beads were added (1 µL) for final concentrations of 20 µg/mL each bead and 1 nM b-rIgG, and plates were read as described above after a 15 min incubation at room temperature in the dark.
RESULTS
Assay Principle and Optimization
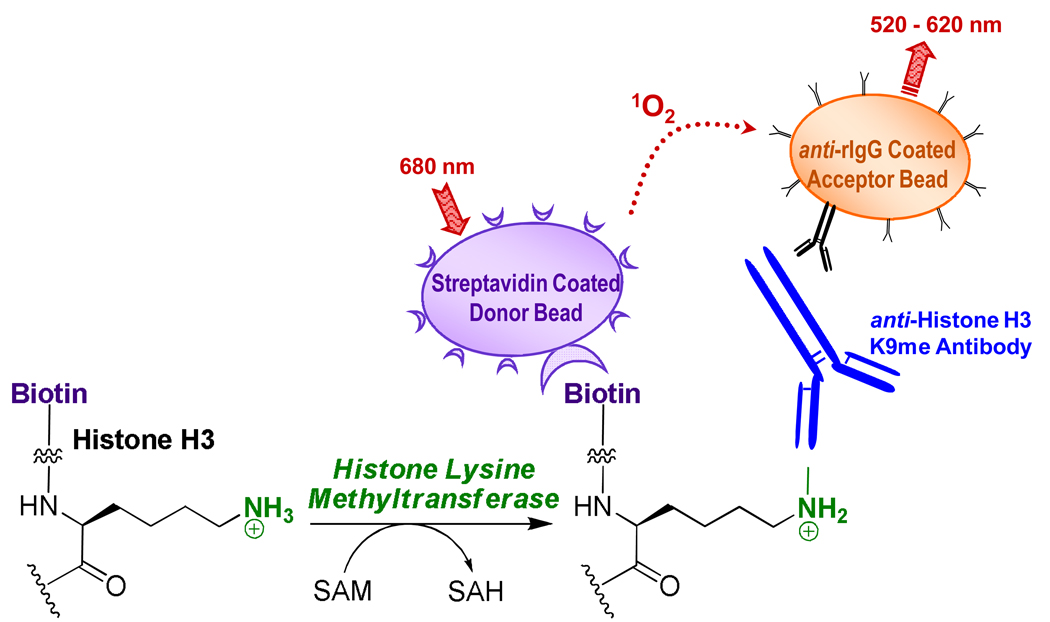
We utilized a chemiluminescence-based detection method to measure HMT activity in 1,536-well plate format using the Amplified Luminescence Proximity Homogenous Assay (AlphaScreen) technology. AlphaScreen was originally designed as a direct homogeneous substitute for the ELISA immunoassay method where the coating, capture and wash steps are replaced by a pair of detection beads capable of recognizing two unique sites on the analyte of interest 21. In our present assay scheme, methylation of a biotinylated-histone peptide substrate results in the formation of a peptide product bearing biotin and methyllysine recognition tags within the same molecule. Accumulation of b-H3K9me is measured through specific methyllysine primary antibody-based interaction, in conjunction with streptavidin-coated donor and secondary antibody-coated acceptor AlphaScreen beads (Scheme 1).
Scheme 1.
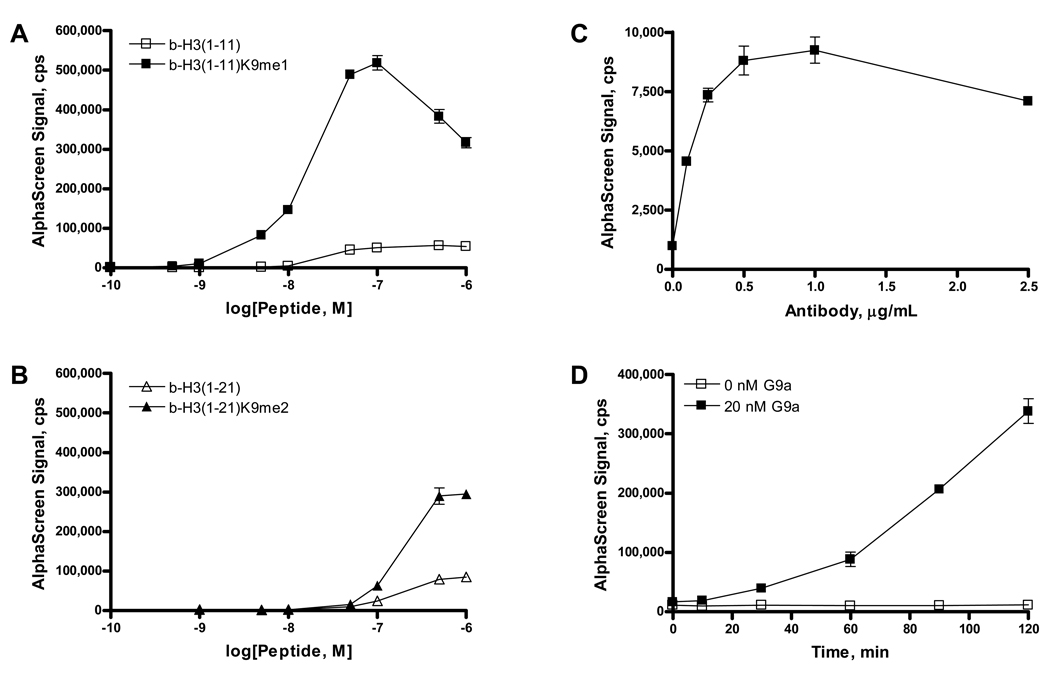
The specificity of polyclonal anti-methyllysine histone H3 antibody was measured for peptides mono- and di-methylated at Lys9 of H3 (H3K9). The antibody displayed affinity for both mono- and di-methylated Lys9 through binding of b-H3(1–11)K9me1 and b-H3(1–21)K9me2 peptides (Fig. 1A,B); however, the response curve for the di-methyl peptide was right-shifted by almost one log unit relative to the mono-methyl epitope, the primary target of said antibody. AlphaScreen signal decreased at peptide concentrations greater than 100 nM b-H3(1–11)K9me1 and 500 nM b-H3(1–21)K9me2. The reduced signal is due to the AlphaScreen “hook effect” where elevated concentration of biotinylated methylated peptide analyte exceeds the binding capacity of the beads and results in reduction of the signal as the proportion of donor and acceptor beads bound to the same analyte molecule decreases. Unmethylated b-H3(1–11) peptide displayed minimal binding to the antibody, and unmethylated b-H3(1–21) exhibited low but appreciable binding. Due to the higher signal-to-background ratio of the 11-mer H3 peptides compared to the 21-mer peptides, we chose to use b-H3(1–11) as the substrate for methyltransferase enzymes during these studies. All subsequent experiments were conducted at 500 nM b-H3(1–11) peptide substrate, so that formation of b-H3K9me1/me2 within the initial rate of the reaction falls below the AlphaScreen hook point.
Figure 1.
Specificity of binding of a polyclonal antibody to human histone H3 methylated at Lys9 was determined with biotinylated peptides of differing methylation states. A, Antibody binding to monomethylated Lys9 was observed as increasing AlphaScreen signal at concentrations up to 100 nM b-H3(1–11)K9me1. B, Binding of antibody to dimethylated Lys9 was also observed, and AlphaScreen signal increased with increasing peptide concentration up to 500 nM b-H3(1–21)me2. Corresponding unmethylated peptides displayed low affinity for antibody binding. C, Optimization of antibody concentration was determined in incubations with 1.0 nM b-H3K9me1 peptide. D, Reaction progress of b-H3(1–11) methylation by 20 nM G9a was measured in 384-well plate format in incubations containing 20 µM SAM and 500 nM peptide substrate. The mean and standard deviation of triplicate measurements are shown.
To further optimize the detection of methylated product, anti-methyllysine histone H3 antibody was titrated in 384-well plate format with b-H3(1–11)K9me1 (Fig. 1C). The signal increased linearly up to 0.5 µg/mL antibody. Subsequent experiments were performed with 0.5 µg/mL antibody for detection of H3K9me1/me2. G9a-catalyzed methylation of b-H3(1–11) was monitored over 2 h in 384-well plates (Fig. 1D), and AlphaScreen signal increased during this time frame. Reactions lacking G9a did not yield a significant background signal (Fig. 1D).
Miniaturized HMT Enzymatic Assay and Counterscreen
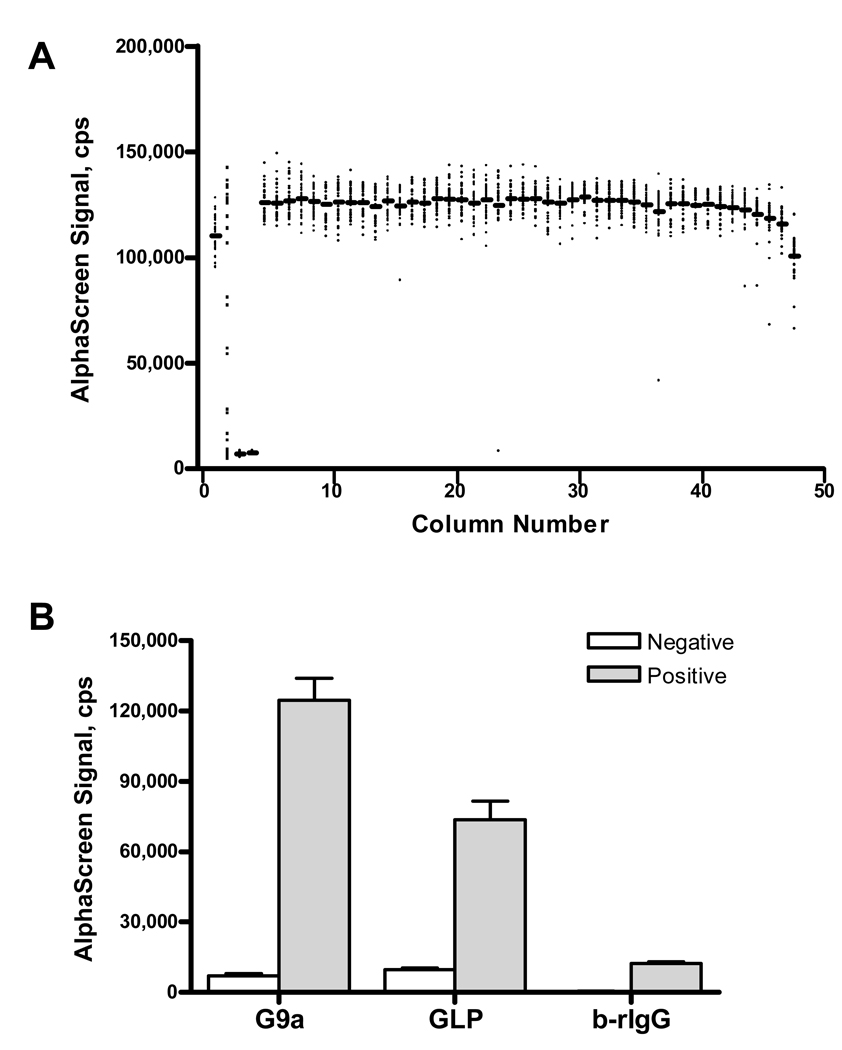
The HMT assay was further miniaturized to a final volume of 4 µL in 1,536-well plates. Reactions were performed with 20 nM enzyme (columns 1, 5–48), no enzyme (columns 3–4), and a 16-point titration of a known G9a inhibitor, BIX-01294 (column 2). AlphaScreen signal was plotted following a two-hour enzymatic reaction with 500 nM b-H3(1–11) followed by a ten-minute incubation with antibody-bead mixture in a scatter plot by column number, as shown in Figure 2A. The assay performed well in 1,536-well format, evidenced by a Z’ value of 0.75, where Z’ is a statistical parameter used to evaluate the quality of high-throughput screening assays, with values over 0.5 being indicative of a robust and HTS-amenable system22, 23. Furthermore, a high degree of signal uniformity was observed across the plate. The slightly lower signal values observed in the first and last columns are characteristic for this white 1,536-well plate type in the AlphaScreen format but are generally mitigated by the use of grey plates. Inhibition data in these columns can be corrected by normalization to a control plate of enzyme with no compound added which is a step routinely performed in HTS 24. The assay integrity was also tested under simulated conditions of overnight unattended screening: the assay components (enzyme, substrate, and antibody-bead mix), formulated as working stocks, were tested when freshly made and after 16-hour storage at 4 °C and were found to retain their activity (Fig. S1).To extend the utility of the new assay, the methyltransferase activity of the G9a-related enzyme, GLP, was similarly tested in 1,536-well format (Fig. 2B). Activity of G9a was higher than that of GLP (125,000 ± 9,000 cps versus 74,000 ± 8,000 cps, respectively). Background signal was low for both G9a and GLP reaction sets, 7,200 ± 800 and 9,800 ± 800 cps, respectively. Z’ was 0.69 for the miniaturized GLP assay test (data not shown).
Figure 2.
The histone methyltransferase assay was performed in miniaturized 1,536-well plate format with G9a and GLP methyltransferase enzymes. A, A scatter plot of AlphaScreen signal by column displays low background and high signal-to-background ratio. The mean value for each column (bars) is shown in addition to the signal in each well (points). Columns 1, 5–48 are G9a reactions, columns 3–4 are no enzyme controls, and column 2 contains a 16-point titration of BIX-01294, a G9a inhibitor. B, Methylation of H3K9 was measured for positive enzymatic or b-rIgG reactions (grey bars) and for negative no enzyme or protein controls (white bars). Values are displayed for the mean and standard deviation of 64 (negative control) and 1,440 (positive control) replicate measurements.
To investigate false positives that may have arisen from interference with the signal or binding of detection reagents, a counterscreen was developed to measure binding of AlphaScreen beads to biotinylated rabbit IgG (b-rIgG), an analyte serving to bind both donor and acceptor beads outside the context of the methyltransferase reaction. Biotinylated-rIgG (1 nM) bound to 20 µg/mL streptavidin-coated donor and anti-rIgG acceptor beads to produce a signal of 12,300 ± 800 cps, versus 460 ± 130 cps in the absence of b-rIgG (Fig. 2B). The high signal achieved with methylation of b-H3(1–11) was not attainable here due to the multiple biotinylation sites of rabbit IgG, which resulted in a suppression of the maximum signal at the AlphaScreen hook point (Fig. S2).
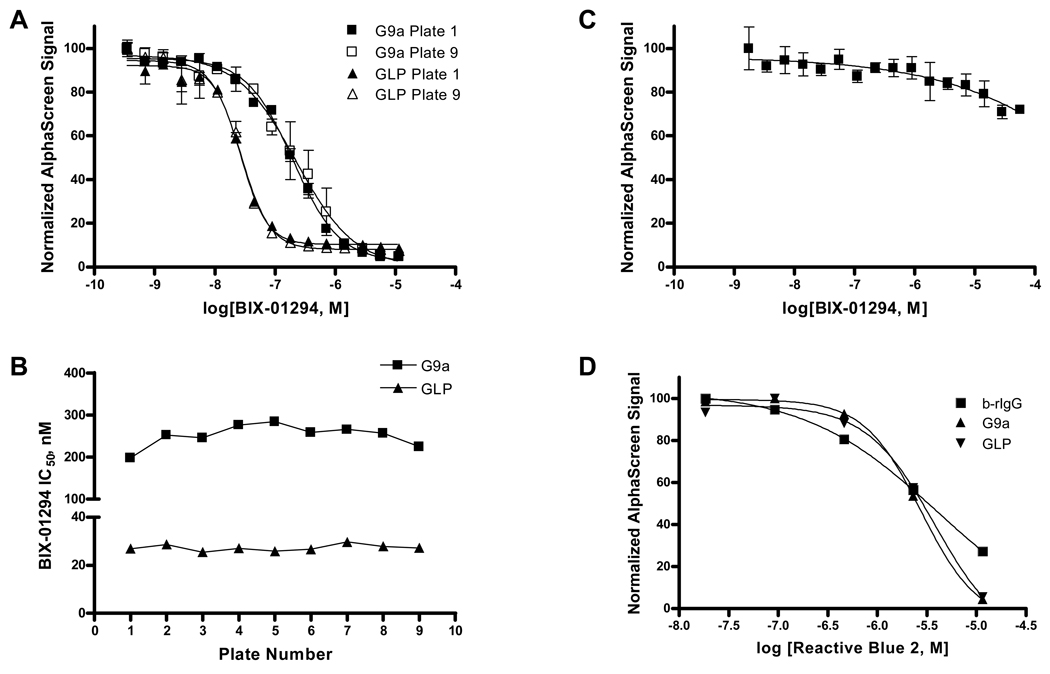
HMT Inhibition with BIX-01294
To evaluate the assay’s robustness and sensitivity, a mock screen was performed of 9 plates for each G9a and GLP methyltransferase. Inhibition of HMT activity with BIX-01294 was used as an intraplate control, with a 16-point titration of compound (Fig. 3A). IC50 values were compared across plates and shown to be highly uniform for each enzyme (Fig. 3B). BIX-01294 was a more potent inhibitor of GLP than of G9a, IC50 = 27 ± 1 nM and 250 ± 30 nM, respectively. To demonstrate that BIX-01294 was not a false positive in the present HMT assays, binding of b-rIgG to AlphaScreen beads was measured in the presence of a 16-point titration of BIX-01294 (Fig. 3C). Signal decreased slightly with the highest concentrations of BIX-01294 tested, but this trend did not significantly contribute to the loss of signal in the HMT enzymatic assays. To validate the b-rIgG counterscreen, we utilized Reactive Blue 2 (Fig. 3D), the anthraquinone dye that constitutes the blue chromophore of blue dextran, a known AlphaScreen color quencher (www.perkinelmer.com). AlphaScreen signal was reduced in the same manner in the G9a and GLP enzymatic assays as in the b-rIgG counterscreen.
Figure 3.
BIX-01294 inhibited G9a and GLP methyltransferases in a mock screen of HMT activity in 9 plates for each enzyme. A, Dose-response curves are shown for BIX-01294 inhibition of G9a (squares) and GLP (triangles) enzymatic activity in the first (solid) and final (empty) plates of the mock screen. B, Reproducibility of IC50 value determinations for BIX-01294 across plates are shown for G9a (squares) and GLP (triangles). C, A counterscreen confirms BIX-01294 as a true HMT inhibitor as minimal loss of signal was observed in a b-rIgG binding assay. D, Reactive Blue 2 is a false positive in AlphaScreen assays and signal interference is observed in both HMT enzymatic reactions and the b-rIgG counterscreen. Data in (A) and (C) represent the mean and standard deviation of duplicate measurements.
DISCUSSION
Here we present an assay that will help identify small molecule inhibitors of protein methyltransferases which will aid in the elucidation of biological roles for the greater than 50 mammalian SET domain enzymes. The assay described here employs AlphaScreen technology 21 that reports on interactions of reaction entities less than 200 nm apart via chemiluminescence process originating from colloidal-size donor and acceptor bead reagents (Scheme 1). Phthalocyanine photosensitizer-containing donor beads are excited by a laser (680 nm) to convert ambient oxygen to singlet oxygen. Acceptor beads (incorporating thioxene, anthracene and rubene derivatives) in close proximity utilize the singlet oxygen in a chemiluminescent reaction to emit light at 520–620 nm. Emission at a shorter wavelength than that of excitation eliminates much of the interference of fluorescent test compounds. Beads are coated with functional groups for specific detection of reaction entities. In the present system, streptavidin-coated donor beads bind biotinylated peptide substrate molecules. Upon HMT-mediated methylation of peptide, a rabbit polyclonal antibody detects methylated lysine residues. Acceptor beads coated with anti-rabbit IgG antibody bring the beads together for measurement of AlphaScreen signal (Scheme 1). Because the addition of antibody and AlphaScreen beads takes place after the methyltransferase reaction has proceeded to a certain desired extent, these detection components do not influence the activity of the target enzyme.
The assay was developed in 384-well format and subsequently miniaturized in 1,536-well plate format. Signal-to-background and Z’ factor were consistently high in both settings and the overall assay protocol was easy to execute due to the use of one combined detection reagent consisting of anti-methyllysine antibody, donor, and acceptor beads. Furthermore, the inhibition by the known G9a inhibitor BIX-01294 observed here was robust, highly reproducible, and in line with previously reported levels, further indicating that the assay can return consistent inhibition data. All assay components, formulated as working stocks, were found to remain stable upon overnight storage at 4 °C (Fig. S1). Thus, the assay can sustain large-scale HTS campaigns. The high sensitivity and very low background of this assay technology make it especially well suited for monitoring enzymatic reactions at low percent of substrate conversion and in the present case the limit of detection of monomethylated product in the presence of unmodified substrate is only restricted by the crossreactivity of the methyllysine antibody available for the particular epigenetic mark. A notable feature of the present HMT assay is the use of the peptide substrate at a concentration below that employed in the current ThioGlo coupled format which sensitizes the reaction to histone substrate-competitive inhibitors, the latter being desirable due to their potential for cross-HMT selectivity optimization. Possible sources of interference in this format include compounds that quench singlet oxygen, intensely colored substances, biotin mimetics, or compounds that interfere with antibody binding to modified peptide. However, the b-rIgG counterscreen demonstrated here can be deployed to immediately interrogate any candidate inhibitors for the above interference before they are considered for further development.
The data presented within indicate that the method can be adopted to a range of HMTs. The assay was initially configured using G9a as a model enzyme but was easily extended to the euchromatic histone-lysine N-methyltransferase 1 (EHMT1, also known as GLP), the only known homolog of G9a which shares 45% amino acid identity with G9a 5. GLP is also a major HMT in euchromatin and exhibits similar functional characteristics as G9a 5, 25. However, these HMTs may have distinct biological roles 7, and their interplay has not been fully elucidated. The assay presented here can measure the activities of both G9a and GLP, providing a platform for the discovery of specific inhibitors of each enzyme.
The control compound tested here, BIX-01294, was identified as an inhibitor of G9a HMT activity and was the first histone methyltransferase inhibitor that was not competitive with the SAM cofactor 26. BIX-01294 was later shown to inhibit GLP methyltransferase activity with greater potency compared to G9a, and was competitive with peptide substrate for binding to the active site 26. Our data obtained using the new assay are in agreement with the more potent inhibition of GLP versus G9a by BIX-01294.
In conclusion, we have developed a simple, sensitive, and miniaturized HMT assay which should be universally applicable to all similar histone-modifying enzymes, with the only changing component being the context-specific antibody for detection of methylated peptide. While we have not investigated these aspects here, we believe that it should be possible to use the present detection strategy in the reverse format, that is, to measure the loss of lysine methyl substituent as a result of the action by demethylase enzymes, and, with the use of an appropriate antibody, to also detect the installment of methyl groups by histone arginine methyltransferases.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Program of NHGRI, NIH. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
REFERENCES
- 1.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 2.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 3.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 4.Hwang KK, Eissenberg JC, Worman HJ. Proc Natl Acad Sci U S A. 2001;98:11423–11427. doi: 10.1073/pnas.211303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Nature chemical biology. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 7.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Cole PA. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Leffler S, Thompson DH, Hrycyna CA. Biochem Biophys Res Commun. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 12.Collazo E, Couture JF, Bulfer S, Trievel RC. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Graves TL, Zhang Y, Scott JE. Anal Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S. J Biol Chem. 2004;279:53248–53258. doi: 10.1074/jbc.M409604200. [DOI] [PubMed] [Google Scholar]
- 15.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 16.Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 17.Eskeland R, Czermin B, Boeke J, Bonaldi T, Regula JT, Imhof A. Biochemistry. 2004;43:3740–3749. doi: 10.1021/bi035964s. [DOI] [PubMed] [Google Scholar]
- 18.Dhayalan A, Dimitrova E, Rathert P, Jeltsch A. J Biomol Screen. 2009;14:1129–1133. doi: 10.1177/1087057109345528. [DOI] [PubMed] [Google Scholar]
- 19.Woo YH, Rajagopalan PT, Benkovic SJ. Anal Biochem. 2005;340:336–340. doi: 10.1016/j.ab.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 20.Capdevila A, Burk RF, Freedman J, Frantzen F, Alfheim I, Wagner C. J Nutr Biochem. 2007;18:827–831. doi: 10.1016/j.jnutbio.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, Pirio M, Pease J, Irvin BR, Singh S, Singh R, Patel R, Dafforn A, Davalian D, Skold C, Kurn N, Wagner DB. Clin Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 22.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JH, Chung TD, Oldenburg KR. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 24.Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, Simeonov A. Assay Drug Dev Technol. 2008;6:637–657. doi: 10.1089/adt.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. Nat Struct Mol Biol. 2009;16:312–317. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.