Abstract
Early embryonic cell cycles in Drosophila consist of rapidly alternating S and M phases. Three genes, pan gu (png), plutonium (plu), and giant nuclei (gnu) coordinate these early S-M cycles by ensuring adequate Cyclin B protein levels. Mutations in any of these genes result in unregulated DNA replication and a lack of mitosis (“giant nuclei” phenotype). png encodes a serine/threonine protein kinase, and plu and gnu encode small, novel proteins. We show that PNG, PLU, and GNU constitute a novel protein kinase complex that specifically regulates S-M cell cycles. All three proteins are required for PNG kinase activity and are phosphorylated by PNG in vitro. Yeast two-hybrid screening revealed a direct interaction between PNG and PLU, and their co-expression is required for physical association and activation of PNG kinase. Artificial dimerization of PLU via fusion to either GST or FK506 binding protein (in the presence of dimerizing agent) abrogates the requirement for GNU to activate PNG kinase. We propose a model in which GNU normally regulates embryonic cell cycles by promoting transient dimerization of a core PNG/PLU complex, thereby stimulating PNG kinase activity.
Keywords: Cell cycle, Drosophila, DNA replication, mitosis, Cyclin B, protein kinase
Protein kinases play crucial regulatory roles in the cell cycle. CDK/cyclin complexes control transitions throughout the cell cycle, and their proper regulation is essential for ensuring the orderly progression of DNA replication and mitosis (for review, see Murray and Hunt 1993). Association of a cyclin subunit with a CDK subunit is needed both for kinase activity and to confer substrate specificity. Thus, one mechanism for control of these kinase complexes involves the accumulation of threshold levels of cyclins via transcription and regulated degradation of cyclin proteins.
Modified cell cycles are used to achieve particular developmental goals. Organisms that must undergo rapid embryogenesis, such as marine invertebrates, amphibians, and insects, utilize a streamlined cell cycle in which DNA replication (S phase) and mitosis (M phase) alternate without intervening gaps. The S-M cycles are driven by maternally provided stockpiles of protein and RNA, eliminating a need for gap phases for gene expression or growth (Foe et al. 1993). Zygotic transcription has not yet begun, so the S-M cell cycles differ from the archetypal cell cycle in that they are regulated solely by posttranscriptional mechanisms. In Drosophila embryos, an additional distinction is that the nuclei divide synchronously in a common cytoplasm (syncytium) during the S-M cycles.
During the first seven cell cycles of Drosophila embryogenesis, levels of the mitotic Cyclins A and B as well as their partner kinase CDK1 are high due to maternal stockpiling, and no detectable fluctuations in their levels or CDK1 activity occur (Edgar et al. 1994). However, localized degradation of Cyclin B during these early cycles has been reported (Huang and Raff 1999), and injection of a stabilized form of Cyclin B into early embryos causes mitotic arrest (Su et al. 1998). Thus, localized oscillations in CDK1 activity likely drive the early embryonic S-M cycles.
Exhaustive screens of Drosophila maternal-effect lethal collections identified three genes needed for proper coupling of S and M phases in early embryos. pan gu (png), plutonium (plu), and giant nuclei (gnu) are first required at the critical transition between completion of meiosis and entry into the first mitotic division (Freeman et al. 1986; Freeman and Glover 1987; Shamanski and Orr-Weaver 1991). Females with mutations in any of these genes produce eggs that complete meiosis, but the embryonic S-M cycles are defective. DNA replication occurs in the absence of mitosis, resulting in large polyploid nuclei (“giant nuclei”). The giant nuclei phenotype is observed even in the absence of fertilization: the four female meiotic products undergo inappropriate DNA replication and become polyploid. In weak alleles of png, a few rounds of S-M cycling occur, but mitosis ultimately ceases and the nuclei become polyploid. Mutations in plu or gnu dominantly enhance this weak png phenotype, indicating that these three genes function in a common pathway to regulate the early embryonic cell cycles.
Several lines of evidence indicate that Cyclin B is a critical target of the giant nuclei class of genes. Cyclin B protein levels are decreased in embryos from png, plu, or gnu mutant females; for png, Cyclin B levels inversely correlate with the severity of the mutant phenotype (Fenger et al. 2000). Mutation of cyclin B dominantly enhances the weak png phenotype, whereas increasing the gene dosage of cyclin B suppresses the giant nuclei phenotype associated with mutations in png, plu, or gnu (Lee et al. 2001). The reduction in Cyclin B protein in the mutants is associated with decreased CDK1 kinase activity (Fenger et al. 2000), accounting for the failure to enter mitosis and the inability to block re-replication of DNA. The mechanism by which the giant nuclei class of genes ensures adequate levels of Cyclin B protein remains to be determined.
png is predicted to encode a serine/threonine protein kinase, and plu and gnu encode small, novel proteins (Axton et al. 1994; Fenger et al. 2000; Renault et al. 2003). The mutations in png that phenotypically appear null cause amino acid substitutions in residues essential for kinase activity, whereas mutations with residual activity resulting in some S-M cycling alter nonconserved amino acids (Fenger et al. 2000). This suggests that PNG kinase activity is required for regulation of early embryonic cell cycles.
In the present study, we took a biochemical approach to characterize both the requirements for PNG kinase activity and the physical interactions between PNG, PLU, and GNU proteins. We found that PNG kinase activity is dependent on the presence of both PLU and GNU. PLU and GNU are not only activators of PNG, but also substrates of PNG kinase in vitro. A yeast two-hybrid screen identified PLU as a specific PNG interactor, indicating a direct interaction between the two proteins. In contrast, GNU is more weakly associated with the PNG/PLU complex and becomes dispensable for PNG kinase activity when the complex is artificially dimerized via fusion of PLU to GST or FK506 binding protein (in the presence of dimerizing agent). These biochemical results provide a molecular explanation for the shared phenotypes and genetic interactions between png, plu, and gnu, three genes required for regulation of early embryonic cell cycles in Drosophila.
Results
New gnu alleles identified by screening a maternal-effect lethal collection
png, plu, and gnu are the only genes identified to date with a giant nuclei mutant phenotype resulting from uncoupled S-M cycles during Drosophila embryogenesis (Freeman et al. 1986; Freeman and Glover 1987; Shamanski and Orr-Weaver 1991). In an effort to identify new genes required for S-M oscillations, we screened a large maternal-effect lethal collection generated in the laboratory of Charles Zuker. Embryos from homozygous mutant females were collected, fixed, stained with DAPI to visualize the DNA, and examined as whole mounts. We screened ∼500 second chromosome lines and ∼1900 third chromosome lines in this manner. We found giant, polyploid nuclei in only two of the ∼2400 lines screened: Z3-0591 and Z3-3770. The giant nuclei phenotype was present in only a subset of embryos from homozygous mutant females of the original Z3-3770 line. A sub-line (Z3-3770A) was derived from a single heterozygous male, and 100% of embryos from homozygous Z3-3770A females were found to have the giant nuclei phenotype.
Given that both Z3-0591 and Z3-3770A are on the third chromosome, we performed complementation testing to determine whether they represented new alleles of gnu. Both Z3-0591 and Z3-3770A were crossed to gnu305, the sole previously described allele of gnu, which contains a premature stop codon and appears to be a protein null (Renault et al. 2003). Z3-0591/gnu305 and Z3-3770A/gnu305 females were found to be sterile, producing embryos with giant, polyploid nuclei (Fig. 1). Thus, Z3-0591 and Z3-3770A represent new alleles of gnu, and no new genes with giant nuclei mutant phenotypes were identified in this screen. gnuZ3-3770A has a strong giant nuclei phenotype characterized by a near absence of nuclear division in embryos from either homozygous (data not shown) or transheterozygous (Fig. 1B) females. gnuZ3-0591 has a weak giant nuclei phenotype with several mitotic divisions occurring in embryos from either homozygous (data not shown) or transheterozygous (Fig. 1C) females. gnuZ3-0591 is one of the weakest giant nuclei alleles identified to date, with a phenotype comparable to that of the previously described png50 (Fenger et al. 2000).
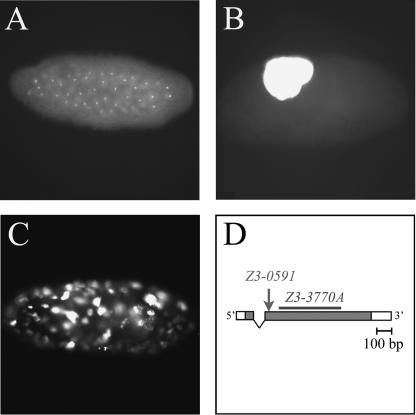
Figure 1.
(A-C) Phenotype of new gnu alleles. Embryos from wild-type or mutant females mated to wild-type males were fixed, stained with propidium iodide to visualize the DNA, and viewed as whole mounts. (A) A fertilized wild-type embryo with diploid nuclei. (B) Embryo from a transheterozygous gnuZ3-3770A/gnu305 female with a single giant nucleus. (C) Embryo from a transheterozygous gnuZ3-0591/gnu305 female with multiple polyploid nuclei. (D) Structure of the wild-type gnu gene and new mutant alleles. Exons are represented by boxes, with shaded regions corresponding to coding sequence. gnuZ3-0591 is a proline-to-leucine change at residue 17 (arrow), and gnuZ3-3770A is a 396-bp deletion in the gnu coding region (bar).
To determine the molecular nature of the weak gnuZ3-0591 and strong gnuZ3-3770A alleles, the gnu gene was PCR-amplified from homozygous mutant flies for sequence analysis (Fig. 1D). gnuZ3-0591 is a missense mutation that causes a proline-to-leucine change at residue 17. gnuZ3-3770A is a 396-bp deletion in the middle of the coding region of gnu with a consequent frame-shift; the resulting predicted fusion protein contains the first 38 amino acids of GNU followed by an additional 97 amino acids of novel sequence. Thus, the molecular nature of the gnuZ3-0591 and gnuZ3-3770A mutations correlates well with the severity of their early embryonic mutant phenotypes (weak and strong, respectively).
PNG kinase activity requires both PLU and GNU
In order to demonstrate that png encodes a functional protein kinase, we generated recombinant PNG protein and developed an in vitro PNG kinase assay. Using an Sf9/baculovirus expression system, a recombinant baculovirus encoding C-terminal Myc-tagged PNG was created. This PNG-Myc fusion protein was shown to be fully functional by its ability to rescue png mutations in transgenic flies (Fenger et al. 2000). Sf9 cells were infected with the PNG-Myc baculovirus, and PNG-Myc protein was immunoprecipitated from cellular lysates using Myc antibody beads. Kinase assays were then performed on these beads, followed by analysis of the reaction products by SDS/PAGE and autoradiography.
In initial PNG-Myc immunoprecipitation (IP) kinase assays, we tested for PNG-Myc autophosphorylation because no PNG-Myc substrates had yet been identified. We observed no autophosphorylation in the IP kinase assay when PNG-Myc was expressed alone (Fig. 2A, lane 2). Common test substrates (for example, myelin basic protein, histone H1, and casein) that can be phosphorylated by other known protein kinases were not phosphorylated by PNG-Myc when added to the IP kinase assay (data not shown).
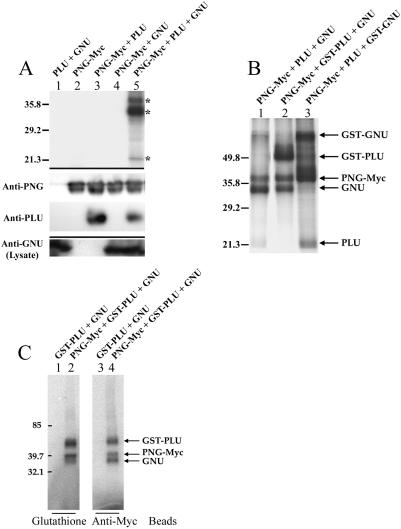
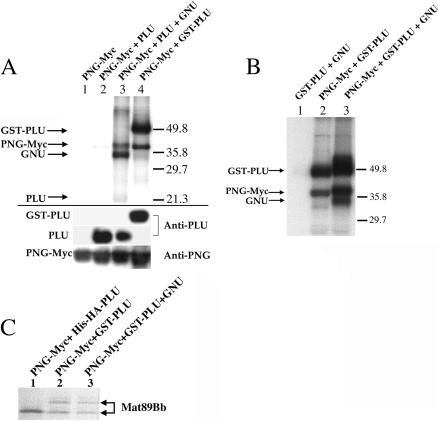
Figure 2.
(A) PNG kinase activity requires both PLU and GNU. (Upper panel) PNG-Myc IP kinase assays. Three major 32P-labeled bands (*) were reproducibly observed following triple infection of Sf9 cells with PNG-Myc, PLU, and GNU. (Middle panel) Corresponding IP immunoblots using anti-PNG and anti-PLU antibodies. (Lower panel) Corresponding immunoblot of cleared lysates using anti-GNU antibodies. (B) PLU and GNU are phosphorylated by PNG kinase. GST fusions of PLU and GNU were used in PNG-Myc IP kinase assays to confirm the identity of two of the three major 32P-labeled bands (marked by * in panel A) as PLU and GNU. An intervening lane between lanes 1 and 2 was removed. (C) Isolation of active PNG kinase complexes using tags on PNG or PLU. GST pull-downs (lanes 1,2) to isolate GST-PLU complexes or Myc immunoprecipitations to isolate PNG-Myc complexes were performed prior to assaying for kinase activity. For all panels, combinations of recombinant baculoviruses used to infect Sf9 cells are shown above each lane.
Given the genetic interactions between png, plu, and gnu and the similarity of their mutant phenotypes, we considered the possibility that PNG might require PLU and/or GNU for its kinase activity. To test this hypothesis, we generated recombinant baculoviruses encoding tagged and untagged versions of PLU and GNU. Sf9 cells were co-infected with various combinations of PNG, PLU, and GNU baculoviruses, and cell lysates were prepared for PNG-Myc IP kinase assays. As shown in the upper panel of Figure 2A, double combinations of PNG-Myc and PLU(lane 3) or PNG-Myc and GNU(lane 4) had no appreciable kinase activity. Only the triple combination of PNG-Myc, PLU, and GNU had a high level of activity in the PNG-Myc IP kinase assay (lane 5). Thus, both PLU and GNU are required for PNG kinase activity.
PLU and GNU form a complex with and are phosphorylated by PNG kinase
Co-infection of Sf9 cells with recombinant baculoviruses encoding PNG-Myc, PLU, and GNU resulted in activity in the PNG-Myc IP kinase assay, and three major 32P-labeled bands were reproducibly observed (Fig. 2A, upper panel, lane 5). Based on their mobilities, we predicted that the three co-expressed recombinant proteins (PNG-Myc, PLU, and GNU) were all present in the immunoprecipitate and were phosphorylated by PNG kinase. PNG and PLU proteins were previously shown to coimmunoprecipitate from Drosophila embryo extracts (Fenger et al. 2000). Immunoblotting of the recombinant PNG-Myc immunoprecipitates revealed that PNG-Myc and PLU co-expressed in Sf9 cells also could be coimmunoprecipitated (Fig. 2A, middle panel, lanes 3,5). Furthermore, recombinant PLU co-immunoprecipitated with PNG-Myc independently of GNU co-expression (Fig. 2A, cf. lanes 3 and 5). When sufficient amounts of Myc antibody beads were used to immunoprecipitate PNG-Myc from Sf9 extracts, essentially all of the co-expressed PLU was co-immunoprecipitated (data not shown). Thus, recombinant PNG and PLU proteins appear to form a tight complex. Trace amounts of recombinant GNU can be specifically detected in the PNG-Myc immunoprecipitates by immunoblotting (data not shown), although it was readily detected in the Sf9 cell lysates (Fig. 2A, lower panel, lanes 1,4,5).
To confirm the identities of the three major phosphorylated bands typically observed in the PNG-Myc IP kinase assay (Fig. 2A, upper panel, lane 5) as PNG-Myc, PLU, and GNU, recombinant baculoviruses encoding tagged versions of these proteins were generated. Addition of a GST tag to the N-terminus of PLU or GNU resulted in large mobility changes on SDS-PAGE (data not shown). Sf9 cells were triply co-infected with various combinations of PNG-Myc and either untagged or GST-tagged versions of PLU and GNU baculoviruses, and the cell lysates were used in PNG-Myc IP kinase assays (Fig. 2B). When GST-PLU was used instead of PLU in a triple co-infection and IP kinase assay, the radiolabeled band with the mobility of PLU disappeared, and a new band with the mobility of GST-PLU appeared (Fig. 2B, cf. lanes 1 and 2). Likewise, when GST-GNU was used instead of GNU in a triple co-infection and IP kinase assay, the radiolabeled band with the mobility of GNU disappeared, and a new band with the mobility of GST-GNU appeared (Fig. 2B, cf. lanes 1 and 3). Thus, detection of 32P incorporation into GNU protein in the IP kinase reaction is a more sensitive assay than immunoblotting for assessing the presence of GNU in the PNG-Myc immunoprecipitates. In similar experiments, substitution of a maltose-binding protein-tagged version of PNG for PNG-Myc resulted in an upward mobility shift of the slowest migrating major radiolabeled band (data not shown), thereby confirming its identity as PNG-Myc. Thus, PNG-Myc, PLU, and GNU form a functional kinase complex, and all three subunits are phosphorylated in the PNG-Myc IP kinase assay.
For the experiments shown in Figure 2A,B, active recombinant PNG kinase complex was isolated from Sf9 cells using the Myc tag on PNG. We have also found that the same active kinase complex can be isolated from Sf9 cells using the GST tag on PLU(Fig. 2C). Sf9 cells were co-infected with a triple combination of PNG-Myc, GST-PLU, and GNU baculoviruses to generate active kinase complex; as a negative control for kinase activity, cells were doubly co-infected with GST-PLU and GNU baculoviruses without PNG-Myc. Cell lysates were prepared, and kinase assays were performed using protein complexes isolated by binding of either GST-PLU to glutathione beads (Fig. 2C, lane 2) or PNG-Myc to Myc antibody beads (Fig. 2C, lane 4). The corresponding negative controls are shown in lanes 1 and 3. The same pattern of radiolabeled bands representing the three recombinant proteins (PNG-Myc, GST-PLU, and GNU) was obtained when kinase assays were performed on the PNG-Myc or GST-PLU beads. Thus, active recombinant PNG kinase complexes can be isolated using tags on either PNG or PLU.
Two-hybrid screening reveals a direct interaction between PNG and PLU
To identify potential downstream molecular targets of PNG, we performed a yeast two-hybrid screen using PNG as the “bait” protein. Screening of 3×106 cDNA clones from a Drosophila ovary library led to the identification of PLU as the only specific PNG interactor. Sixty-three cDNA clones encoding PLU were isolated in this screen. We previously reported that PNG and PLU did not interact in a yeast two-hybrid assay (Fenger et al. 2000). In the present study, we found it necessary to grow the yeast on selective media for relatively long periods (>7 d) in order to detect interaction between PNG and PLU. Furthermore, this interaction was only marginally detected in a secondary screen involving generation of blue color on X-gal plates. PNG and PLU proteins have been shown to co-immunoprecipitate from both Drosophila embryo extracts (Fenger et al. 2000) and as recombinant proteins from Sf9 extracts (this study); the PNG yeast two-hybrid screen results indicate that this interaction between PNG and PLU proteins is direct.
In an effort to define subdomains of PNG and PLU required for their interaction in the yeast two-hybrid assay, the PNG “bait” protein was divided into three regions of roughly equal size. Deletion constructs containing N-terminal, central, and C-terminal regions of PNG bait in various combinations were generated. None of these PNG deletion mutants interacted with full-length PLU“prey” in the yeast two-hybrid assay (data not shown). Likewise, deletion constructs containing N-terminal, central (containing the three ankyrin repeats of PLU), and C-terminal regions of PLU prey in various combinations were generated, and none of these PLU deletion mutants interacted with full-length PNG bait (data not shown). The shortest plu cDNA clone isolated in the yeast two-hybrid screen lacked coding region for only the first five amino acids of PLU. The failure to define subdomains of PNG and PLU required for their interaction in the yeast two-hybrid assay suggests that multiple regions across the primary structures of the proteins are needed for binding and/or that protein folding is compromised.
Co-expression of PNG and PLU is required for both physical association and activation of PNG kinase
To generate the active PNG kinase complex shown in Figure 2, Sf9 cells were triply co-infected with PNG, PLU, and GNU baculoviruses such that the three proteins were co-expressed in the same cells. To determine whether or not co-expression of recombinant PNG with PLU and/or GNU was necessary for its kinase activity, a mixing experiment was performed (Fig. 3). When PNG-Myc, PLU, and GNU were co-expressed in Sf9 cells, PNG-Myc IP kinase activity (as measured by 32P incorporation into GNU) was easily detected (Fig. 3, upper panel, lane 1). To test PNG's requirement for GNU co-expression, PNG-Myc and PLU were co-expressed, but GNU was expressed separately; cell extracts were mixed prior to the immunoprecipitation and kinase assay. PNG kinase activity was identical whether GNU was co-expressed with the PNG/PLU complex or separately expressed (Fig. 3, upper panel, cf. lanes 1 and 2).
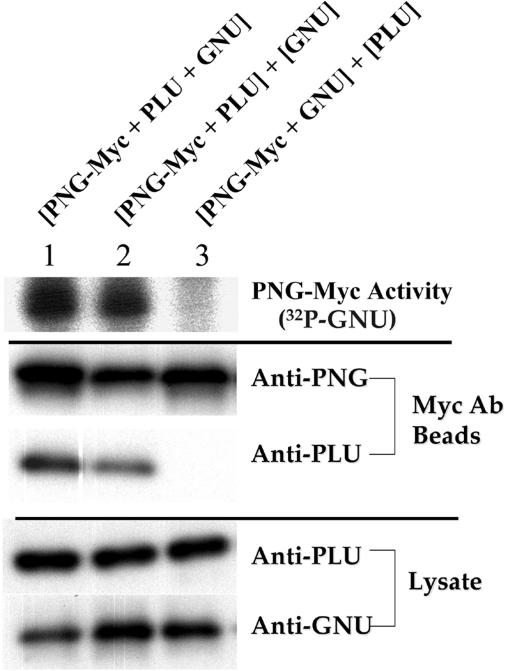
Figure 3.
PNG kinase activity requires PLU co-expression. Combinations of recombinant baculoviruses used to infect Sf9 cells are shown above each lane. Brackets indicate whether proteins were co-expressed in Sf9 cells (shared brackets) or expressed separately (different brackets). When one of the proteins was separately expressed (lanes 2,3), cell extracts were mixed prior to performing PNG-Myc immunoprecipitations and kinase assays. (Upper panel) PNG-Myc IP kinase assays. Incorporation of 32P label into GNU protein was used as a measure of PNG kinase activity. (Middle panel) Corresponding IP immunoblots using anti-PNG and anti-PLU antibodies. (Lower panel) Corresponding immunoblots of cleared lysates using anti-PLU and anti-GNU antibodies. An intervening lane between lanes 1 and 2 was removed.
To test PNG's requirement for PLU co-expression, PNG-Myc and GNU were co-expressed, but PLU was expressed separately; cell extracts were mixed prior to the immunoprecipitation and kinase assay. No PNG kinase activity was detected when PLU was expressed separately from PNG and GNU(Fig. 3, upper panel, cf. lanes 1and 3). Similarly, when PLU and GNU were co-expressed separately from PNG-Myc, PNG kinase activity was not reconstituted by mixing extracts (data not shown). Immunoblotting of the immunoprecipitates revealed that PLU no longer co-immunoprecipitated with PNG-Myc when the proteins were separately expressed (Fig. 3, middle panel, lane 3), although full-length PLU protein was present at comparable levels in the cellular lysates (Fig. 3, lower panel, lane 3). Thus, co-expression of PNG and PLU is required for their physical association, which is necessary for PNG kinase activity.
PNG and PLU exist in a large complex in Drosophila embryo extracts
Although PNG and PLU proteins can be co-immunoprecipitated from Drosophila embryo extracts, GNU does not co-immunoprecipitate with the PNG/PLU complex from Drosophila embryo extracts (Fenger et al. 2000; L. Vardy and T. Orr-Weaver, unpubl.). The failure to detect a complex with GNU in vivo is consistent with the weak/transient interaction we have observed between recombinant PNG/PLU complexes and GNU. To determine whether the core PNG/PLU complex exists as a heterodimer or as a large-molecular-weight complex in vivo, a sucrose density gradient was performed using Drosophila embryo extracts, followed by immunoblotting for PNG and PLU(Fig. 4A). PNG had a profile almost identical to that of aldolase, the 158-kD standard. The PLU profile was slightly shifted to the left (indicating a smaller size), but generally overlapped well with that of PNG. PNG is a 33-kD protein, and PLU is a 19-kD protein. A complex containing one molecule each of PNG and PLU would be ∼52 kD, which is significantly smaller than their apparent size of ∼158 kD in embryo extracts by sucrose density gradient analysis.
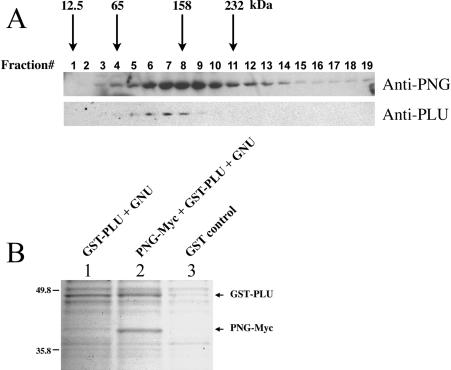
Figure 4.
(A) PNG and PLU exist in a large complex in Drosophila embryo extracts. Wild-type embryo extracts were fractionated on a sucrose density gradient, and fractions were analyzed by immunoblotting using anti-PNG and anti-PLU anti-bodies. Arrows indicate peak fractions of various molecular weight standards run in parallel. (B) PNG and PLU interact with a 1:1 stoichiometry. Combinations of recombinant baculoviruses used to infect Sf9 cells are shown above each lane. GST-PLU complexes were purified from Sf9 extracts using glutathione beads. Eluted proteins were resolved by SDS-PAGE and visualized by staining with GelCode Blue. As a negative control, recombinant GST (lane 3) was also purified from Sf9 extracts.
To determine the stoichiometry of the interaction between PNG and PLU, active recombinant PNG kinase complex was isolated and resolved by SDS-PAGE, followed by staining to visualize proteins (Fig. 4B). GNU was not detected in the PNG/PLU complexes by this method. Densitometry of the PNG-Myc and GST-PLU bands revealed a ratio of 0.9, respectively, indicating that they interact with a 1:1 stoichiometry.
Recombinant PNG kinase complex phosphorylates endogenous GNU from Drosophila embryo extracts
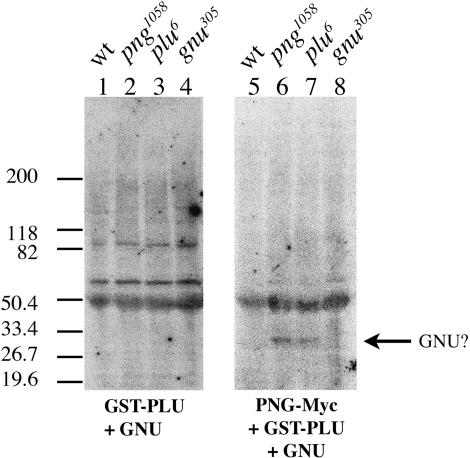
To test for the presence of PNG kinase complex substrates in Drosophila embryo extracts, a filter kinase assay was performed. Extracts prepared from embryos collected from wild-type or homozygous giant nuclei mutant females were resolved by SDS-PAGE and transferred to Immobilon. The filters were incubated with either purified recombinant PNG kinase complex or a negative control under kinase assay conditions, and 32P incorporation into embryonic proteins on the filters was assessed. A prominent radiolabeled band with mobility similar to that of GNU was detected in png and plu mutant embryo extracts following treatment with active recombinant PNG kinase complex, but not with the negative control (Fig. 5, cf. lanes 6-7 and lanes 2-3, respectively). This 32P-labeled band was not detected in wild-type or gnu mutant embryo extracts following treatment with active PNG kinase (Fig. 5, lanes 5 and 8, respectively).
Figure 5.
Phosphorylation of endogenous GNU by recombinant PNG kinase complex. Extracts of embryos from wild-type or mutant females were separated by SDS-PAGE and transferred to Immobilon. Maternal genotypes are indicated above each lane. Filter kinase assays were performed using active recombinant PNG kinase complex (right panel) or a negative control lacking the PNG subunit (left panel). 32P incorporation into embryonic proteins on the filter was assessed by autoradiography.
We believe that this radiolabeled protein represents phosphorylated GNU, for several reasons. Subsequent immunoblotting of the filters (after decay of the 32P signal) using anti-GNU antibodies confirmed that the radiolabeled protein and GNU had identical mobilities (data not shown). PNG kinase activity in vitro requires PNG, PLU, and GNU. If the same were true in vivo, then PNG kinase substrates (for example, GNU) in png, plu, or gnu mutant embryos should be underphosphorylated relative to wild-type embryos. As a consequence of this underphosphorylated state, GNU from png and plu mutant embryo extracts would be expected to incorporate higher amounts of 32P in the PNG filter kinase assay relative to GNU from wild-type embryos that has presumably already been phosphorylated by the PNG kinase complex. The gnu305 allele contains a premature stop codon (Renault et al. 2003). Typically, no GNU protein is detected in embryo extracts from homozygous gnu305 females by immunoblotting, although GNU from wild-type embryos is readily detected (data not shown; Renault et al. 2003); we rarely observe trace amounts of truncated GNU protein in gnu305 embryos (data not shown). These observations indicate that the truncated GNU protein is unstable in embryo extracts. Thus, lack of a radiolabeled GNU band in gnu305 embryo extracts in the filter kinase assay is consistent with the observation that gnu305 is essentially a protein null allele. The results from the filter kinase assay suggest that the recombinant PNG kinase complex can phosphorylate endogenous GNU from Drosophila embryo extracts.
GST tagging of PLU abrogates the requirement for GNU in activation of PNG kinase
We have shown that PNG kinase activity in vitro requires both PLU and GNU(Fig. 2A). However, when a GST-tagged version of PLU was substituted for PLU in the PNG kinase assays, an unexpected result was obtained. As controls, the double combination of PNG-Myc and PLU had no activity in the PNG-Myc IP kinase assay, whereas the triple combination of PNG-Myc, PLU, and GNU was active (Fig. 6A, upper panel, lanes 2 and 3, respectively). Surprisingly, we found that the double combination of PNG-Myc and GST-PLU also had a high level of activity in the PNG-Myc IP kinase assay (Fig. 6A, upper panel, lane 4). This difference in kinase activity between PNG-Myc/PLU and PNG-Myc/GST-PLU complexes (Fig. 6A, upper panel, cf. lanes 2 and 4) was not due to differences in the levels of PNG-Myc or PLU(tagged or untagged) proteins in the immunoprecipitates (Fig. 6A, lower panel, cf. lanes 2 and 4). When PNG kinase complexes were isolated using the GST tag on PLU instead of the Myc tag on PNG, a similar result was obtained: PNG-Myc/GST-PLU complexes bound to glutathione beads also had a high level of PNG kinase activity (Fig. 6B). Thus, addition of a GST tag to PLU abrogates the requirement for GNU in activation of the PNG kinase complex as assessed by 32P incorporation into components of the complex.
Figure 6.
GST-PLU activates PNG kinase in the absence of GNU. (A) (Upper panel) PNG-Myc IP kinase assays. (Lower panel) Corresponding IP immunoblots using anti-PNG and anti-PLU antibodies. An intervening lane between lanes 2 and 3 was removed. (B) GST pull-downs and kinase assays. (C) PNG kinase complexes were purified on nickel beads (lane 1) or glutathione beads (lanes 2,3) using affinity-tagged PLU. Eluted PNG kinase complexes were tested for the ability to shift the SDS-PAGE mobility of the 35S-labeled PNG kinase substrate Mat89Bb. This mobility shift is due to phosphorylation, as it is eliminated by treatment with λ-phosphatase (data not shown). Combinations of recombinant baculoviruses used to infect Sf9 cells are shown above each lane.
To test further the ability of GST-PLU to activate PNG kinase in the absence of GNU, we used an exogenous substrate of PNG kinase that was recently identified in a genome-wide biochemical screen (data not shown). Mat89Bb is a novel maternal protein of unknown function that undergoes a mobility shift on SDS-PAGE due to phosphorylation following treatment with active recombinant PNG kinase complex. We tested the ability of various PNG kinase complexes that were purified using affinity-tagged PLU to phosphorylate 35S-labeled Mat89Bb (Fig. 6C). We found that PNG-Myc/GST-PLU complexes recognized exogenous Mat89Bb as a substrate (Fig. 6C, lane 2). Interestingly, addition of other affinity tags to the amino terminus of PLU, including hexa-histidine-hemagglutinin (Fig. 6C, lane 1) or maltose-binding protein (data not shown), was not sufficient to activate PNG kinase activity in the absence of GNU. His-HA-PLU and MBP-PLU were expressed in Sf9 cells at levels comparable to that of GST-PLU, and both could activate PNG kinase in the presence of GNU(data not shown). Furthermore, the kinase activity of PNG-Myc/GST-PLU complexes towards the exogenous substrate Mat89Bb was not further enhanced by the presence of GNU(Fig. 6C, cf. lanes 2 and 3), indicating that PNG-Myc/GST-PLU represents the fully active form of PNG kinase.
Inducible dimerization of PNG/PLU eliminates the requirement for GNU in activation of PNG kinase
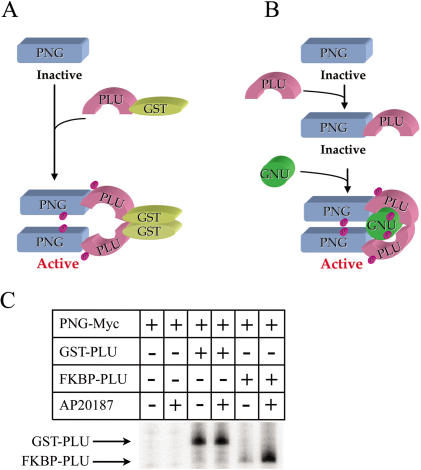
The ability of the GST-PLU fusion protein to maximally stimulate PNG kinase activity even in the absence of GNU can be explained by a model in which activation of PNG kinase involves dimerization of PNG/PLU complexes (Fig. 7A). Under normal conditions, PLU is necessary (but not sufficient) for PNG kinase activity, and PNG and PLU form a tight complex with an ∼1:1 stoichiometric ratio (Fig. 7B). GST protein can form homodimers (Lim et al. 1994), and addition of a GST tag to a variety of proteins has been found to promote dimerization of the resulting fusion proteins (E. Lee, pers. comm.). According to our model, fusion of GST to PLU results in activation of PNG kinase by artificially forcing dimerization of PNG/PLU complexes (Fig. 7A), which is normally a transient state mediated by GNU(Fig. 7B). Although in our model we indicate that there is one GNU molecule for every two molecules of PNG/PLU, the actual number of GNU molecules required to interact transiently with and activate the PNG/PLU complex may be greater. As predicted, addition of a GST tag to PLU results in a shift in the mobility of PNG-PLU complexes on gel filtration, consistent with the formation of multimeric complexes (data not shown).
Figure 7.
Activation of PNG kinase by dimerization. Panels (A) and (B) present a model accounting for our experimental observations. (A) Fusion of GST to PLU, which forms a stable complex with PNG, promotes dimerization of PNG/GST-PLU complexes via the GST moiety. This artificial dimerization leads to activation of PNG kinase in a GNU-independent manner. (B) PNG kinase activity normally requires both GNU and PLU. Abrogation of the requirement for GNU when the PNG/PLU complex is artificially dimerized via GST implies that GNU normally performs this function. (C) Experimental test of the dimerization model. Sf9 cells were infected with PNG-Myc baculovirus alone or in combination with either GST-PLU or FKBP-PLU baculoviruses as indicated. Cleared lysates were either untreated or treated with the FKBP dimerizing agent AP20187 prior to performing PNG-Myc immunoprecipitations and kinase assays. Incorporation of 32P label into GST-PLU or FKBP-PLU proteins was used as a measure of PNG kinase activity.
In the experiments shown in Figure 6, dimerization of PNG/PLU complexes was driven by a GST-PLU fusion protein. It is not clear whether PNG kinase alone can be activated via artificial dimerization in the absence of PLU. Attempts to test this possibility failed because a GST-tagged version of PNG was found to have very little kinase activity, even in the presence of both PLU and GNU(data not shown).
To test our model of multimerization as a mechanism for PNG kinase activation, we created a fusion between PLU and FK506 binding protein (FKBP) that can be dimerized in a regulated manner by addition of the compound AP20187 (Amara et al. 1997; Clackson et al. 1998). As shown in Figure 7C, extracts from Sf9 cells infected with PNG-Myc and FKBP-PLU exhibited a low level of PNG kinase activity that was substantially increased by addition of the dimerizer AP20187. This AP20187-induced PNG kinase activity was similar to that of PNG-Myc plus GST-PLU, and immunoblotting revealed comparable levels of FKBP-PLU and GST-PLU as well as PNG-Myc proteins in the immunoprecipitates (data not shown). The low level of kinase activity associated with FKBP-PLU in the absence of the dimerizer AP20187 is consistent with reports of low levels of dimerization observed for other FKBP fusion proteins when expressed at high levels (Clackson et al. 1998). AP20187 had no effect on the kinase activities of the negative (PNG-Myc) or positive (PNG-Myc plus GST-PLU) controls. The fact that two different fusions of PLU that induce dimerization (GST-PLU and FKBP-PLU in the presence of AP20187) are sufficient to activate PNG kinase in the absence of GNU provides strong evidence for a dimerization model of PNG kinase activation.
cyclin B transcript levels are normal in png and plu mutants
Cyclin B protein levels are decreased in embryos from females with mutations in png, plu, or gnu (Fenger et al. 2000). In addition to their effects on the S-M cycles, png, plu, and gnu are needed for the destabilization of certain maternal transcripts that occurs at the midblastula transition of embryogenesis (Tadros et al. 2003). This observation raised the possibility that the PNG kinase complex might regulate cyclin B transcripts, possibly ensuring adequate protein levels by stabilizing the message. To help elucidate the mechanism by which the giant nuclei class of genes regulates Cyclin B protein levels, we measured cycB transcript levels in embryos from wild-type and mutant females by RT-PCR (Fig. 8A). Embryos from wild-type, png, and plu females were found to have comparable cycB mRNA levels after normalizing to Actin 42A. Normal cycB mRNA levels were similarly observed in embryos from gnu females (data not shown). Thus, decreased Cyclin B protein levels associated with the giant nuclei mutant phenotype are not a consequence of decreased cycB transcripts.
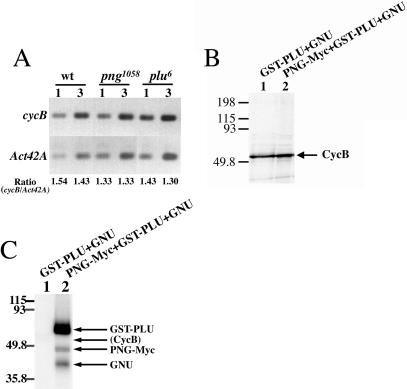
Figure 8.
(A) cyclin B mRNA levels are normal in png and plu mutants. RT-PCR was used to assess cycB mRNA levels in embryos from wild-type, png1058, and plu6 females. The number of embryo equivalents of reverse transcription reaction products used as input for PCR is shown above each lane. The ratio of cycB to Act42A RT-PCR products is shown below each lane. An intervening lane between lanes 4 and 5 was removed. (B,C) Cyclin B is not a PNG kinase substrate. (B) 35S-labeled Cyclin B fails to undergo a mobility shift on SDS-PAGE following treatment with active recombinant PNG kinase complex (lane 2). (C) Cyclin B fails to incorporate 32P when incubated with active recombinant PNG kinase complex (lane 2). Combinations of recombinant baculoviruses used to infect Sf9 cells are shown above each lane.
Cyclin B is not a direct substrate of the PNG kinase complex
We also tested the possibility that Cyclin B protein could be stabilized by direct phosphorylation of Cyclin B by the PNG kinase complex. Treatment of 35S-labeled Cyclin B with active recombinant PNG kinase complex failed to result in a mobility shift on SDS-PAGE that often occurs as a consequence of protein phosphorylation (Fig. 8B). Furthermore, treatment of purified Cyclin B with active PNG kinase complex does not result in detectable incorporation of 32P into Cyclin B protein (Fig. 8C). Thus, Cyclin B does not appear to be a direct substrate of the PNG kinase complex.
Discussion
We demonstrate a novel cell-cycle protein kinase complex composed of the PNG kinase and its associated subunits PLU and GNU. The PNG kinase complex regulates the early embryonic S-M cycles in Drosophila, promoting mitosis and inhibiting DNA replication. PNG, PLU, and GNU proteins are present only in ovaries and early embryos, and null mutations in their genes reveal that they play essential developmental roles exclusively during the early embryonic divisions following completion of meiosis (Axton et al. 1994; Fenger et al. 2000; Renault et al. 2003). The biochemical analysis of the PNG kinase complex described here defines the requirement for png, plu, and gnu during the S-M cycles and explains their shared phenotypes and genetic interactions by establishing that all three proteins are essential for PNG kinase activity.
Cell-cycle regulatory kinases must be precisely controlled to ensure the orderly progression of cell-cycle events. Parallels can be drawn between the PNG kinase complex and CDK/cyclin complexes. CDK kinases require association of an activating cyclin subunit. Similarly, the PLU and GNU proteins are activating subunits of the PNG kinase. Each CDK molecule partners with a single cyclin subunit (Jeffrey et al. 1995), and we find that the PLU subunit is stably associated with PNG in a 1:1 stoichiometry. GNUis also needed for PNG kinase activity, but it is much less stably associated with the PNG/PLU complex, possibly serving to control when and/or where PNG kinase is active. Both CDK1/cyclin complexes and the PNG kinase complex promote entry into mitosis and block DNA re-replication.
The PNG and PLU subunits are tightly and directly associated. In addition to the interaction we observe following their co-expression in baculovirus-infected cells, we find they directly bind each other in yeast cells in a two-hybrid assay. Furthermore, they form a complex in vivo and can be co-immunoprecipitated from Drosophila ovary or embryo extracts (Fenger et al. 2000).
It is striking that PNG and PLU must be co-expressed to associate with each other and to form an active kinase with GNU. A similar observation has been reported for the β and γ subunits of heterotrimeric G proteins (Fung 1983; Hildebrandt et al. 1984). Physical interaction between β and γ subunits to form a functional heterodimer requires co-expression. In both cases, the requirement for co-expression likely reflects the fact that the physical association between the two proteins is very tight, and folding of the proteins may be coordinated. Consistent with this explanation, attempts to dissect the interaction domains between PNG and PLU using the two-hybrid assay failed, suggesting that multiple regions on each protein are needed for interaction and/or that the truncated proteins did not fold correctly. Interestingly, PNG mutant proteins predicted to have diminished or no kinase activity fail to co-immunoprecipitate with PLU-Myc from Drosophila embryo extracts, despite their presence at nearly normal levels (Fenger et al. 2000). Our results demonstrate that PNG and PLU association can occur in the absence of GNU; thus, functional kinase activity is not required for this association. Instead, mutations that affect PNG kinase activity also affect its interaction with PLU, possibly via alteration of its three-dimensional structure.
The intimate association between PNG and PLU suggests that the sole function of PLU is to form a complex with and activate PNG. Co-expression of PNG and PLU may permit PNG to fold into an active conformation. It is also possible that PLU provides substrate specificity to the PNG kinase by analogy to cyclin subunits and their CDKs. It remains to be determined whether phosphorylation of PLU affects its activity or stability. It is only weakly phosphorylated by PNG in IP kinase assays. Thus, it is possible that PLU is only “incidentally” phosphorylated as a consequence of its proximity to PNG in the complex. Density gradient analysis suggests that PNG and PLU proteins are part of a larger-molecular-weight complex in vivo, representing either a multimeric complex of the PNG/PLU heterodimer or a complex containing additional, as yet unidentified, proteins. Future identification of other regulators and/or substrates of the PNG kinase complex will further our understanding of how S-M cycling is regulated and help to identify the mechanism by which the PNG kinase complex controls transcript stability (Tadros et al. 2003).
In contrast to PLU, GNU is not tightly associated with PNG. GNU is present in active recombinant PNG kinase complexes as assessed by 32P incorporation in PNG-Myc IP kinase assays. A very low level of GNU protein is detected in these PNG-Myc immunoprecipitates by immunoblotting, and the interaction between GNU and the PNG/PLU core complex appears to be transient and/or weak. PNG and GNU fail to interact in a yeast two-hybrid assay. Furthermore, endogenous GNU (in contrast to PNG) does not co-immunoprecipitate with PLU-Myc from Drosophila embryo extracts (L. Vardy and T. Orr-Weaver, unpubl.).
GNU is required for PNG kinase activity, and we propose that GNU normally activates a core PNG/PLU complex by facilitating its multimerization (Fig. 7B). This model was initially suggested by the observation that a GST-PLU fusion protein can fully activate PNG kinase in the absence of GNU (Fig. 7A). We demonstrate the role of dimerization in activating PNG kinase directly by using an FK506-binding protein fused to PLU and the AP20187 compound to dimerize the FKBP domain. Inducible dimerization of the FKBP domain greatly stimulated the kinase activity of the PNG/FKBPPLU complex to levels comparable to that of the PNG/PLU/GNU and PNG/GST-PLU complexes. There is precedence for the regulation of protein kinases via dimerization. Receptor tyrosine kinases dimerize upon ligand binding, and this dimerization activates their intracellular kinase domains (Heldin 1995). In another type of mechanism, dimerization of the MAP kinase ERK2 permits its entry into the nucleus (Khokhlatchev et al. 1998).
The weak association between GNU and the PNG/PLU complex raises the possibility that GNU has other as yet unidentified functions in addition to activation of PNG kinase. GNU could interact with other proteins in addition to the PNG/PLU complex to potentially regulate other processes. However, the genetic phenotypes and developmental expression of GNU show that, like PNG and PLU, its activity is restricted to early embryogenesis. Ectopic expression of GNU in ovarian nurse cells caused female sterility (Renault et al. 2003). This effect was dependent on having functional PNG and PLU, suggesting that at least some of GNU's in vivo activity is mediated through the PNG kinase complex. This observation is consistent with our biochemical results showing that GNU is an activator of a core PNG/PLU complex, and suggests that the female sterility associated with ectopic GNU expression is a consequence of ectopic activation of PNG kinase.
GNU is strongly phosphorylated by PNG in IP kinase assays using recombinant proteins. The results from our PNG filter kinase assay indicate that endogenous GNU (from embryo extracts) also is phosphorylated by recombinant PNG kinase complex in vivo. Our filter kinase assay results are consistent with our previous observation that an ∼30-kD protein present in Drosophila embryo extracts is phosphorylated in a PNG-dependent manner (Fenger et al. 2000). Although not yet identified, this ∼30-kD 32P-labeled band likely represents GNU.
Phosphorylation of proteins on particular residues can sometimes result in an upward mobility shift on SDS-PAGE. Incubation of 35S-labeled GNU (generated in a coupled in vitro transcription/translation reaction using reticulocyte lysate) with active recombinant PNG kinase complex resulted in an easily detectable upward shift in its mobility on SDS-PAGE (L. Lee and T. Orr-Weaver, unpubl.). Therefore, we compared the mobility of GNU in embryo extracts from wild-type, png, and plu mutant females. We observed no differences in the mobility of GNU from these different genetic backgrounds (L. Lee and T. Orr-Weaver, unpubl.); similar observations have been reported by others (Renault et al. 2003). Possible explanations for the failure to detect mobility differences in GNU from wild-type versus png and plu mutant backgrounds is that only a small percentage of the total pool of GNU is phosphorylated by PNG, the modification is transient, and/or phosphorylated GNU is unstable. Another possible explanation is that the in vivo PNG phosphorylation sites differ from the in vitro phosphorylation sites, and they do not result in a mobility shift of GNU (that is, GNU is relatively “hyperphosphorylated” by PNG in vitro). In addition to possibly being a PNG substrate in vivo, Renault et al. (2003) found that GNU is phosphorylated by a different kinase during oogenesis, and this phosphorylation is removed at egg activation. The functional consequences of phosphorylation of GNU await elucidation.
Our previous studies revealed that the giant nuclei class of genes is required to maintain adequate levels of Cyclin B protein normally needed to block DNA re-replication and to promote the onset of mitosis (Fenger et al. 2000; Lee et al. 2001). This study indicates that png, plu, and gnu do not affect the steady-state level of cyclin B transcripts. Furthermore, Cyclin B protein does not appear to be a direct substrate of the PNG kinase complex. In an effort to determine the mechanism by which the giant nuclei class of genes regulates Cyclin B protein levels, we recently performed a genome-wide biochemical screen to identify PNG kinase substrates (L.A. Lee, E. Lee, H. Kashevsky, M. Benasutti, M.W. Kirschner, and T. Orr-Weaver, unpubl.). Seven substrates of the PNG kinase complex (including GNU) were identified in this screen. Thus, regulation of Cyclin B protein levels may occur further downstream of these direct targets of the PNG kinase complex.
png, plu, and gnu are the only genes known to be essential for S-M oscillations in the early embryo, and our biochemical results suggest that activation of PNG kinase is a major function of both PLU and GNU. We previously reported that archetypal cell cycles in developing eye imaginal discs were not perturbed by ectopic co-expression of PNG and PLU(Fenger et al. 2000). Likewise, misexpression of GNU in polytene salivary glands and ovarian follicle cells had no obvious effects on the cell cycles in these tissues (Renault et al. 2003). Given our finding that PNG kinase activity requires both PLU and GNU, ectopic expression of only one or two components of the complex would likely have no effect on cell cycles in other tissues. In screens of maternal-effect lethal collections (including a large, likely saturating collection generated in the laboratory of Charles Zuker), png, plu, and gnu are the only mutants identified in which S phase is uncoupled from mitosis in the early embryo (this study; Freeman et al. 1986; Freeman and Glover 1987; Shamanski and Orr-Weaver 1991). Thus, the PNG/PLU/GNU kinase complex appears to be the crucial regulatory switch unique to the S-M cycles of early Drosophila embryogenesis.
Materials and methods
Fly stocks
Flies were maintained at 25°C using standard techniques (Greenspan 1997). The wild-type stocks used were Oregon R and yw. png1058 (Shamanski and Orr-Weaver 1991), plu6 (Shamanski and Orr-Weaver 1991; Axton et al. 1994), and gnu305 (Freeman et al. 1986; Freeman and Glover 1987) alleles have been previously described.
Embryo fixation, staining, and microscopy
Homozygous mutant females from a large maternal-effect lethal collection (a gift from Charles Zuker,) were fattened on yeast for several days. Embryos were collected overnight, dechorionated, fixed, 4′,6-diamidino-2-phenylindole (DAPI)-stained, and prepared for microscopy as described by Shamanski and Orr-Weaver (1991). Zeiss Axiophot and Axioskop microscopes with Pan-neofluar 10×, 20×, and 40× objectives were used to examine fluorescence staining. In subsequent analyses of mutants with giant nuclei phenotypes, 0-3-h embryos were stained with propidium iodide instead of DAPI to visualize the DNA (Fenger et al. 2000).
Sf9 cell/baculovirus expression
Sf9 cells were routinely grown in suspension at 27°C. cDNAs encoding PLU and GNU and genomic DNA encoding PNG (which lacks introns) were subcloned into the untagged pFastBac1 vector (GIBCO BRL) and/or derivatives of pFastBac1 containing N-terminal glutathione S-transferase (GST), maltose-binding protein, hexa-histidine, or hexa-histidine-hemagglutinin (His-HA) affinity tags for protein purification (a gift from Ethan Lee, Harvard Medical School). An FspI/NheI fragment containing genomic DNA encoding PNG with three C-terminal Myc epitope repeats was removed from a transgenic rescue construct (Fenger et al. 2000) and subcloned into pFast-Bac. Recombinant baculoviruses were generated following protocols from the manufacturer (GIBCO BRL). Infections were performed by addition of amplified baculovirus stock (at 1:100 (v/v) virus/cells) to growing Sf9 cells (1 × 106 cells/ml). For co-infections with 2-3 baculovirus stocks, each amplified baculovirus stock was added at 1:100 (v/v) virus/cells. Cells were harvested at 48 h p.i., and cell pellets were snap frozen and stored at -80°C.
PNG-Myc IP kinase assays and GST pull-downs/kinase assays
Frozen Sf9 cell pellets (from 25-mL infected cultures) were resuspended in 150 μL of ice-cold IP Buffer (50 mM HEPES pH 7.7, 100 mM NaCl, 5 mM MgCl2, 1 mM EGTA, 0.1% Triton X-100, 10% glycerol, protease inhibitors), briefly sonicated, and cleared by microcentrifugation at 4°C for 10 min. Cleared lysates (150 μL) were added to 25 μL Myc antibody beads (Santa Cruz Bio-technology), and samples were rotated at 4°C for 2 h to allow binding. The beads were washed 3× as follows: ice-cold IP Buffer, IP Buffer with 350 mM NaCl, IP Buffer (250 μL each). Washed beads (5 μL) were incubated with 10 μL Kinase Buffer (50 mM HEPES pH 7.7, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 1 mM EDTA, 10 mM n-octylglucoside, 2 mM DTT, 0.1 mM cold ATP, and 10 microcuries γ[32P]ATP (10 mCi/mL, 3,000 Ci/mmol, Amersham) at 30°C for 30 min. Reactions were stopped by addition of 5 μL 6X SDS-PAGE sample buffer. Samples were briefly microfuged prior to analysis by SDS-PAGE and autoradiography (7.5 μL/lane). In some experiments, PNG kinase complexes were isolated by binding of GST-PLU to glutathione beads (Pharmacia) followed by kinase assays using protocols identical to those described above for the PNG-Myc IP kinase assays. To assess the stoichiometry of PNG and PLU in the complexes, proteins were eluted from glutathione beads with IP Buffer containing 5 mM glutathione, separated by SDS-PAGE, and stained with GelCode Blue (Pierce). The dried gel was scanned, and the bands corresponding to PNG-Myc and GST-PLU were quantitated using NIH Image 1.63. The ratio of PNG-Myc to GST-PLU protein was determined after subtracting the background from the corresponding regions in the GST control lane.
PNG kinase assays using exogenous substrates
35S-labeled proteins (Mat89Bb and Cyclin B) were synthesized in a coupled transcription-translation system using reticulocyte lysate according to the manufacturer's protocol (TNT system from Promega). Recombinant PNG kinase complexes containing GST-PLU or His-HA-PLU were isolated by binding to glutathione beads (Pharmacia) or Ni-NTA beads (QIAGEN), respectively, and washed as described above for the PNG-Myc IP kinase assays except that EGTA was omitted when using Ni-NTA. Proteins were eluted from glutathione beads with Kinase Buffer B (50 mM HEPES pH 7.7, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 1 mM EDTA, 10 mM n-octylglucoside, 2 mM DTT, 1 mM ATP) supplemented with 50 mM glutathione or from Ni-NTA beads with 50 mM HEPES pH 7.7, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 10 mM n-octylglucoside, 1 mM ATP, and 300 mM imidazole pH 8.0. Three rounds of elution (100 μL each) were performed, and eluates were combined. Eluted proteins were dialyzed against Kinase Buffer B. Kinase assays were performed at 30°C for 30 min following addition of 1 μL of 35S-labeled protein to 10 μL of purified PNG kinase complex. Reactions were stopped by addition of 5 μL of 6X SDS-PAGE sample buffer and analyzed by SDS-PAGE and autoradiography (7.5 μL/lane).
To test Cyclin B as a PNG kinase substrate by 32P incorporation, an MBP-Cyclin B fusion was generated by subcloning Drosophila cycB cDNA into a derivative of pMAL containing a TEV protease site C-terminal to MBP (a gift from Ethan Lee). Recombinant MBP-Cyclin B was purified from bacterial lysates using amylose beads (New England Biolabs) and treated with TEV protease (Life Technologies) so as to cleave the MBP tag from Cyclin B. Purified Cyclin B (0.5 μg) was added to PNG kinase complexes bound to glutathione beads (via GST-PLU), and 32P incorporation into Cyclin B was assessed as described above (GST pull-downs/kinase assays).
Inducible dimerization of FKBP-PLU
The FKBP-PLU fusion was generated by subcloning plu cDNA into a derivative of pFastBac1 containing FK506 binding protein (F36V mutant) at the amino terminus (a gift from Ethan Lee). Generation of recombinant baculovirus and infections were performed as described above. Cleared lysates were prepared as described above and divided into two equal portions. Dimerizing agent AP20187 (a gift from ARIAD Pharmaceuticals; www.ariad.com/regulationkits) was added to one of the portions (f.c. 200 nM), and PNG-Myc immunoprecipitations and kinase assays were performed as described above.
Polyclonal antibodies against GNU
A fusion between GST and 112 amino acids from the amino terminus of GNUwas used to produce antibodies in guinea pigs. The construct encoding the GST-GNU fusion protein was generated by PCR amplification of gnu cDNA (LD12084) followed by subcloning into the pGEX-4T-1 expression vector (Pharmacia). GST-GNU was expressed in BL21 bacterial cells by IPTG induction, purified from the soluble fraction using glutathione beads, and injected into guinea pigs for antibody production (Covance). In immunoblots of Drosophila embryo extracts, the anti-GNU antibody recognizes a cluster of bands slightly larger than the 27 kD predicted by the sequence of GNU.
Immunoblotting
For IP immunoblots, PNG kinase complexes were immunoprecipitated using Myc antibody beads, and beads were washed as described above. Proteins were eluted from washed beads (1 μL) by incubation at 65°C for 1 min in IP Buffer with 1% SDS (8 μL). 6X SDS-PAGE sample buffer (2 μL) was added to the eluates, and proteins were resolved by SDS-PAGE. For immunoblots of cleared lysates, 1.5 μL of sample/lane was resolved by SDS-PAGE. Proteins were transferred to Immobilon for immunoblotting as described (Tang et al. 1998). Antibodies were used at the following dilutions: guinea pig anti-PNG serum (1:2000; Fenger et al. 2000), affinity-purified rabbit anti-PLU (1:200; Elfring et al. 1997), and guinea pig anti-GNU serum (1:5000).
Sucrose density gradient analysis
Embryos (0-2-h) collected from Oregon R females were dechorionated in 50% bleach, washed with 0.9% NaCl/0.01% Triton X-100, and homogenized at 1:2 (v/v) embryo/Homogenization Buffer (50 mM HEPES pH 7.6, 100 mM NaCl, 1 mM EGTA, 1 mM DTT, 0.002% NP40, protease inhibitors). The sample was cleared by microcentrifugation at 4°C for 15 min. The cleared supernatant (300 μL) was loaded onto a sucrose density gradient (5 mL, 5%-20% in Homogenization Buffer) and centrifuged at 4°C for 15 h (SW 50.1 rotor, 30K rpm). The following standards (Pharmacia, 50 μg each in 300 μL Homogenization Buffer) were run in parallel: cytochrome c (12.5 kD), albumin (65 kD), aldolase (158 kD), and catalase (232 kD). Following centrifugation, fractions (200-μL) were withdrawn from the top of the gradient, and aliquots (20-μL) were analyzed by SDS-PAGE and immunoblotting for PNG and PLU as described above.
Filter kinase assay
Extracts were prepared from embryos (0-2-h) collected from females with the following genotypes: yw (wild-type), png1058, plu6, and gnu305. Embryos were dechorionated with 50% bleach, washed with 0.9% NaCl/0.01% Triton X-100, and homogenized at 1:4 (v/v) embryo/urea sample buffer. Proteins were resolved by SDS-PAGE followed by transfer to Immobilon filters (Tang et al. 1998). Filters were blocked by incubation in TBS-TB (Tris-buffered saline, 0.1% Tween, 1% BSA) at room temperature for 1 h.
Lysates were prepared from frozen Sf9 cell pellets (25-mL cultures), and proteins were bound to and eluted from glutathione beads (25-μL) as described above. Eluates (200-μL each) were diluted into 850 μL Filter Kinase Buffer (50 mM HEPES pH 7.7, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 1 mM EDTA, 10 mM n-octylglucoside, 2 mM DTT, 0.1 mM cold ATP, and 100 microcuries γ[32P]ATP [10 mCi/mL, 3,000 Ci/mmol, Amersham]). Blocked Immobilon filters in Seal-A-Meal bags were incubated with the eluates in Filter Kinase Buffer at room temperature for 1.5 h, washed with TBS-TB for 5 min, then washed twice with phosphate-buffered saline for 5 min each (all washes at room temperature). Filters were air dried, and 32P-labeled proteins were detected using Kodak MR film.
Yeast two-hybrid screen
A yeast two-hybrid screen to identify PNG interactors was performed using a Gal4/LexA system developed by the Brent lab (Golemis et al. 1997). An EcoRI/NotI fragment containing genomic DNA encoding PNG (which lacks introns) was subcloned into the HIS3 2-μm bait vector pEG202 in-frame with the last codon of LexA to produce a bait construct. The yeast strain EGY48 carrying the PNG bait construct was transformed with DNA from an amplified Drosophila ovary cDNA library (ovoIb) cloned in the vector pJG4-5 (Großhans et al. 1999), and galactose-dependent Leu+ colonies were recovered. The yeast strain used for screening also contained the lacZ reporter pSH18-34. As a secondary screen, galactose-dependent Leu+ colonies were restreaked and tested for β-galactosidase expression. Library plasmids from all of the galactose-dependent Leu+ colonies were recovered in E. coli using standard techniques, and the cDNA inserts were sequenced. Specificity was determined by testing for yeast two-hybrid interactions between positives from the screen and the following “irrelevant” baits: MEI-S332 (Tang et al. 1998) and YAN (a gift from Ilaria Rebay, Whitehead Institute).
RT-PCR
First, 225 embryos (0-2-h) collected from wild-type, png, or plu mutant females were snap-frozen in liquid nitrogen, and RNA was isolated using RNAzol B (Tel-Test) according to the manufacturer's protocols. Reverse transcription (25-μL reactions) was performed using oligo-dT and MMLV-RT (Promega), and PCR was subsequently performed using 1/9 or 1/3 μL of reaction products (1 or 3 embryo equivalents, respectively) as input. A 261-bp sequence in the 3′ coding region of cyclin B (cycB) cDNA that spans a 70-bp intron in the genomic sequence was amplified, and a 240-bp sequence in the 3′ coding region of Actin 42A (Act42A) was amplified in separate reactions as a control. Reaction products were analyzed on a 2% agarose gel, the photographed gel was scanned, and bands corresponding to cycB and Act42A PCR products were quantitated using NIH Image 1.63.
Acknowledgments
We thank Ethan Lee for helpful ideas and discussions throughout the course of this work, and for the Sf9/baculovirus expression vectors. Both Ethan Lee and Marc Kirschner provided enthusiastic support for our biochemical endeavors. The maternal-effect mutant collection was kindly provided by Charles Zuker's laboratory, University of California, San Diego. We thank Edmund Koundakjian, David Cowan, and Robert Hardy of the Zuker lab for establishing the collection of F3 viable lines, and Barbara Wakimoto, Dan Lindsley, and Mike McKeon for identifying the subset of female-sterile lines. We gratefully acknowledge the work of fellow lab members in jointly screening this collection: Kimberly Dej, Irena Ivanovska, Dmitry Epstein, Helena Kashevsky, and Giovanni Bosco. We thank ARIAD Pharmaceuticals for expeditiously providing the AP20187 dimerizing agent; Christiane Nusslein-Volhard for the Drosophila ovary cDNA library; Helena Kashevsky for generating the PNG-Myc construct; Ilaria Rebay for the YAN bait strain; and Giovanni Bosco, Colleen Raymond, Deborah Burney-Sigman, and Lynn Young for help with the PNG yeast two-hybrid screen. Leah Vardy provided helpful comments on the manuscript. L.A.L. was supported by postdoctoral fellowships from the NIH and the Charles A. King Trust (The Medical Foundation). This work was supported by NIH grant GM39341 to T.O.-W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1132603.
References
- Amara J.F., Clackson, T., Rivera, V.M., Guo, T., Keenan, T., Natesan, S., Pollock, R., Yang, W., Courage, N.L., Holt, D.A., et al. 1997. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc. Natl. Acad. Sci. 94: 10618-10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axton J.M., Shamanski, F.L., Young, L.M., Henderson, D.S., Boyd, J.B., and Orr-Weaver, T.L. 1994. The inhibitor of DNA replication encoded by the Drosophila gene plutonium is a small, ankyrin repeat protein. EMBO J. 13: 462-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T., Yang, W., Rozamus, L.W., Hatada, M., Amara, J.F., Rollins, C.T., Stevenson, L.F., Magari, S.R., Wood, S.A., Courage, N.L., et al. 1998. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. 95: 10437-10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Sprenger, F., Duronio, R.J., Leopold, P., and O'Farrell, P.H. 1994. Distinct molecular mechanisms regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes & Dev. 8: 440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring L.K., Axton, J.M., Fenger, D.D., Page, A.W., Carminati, J., and Orr-Weaver, T.L. 1997. The Drosophila PLUTONIUM protein is a specialized cell cycle regulator required at the onset of development. Mol. Biol. Cell 8: 583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger D.D., Carminati, J.L., Burney-Sigman, D.L., Kashevsky, H., Dines, J.L., Elfring, L.K., and Orr-Weaver, T.L. 2000. PAN GU: A protein kinase that inhibits S phase and promotes mitosis in early Drosophila development. Development 127: 4763-4774. [DOI] [PubMed] [Google Scholar]
- Foe V.E., Odell, G.M., and Edgar, B.A. 1993. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. In The development of Drosophila melanogaster (eds. M. Bate and A. Martinez Arias), pp. 149-300. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Freeman M. and Glover, D. 1987. The gnu mutation of Drosophila causes inappropriate DNA synthesis in unfertilized and fertilized eggs. Genes & Dev. 1: 924-930. [DOI] [PubMed] [Google Scholar]
- Freeman M., Nusslein-Volhard, C., and Glover, D. 1986. The dissociation of nuclear and centrosomal division in gnu, a mutation causing giant nuclei in Drosophila. Cell 46: 457-468. [DOI] [PubMed] [Google Scholar]
- Fung B.K. 1983. Characterization of transducin from bovine retinal rod outer segments. I. Separation and reconstitution of the subunits. J. Biol. Chem. 258: 10495-10502. [PubMed] [Google Scholar]
- Golemis E.A., Serebriiskii, I., Gyuris, J., and Brent, R. 1997. Interaction trap/two-hybrid system of identify interacting proteins. In Current protocols in molecular biology (eds. F. Ausubel et al.), pp. 20.21.21-20.21.35. J. Wiley, New York.
- Greenspan R.J. 1997. Fly pushing: The theory and practice of Drosophila genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Großhans J., Schnorrer, F., and Nusslein-Volhard, C. 1999. Oligomerisation of Tube and Pelle leads to nuclear localisation of Dorsal. Mech. Dev. 81: 127-138. [DOI] [PubMed] [Google Scholar]
- Heldin C.H. 1995. Dimerization of cell surface receptors in signal transduction. Cell 80: 213-223. [DOI] [PubMed] [Google Scholar]
- Hildebrandt J.D., Codina, J., Risinger, R., and Birnbaumer, L. 1984. Identification of a γ subunit associated with the adenylyl cyclase regulatory proteins Ns and Ni. J. Biol. Chem. 259: 2039-2042. [PubMed] [Google Scholar]
- Huang J. and Raff, J.W. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18: 2184-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey P.D., Russo, A.A., Polyak, K., Gibbs, E., Hurwitz, J., Massague, J., and Pavletich, N.P. 1995. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313-320. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A.V., Canagarajah, B., Wilsbacher, J., Robinson, M., Atkinson, M., Goldsmith, E., and Cobb, M.H. 1998. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93: 605-615. [DOI] [PubMed] [Google Scholar]
- Lee L.A., Elfring, L.K., Bosco, G., and Orr-Weaver, T.L. 2001. A genetic screen for suppressors and enhancers of the Drosophila PAN GUcell cycle kinase identifies Cyclin B as a target. Genetics 158: 1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K., Ho, J.X., Keeling, K., Gilliland, G.L., Ji, X., Ruker, F., and Carter, D.C. 1994. Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 3: 2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. and Hunt, T. 1993. The cell cycle: An introduction. Freeman, New York.
- Renault A.D., Zhang, X.H., Alphey, L.S., Frenz, L.M., Glover, D.M., Saunders, R.D., and Axton, J.M. 2003. giant nuclei is essential in the cell cycle transition from meiosis to mitosis. Development 130: 2997-3005. [DOI] [PubMed] [Google Scholar]
- Shamanski F. and Orr-Weaver, T. 1991. The Drosophila plutonium and pan gu genes regulate entry into S phase at fertilization. Cell 66: 1289-1300. [DOI] [PubMed] [Google Scholar]
- Su T.T., Sprenger, F., DiGregorio, P.J., Campbell, S.D., and O'Farrell, P.H. 1998. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes & Dev. 12: 1495-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Houston, S.A., Bashirullah, A., Cooperstock, R.L., Semotok, J.L., Reed, B.H., and Lipshitz, H.D. 2003. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics 164: 989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T.T.-L., Bickel, S.E., Young, L.M., and Orr-Weaver, T.L. 1998. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes & Dev. 12: 3843-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]