Abstract
The α7 nicotinic acetylcholine receptor (nAChR) is well established as the principal high-affinity α-bungarotoxin-binding protein in the mammalian brain. We isolated carbachol-sensitive α-bungarotoxin-binding complexes from total mouse brain tissue by affinity immobilization followed by selective elution, and these proteins were fractionated by SDS-PAGE. The proteins in subdivided gel lane segments were tryptically digested, and the resulting peptides were analyzed by standard mass spectrometry. We identified 55 proteins in wild-type samples that were not present in comparable brain samples from α7 nAChR knockout mice that had been processed in a parallel fashion. Many of these 55 proteins are novel proteomic candidates for interaction partners of the α7 nAChR, and many are associated with multiple signaling pathways that may be implicated in α7 function in the central nervous system. The newly identified potential protein interactions, together with the general methodology that we introduce for α-bungarotoxin-binding protein complexes, form a new platform for many interesting follow-up studies aimed at elucidating the physiological role of neuronal α7 nAChRs.
Keywords: α-bungarotoxin, α7 nAChR, membrane protein, mass spectrometry, nicotinic receptor
INTRODUCTION
Nicotinic acetylcholine receptors (nAChRs) comprise a diverse family of ligand-gated ion channels (LGICs) that are expressed throughout the vertebrate central and peripheral nervous systems. nAChRs are composed of five subunits that assemble symmetrically around an axis perpendicular to the plasma membrane.1 Each subunit consists of a large N-terminal ligand-binding extracellular domain and four transmembrane helices, M1 through M4.2 Twelve neuronal nAChR subunits have been identified (α2–α10 and β2–β4). Subunit composition determines the localization and the physiological and pharmacological properties of each receptor subtype.3 Neuronal nAChR subtypes may be classified into two major subclasses based on sensitivity to the nicotinic antagonist α-bungarotoxin (α-bgtx). The neurotoxin, α-bgtx, has been used extensively to identify, localize, and purify neuronal α-bgtx-sensitive nicotinic acetylcholine receptors with α7, α8, α9, and α10 subunits, and the skeletal muscle-type nAChR.4
The α7 nAChR is the only known endogenous receptor in the central nervous system (CNS) that binds α-bgtx with high affinity.5 α7 subunit-containing receptors are abundant in the mammalian hippocampus and the cerebral cortex, brain regions associated with learning and memory.6 Located pre-and peri-synaptically, these receptors can modulate neurotransmitter release, while post-synaptic α7 nAChRs can mediate excitatory currents and are involved in complex signal transduction as a consequence of the intrinsic channel’s high permeability to calcium.5 Similarly calcium entry through α7 nAChRs has been proposed to affect the initiation of secondary messenger systems during neuronal development.7 Although most abundantly expressed in neuronal tissue, recent evidence suggests that the α7 nAChR is found in mammalian macrophages,8 microglia,9, 10 keratinocytes,11 smooth muscle cells,12 astrocytes,13 endothelial cells,14, 15 and lymphocytes.16, 17
Additionally, the α7 nAChR is thought to interact with proteins involved in trafficking and targeting,18–20 scaffolding,21, 22 as well as with kinases and other signaling proteins.23, 24 The intracellular region coupling the third and fourth transmembrane helices of the α7 nAChR, the M3-M4 linker, is potentially a major site for intracellular protein interaction. The bulk of the intracellular domain of nAChRs is comprised of the M3-M4 linker, which displays substantial variability in both sequence and length among nAChRs. The M3-M4 linker region has been implicated in receptor assembly,25 folding,5, 6 and trafficking.22, 26–29 Several potential binding partners of the α7 nAChR have been reported in studies using a variety of methods including immunolabeling,22 co-immunoprecipitations,23, 24 yeast two-hybrid assays,18 and Western blotting.20, 21 However, our study is the first large-scale proteomics-based analysis of the α7 nAChR interactome.
As is well documented, mass spectrometry is a highly effective methodology for elucidating protein-protein interaction networks.30, 31 Following enzymatic digestion, peptides are fractionated and subjected to nano-electrospray ionization (nano-ESI). Mass measurement of the peptide ions followed by further fragmentation analysis allows for the determination of the primary sequences of the peptides, ultimately resulting in protein identification.32 We utilized mass spectrometry as a high-throughput method for identifying proteins that remain associated with the α7 nAChR following detergent solubilization from native neuronal tissue. Using α-bgtx-conjugated beads and specific competitive elution with carbachol (a low molecular weight cholinergic agonist of the α7 nAChR), we isolated protein complexes from murine brain membranes solubilized with the detergent, Triton X-100. Brain tissue from α7 nAChR knockout mice, lacking high-affinity binding sites, was used in our studies to control for proteins binding nonspecifically to the affinity bead matrix. The α7 nAChR knockout mice appear to be largely normal in development and in gross neurological function, although it has been observed that homozygotes may not breed reliably.33 Following SDS-PAGE fractionation and in-gel tryptic digestion of the selectively eluted binding proteins, the peptides were characterized by nano-ESI tandem mass spectrometry (MS/MS), and the SEQUEST algorithm was applied to the data analysis to determine the identity of the eluted proteins. Using these techniques, 55 potential members of the α7 nAChR interactome were identified reproducibly in at least two of the three independent mass spectrometric analyses performed as part of this study. Further validation by orthogonal methodologies of this preliminary list of proteins is necessary before these proteins can be deemed as bona fide interacting partners of the α7 nAChR. The α7 nAChR is thought to be involved in many cognitive and behavioral functions. Recent studies have shown a correlation between altered expression of the α7 nAChR and human cognitive disorders, such as epilepsy,34, 35 schizophrenia,36, 37 autism,38, 39 and attention-deficit / hyperactivity disorder.40, 41 Understanding the interplay of the α7 nAChR in various signaling pathways may have important implications in the diagnosis, treatment, and prevention of a variety of neuropsychiatric and neurodegenerative diseases.42
EXPERIMENTAL PROCEDURES
Materials
Trypsin Gold, mass spectrometry grade (V5280) was bought from Promega Corp. (Madison, WI). Triton X-100 (807426) was purchased from MP Biomedicals (Solon, OH). Complete, Mini protease inhibitor cocktail was purchased from Roche (Mannheim, Germany). Rabbit anti-α7 nAChR antibody (ab10096) was bought from Abcam (Cambridge, MA). Goat anti-α7 nAChR antibody (SC-1477) was from Santa Cruz Biotechnology (Santa Cruz, CA). CL-XPosure Film (34090) and Supersignal West Pico Chemiluminescence substrate (34080) were purchased from Pierce (Rockford, IL). BenchMark Pre-Stained (10748–010) and MagicMark XP (LC5602) protein standards were bought from Invitrogen (Carlsbad, CA). Strong cation exchange ZipTips (ZipTipSCX, ZTSCXS096) were purchased from Millipore (Billerica, MA). Cyanogen bromide (CNBr) activated sepharose beads (C9142), Brilliant Blue G-Colloidal Coomassie stain (B2025), horse-radish peroxidase-conjugated goat anti-rabbit antibody (A9169), horse-radish peroxidase-conjugated rabbit anti-goat antibody (A5420), carbachol (C4382), α-bungarotoxin (T3019), and the remaining general chemicals used were purchased from Sigma-Aldrich (St. Louis, MO).
Preparation of ligand affinity beads
CNBr-activated sepharose 4B beads (1.5 g) were hydrated in 5 ml of 1 mM HCl for 30 minutes and washed on a coarse glass filter with 500 ml of 1 mM HCl. The beads were added to 7.5 ml of coupling buffer (0.25 M NaHCO3, 0.5 M NaCl, pH 8.3) and centrifuged for 5 minutes at 1,500 × g. The beads were pelleted and resuspended in 15 ml of coupling buffer containing 3 mg of α-bgtx prior to gentle rotation overnight at 4°C. The beads were then pelleted, resuspended in 15 ml of 0.2 M glycine in 80% coupling buffer and gently rotated overnight at 4°C. This mixture was washed on a fritted coarse glass filter, first with 100 ml of 0.1 M NaHCO3, 0.5 M NaCl, pH 8.0, next with 100 ml of 0.1 M NaCH3CO2, 0.5 M NaCl, pH 4.0, then again with 100 ml of 0.1 M NaHCO3, 0.5 M NaCl, pH 8.0, followed by a wash in coupling buffer. The beads were finally washed twice with Tris-buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.6), and resuspended in TBS supplemented with 0.1% Triton X-100 and 0.02% sodium azide.
Membrane protein solubilization
Frozen whole mouse brains of wild-type or α7 nAChR knockout mice (C57 strain genetic background) were thawed and homogenized in ice-cold TBSp (TBS supplemented with protease inhibitors) with 20–25 strokes of a Potter-Elvehjem glass homogenizer. One milliliter of buffer was added per 400 mg of mouse brain tissue. The homogenate was ultracentrifuged at 100,000 × g for 60 minutes at 4°C. The pellet (containing the cell membrane fraction) was reconstituted in an ice-cold TBSp plus 1% Triton X-100 solution. The resuspended pellet was homogenized with 20–25 strokes of a Potter-Elvehjem glass homogenizer and incubated on ice with gentle agitation for 2 hours. Detergent-solubilized proteins were separated from the insoluble proteins and cellular debris by ultracentrifugation at 100,000 × g for 60 minutes at 4°C.
α-Bgtx complex isolation
Detergent-solubilized proteins were incubated with 100 µl of α-bungarotoxin-conjugated sepharose bead slurry overnight at 4°C with gentle rotation. Following the incubation, the beads were centrifuged at 2,000 × g for 5 minutes and unbound proteins were removed. The beads were washed three times with 1.5 ml of ice-cold TBSp plus 1% Triton X-100 to reduce nonspecific binding. The α7 nAChR and associated proteins were selectively eluted by incubation with 50 µl of a 1 M carbamylcholine chloride (carbachol) solution, and subsequently precipitated with acetone. In brief, four times the sample volume of cold (−20°C) 100% acetone was added to each sample, which was mixed by vortexing and then incubated for 60 minutes at −20°C. The samples were centrifuged for 10 minutes at 13,000 × g, after which the supernatants were aspirated and the pellets were air-dried.
Sample preparation, SDS-PAGE, and Western blotting
Acetone-precipitated samples were resuspended in 10 µl of SDS-loading buffer (100 mM Tris-HCl, pH 8, 2% SDS, 100 mM DTT, 20% glycerol, 0.002% bromophenol blue) and incubated at 55°C for 30 minutes. One microliter of 1 M iodoacetamide was added to the samples which were then incubated at room temperature in darkness for 30 minutes. Samples were loaded onto a 15% polyacrylamide gel and SDS-PAGE was performed at 125 V for approximately 45 minutes. Proteins were visualized by colloidal Coomassie staining following the manufacturer’s protocol.
Western blotting confirmed that the α7 subunit was captured by the α-bgtx-conjugated sepharose bead procedure. Proteins were transferred from polyacrylamide gels onto a nitrocellulose membrane at 100 V for 90 minutes at 4°C in electroblotting buffer (25 mM Tris-HCl, 0.2 M glycine, 20% methanol). The membrane was blocked for 1 hour at room temperature with 5% nonfat dry milk dissolved in TBS-T (TBS supplemented with 0.1% Tween-20). The membrane was then incubated with 10 ml of a 1:500 dilution of anti-α7 nAChR antibody in TBS at room temperature for 2 hours. After two 5-minute washes in both TBS-T and TBS, a 1:20,000 dilution of the appropriate horseradish peroxidase-conjugated secondary antibody in TBS was added to the membrane, which was incubated for 1 hour at room temperature. After another set of washes in TBS-T and TBS, the membrane was incubated with the chemiluminescence substrate for 5 minutes following the manufacturer’s protocol. Finally, the membrane was exposed to X-ray film for 15–30 seconds and the film was developed.
In-gel digestion
Gel slices were excised from the Coomassie-stained polyacrylamide gel and washed three times with 200 µl of 50% acetonitrile, 50% 50 mM ammonium bicarbonate solution for 30 minutes at 37 °C. Gel slices were then dehydrated by adding 200 µl of neat acetonitrile and incubated at room temperature for 15 minutes. After removing the acetonitrile and drying for 30 minutes at 37 °C, the gel slices were rehydrated with the addition of 10 µl of 25 mM ammonium bicarbonate, pH 8.0 containing 100 ng of trypsin. Following a 30-minute room temperature incubation, an additional 50 µl of 25 mM ammonium bicarbonate was added to completely immerse the gel slices in buffer. After an overnight incubation at 37°C, the peptides were extracted with 40 µl of 0.1% TFA (trifluoroacetic acid) in increasing concentrations of acetonitrile (0%, 25%, 50%, and 75%). These fractions were combined in a single low-retention microcentrifuge tube and vacuum centrifuged to dryness. The peptides were desalted with a ZipTipSCX prior to analysis by mass spectrometry following the manufacturer’s instructions. Briefly, the lyophilized peptides were resuspended in 10 µl of 0.1% TFA and the ZipTipSCX resin was equilibrated with 0.1% TFA. The peptides were bound to the ZipTip resin by trituration and washed in 0.1% TFA, 30% methanol. Finally, the peptides were eluted with 5 µl of 5% ammonium hydroxide, 30% methanol and the sample was vacuum centrifuged to dryness.
Mass spectrometry
One of the data sets (Data Set A) was obtained using an LTQ-Orbitrap at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School (Boston, MA). The samples were reconstituted in 10 µl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nano-scale reversed-phase HPLC capillary was prepared with 5 µm C18 spherical silica beads packed into a fused silica capillary (100 µm inner diameter and approximately 12 cm in length) with a flame-drawn tip. After equilibrating the column, the sample was loaded via a Famos autosampler (LC Packings, San Francisco, CA). A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). The eluted peptides were subjected to nano-ESI and the resulting ions were detected, isolated, and fragmented to produce MS/MS spectra of specific fragment ions using an LTQ Orbitrap mass spectrometer (ThermoFinnigan, San Jose, CA). The MS/MS spectra were automatically searched against the mouse NCBI non-redundant protein database (November 2007) using the SEQUEST algorithm.43
The other two data sets (Data Sets B and C) were obtained using an LTQ mass spectrometer at the NSF/EPSCoR Proteomics Facility at Brown University (Providence, RI). The peptides were reconstituted in 20 µl of 0.1 M acetic acid. A nano-scale reversed-phase HPLC capillary was prepared with 5 µm Monitor C18 spherical silica beads packed into a fused silica capillary (75 µm inner diameter and approximately 12 cm in length) with an integrated ESI emitter tip having a 4 µm opening and fritted with 3 µm silica particles (Bangs Labs, Fishers, IN). A gradient was formed and peptides were eluted with increasing concentrations of solvent B (90% acetonitrile, 0.1 M acetic acid). The eluted peptides were then scanned by an LTQ mass spectrometer (data-dependant scanning with 1 MS scan followed by 5 MS/MS scans). MS/MS spectra were generated and automatically searched against the mouse NCBI non-redundant protein database using the SEQUEST algorithm provided with BioWorks 3.2 SR (ThermoFinnigan, San Jose, CA). The list of predicted proteins was imported into a custom-made FileMaker Pro (FileMaker, Santa Clara, CA) relational database for data analysis.
Each data set (A through C) represents a different preparation of mouse brain tissue. The murine brain tissue samples were processed and in-gel digestions were preformed in parallel for Data Sets A and B. Data Set C samples were prepared several weeks later using identical protocols.
Data processing
Peak list files were created by the program “extract_msn.exe” that was installed with BioWorks version 3.2 SR using the following parameters: The mass must fall within the range of 600 to 4500 Daltons and the minimum total ion current for the scan must exceed 1000. The precursor tolerance for grouping was 1.5 Da, with no differing intermediate scans allowed and only a single scan required to create a peak file. The minimum signal-to-noise for a peak to be written to the peak file was 3, and 25 such peaks must be found for a peak file to be created. The program calculated charge states, however, in cases of ambiguity, peak files for both the +2 and +3 charge states were created. No smoothing or de-isotoping was performed. The precursor-ion tolerance was 2.0 Da and the fragment-ion tolerance was 0.8 Da. The modifications allowed were iodoacetamide alkylation of cysteine and oxidation of methionine. Enzymatic digestion was specified as trypsin, with up to 2 missed cleavages allowed. Searches were performed using the NCBI non-redundant mouse database (November, 2007). A list of reversed-sequences was created from these entries and appended to the original database for searching so that false positive rates could be estimated. Final false positive rates were estimated at approximately 1%. The mass tolerance parameter cut-offs were as follows: Peptides with charge of +1, +2 and +3 must have Xcorr values that are greater than 1.5, 2.0 and 2.5, respectively.
RESULTS
Affinity sedimentations, immunoblotting, and Coomassie staining
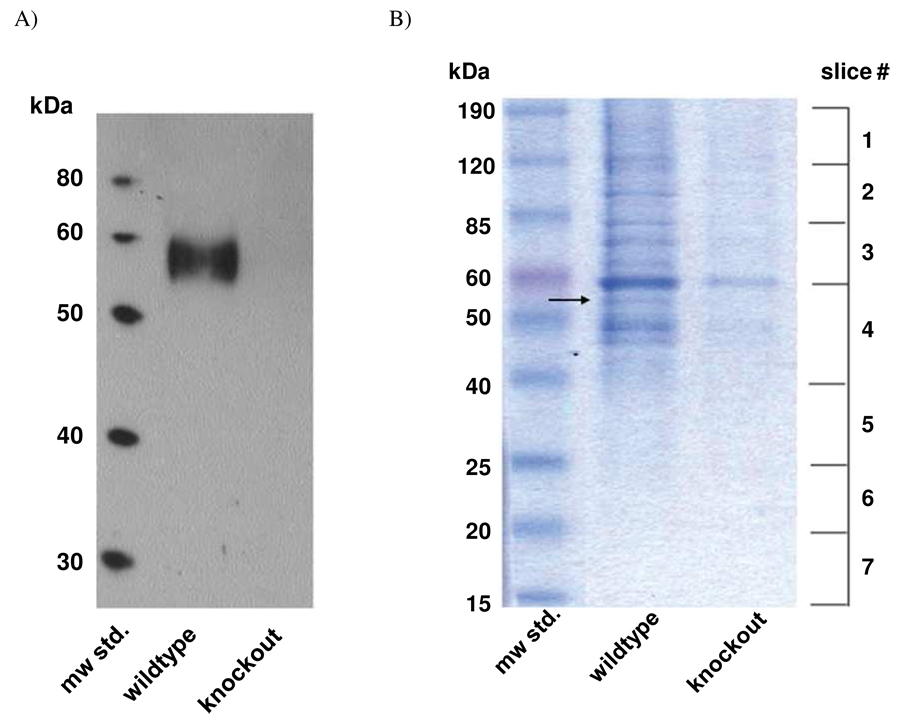
To achieve greater specificity, we used the nicotinic agonist, carbachol, to elute from α-bgtx-conjugated affinity beads those complexes bound in a manner consistent with the nicotinic pharmacology of the α7 nAChR. Carbachol eluants of α-bgtx bead isolations from wild-type and α7 knockout mouse brain tissue were analyzed on SDS-PAGE. Western blotting performed with antibodies against the α7 nAChR (Figure 1A) verified that the α7 receptor was successfully sedimented with the α-bgtx beads and released upon carbachol incubation. In these Western blots, no band was observed in the lane of samples originating from the α7 nAChR knockout mouse, confirming that the α7 nAChR is not present in the control mouse samples.
Figure 1.
Western blotting and SDS-PAGE analysis of mouse brain-derived samples following affinity isolation. A) Western blotting confirms that the α7 subunit was present in a lane loaded with the carbachol eluant from an α-bgtx affinity bead preparation of solubilized, wild-type mouse brain tissue. No corresponding band was present in the lane corresponding to a sample from the α7 nAChR knockout mouse. The nitrocellulose membrane was probed with anti-α7 nAChR antibody. B) Coomassie-stained SDS-PAGE gel illustrating the profile of proteins isolated by an α-bgtx affinity bead sedimentation from wild-type and α7 nAChR knockout mouse brain tissue. The approximate molecular weights of the protein standards and gel slice regions are denoted to the left and right of the gel, respectively.
Affinity-isolated proteins were visualized on polyacrylamide gels stained with colloidal Coomassie blue. The protein staining showed multiple bands from the α-bgtx affinity sedimentations of wild-type samples (Figure 1B). These protein bands confirm the isolation of a diverse array of candidate interacting partners of the α7 nAChR. Protein bands in the sample lanes from the knockout mouse are most likely due to nonspecific binding, or could be novel non-α7 α-bgtx-binding proteins. Notably, protein bands from the wild-type mouse samples were significantly more intense (greater than two-fold) than those from the knockout mouse samples, although identical masses of brain tissue were homogenized in both cases. In our preliminary studies, we targeted specific protein bands for detailed MALDI MS/MS analysis which readily identified the α7 subunit at the correct molecular weight in the wild-type gel lane (Figure 1B, arrow). A similar MALDI analysis of the corresponding gel region in the α7 nAChR knockout mouse lane did not detect any α7 protein. In addition, we identified the most intensely stained band in the knockout sample lane as the highly abundant protein, tubulin.
Mass spectrometric analysis of α-bgtx affinity-isolated proteins
For a more thorough analysis of the proteins isolated by our methodology, SDS-PAGE gel lanes of α-bgtx isolations from wild-type and knockout mice were divided into 7 slices (Figure 1B) corresponding to the molecular weights: (1) 190–120 kDa, (2) 120–85 kDa, (3) 85–60 kDa, (4) 60–40 kDa, (5) 40–25 kDa, (6) 25–20 kDa, and (7) 20–15 kDa. Tryptic peptides prepared from each gel slice were separated by reversed-phase high-performance liquid chromatography (RP-HPLC) prior to nano-ESI mass spectrometric analysis. Separation at the peptide level resulted in the identification of hundreds of proteins from a single gel slice. The use of ZipTipSCX sample preparation pipette tips also improved spectra quality by removing salts and residual detergent from the eluted peptides.
Proteins identified from wild-type mouse brain tissue, but absent in corresponding isolations from α7 nAChR knockout mice, and which also appear in at least two of the three independent data sets are listed in Table 1. These proteins were broadly categorized by their cellular functions by means of gene ontology queries.44 Exclusively from wild-type mouse brain tissue, we identified 22 proteins that provide structural support and/or play a role in trafficking, 19 potentially interacting proteins involved in signal transduction, 7 proteins related to basic metabolism, 5 chaperone proteins, 2 proteins associated with proteolysis, for a total of 55 proteins. Supplementary Table 1 lists the identified peptides corresponding to the proteins listed in Table 1. An additional 153 proteins were identified in only one of the three acquired mass spectrometry data sets (Supplemental Table 2). In addition, a total of 129 proteins were identified by MS/MS as being represented in both the wild-type and the α7 nAChR knockout mouse samples (Supplemental Table 3). Proteins listed in Supplementary Table 2 and Table 3 were not included in our list of potential interacting partners of the α7 nAChR. Further repetitions would be necessary to determine whether the proteins in Supplementary Table 2 are due to physiologically significant interactions with the network of proteins interacting with the α7 nAChR. Likewise, without further supporting data, the proteins listed in Supplemental Table 3 are most likely products of nonspecific binding, novel non-α7 nAChR α-bgtx-binding proteins, or false positive protein identifications resulting from the data analysis algorithm. Supplementary Table 4 lists additional information on the sub-cellular location and function of those candidate interacting proteins that are unique to the wild-type samples (i.e., those in Table 1).
Table 1. Proteins from wild-type mouse brain tissue identified by nano-ESI-MS/MS of peptides originating from in-gel digested proteins isolated by selective elution from α-bgtx-conjugated affinity beads.
Only proteins that appear in wild-type samples and are absent in knockout samples are listed. This table lists proteins that were identified in at least two of three data sets from α-bgtx isolations of different mouse brain preparations. Supplemental Table 1 lists all those proteins from wild-type mouse brain tissue that appear in at least one of the three data sets associated with selective elution from α-bgtx-conjugated beads. Each Data Set (A through C) represents a separate preparation of the same amount of mouse brain tissue. The table lists the name of the protein, the Swiss-Prot accession number, protein mass in Daltons, as well as the total number of matched peptides per protein, and the gel slice from which these peptides were derived.
| Protein | Swiss-Prot acc. no. |
protein mass (Da) |
Data Set A | Data Set B | Data Set C | |||
|---|---|---|---|---|---|---|---|---|
| matched peptides |
slice no. | matched peptides |
slice no. | matched peptides |
slice no. | |||
| Basic metabolic proteins | ||||||||
| Adenylyl cyclase-associated protein 2 | Q9CYT6 | 52,862 | 1 | 4 | 1 | 4 | 1 | 4 |
| Aldehyde dehydrogenase, mitochondrial | P47738 | 56,538 | 2 | 4 | 1 | 4 | ||
| Glutathione S-transferase Mu 1 | P10649 | 25,970 | 1 | 5 | 1 | 5 | ||
| L-lactate dehydrogenase A chain | P06151 | 36,499 | 2 | 4 | 1 | 4 | ||
| Myristoylated alanine-rich C-kinase substrate | P26645 | 29,661 | 1 | 2 | 3 | 2 | ||
| Probable oxidoreductase KIAA1576 | Q80TB8 | 45,817 | 1 | 4 | 1 | 4 | ||
| Vacuolar proton translocating ATPase A1 | Q9Z1G4 | 96,501 | 1 | 1 | 2 | 2 | 2 | 2 |
| Cell structure and protein trafficking | ||||||||
| Alpha-catenin 2 | Q61301 | 105,286 | 1 | 1 | 1 | 1 | ||
| Ankyrin 2, isoform 1 | Q6PCN2 | 117,436 | 4 | 1 | 3 | 1 | 1 | 1 |
| Ankyrin 2, isoform 2 | Q8C8R3 | 131,202 | 2 | 1 | 2 | 1 | 1 | 1 |
| AP-2 complex subunit beta-1 | Q9DBG3 | 104,583 | 2 | 1 | 1 | 1 | ||
| AP-3 complex subunit beta-2 | Q9JME5 | 119,193 | 1 | 1 | 1 | 1 | ||
| Brain acid soluble protein 1; BASP1, NAP-22 | Q91XV3 | 22,087 | 7 | 4 | 2 | 4 | ||
| Protein disulfide-isomerase A3 | P27773 | 56,678 | 3 | 4 | 1 | 4 | ||
| Endophilin-1 | Q62420 | 39,877 | 2 | 4 | 1 | 4 | ||
| Endophilin-2 | Q62419 | 41,518 | 1 | 3 | 1 | 3 | ||
| Fascin | Q61553 | 54,508 | 2 | 3 | 5 | 3 | ||
| Gamma-soluble NSF attachment protein | Q9CWZ7 | 34,732 | 1 | 4 | 1 | 4 | ||
| Homer protein homolog 1 | Q9Z2Y3 | 41,413 | 1 | 4 | 1 | 4 | ||
| Kinesin-like protein KIF2 | P28740 | 79,756 | 1 | 2 | 1 | 2 | ||
| Microtubule-associated protein tau | P10637 | 76,243 | 2 | 4 | 1 | 4 | 5 | 4 |
| Myosin-VI | Q64331 | 146,409 | 1 | 1 | 3 | 1 | ||
| Nck-associated protein 1 | P28660 | 128,784 | 1 | 1 | 1 | 2 | ||
| Spectrin beta chain | P15508 | 245,250 | 1 | 1 | 2 | 1 | ||
| Synaptogyrin-1 | O55100 | 25,653 | 1 | 5 | 1 | 5 | ||
| Synaptojanin-1 | Q8CHC4 | 172,617 | 1 | 1 | 1 | 1 | ||
| Syntaxin-1 A | O35526 | 33,054 | 1 | 6 | 1 | 4 | ||
| Syntaxin-1 B | O35525 | 33,079 | 1 | 4 | 1 | 4 | ||
| Tropomodulin-2 | Q9JKK7 | 39,511 | 1 | 4 | 1 | 4 | ||
| Chaperones | ||||||||
| 14-3-3 protein beta/alpha | Q9CQV8 | 28,086 | 1 | 6 | 2 | 6 | ||
| 14-3-3 protein eta | P68510 | 28,212 | 1 | 5 | 1 | 5 | ||
| 14-3-3 sigma | O70456 | 27,713 | 1 | 4 | 1 | 4 | ||
| Gelsolin | P13020 | 85,942 | 1 | 2 | 3 | 2 | ||
| Heat shock protein HSP 90-beta | P11499 | 83,325 | 2 | 2 | 2 | 2 | ||
| Proteolytic pathway proteins | ||||||||
| Ubiquitin carboxyl-terminal hydrolase 5 | P56399 | 95,833 | 1 | 1 | 2 | 2 | ||
| Ubiquitin-activating enzyme E1 | Q02053 | 117,809 | 2 | 1 | 2 | 1 | 3 | 1 |
| Signal transduction | ||||||||
| Calmodulin | P62204 | 16,838 | 1 | 6 | 1 | 6 | 2 | 6 |
| cAMP-dependent protein kinase, PKA | P68181 | 40,708 | 1 | 3 | 1 | 3 | ||
| Clathrin coat assembly protein AP180 (SNAP91) | Q61548 | 91,851 | 1 | 2 | 2 | 2 | ||
| Contactin-1 | P12960 | 113,388 | 1 | 1 | 3 | 1 | ||
| Guanine nucleotide-binding protein alpha-12 | P27600 | 44,095 | 2 | 4 | 2 | 4 | ||
| Guanine nucleotide-binding protein G(i), alpha-2 | P08752 | 40,471 | 1 | 4 | 2 | 4 | ||
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) beta 3 | Q61011 | 37,240 | 1 | 4 | 1 | 4 | ||
| Guanine nucleotide-binding protein G(o) alpha 1 | P18872 | 40,085 | 7 | 4 | 2 | 4 | 6 | 4 |
| Guanine nucleotide-binding protein G(q) alpha | P21279 | 41,483 | 1 | 4 | 1 | 4 | ||
| Mitsugumin-23 | Q3UBX0 | 26,306 | 1 | 6 | 1 | 6 | ||
| Neuromodulin (GAP-43) | P06837 | 23,632 | 3 | 5 | 1 | 5 | ||
| Phosphatase 2A inhibitor, TAF-1, SET | Q9EQU5 | 33,378 | 1 | 4 | 2 | 4 | ||
| Protein kinase C gamma type | P63318 | 78,358 | 1 | 2 | 1 | 2 | ||
| Protein phosphatase 1 regulatory subunit 7 | Q3UM45 | 41,262 | 2 | 4 | 1 | 3 | ||
| Ras-related protein Rab-2A | P53994 | 23,548 | 1 | 5 | 1 | 5 | ||
| Serine/threonine-protein phosphatase 2B | P63328 | 58,644 | 2 | 3 | 1 | 3 | ||
| Serine/threonine-protein phosphatase PP1-alpha | P62137 | 37,540 | 2 | 4 | 1 | 4 | ||
| Tenascin-R | Q8BYI9 | 149,563 | 1 | 1 | 1 | 1 | ||
| Transgelin-3 | Q9R1Q8 | 22,471 | 4 | 5 | 3 | 5 | ||
DISCUSSION
The combination of affinity isolation and tandem mass spectrometry is a powerful method for revealing many potential interacting proteins associated directly or indirectly with a protein of interest. The isolation of a protein target is one of the major challenges of a proteomics experiment. Using antibodies coupled to a bead support for immunoprecipitations is a common technique for isolating proteins from their native environment. Antibodies, however, can lead to undesired sample complexity in mass spectrometry-based experiments. Anti-peptide antibodies against the α7 nAChR, for example, have been shown to cross-react with non-α7 nAChR proteins and are unreliable for use in many in vivo and in situ studies.45, 46 In addition, antibodies binding to certain functionally relevant epitopes may perturb the conformation of the receptor and compromise important protein-protein interactions. Moreover, antibodies are not easily dissociated from their target, and an abundance of antibody protein in a sample may suppress the signal of peptides of interest.47 The use of α-bgtx for affinity purification resolves many of these issues. In wild-type brain tissue, α-bgtx binds selectively to the α7 nAChR with high affinity, and brain sections from α7 receptor knockout mice show little evidence of α-bgtx binding to other sites.33
The interactions of various neuronal membrane receptors have been investigated via immunoprecipitations and/or affinity precipitations.48 These studies have both confirmed several previously suspected binding partners and yielded important information on novel interacting partners. Several such studies have investigated the post-synaptic density, which is a protein dense region at the post-synaptic plasma membrane of the neuronal synaptic cleft. Proteins in this region include glutamate receptors that are present at the major excitatory synapses in the mammalian CNS.49–51 Recently, 698 proteins associated with post-synaptic terminals of the murine CNS have been identified via immunoprecipitations of the glutamate receptors, AMPAR (α-amino-3-hydroxy-5-methylisoxazole-4-propionate receptor) and NMDAR (N-methyl-D-aspartic acid receptor), as determined by LC-MS/MS.52
Our mass spectrometric analysis of proteins isolated by α-bgtx-affinity methods has identified several proteins that were among the 698 proteins listed by Collins, et al.52 Twenty-six of the 55 proteins isolated using our α-bgtx binding strategy and identified only in the wild-type mouse sample are among the previously-described post-synaptic density proteins (listed in Table 2), including cell structure and trafficking proteins, molecular chaperones, and proteins involved in signal transduction. The α7 nAChR itself was not identified in the study by Collins, et al., and, similarly, we also did not identify the NMDAR or the AMPAR subunits in our interactome study. These data suggest that α7 nAChRs are not directly complexed with glutamate receptors, although indirect interactions that are detergent sensitive remain a possibility.53 It is certainly possible that the 26 proteins identified in common by both Collins, et al. and our study may interact with two different sets of receptor associated proteins, the α7 receptor interactome in one case, and the NMDAR/AMPAR post-synaptic density interactome in the other case. Further studies would be needed to determine whether any of the overlapping 26 proteins are due to NMDAR/AMPAR protein complexes that include post-synaptic α7 nAChRs.
Table 2. Proteins from Table 1 that have also been identified in the post-synaptic density proteome.
These proteins are a subset of those in Table 1 that are among the 698 proteins identified in the study of the post-synaptic density proteome by Collins et al.52
| Protein | Swiss-Prot acc. no. |
|---|---|
| Basic metabolic proteins | |
| L-lactate dehydrogenase A chain | P06151 |
| Vacuolar proton translocating ATPase A isoform 1 | Q9Z1G4 |
| Cell structure and protein trafficking | |
| Alpha-catenin 2 | Q61301 |
| AP-2 complex subunit beta-1 | Q9DBG3 |
| Brain acid soluble protein 1; BASP1, NAP-22 | Q91XV3 |
| Fascin | Q61553 |
| Gamma-soluble NSF attachment protein | Q9CWZ7 |
| Homer protein homolog 1 | Q9Z2Y3 |
| Kinesin-like protein KIF2 | P28740 |
| Microtubule-associated protein tau | P10637 |
| Myosin-VI | Q64331 |
| Synaptogyrin-1 | O55100 |
| Syntaxin-1 A | O35526 |
| Syntaxin-1 B | O35525 |
| Tropomodulin-2 | Q9JKK7 |
| Chaperones | |
| 14-3-3 protein beta/alpha | Q9CQV8 |
| 14-3-3 protein eta | P68510 |
| Gelsolin; Actin-depolymerizing factor Brevin | P13020 |
| Heat shock protein HSP 90-beta | P11499 |
| Signal transduction | |
| cAMP-dependent protein kinase, PKA | P68181 |
| Contactin-1 | P12960 |
| Guanine nucleotide-binding protein G(i), alpha-2 subunit | P08752 |
| Guanine nucleotide-binding protein G(o) subunit alpha 1 | P18872 |
| Guanine nucleotide-binding protein G(q) subunit alpha | P21279 |
| Protein kinase C gamma type | P63318 |
| Ras-related protein Rab-2A | P53994 |
We also have identified potential α7 nAChR binding partners that may be associated with pre-synaptically localized α7 nAChRs, including the proteins, BASP1/NAP-22, tenascin-R, neuromodulin (GAP-43), and synaptogyrin-1. Of these identifications, BASP1/NAP-22 is particularly interesting as this protein binds cholesterol and has been associated with microdomains (lipid rafts) at axonal termini.54, 55 Coincidently, several studies have suggested that α7 nAChRs may also be found in lipid rafts (as well as pre-synaptically), strengthening the possibility of an interaction between the two proteins.23, 56, 57 The identification of both pre- and post-synaptic proteins in our proteomics study is not surprising considering that there exists much evidence that α7 nAChRs are associated with both presynaptic and postsynaptic membranes.58, 59
The intrinsic ion channel of the α7 nAChR is highly permeable to calcium ions, as has been established through electrophysiological recordings of heterologously-expressed receptors.60, 61 In addition, the α7 nAChR has been implicated in calcium entry-dependent events, including the regulation of secondary messenger cascades and of neurotransmitter release.13, 61 Phosphorylation of the receptor by protein kinase A (PKA) and/or protein kinase C (PKC), which play prominent roles in cellular regulation and signaling cascades, may also regulate and be regulated by α7 nAChR-mediated calcium modulation.62–64 Interestingly, we have identified both PKA and PKC in our proteomics study as two of the 55 candidate interacting proteins present in samples prepared from wild-type mice, but absent in samples originating from α7 nAChR knockout mice (Table 1).
Calcium regulation by the α7 nAChR is thought to influence both neurite outgrowth and secondary messenger systems in the fetal and adult hippocampus.65 The relatively high calcium permeability of the α7 nAChR may lead to the efficient co-localization of calcium-binding or calcium-dependent proteins in neurons. In the developing nervous system, an increase in calcium concentration in regions of neurite growth is directly associated with neurotransmitter release and neurite extension or retraction.66 Terminal plasticity of neurites persists following synapse development.67 In our interactome study, we identified several proteins that are known to be active in neurite outgrowth and maintenance, including alpha-catenin 2,68 BASP1/NAP-22,69 gelsolin,70 MARCKS,71 homer 1,72 neuromodulin,71 and tenascin C.73 Many of the aforementioned proteins also interact with calmodulin, PKC, and protein phosphatase 2B, which have roles in the modulation of calcium-dependent processes and may indirectly affect the rate of elongation and branching of growing neurites. Tubulin, actin, and other cell structure proteins, such as fascin and tau, provide essential structural support to neurites.74, 75 Moreover, actin has been shown, using confocal fluorescence microscopy and immunogold labeling in electron microscopic studies, to co-localize with the α7 nAChR.22 In chick ciliary ganglion, the α7 nAChR has been shown to influence actin filament and/or microtubule assembly through calcium regulation.76 Although actin and tubulin may interact with α7 nAChRs at some level in some tissues and regions, as has been suggested by the correlated presence of these proteins in neurite growth cones,77 these two very abundant cytoskeletal proteins were eliminated from our list of potential interacting proteins because they were also found in samples originating from α7 nAChR knockout mice.
We have identified several proteins, as mentioned above, that are known to contribute to developmentally important regulatory mechanisms of the nervous system. Correspondingly, previous studies have shown that [125I]-α-bgtx-binding is developmentally regulated in postnatal mice.78 It would be a reasonable extension of our work to apply our α-bgtx-based isolation strategy to the identification of binding partners at various stages of postnatal mouse development to search for novel developmental stage-specific α7 nAChR interactions. Of particular interest may be to further investigate the interaction of the α7 nAChR with chaperone proteins which we have identified as potential interacting partners. For example, 14-3-3η, a member of family of chaperone/ scaffolding/ adaptor proteins, has been linked with regulation of expression and subunit stabilization in the α4-subunit containing nAChR.79 We have identified 14-3-3η, as well as14-3-3α/β and 14-3-3σ, exclusively in α-bgtx affinity isolations from adult wild-type mice (Table 1), but the characterization of their interaction with α7 nAChR in both a global and developmental context remains undefined. Another example of a protein that we identified that may also have a significant role in α7 nAChR-related nervous system development is protein disulfide-isomerase A3, which is a member of an enzyme family that catalyzes the rearrangement of disulfide bonds in proteins.80 Although we present the first evidence that this enzyme may interact with the disulfide-bind containing α7 nAChR, there are reports in the literature of the function of protein disulfide-isomerases in developmental regulation.81–83 These proteins, and others that we have identified, may have developmentally regulated roles for the localization and/or folding α7 nAChR. Further proteomic experiments at different mouse developmental stages could help clarify the roles and interactions of the α7 nAChR and its binding partners in the developing nervous system.
As is well known, the G protein (guanine nucleotide-binding protein) family functions in numerous important signal transduction and secondary messenger cascades. These signaling cascades influence embryonic development, learning and memory, as well as organismal homeostasis.84 As is the case with α7 nAChRs, G proteins have been prominently implicated in neurite development 85 and in calcium-dependent signaling.86, 87 Several G proteins (Table 1) met the identification criteria of our mass spectrometric analysis of proteins enriched through α-bgtx affinity isolation. The likelihood of a physiologically important interaction between the α7 nAChR and G proteins is strengthened by the report of an association of the α7 nAChR with adenylyl cyclase. Adenylyl cyclase, an enzyme catalyzing the conversion of ATP to 3',5'-cyclic AMP (cAMP) and pyrophosphate, is known to be stimulated directly by G proteins.88 Similarly, immunoprecipitation and Western blotting data suggest that adenylyl cyclase interacts with the α7 nAChR23 Furthermore, although we did not identify the multi-transmembrane spanning protein adenylyl cyclase, we did identify, an adenylyl cyclase-associated protein, adenylyl cyclase-associated protein 2, as present only in those samples originating from wild-type mice. In addition, neuromodulin (GAP-43) is thought to bind G proteins and to stimulate binding of GTP to the Go subunit.89 In a related interaction, neuromodulin binds and inactivates calmodulin, which is then released in response to PKC activation.90 Notably, neuromodulin, the Go subunit, calmodulin, and PKC are all candidates for interacting partners with α7 nAChRs as they were identified only in samples originating from wild-type mice (Table 1).
Our results with respect to the Go subunit differ from those of a previous study that failed to detect interaction between the α7 nAChR and the Go subunit.91 However, these researchers based their conclusions on Western blots of pull-downs performed using a GST-fusion protein of the α7 nAChR M3-M4 intracellular linker region. As a consequence of the prokaryotic expression of this GST-fusion protein, suboptimal folding, and/or deficiencies in post-translational modifications may not have permitted the association of the M3-M4 linker with all of its native binding partners in that study. Our data together with conclusions drawn from the available literature about common potential binding partners argue that associations between the α7 nAChR and G proteins are highly probable and clearly merit further investigation. For all proteins identified in our study, additional validation using independent methodologies is necessary to confirm that these proteins are bona fide interacting partners of the α7 nAChR in vivo.
CONCLUSION
We have successfully solubilized and affinity-isolated the α7 nAChR and associated proteins from murine brain tissue. Mass spectrometric analysis of in-gel, tryptically-digested peptides identified 55 proteins as members of the neuronal α7 nAChR interactome (Table 1). With the exception of tau, most of the proteins listed in Table 1 have not been previously identified as interaction partners of the α7 nAChR.92 To our knowledge, of the proteins listed in Table 1, only PKA,93, 94 PKC,94 and 14-3-3η93 have been shown previously to be associated with non-α7 nAChRs. The compiled list of proteins in the interaction network obtained from this initial study is not meant to be comprehensive, nor have we validated these identified proteins via orthogonal assays. Verification of the proteins that we identified as potential members of the α7 nAChR interactome will need to be performed via traditional biochemical methods, such as Western blotting and co-immunoprecipitations, before more concerted efforts are devoted to the detailed study of these interactions. Nevertheless, our work has laid a solid foundation for further exploration of the α7 nAChR interactome in both neuronal and non-neuronal tissues. Furthermore, the affinity purification with α-bgtx-conjugated beads together with the proteomic analysis procedure that we have developed could also be applied to the investigation of the interactomes of other receptors. The use of a pharmatope tag consisting of a short amino acid sequence to confer α-bgtx sensitivity to a target protein has been well described in cell culture95 and oocyte studies.96, 97 More specifically, such a tag may be inserted into a receptor for which suitable antibodies are not available and/or for which affinity-ligand beads are unavailable or impractical.98 Coupling pharmatope-tagging with our methodology of α-bgtx isolation in an α7 nAChR null background may facilitate the proteomic analyses of other functionally important membrane proteins.
Supplementary Material
Tables of proteins and peptides identified by nano-ESI-MS/MS that are exclusive to wild-type mouse brain tissue samples and which appear in at least two of the three data sets; Proteins identified by nano-ESI-MS/MS that are exclusive to wild-type mouse brain tissue samples, but which appear in only one data set; Proteins identified by nano-ESI-MS/MS in samples originating from both wild-type and α7 nAChR knockout mice; Sub-cellular location and function of proteins listed in Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENT
This work was supported by National Institutes of Health grants #GM32629 and DA021765. Some of the work presented here was done in partial fulfillment of the requirements for a Ph.D. degree (J.A.P.) from Brown University. Some of the instrumentation used in this study was made available through an NSF/EPSCoR award (#0554548) and by an award from the Rhode Island Research Alliance. We thank Dr. P. Caffery for critical reading of this manuscript, and Dr. James Clifton and Dr. T. Shen for their assistance and input on the theory and techniques of mass spectrometry.
References
- 1.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346(4):967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 2.Lindstrom J, Anand R, Gerzanich V, Peng X, Wang F, Wells G. Structure and function of neuronal nicotinic acetylcholine receptors. Prog Brain Res. 1996;109:125–137. doi: 10.1016/s0079-6123(08)62094-4. [DOI] [PubMed] [Google Scholar]
- 3.Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- 4.Moise L, Piserchio A, Basus VJ, Hawrot E. NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor. J Biol Chem. 2002;277(14):12406–12417. doi: 10.1074/jbc.M110320200. [DOI] [PubMed] [Google Scholar]
- 5.Berg DK, Conroy WG. Nicotinic alpha 7 receptors: synaptic options and downstream signaling in neurons. J Neurobiol. 2002;53(4):512–523. doi: 10.1002/neu.10116. [DOI] [PubMed] [Google Scholar]
- 6.Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochem Pharmacol. 2007 doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 7.Role LW, Berg DK. Nicotinic receptors in the development and modulation of CNS synapses. Neuron. 1996;16(6):1077–1085. doi: 10.1016/s0896-6273(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, Li JH, Yang H, Ulloa L, Al-Abed Y, Czura CJ, Tracey KJ. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. 2003;421(6921):384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 9.De Simone R, Ajmone-Cat MA, Carnevale D, Minghetti L. Activation of alpha7 nicotinic acetylcholine receptor by nicotine selectively up-regulates cyclooxygenase-2 and prostaglandin E2 in rat microglial cultures. J Neuroinflammation. 2005;2(1):4. doi: 10.1186/1742-2094-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shytle RD, Mori T, Townsend K, Vendrame M, Sun N, Zeng J, Ehrhart J, Silver AA, Sanberg PR, Tan J. Cholinergic modulation of microglial activation by alpha 7 nicotinic receptors. J Neurochem. 2004;89(2):337–343. doi: 10.1046/j.1471-4159.2004.02347.x. [DOI] [PubMed] [Google Scholar]
- 11.Zia S, Ndoye A, Lee TX, Webber RJ, Grando SA. Receptor-mediated inhibition of keratinocyte migration by nicotine involves modulations of calcium influx and intracellular concentration. J Pharmacol Exp Ther. 2000;293(3):973–981. [PubMed] [Google Scholar]
- 12.Wada T, Naito M, Kenmochi H, Tsuneki H, Sasaoka T. Chronic nicotine exposure enhances insulin-induced mitogenic signaling via up-regulation of alpha7 nicotinic receptors in isolated rat aortic smooth muscle cells. Endocrinology. 2007;148(2):790–799. doi: 10.1210/en.2006-0907. [DOI] [PubMed] [Google Scholar]
- 13.Sharma G, Vijayaraghavan S. Nicotinic cholinergic signaling in hippocampal astrocytes involves calcium-induced calcium release from intracellular stores. Proc Natl Acad Sci U S A. 2001;98(7):4148–4153. doi: 10.1073/pnas.071540198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maus AD, Pereira EF, Karachunski PI, Horton RM, Navaneetham D, Macklin K, Cortes WS, Albuquerque EX, Conti-Fine BM. Human and rodent bronchial epithelial cells express functional nicotinic acetylcholine receptors. Mol Pharmacol. 1998;54(5):779–788. doi: 10.1124/mol.54.5.779. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Pereira EF, Maus AD, Ostlie NS, Navaneetham D, Lei S, Albuquerque EX, Conti-Fine BM. Human bronchial epithelial and endothelial cells express alpha7 nicotinic acetylcholine receptors. Mol Pharmacol. 2001;60(6):1201–1209. doi: 10.1124/mol.60.6.1201. [DOI] [PubMed] [Google Scholar]
- 16.Benhammou K, Lee M, Strook M, Sullivan B, Logel J, Raschen K, Gotti C, Leonard S. [(3)H]Nicotine binding in peripheral blood cells of smokers is correlated with the number of cigarettes smoked per day. Neuropharmacology. 2000;39(13):2818–2829. doi: 10.1016/s0028-3908(00)00153-2. [DOI] [PubMed] [Google Scholar]
- 17.Toyabe S, Iiai T, Fukuda M, Kawamura T, Suzuki S, Uchiyama M, Abo T. Identification of nicotinic acetylcholine receptors on lymphocytes in the periphery as well as thymus in mice. Immunology. 1997;92(2):201–205. doi: 10.1046/j.1365-2567.1997.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baer K, Burli T, Huh KH, Wiesner A, Erb-Vogtli S, Gockeritz-Dujmovic D, Moransard M, Nishimune A, Rees MI, Henley JM, Fritschy JM, Fuhrer C. PICK1 interacts with alpha7 neuronal nicotinic acetylcholine receptors and controls their clustering. Mol Cell Neurosci. 2007;35(2):339–355. doi: 10.1016/j.mcn.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, Millar NS. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol Pharmacol. 2005;68(5):1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- 20.Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, Aiyar J. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor alpha7 subunit in mammalian cells. J Biol Chem. 2005;280(2):1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- 21.Dasgupta P, Rastogi S, Pillai S, Ordonez-Ercan D, Morris M, Haura E, Chellappan S. Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J Clin Invest. 2006;116(8):2208–2217. doi: 10.1172/JCI28164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoop RD, Yamada N, Berg DK. Cytoskeletal links of neuronal acetylcholine receptors containing alpha 7 subunits. J Neurosci. 2000;20(11):4021–4029. doi: 10.1523/JNEUROSCI.20-11-04021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshikawa J, Toya Y, Fujita T, Egawa M, Kawabe J, Umemura S, Ishikawa Y. Nicotinic acetylcholine receptor alpha 7 regulates cAMP signal within lipid rafts. Am J Physiol Cell Physiol. 2003;285(3):C567–C574. doi: 10.1152/ajpcell.00422.2002. [DOI] [PubMed] [Google Scholar]
- 24.Kihara T, Shimohama S, Sawada H, Honda K, Nakamizo T, Shibasaki H, Kume T, Akaike A. alpha 7 nicotinic receptor transduces signals to phosphatidylinositol 3-kinase to block A beta-amyloid-induced neurotoxicity. J Biol Chem. 2001;276(17):13541–13546. doi: 10.1074/jbc.M008035200. [DOI] [PubMed] [Google Scholar]
- 25.Yu XM, Hall ZW. A sequence in the main cytoplasmic loop of the alpha subunit is required for assembly of mouse muscle nicotinic acetylcholine receptor. Neuron. 1994;13(1):247–255. doi: 10.1016/0896-6273(94)90473-1. [DOI] [PubMed] [Google Scholar]
- 26.Bencherif M, Lukas RJ. Cytochalasin modulation of nicotinic cholinergic receptor expression and muscarinic receptor function in human TE671/RD cells: a possible functional role of the cytoskeleton. J Neurochem. 1993;61(3):852–864. doi: 10.1111/j.1471-4159.1993.tb03596.x. [DOI] [PubMed] [Google Scholar]
- 27.Colledge M, Froehner SC. Tyrosine phosphorylation of nicotinic acetylcholine receptor mediates Grb2 binding. J Neurosci. 1997;17(13):5038–5045. doi: 10.1523/JNEUROSCI.17-13-05038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Temburni MK, Blitzblau RC, Jacob MH. Receptor targeting and heterogeneity at interneuronal nicotinic cholinergic synapses in vivo. J Physiol. 2000;525(Pt 1):21–29. doi: 10.1111/j.1469-7793.2000.00021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams BM, Temburni MK, Levey MS, Bertrand S, Bertrand D, Jacob MH. The long internal loop of the alpha 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nat Neurosci. 1998;1(7):557–562. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- 30.Grant SG, Husi H. Proteomics of multiprotein complexes: answering fundamental questions in neuroscience. Trends Biotechnol. 2001;19(10 Suppl):S49–S54. doi: 10.1016/S0167-7799(01)01799-1. [DOI] [PubMed] [Google Scholar]
- 31.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17(7):676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 32.Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5(9):699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 33.Orr-Urtreger A, Goldner FM, Saeki M, Lorenzo I, Goldberg L, De Biasi M, Dani JA, Patrick JW, Beaudet AL. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J Neurosci. 1997;17(23):9165–9171. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnaiz-Cot JJ, Gonzalez JC, Sobrado M, Baldelli P, Carbone E, Gandia L, Garcia AG, Hernandez-Guijo JM. Allosteric modulation of alpha 7 nicotinic receptors selectively depolarizes hippocampal interneurons, enhancing spontaneous GABAergic transmission. Eur J Neurosci. 2008;27(5):1097–1110. doi: 10.1111/j.1460-9568.2008.06077.x. [DOI] [PubMed] [Google Scholar]
- 35.Feuerbach D, Lingenhoehl K, Olpe HR, Vassout A, Gentsch C, Chaperon F, Nozulak J, Enz A, Bilbe G, McAllister K, Hoyer D. The selective nicotinic acetylcholine receptor alpha7 agonist JN403 is active in animal models of cognition, sensory gating, epilepsy and pain. Neuropharmacology. 2008 doi: 10.1016/j.neuropharm.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 36.Fan JB, Ma J, Li XW, Zhang CS, Sun WW, He G, Gu NF, Feng GY, St Clair D, He L. Population-based and family-based association studies of an (AC)n dinucleotide repeat in alpha-7 nicotinic receptor subunit gene and schizophrenia. Schizophr Res. 2006;84(2–3):222–227. doi: 10.1016/j.schres.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Martin LF, Freedman R. Schizophrenia and the alpha7 Nicotinic Acetylcholine Receptor. Int Rev Neurobiol. 2007;78:225–246. doi: 10.1016/S0074-7742(06)78008-4. [DOI] [PubMed] [Google Scholar]
- 38.Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, Hegde V, Hundley R, Khwaja O, Kothare S, Luedke C, Nasir R, Poduri A, Prasad K, Raffalli P, Reinhard A, Smith SE, Sobeih M, Soul J, Stoler J, Takeoka M, Tan WH, Thakuria J, Wolff P, Yusupov R, Gusella JF, Daly MJ, Wu BL. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J Med Genet. 2008 doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray MA, Graham AJ, Lee M, Perry RH, Court JA, Perry EK. Neuronal nicotinic acetylcholine receptor subunits in autism: an immunohistochemical investigation in the thalamus. Neurobiol Dis. 2005;19(3):366–377. doi: 10.1016/j.nbd.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 40.Weiss S, Tzavara ET, Davis RJ, Nomikos GG, Michael McIntosh J, Giros B, Martres MP. Functional alterations of nicotinic neurotransmission in dopamine transporter knock-out mice. Neuropharmacology. 2007;52(7):1496–1508. doi: 10.1016/j.neuropharm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Weiss S, Nosten-Bertrand M, McIntosh JM, Giros B, Martres MP. Nicotine improves cognitive deficits of dopamine transporter knockout mice without long-term tolerance. Neuropsychopharmacology. 2007;32(12):2465–2478. doi: 10.1038/sj.npp.1301385. [DOI] [PubMed] [Google Scholar]
- 42.Paterson D, Nordberg A. Neuronal nicotinic receptors in the human brain. Prog Neurobiol. 2000;61(1):75–111. doi: 10.1016/s0301-0082(99)00045-3. [DOI] [PubMed] [Google Scholar]
- 43.Eng JK, Mccormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moser N, Mechawar N, Jones I, Gochberg-Sarver A, Orr-Urtreger A, Plomann M, Salas R, Molles B, Marubio L, Roth U, Maskos U, Winzer-Serhan U, Bourgeois JP, Le Sourd AM, De Biasi M, Schroder H, Lindstrom J, Maelicke A, Changeux JP, Wevers A. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J Neurochem. 2007;102(2):479–492. doi: 10.1111/j.1471-4159.2007.04498.x. [DOI] [PubMed] [Google Scholar]
- 46.Shelukhina IV, Kryukova EV, Skok MV, Lykhmus EY, Zhmak MN, Mordvintsev DY, Kasheverov IE, Tsetlin VI. Analysis of specificity of antibodies against synthetic fragments of different neuronal nicotinic acetylcholine receptor subunits. Biochemistry (Mosc) 2006;71(7):749–758. doi: 10.1134/s0006297906070078. [DOI] [PubMed] [Google Scholar]
- 47.Sterner JL, Johnston MV, Nicol GR, Ridge DP. Signal suppression in electrospray ionization Fourier transform mass spectrometry of multi-component samples. J Mass Spectrom. 2000;35(3):385–391. doi: 10.1002/(SICI)1096-9888(200003)35:3<385::AID-JMS947>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 48.Andrade EC, Krueger DD, Nairn AC. Recent advances in neuroproteomics. Curr Opin Mol Ther. 2007;9(3):270–281. [PMC free article] [PubMed] [Google Scholar]
- 49.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3(7):661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 50.Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem. 2004;279(2):987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 51.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J Neurochem. 2004;88(3):759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 52.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97 Suppl 1:16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 53.Barik J, Wonnacott S. Indirect modulation by alpha7 nicotinic acetylcholine receptors of noradrenaline release in rat hippocampal slices: interaction with glutamate and GABA systems and effect of nicotine withdrawal. Mol Pharmacol. 2006;69(2):618–628. doi: 10.1124/mol.105.018184. [DOI] [PubMed] [Google Scholar]
- 54.Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int Rev Cytol. 2005;245:245–325. doi: 10.1016/S0074-7696(05)45007-X. [DOI] [PubMed] [Google Scholar]
- 55.Mosevitsky MI, Capony JP, Skladchikova G, Novitskaya VA, Plekhanov A, Zakharov VV. The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physico-chemical properties. Biochimie. 1997;79(6):373–384. doi: 10.1016/s0300-9084(97)80032-6. [DOI] [PubMed] [Google Scholar]
- 56.Epand RM, Epand RF, Maekawa S. The arrangement of cholesterol in membranes and binding of NAP-22. Chem Phys Lipids. 2003;122(1–2):33–39. doi: 10.1016/s0009-3084(02)00176-7. [DOI] [PubMed] [Google Scholar]
- 57.Maekawa S, Taguchi K. Localization of the Cl(-)-ATPase activity on NAP-22 enriched membrane microdomain (raft) of rat brain. Neurosci Lett. 2004;362(2):158–161. doi: 10.1016/j.neulet.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 58.Grilli M, Raiteri L, Patti L, Parodi M, Robino F, Raiteri M, Marchi M. Modulation of the function of presynaptic alpha7 and non-alpha7 nicotinic receptors by the tryptophan metabolites, 5-hydroxyindole and kynurenate in mouse brain. Br J Pharmacol. 2006;149(6):724–732. doi: 10.1038/sj.bjp.0706914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pakkanen JS, Jokitalo E, Tuominen RK. Up-regulation of beta2 and alpha7 subunit containing nicotinic acetylcholine receptors in mouse striatum at cellular level. Eur J Neurosci. 2005;21(10):2681–2691. doi: 10.1111/j.1460-9568.2005.04105.x. [DOI] [PubMed] [Google Scholar]
- 60.Bertrand D, Galzi JL, Devillers-Thiery A, Bertrand S, Changeux JP. Mutations at two distinct sites within the channel domain M2 alter calcium permeability of neuronal alpha 7 nicotinic receptor. Proc Natl Acad Sci U S A. 1993;90(15):6971–6975. doi: 10.1073/pnas.90.15.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seguela P, Wadiche J, Dineley-Miller K, Dani JA, Patrick JW. Molecular cloning, functional properties, and distribution of rat brain alpha 7: a nicotinic cation channel highly permeable to calcium. J Neurosci. 1993;13(2):596–604. doi: 10.1523/JNEUROSCI.13-02-00596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Papke RL, He YJ, Millard WJ, Meyer EM. Characterization of the neuroprotective and toxic effects of alpha7 nicotinic receptor activation in PC12 cells. Brain Res. 1999;830(2):218–225. doi: 10.1016/s0006-8993(99)01372-4. [DOI] [PubMed] [Google Scholar]
- 63.Dajas-Bailador FA, Soliakov L, Wonnacott S. Nicotine activates the extracellular signal-regulated kinase 1/2 via the alpha7 nicotinic acetylcholine receptor and protein kinase A, in SH-SY5Y cells and hippocampal neurones. J Neurochem. 2002;80(3):520–530. doi: 10.1046/j.0022-3042.2001.00725.x. [DOI] [PubMed] [Google Scholar]
- 64.Quik M, Philie J, Choremis J. Modulation of alpha7 nicotinic receptor-mediated calcium influx by nicotinic agonists. Mol Pharmacol. 1997;51(3):499–506. [PubMed] [Google Scholar]
- 65.Ospina JA, Broide RS, Acevedo D, Robertson RT, Leslie FM. Calcium regulation of agonist binding to alpha7-type nicotinic acetylcholine receptors in adult and fetal rat hippocampus. J Neurochem. 1998;70(3):1061–1068. doi: 10.1046/j.1471-4159.1998.70031061.x. [DOI] [PubMed] [Google Scholar]
- 66.Funte LR, Haydon PG. Synaptic target contact enhances presynaptic calcium influx by activating cAMP-dependent protein kinase during synaptogenesis. Neuron. 1993;10(6):1069–1078. doi: 10.1016/0896-6273(93)90055-v. [DOI] [PubMed] [Google Scholar]
- 67.Wong WT, Wong RO. Rapid dendritic movements during synapse formation and rearrangement. Curr Opin Neurobiol. 2000;10(1):118–124. doi: 10.1016/s0959-4388(99)00059-8. [DOI] [PubMed] [Google Scholar]
- 68.Lee HS, Tomarev SI. Optimedin induces expression of N-cadherin and stimulates aggregation of NGF-stimulated PC12 cells. Exp Cell Res. 2007;313(1):98–108. doi: 10.1016/j.yexcr.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bomze HM, Bulsara KR, Iskandar BJ, Caroni P, Skene JH. Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat Neurosci. 2001;4(1):38–43. doi: 10.1038/82881. [DOI] [PubMed] [Google Scholar]
- 70.Furnish EJ, Zhou W, Cunningham CC, Kas JA, Schmidt CE. Gelsolin overexpression enhances neurite outgrowth in PC12 cells. FEBS Lett. 2001;508(2):282–286. doi: 10.1016/s0014-5793(01)03078-2. [DOI] [PubMed] [Google Scholar]
- 71.Laux T, Fukami K, Thelen M, Golub T, Frey D, Caroni P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149(7):1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ango F, Robbe D, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L. Homer-dependent cell surface expression of metabotropic glutamate receptor type 5 in neurons. Mol Cell Neurosci. 2002;20(2):323–329. doi: 10.1006/mcne.2002.1100. [DOI] [PubMed] [Google Scholar]
- 73.Liu HY, Nur EKA, Schachner M, Meiners S. Neurite guidance by the FnC repeat of human tenascin-C: neurite attraction vs. neurite retention. Eur J Neurosci. 2005;22(8):1863–1872. doi: 10.1111/j.1460-9568.2005.04383.x. [DOI] [PubMed] [Google Scholar]
- 74.Kiddie G, McLean D, Van Ooyen A, Graham B. Biologically plausible models of neurite outgrowth. Prog Brain Res. 2005;147:67–80. doi: 10.1016/S0079-6123(04)47006-X. [DOI] [PubMed] [Google Scholar]
- 75.Kraft R, Escobar MM, Narro ML, Kurtis JL, Efrat A, Barnard K, Restifo LL. Phenotypes of Drosophila brain neurons in primary culture reveal a role for fascin in neurite shape and trajectory. J Neurosci. 2006;26(34):8734–8747. doi: 10.1523/JNEUROSCI.2106-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pugh PC, Berg DK. Neuronal acetylcholine receptors that bind alpha-bungarotoxin mediate neurite retraction in a calcium-dependent manner. J Neurosci. 1994;14(2):889–896. doi: 10.1523/JNEUROSCI.14-02-00889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kater SB, Mills LR. Regulation of growth cone behavior by calcium. J Neurosci. 1991;11(4):891–899. doi: 10.1523/JNEUROSCI.11-04-00891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu WF, Guan ZZ, Nordberg A. Postnatal upregulation of alpha4 and alpha3 nicotinic receptor subunits in the brain of alpha7 nicotinic receptor-deficient mice. Neuroscience. 2007;146(4):1618–1628. doi: 10.1016/j.neuroscience.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 79.Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem. 2001;276(30):28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- 80.Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol. 2002;193(2):154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- 81.Miyaishi O, Kozaki K, Iida K, Isobe K, Hashizume Y, Saga S. Elevated expression of PDI family proteins during differentiation of mouse F9 teratocarcinoma cells. J Cell Biochem. 1998;68(4):436–445. doi: 10.1002/(sici)1097-4644(19980315)68:4<436::aid-jcb4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 82.Liu Y, Hogan BL. Differential gene expression in the distal tip endoderm of the embryonic mouse lung. Gene Expr Patterns. 2002;2(3–4):229–233. doi: 10.1016/s1567-133x(02)00057-1. [DOI] [PubMed] [Google Scholar]
- 83.Shan SW, Tang MK, Cai DQ, Chui YL, Chow PH, Grotewold L, Lee KK. Comparative proteomic analysis identifies protein disulfide isomerase and peroxiredoxin 1 as new players involved in embryonic interdigital cell death. Dev Dyn. 2005;233(2):266–281. doi: 10.1002/dvdy.20404. [DOI] [PubMed] [Google Scholar]
- 84.Neves SR, Ram PT, Iyengar R. G Protein Pathways. Science. 2002;296(5573):1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 85.Hansen RK, Christensen C, Korshunova I, Kriebel M, Burkarth N, Kiselyov VV, Olsen M, Ostergaard S, Holm A, Volkmer H, Walmod PS, Berezin V, Bock E. Identification of NCAM-binding peptides promoting neurite outgrowth via a heterotrimeric G-protein-coupled pathway. J Neurochem. 2007;103(4):1396–1407. doi: 10.1111/j.1471-4159.2007.04894.x. [DOI] [PubMed] [Google Scholar]
- 86.Dolphin AC. G protein modulation of voltage-gated calcium channels. Pharmacol Rev. 2003;55(4):607–627. doi: 10.1124/pr.55.4.3. [DOI] [PubMed] [Google Scholar]
- 87.Mirotznik RR, Zheng X, Stanley EF. G-Protein types involved in calcium channel inhibition at a presynaptic nerve terminal. J Neurosci. 2000;20(20):7614–7621. doi: 10.1523/JNEUROSCI.20-20-07614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gilman AG. G Proteins: Transducers of Receptor-Generated Signals. Annual Review of Biochemistry. 1987;56(1):615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 89.Sternweis PC, Robishaw JD. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J Biol Chem. 1984;259(22):13806–13813. [PubMed] [Google Scholar]
- 90.Skene JH. GAP-43 as a 'calmodulin sponge' and some implications for calcium signalling in axon terminals. Neurosci Res Suppl. 1990;13:S112–S125. doi: 10.1016/0921-8696(90)90040-a. [DOI] [PubMed] [Google Scholar]
- 91.Fischer H, Liu DM, Lee A, Harries JC, Adams DJ. Selective modulation of neuronal nicotinic acetylcholine receptor channel subunits by Go-protein subunits. J Neurosci. 2005;25(14):3571–3577. doi: 10.1523/JNEUROSCI.4971-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang HY, Li W, Benedetti NJ, Lee DH. Alpha 7 nicotinic acetylcholine receptors mediate beta-amyloid peptide-induced tau protein phosphorylation. J Biol Chem. 2003;278(34):31547–31553. doi: 10.1074/jbc.M212532200. [DOI] [PubMed] [Google Scholar]
- 93.Exley R, Moroni M, Sasdelli F, Houlihan LM, Lukas RJ, Sher E, Zwart R, Bermudez I. Chaperone protein 14-3-3 and protein kinase A increase the relative abundance of low agonist sensitivity human alpha 4 beta 2 nicotinic acetylcholine receptors in Xenopus oocytes. J Neurochem. 2006;98(3):876–885. doi: 10.1111/j.1471-4159.2006.03915.x. [DOI] [PubMed] [Google Scholar]
- 94.Pollock VV, Pastoor TE, Wecker L. Cyclic AMP-dependent protein kinase (PKA) phosphorylates Ser362 and 467 and protein kinase C phosphorylates Ser550 within the M3/M4 cytoplasmic domain of human nicotinic receptor alpha4 subunits. J Neurochem. 2007;103(2):456–466. doi: 10.1111/j.1471-4159.2007.04853.x. [DOI] [PubMed] [Google Scholar]
- 95.McCann CM, Bareyre FM, Lichtman JW, Sanes JR. Peptide tags for labeling membrane proteins in live cells with multiple fluorophores. Biotechniques. 2005;38(6):945–952. doi: 10.2144/05386IT02. [DOI] [PubMed] [Google Scholar]
- 96.Levandoski MM, Lin Y, Moise L, McLaughlin JT, Cooper E, Hawrot E. Chimeric analysis of a neuronal nicotinic acetylcholine receptor reveals amino acids conferring sensitivity to alpha-bungarotoxin. J Biol Chem. 1999;274(37):26113–26119. doi: 10.1074/jbc.274.37.26113. [DOI] [PubMed] [Google Scholar]
- 97.Sanders T, Hawrot E. A novel pharmatope tag inserted into the beta4 subunit confers allosteric modulation to neuronal nicotinic receptors. J Biol Chem. 2004;279(49):51460–51465. doi: 10.1074/jbc.M409533200. [DOI] [PubMed] [Google Scholar]
- 98.Moise L, Zeng H, Caffery P, Rogowski RS, Hawrot E. Structure and function of alpha-bungarotoxin. Journal of Toxicology-Toxin Reviews. 2002;21(3):293–317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables of proteins and peptides identified by nano-ESI-MS/MS that are exclusive to wild-type mouse brain tissue samples and which appear in at least two of the three data sets; Proteins identified by nano-ESI-MS/MS that are exclusive to wild-type mouse brain tissue samples, but which appear in only one data set; Proteins identified by nano-ESI-MS/MS in samples originating from both wild-type and α7 nAChR knockout mice; Sub-cellular location and function of proteins listed in Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.