Abstract
Advances in x-ray light sources and detectors have created opportunities for advancing our understanding of structure and structural dynamics for supramolecular assemblies in solution by combining x-ray scattering measurement with coordinate-based modeling methods. In this review the foundations for x-ray scattering are discussed and illustrated with selected examples demonstrating the ability to correlate solution x-ray scattering measurements to molecular structure, conformation, and dynamics. These approaches are anticipated to have a broad range of applications in natural and artificial photosynthesis by offering possibilities for structure resolution for dynamic supramolecular assemblies in solution that can not be fully addressed with crystallographic techniques, and for resolving fundamental mechanisms for solar energy conversion by mapping out structure in light-excited reaction states.
Keywords: x-ray scattering, photosynthesis, artificial photosynthesis, solution structure, supramolecular chemistry, molecular dynamics, structure-function
Introduction
Key design features of natural photosynthesis are the hierarchal, modular architectures with module-specific protein host-cofactor guest chemistries. This partitioning of photochemical function allows the individual tasks of solar energy conversion such as light-harvesting, primary charge separation, charge accumulation, and terminal water-splitting reductive chemistries to be optimized within individual functional modules, which can then be integrated, regulated, and repaired for overall optimal solar energy conversion throughput. Advances in supramolecular chemistry are allowing synthetic molecular devices to be made that mimic aspects of the hierarchal, modular designs found in biology. (Gust et al. 2001; Wasielewski 2006) Central challenges in solar energy research lie in deciphering the fundamental structural bases for photosynthetic function in biological and biomimetic modular architectures.
Three dimensional structure determinations of proteins are generally obtained through x-ray crystallography or NMR techniques. While crystallography provides the most detailed and accurate measurement of molecular structure, structural information is necessarily confined to the crystalline state. Conformational dynamics and preferred conformations for proteins in crystals can differ from those in non-crystalline states. The differences in conformational preference between liquid and solid state are particularly critical for problems concerning protein-protein interactions. NMR approaches are employed for determination of biomolecular structures in solution, but as discussed below structural determination can frequently be limited by the number of atom pair distance constraints resolved by NMR measurements.
X-ray scattering approaches offer additional opportunities to resolve links between the structural hierarchies and function in natural and artificial photosynthesis that are complementary to crystallographic and NMR measurements. Advantages of scattering technique for structure characterization are that they provide tools for measuring atomic pair distances for molecules in solution and other non-crystalline media that are directly relevant to photosynthetic function, and with a resolution range that extends from single bond distances to the submicron range. Further, developments in synchrotron and pulsed x-ray facilities are offering x-ray light sources of exceptional brilliance, energy tunability, and stability(Hellemans 1997; Riekel et al. 1996; Winick 1998). These extended capabilities include opportunities to record scattering patterns to high spatial resolution (Fischetti et al. 2004; Makowski et al. 2008; O’Donnell et al. 2007; Zuo et al. 2006), with element-specific sensitivity (anomalous scattering), and with ultrafast time resolution using laser pump x-ray probe scattering methods (Cammarata et al. 2008; Ihee et al. 2005; Plech et al. 2004).
An inherent limitation in scattering is that the measurement provides information only on the distribution of atom pair distances but does not directly provide information on three dimensional structure. Emerging techniques are demonstrating opportunities to achieve solution-state biomolecular structure refinement using scattering data as a fitting constraint in conjunction with model-based conformational searches (Bernadó et al. 2007; Kojima et al. 2004; Petoukhov et al. 2002; Petoukhov and Svergun 2005), and in combination with crystallographic data (Putnam et al. 2007), and NMR distance constraints (Gabel et al. 2006; Grishaev et al. 2005a; Schwieters and Clore 2006; Zuo et al. 2008). Used in this way, the x-ray scattering measurement when combined with complementary structural information and coordinate-based modeling provides a method to achieve atomic-scale insight into the structure and conformational dynamics of complex supramolecular architectures in solution. In this educational review we provide examples demonstrating opportunities for photosynthetic supramolecular structure resolution using coordinate-based analyses of solution x-ray scattering data.
Methods
The X-ray scattering experiment
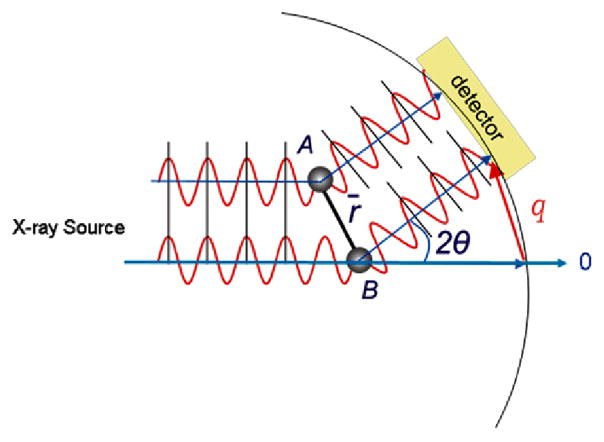
An outline of the x-ray scattering experiment is shown in Figure 1. A solution sample in capillary flow cell is placed in the path of a collimated or long focused monochromatic X-ray beam from a synchrotron light source that typically has a cross sectional dimensions of 0.3 mm × 0.5 mm or less (Seifert et al. 2000). The transmitted beam is blocked by a beam stop, while the 2-dimensional scattering pattern is measured by an area detector. For solution samples the isotropic 2-dimensional scattering image is radially averaged as a function of the scattering angle 2Θ with respect to the transmitted beam to produce a 1-dimensional reciprocal space scattering pattern of scattered X-ray intensity plotted as a function of angle. Typically thin-walled capillaries with X-ray pathlengths of 1.0 mm to 1.5 mm are used. The pathlength combined with the X-ray cross-section results in a sample irradiated volume on the order of 0.25 μl or less. At an undulator beam line at the Advanced Photon Source (Argonne National Laboratory), we find that high quality scattering patterns can be recorded for photosynthetic samples with data acquisition times that range from a few seconds to minutes (Svensson et al. 2004; Tiede et al. 2002; Zuo et al. 2006), depending upon the sample concentration and the spatial resolution of the measurement as discussed below. In addition, we have found that radiation damage can be avoided by flowing samples at a rate of 50 μl/min. Hence, high quality synchrotron X-ray scattering patterns can typically be recorded for samples with a total volume of 0.1 ml or less, and with protein or photosynthetic complex concentrations in the 0.01 mM to 10 mM range. More in-depth descriptions of x-ray scattering experiments, data processing, and analyses can be found in several excellent recent reviews (Koch 2006; Lipfert and Doniach 2007; Putnam et al. 2007; Tsuruta and Irving 2008).
Figure 1.
Schematic diagram of the x-ray scattering experiment. A synchrotron light source is used to provide a narrow collimated or long-focused beam targeted on a solution sample. A 2-dimensional scattering image centered on the incident beam direction is measured by an area detector, and the scattered x-ray intensity is azimuthally averaged as a function of the scattering vector, q, and the scattering angle 2Θ to yield the scattered intensity pattern as a function of q.
Atomic coordinate-based X-ray scattering analysis
Fundamental aspects of x-ray scattering can be understood by considering X-ray scattering for a hypothetical diatomic molecule with atomic spacing, r, shown in Figure 2. As a result of the interactions between the oscillating electric fields in the X-rays and electrons surrounding each atom, a small fraction of the impinging X-ray intensity is scattered coherently. The magnitude and angular dependence of this scattering are described by the atomic scattering factors that are in turn dependent upon the atomic number and ionization state of each atom (Warren 1990). Coherent scattering changes only the direction of the X-rays, not the phase or energy. However, the pathlengths taken by the X-rays scattered from each atom to arrive at a detector positioned at an angle 2θ with respect to the transmitted beam will differ, as illustrated in Figure 2. As a result, the combined X-rays scattered from each atom will vary between constructive (in-phase) and destructive (out-of-phase) interference as a function of the measured scattering angle, usually defined in terms of the vector q= (4π/λ)sinθ, where λ is the X-ray wavelength and θ is the half scattering angle. The function describing the pair-wise interference from N atoms in a molecule is (Svergun et al. 1995):
| (Eq. 1) |
where Aj is the atomic scattering amplitude for the jth atom, rj,k is the distance between the jth and kth atoms. Eq. 1 has the format of a Fourier transform indicating that information on inter-atomic distance, r, is converted to a frequency pattern in scattered X-ray intensity when measured as a function of the scattering vector, q, often referred to as the reciprocal space parameter where q has the units of inverse distance. For randomly oriented molecules in solution, Eq. 1 must be averaged over all orientations of the vectors q and r, and the atomic scattering amplitudes must include methods to account for the electron density background of the solvent and the solvent excluded volumes occupied by each of the atoms (Svergun et al. 1995; Zhang and Friesner 1998). Programs CRYSOL (Svergun et al. 1995) and solX (Zhang and Friesner 1998; Zuo et al. 2006) have been developed for calculation of solution scattering patterns based on coordinate molecules and are available on request.
Figure 2.
Scattering paths for X-rays striking a pair of atoms, A and B, separated by a distance, r, and detected at a scattering angle 2θ with respect to the direction of the incident beam.
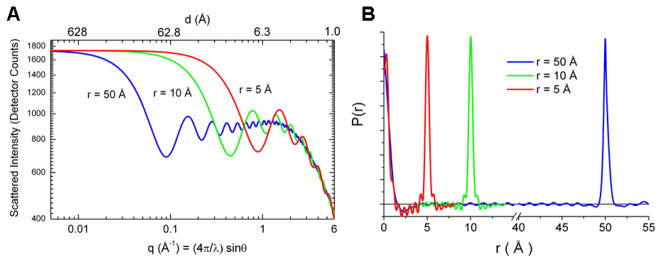
Molecular scattering patterns can be understood to arise from the sum of scattering from all pairs of atoms. Figure 3A illustrates how the reciprocal space solution scattering patterns change for individual atom pairs with inter-atomic spacings of 5 Å, 10 Å, and 50 Å. Figure 3B shows the corresponding inverse Fourier transforms for each or the scattering curves to produce pair distribution functions, PDF, (Putnam et al. 2007; Semenyuk and Svergun 1991) that reconstruct the originating real-space atom pair distances. Corresponding scattering and PDF patterns for multi-atom molecules would sum traces for all of the composite individual atom pairs.
Figure 3.
Scattering patterns calculated for atom pairs having different interatomic distances. Part A shows the reciprocal space scattering patterns and Part B shows the corresponding PDF functions. The curves in red, green, and blue correspond to atom pairs with separations of 5 Å, 10 Å, and 50 Å, respectively.
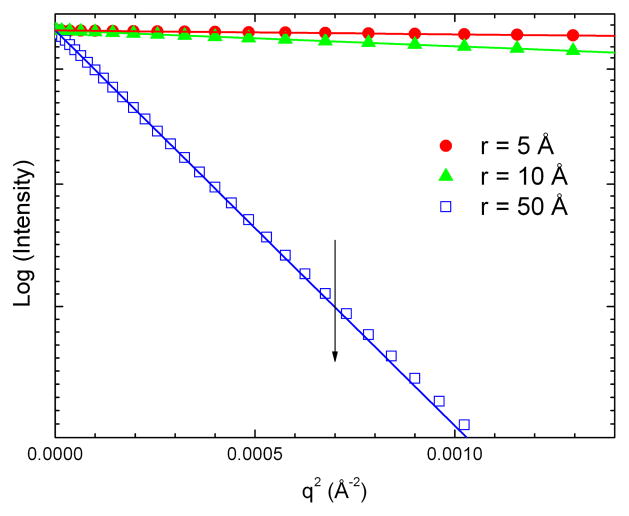
The individual atom-pair scattering patterns demonstrate the angle dependent spatial resolution contained in a scattering measurement. At small angle the pathlengths for scattering from each atom converge, causing interference and spatial resolution (d = 2π/q) to be lost, and the amplitude of the scattered X-rays asymptotically approaches the value at zero scattering angle, I(0). The scattered intensity at zero angle is proportional to the concentration weighted square of the electron density contributed by all of the atoms in the complex: I(0)= conc*V2 (ρ – ρo)2 where V, the total solvent excluded volume for the molecule, ρ, the average electron density of the molecule, and ρo, the average electron density of the solvent (Guinier and Fournet 1955) Measurement of I(0) for a molecule can be used to determine the molecular weight (Lipfert and Doniach 2007). In the small angle region, the asymptotic approach of scattered intensity to I(0) follows the Guinier relationship(Guinier and Fournet 1955; Lipfert and Doniach 2007; Putnam et al. 2007): , where Rg is the radius of gyration corresponding to the mean electron density-weighted atomic distance from the center of mass. In this region, a plot of the logarithm of the scattered intensity versus q2 has a linear format as shown in Figure 4, illustrated by Guinier plots for the calculated diatomic scattering curves. The Guinier plots have slopes corresponding to the radii of gyration, in this case 2.5 Å, 5 Å, and 25 Å, respectively, and a common I(0) intercept. In photosynthesis research, examples of the use of measurements of I(0) and Rg include the determination of the aggregation state of recombinantly expressed manganese stabilizing protein from PSII (Svensson et al. 2002; Svensson et al. 2004), characterization of the aggregation state of reaction centers in crystallization mixtures (Marone et al. 1998), and determine the stoichiometry of cytochrome c2 docking in electron transfer complexes with the bacterial reaction center (Tiede et al. 2000).
Figure 4.
Small angle Guinier plots for the scattering patterns calculated for atom pairs having different inter-atomic distances. The symbols re-plot the calculated patterns shown in Figure 3, with the atom pair spacing of 5 Å ( ), 10 Å (
), 10 Å ( ), and 50 Å (
), and 50 Å ( ). The solid lines are fits using the Guinier equation described in the text. The Guinier relationship holds under the condition that q is small compared to the dimension of the molecule. Figure 3 shows that this condition is achieved in different q-ranges depending on the dimensions of the molecule. A rule of thumb suggests that the Guinier condition will be met when q < 1/1.5*Rg. The vertical line marks this position for atom pair with a 50 Å separation. The data below this position are fit by the Guinier approximation. Deviations are seen at higher q.
). The solid lines are fits using the Guinier equation described in the text. The Guinier relationship holds under the condition that q is small compared to the dimension of the molecule. Figure 3 shows that this condition is achieved in different q-ranges depending on the dimensions of the molecule. A rule of thumb suggests that the Guinier condition will be met when q < 1/1.5*Rg. The vertical line marks this position for atom pair with a 50 Å separation. The data below this position are fit by the Guinier approximation. Deviations are seen at higher q.
At high angles an oscillatory interference pattern can be seen to develop in the model atom pair scattering shown in Figure 3A. The onset of the oscillatory interference and the corresponding frequencies provide a precise measure of interatomic spacings that can be obtained by Fourier transform. The high angle scattering features are of interest since they provide an atomic scale measurement of molecular structure that can be carried out in solution. However, the ability to utilize high-resolution scattering data for structural analysis depends on a combination of factors, including the size, complexity, and dynamics of the molecular system. In the following sections, we will give examples of scattering characteristics for molecular systems that demonstrate how characteristics of hierarchical molecular structure are reflected in wide angle X-ray scattering data, and describe progress and prospects for using scattering data for solution structure resolution.
Small Molecule Scattering: A Transition Metal Complex Example
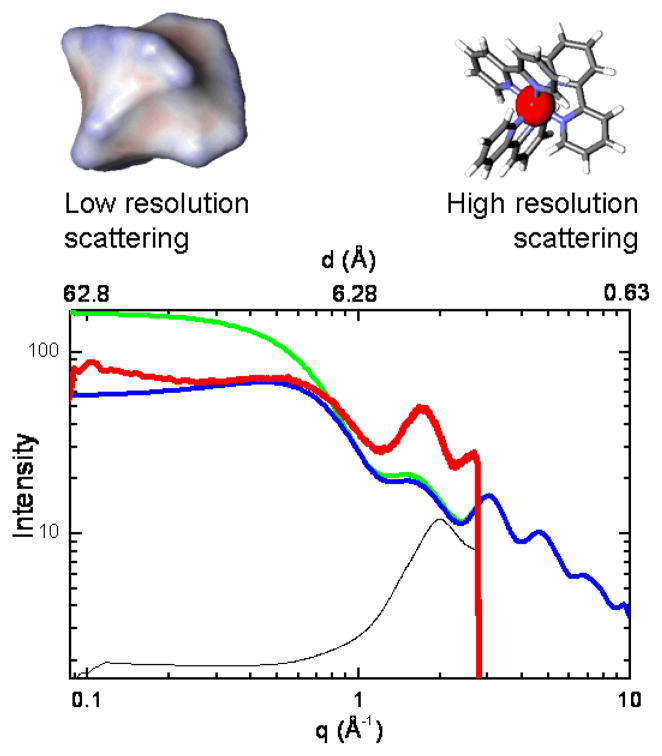
Features of atomic coordinate-based scattering for a small molecule transition metal complex can be illustrated by comparing experimental and calculated x-ray scattering for an aqueous solution of ruthenium-tris(2,2′-bipyridyl) dichloride, Ru(bpy)32+, in Figure 5. Ru(bpy)32+ is a light-absorbing molecule used widely as a photosensitizer in dye-sensitized solar cells and artificial photosynthesis. Calculated scattering patterns are shown using two different sets of atomic volumes. The trace in blue shows a calculation using the atomic volumes use typically used for calculation of x-ray scattering for proteins in aqueous solution (Fraser et al. 1978; Svergun et al. 1995), while the trace in green shows a calculation using volumes that we have found empirically to be useful for simulation of scattering for a variety of molecules in organic solvents (Lee et al. 2008; O’Donnell et al. 2007; Tiede et al. 2004). The atomic volumes used for molecules in non-aqueous solvents have volumes for aromatic carbon atoms reduced by 25% from the values used for aqueous solvent, and expansion of the volumes around polar and charged groups, analogous to the atomic volumes found in the inner, anhydrous core of proteins (Pontius et al. 1996). The experimental scattering pattern for an aqueous solution of 10 mM Ru(bpy)32+ containing 0.1 M KH2PO4 is shown by the red trace in Figure 5. The q-range measured in this experiment was limited by the dimensions of the detector (Mar CCD 165) and the single fixed distance used between the sample and the detector. This causes the sudden truncation of scattering data at q~ 2.8 Å−1. The small angle limit is determined by the size and position of the X-ray beam stop.
Figure 5.
Comparison of calculated and experimental x-ray scattering for tris(bipyridine)ruthenium(II) dichloride in solution. Solution scattering patterns for this compound were calculated in two ways. First using atomic van der Waals volumes (blue curve) and second with the atomic volumes for the aromatic carbon atoms in the bipyridyl ligands reduced by 25% (green curve) in order to illustrate the effect of the atomic solvent excluded volume on scattering, as described in the text. Experimental solution scattering for the Ru(II)tris(bipyridyl) compound was measured as the difference (red curve) between scattering for a 10 mM Ru(II)tris(bipyridyl)Cl2 solution in 0.1 M K2HPO4 and the 0.1 M K2HPO4 buffer solution alone. The scattering for the 0.1 M K2HPO4 buffer solution is shown in lower trace (black line), reduced in scale by a factor of 340 for illustration. Note that the residual solvent layer peak in the difference pattern has a peak position slightly different than that measured for bulk water, and is suggestive of scattering originating from a hydration layer with slightly expanded volume. The insets at the top of the figure illustrate the low resolution size shape and net electron density detected in the small angle scattering region on the left, and the interference pattern arising form coordinate-based structure at higher angle on the right.
A comparison of the two calculated curves shows that adjustments in the atomic volumes have the greatest effect in the small angle region where the loss of spatial resolution makes the scattering insensitive to the details of atomic structure, but is primarily sensitive to the size, shape, and the average electron density contrast of the solute molecule with the solvent. A characteristic feature of the experimental scattering pattern is the bend, or broad peak, seen in the small angle region (q ~ 0.7 Å−1). Maximum of this type in small angle scattering can arise from repulsive interactions that establish a minimum spacing between charged molecules in solution (Ducruix et al. 1996; Tiede et al. 2002). However, inter-molecular electrostatic interactions and their effects on scattering patterns are modulated by changing solution ionic strength and molecular concentration. The Ru(bpy)32+ scattering was found to be insensitive to these changes, indicating that the peak in the small angle scattering is a property of intra-molecular scattering.
The calculated scattering patters show that low angle peaks can be understood to arise from a partial electron density match between the solvent and the Ru(bpy) 32+. The reduction of atomic volumes for the aromatic carbon atoms causes all parts of the Ru(bpy) 32+ molecule to have a higher electron density compared to the solvent water, the I(0) calculated for the molecule increases, and the calculated scattering pattern follows a straightforward Guinier relationship in the small angle region. With the standard atomic volumes, the bipyridyl groups have a lower contrast with the water solvent, and amplitude of the small angle scattering, I(0), is reduced. Both curves converge to the same pattern at high angle, exhibiting an oscillatory pattern that is the reciprocal space representation of the sum of the atom pair distances associated with the bipyridyl ligands. Measurement of the interference pattern can be extended to higher angle either by changing the X-ray wavelength or by re-positioning the area detector subtend a larger portion of the high angle scattering. Because of the fixed dimensions of the beam stop and detector, the shift to record high angle scattering data will occur with loss of the small angle scattering data. Recording of scattering patterns across an extended q-range requires multiple measurements with different data acquisition geometries. In cases where the scattering patterns have been measured to q ≥ 6 Å−1, corresponding to a spatial resolution of better than 1 Å, PDF analysis allows the individual metal-ligand atom pair distances to be resolved, and structure of the transition metal complex to be determined in solution (Megyes et al. 2008; O’Donnell et al. 2007; Vaughan et al. 1950).
The scattering calculations for Ru(bpy) 32+ illustrate that atomic coordinate structure and the solvent excluded volumes make distinguishable, angle-dependent contributions to solution scattering. In addition, the experimental data shows a peak near q=1.8 Å−1 that falls within the solvent envelope. The experimental Ru(bpy) 32+ scattering pattern is measured as the difference between scatting for the Ru(bpy) 32+ solution and the aqueous buffer background (O’Donnell et al. 2007). The normalized subtraction of the buffer removes contributions of bulk solvent from the Ru(bpy) 32+ solution scattering. The residual peak has a position and shape different than the bulk solvent, and is suggestive of structure within a solvation layer associated with Ru(bpy)32+ or the accompanying counter ions. This kind of solvation layer structure can often be resolved in transition metal complex (Megyes et al. 2008; Megyes et al. 2005) and supramolecular (Lee et al. 2006; O’Donnell et al. 2007) scattering.
Macromolecule Scattering: A Protein Example
Wide-angle x-ray scattering patterns for proteins can be expected to be complicated by the much larger ensemble of atom pair distance correlations than is seen in photosynthetic model compounds. Scattering for a protein can generally be resolved into three regions. As discussed above, scattering in the small angle Guinier region provides information on the size, shape, and organization of domains in a multimeric protein (Koch 2006; Lipfert and Doniach 2007; Petoukhov and Svergun 2005; Putnam et al. 2007). Scattering in the q-region with a spatial resolution of 20 Å to 6 Å (0.2 Å−1 < q < 1.0 Å−1) provides a measure of the tertiary or protein fold structure, while scattering with a spatial resolution of 6 Å to 2 Å (1.0 Å−1 < q < 3.0 Å−1) provides a measure of secondary protein structure as reflected by characteristic hydrogen bonding and near-neighbor repeat distances in α-helix and β-sheet structures. Numerous groups have noted these scattering features and the fact that they provide a fingerprint for protein domain structure (Fedorov and Denesyuk 1978; Hirai et al. 2004; Makowski et al. 2008; Svergun et al. 2001; Tiede et al. 2002). For example, we have used x-ray scattering in combination with FTIR measurements to identify the β-sheet domain structure of the isolated manganese stabilizing protein from photosystem II, and demonstrated the consistency of the structure within crystalline photosystem II and in solution (Svensson et al. 2002; Svensson et al. 2004).
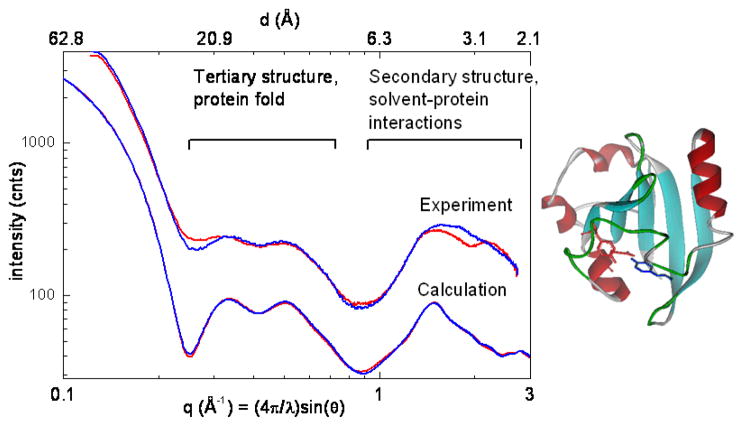
An example of x-ray scattering for a protein is illustrated in Figure 6 showing a comparison of experimental (in solution; top) and calculated (from crystals; bottom) scattering patterns for the photoactive yellow protein, PYP (Philip et al. 2008). Scattering patterns are shown measured for PYP samples in the dark (blue lines; also see (Kim et al. 2004)) and during continuous blue light illumination during a 10 second scattering measurement (red lines D. M. Tiede and W.D. Hoff, unpublished). The experimental light and dark scattering patterns show readily reversible changes (see e.g. at q = 0.25 Å-1 and q = 1.8) reflecting significant changes in secondary and tertiary protein structures. Comparable light-induced solution scattering results were also published previously with a lower 6 Å spatial resolution (Kamikubo et al. 2007).
Figure 6.
Comparison of experimental and calculated solution scattering patterns for the photochemical yellow protein (PYP). The top two traces show experimental traces for the wild-type PYP from Halorhodospira halophila (10 mg/ml) in Tris buffer (pH 7.5) (Philip et al. 2008) measured in the dark (blue line) and while illuminated (red line) with light from a 75 W xenon arc lamp filter through a broad bandpass 460 nm glass filter. The bottom two traces show scattering calculations from x-ray crystal structures 2PHY (Borgstahl et al. 1995) (blue line) and 2PYP (Genick et al. 1997) (red line) from the Protein Data Bank (Bernstein et al. 1977) that are dark adapted and laser-excited photo-stationary state I2, respectively.
The calculated scattering patterns used crystal coordinates obtained for the dark adapted crystal (Borgstahl et al. 1995) and for crystals in which the photo-stationary state (50% population) was produced on the milliseconds time-scale following laser excitation (Genick et al. 1997). Calculations from both crystal structures produce nearly identical scattering patterns, indicating that the light-induced structural changes are much smaller in crystals than in solution. The calculated scattering patterns show a pair of peaks in the protein tertiary structure region that correspond nicely to those observed for the dark-adapted solution sample, although the solution experiment shows “flatter” features possibly reflective of a dynamic conformational ensemble as described below. In the protein secondary region scattering features measured in experiment with the dark-adapted sample also align with features calculated from crystal structures, but the experiment shows a broader peak, particularly near the 3.1 Å spatial resolution position. This may be indicative of the presence of a solvation layer that is measured in experiment but is absent in the present model calculation, akin to the interactions observed in the Ru(bpy)32+ described above.
The light-generated state in solution (red line) produces changes throughout the PYP scattering pattern, including small angle, Rg region, a substantial flattening of the tertiary scattering peak, and changes in the tertiary protein structure scattering peak (see also (Kamikubo et al. 2007)), and in the secondary structure and protein-solvent scattering peak. These multiple length-scale changes are indicative of the light-induced “protein-quake” that propagates throughout the protein (Xie et al. 2001). The scattering data are a direct measure of the amplitude of this quake and demonstrate that the quake is much larger in solution than in crystals (small differences are seen between scattering patterns calculated for the light and dark crystal structures, but these differences are not easily seen with the scale of the plots in Figure 6), and possibly correlates with the altered photocycle function observed in crystals (Yeremenko et al. 2006). Laser-initiated, time-resolved wide-angle solution scattering has been extended to the ultrafast timescale using synchrotron-based pump-probe techniques allowing the structural cascade initiated byphoto-deligation of CO to be followed for hemoglobin in solution, and quantitatively compared to crystal coordinated data (Cammarata et al. 2008). Time-resolved scattering studies spanning the ultrafast to seconds time scales have clear applications for resolving the connections between light-induced structural dynamics and photosynthetic function.
Modeling Solution X-ray Scattering Based on Molecular Dynamics Simulations
The determination of the structure and dynamics of macromolecules in non-crystalline media is critical for understanding in-situ function. In solution, macromolecules exist in a number of different conformational states governed by a typically complex potential energy surface. This ensemble of states is in contrast to the picture of the single, rigid structure produced by x-ray crystallography. One of the strengths of the x-ray scattering technique is that it provides a direct, all-atom measure of the configurational ensemble present in solution that can be quantitatively compared to coordinate models of the static crystal structure and dynamics simulations of the ensemble. Multiple computational methods are available for generating molecular conformations.(Christen and van Gunsteren 2008; Howard and Kollman 1988; Tama and Brooks 2006; Vengadesan and Gautham 2005) Of these, when dynamic evolution of molecular motions is of interest, molecular dynamics is the method of choice. Molecular dynamics (MD) is capable of producing weighted ensembles with accompanying dynamic information, provided that empirical force fields and simulation times are adjusted to representatively sample thermally accessible conformational states. However, the validation of MD simulation results has been a long-standing problem.(van Gunsteren et al. 2006)
Several studies have demonstrated the ability to use experimental X-ray scattering as a discriminating benchmark for evaluating the accuracy of molecular simulation to represent real world configurational ensembles. For example, small angle scattering data parameters have been tested against those calculated from the simulated ensembles produced byMD simulation (Kim et al. 2008; Sivaramakrishnan et al. 2008; Vigil et al. 2001; von Ossowski et al. 2005) and other configurational search methods (Bernadó et al. 2007; Petoukhov et al. 2002; Petoukhov and Svergun 2005). The sensitivity of scattering data to conformations contained within the simulated ensembles have led to the development of structural refinement algorithms that use small angle scattering data in restrained MD simulations (Kojima et al. 2008; Kojima et al. 2004). The experimental evaluation of MD simulations have been further extended by comparing simulated ensembles with wide-angle scattering features for DNA (Zuo et al. 2006), a transition metal complex (Megyes et al. 2008), and light-harvesting porphyrin supramoleuclar assembly (Mardis et al. 2009). The extension of the combined MD and x-ray scattering analyses to wide angle permits a higher resolution, atomic-scale comparison between experimental and simulated ensembles.
As an example of a solution state conformational ensemble characterization by combined x-ray scattering and MD analyses, we summarize here the results for the light-harvesting, hexameric, diphenylethyne-linked porphyrin array in solution (Mardis et al. 2009). This specific example will be used to illustrate the synergies that exist between MD simulation and solution-phase x-ray scattering.
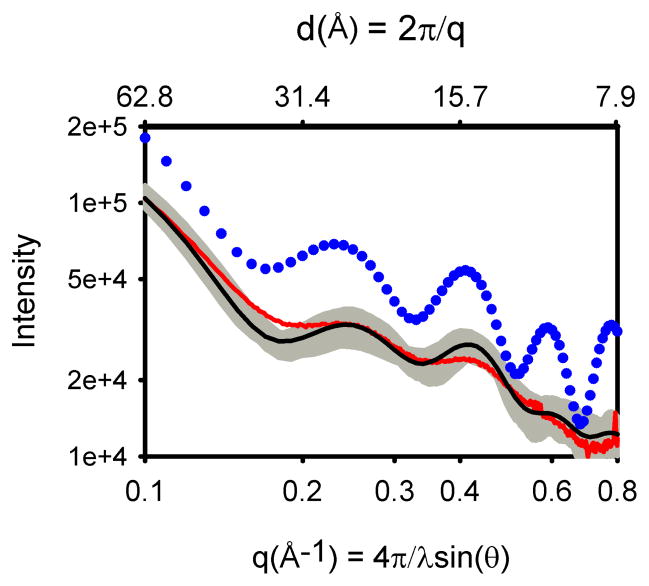
X-ray scattering measured for the porphyrin array in dilute toluene solutions at room temperature were found to exhibit broadening of scattering features arising from a distribution of the solution conformers (Tiede et al. 2004). An example is shown in Figure 7 that compares part of an experimentally measured scattering pattern (red trace) to a scattering pattern (blue dots) calculated from a single energy minimized structure (Figure 8A and B). The portion of the scattering pattern shown in Figure 7 covers the 0.1 Å−1 < q < 1.0 Å−1 region, and probes structure across the 60 Å to 6 Å spatial resolution range (d = 2π/q). This distance range provides a direct measure of the hexameric ring conformation. While a correspondence is seen between the number and approximate positions of peaks in the experimental and calculated patterns, the amplitude of the oscillations measured in experiment is markedly damped compared to that seen in patterns calculated for single supramolecular conformers. In addition, slight differences in the oscillatory peak patterns can be seen between the patterns calculated for the energy-minimized structure and experiment. This has been shown to correspond to a 1.4 Å compression of the overall dimension of the hexameric ring measured in experiment compared to the energy-minimized structure (Tiede et al. 2004). Observation that small distortions in the porphyrin array conformers produce significant shifts in calculated inference peak positions and amplitudes in scattering patterns suggests that the dampening of experimental scattering features could arise from a distribution of porphyrin array conformers in solution, and that dampening in experimental scattering data provides a monitor of the magnitude of this configurational dispersion.
Figure 7.
Experimental and calculated scattering patterns for the hexameric, diphenylethyne-linked porphyrin array composed of a cyclic array of alternating zinc and free-base porphyrins. Experimental data (red) are compared with the calculated scattering pattern for an energy-minimized structure (blue dots) and the MD ensemble-average (black). The scattering patterns calculated for individual snapshots taken every 1 ps during the production phase of the simulation are plotted in grey.
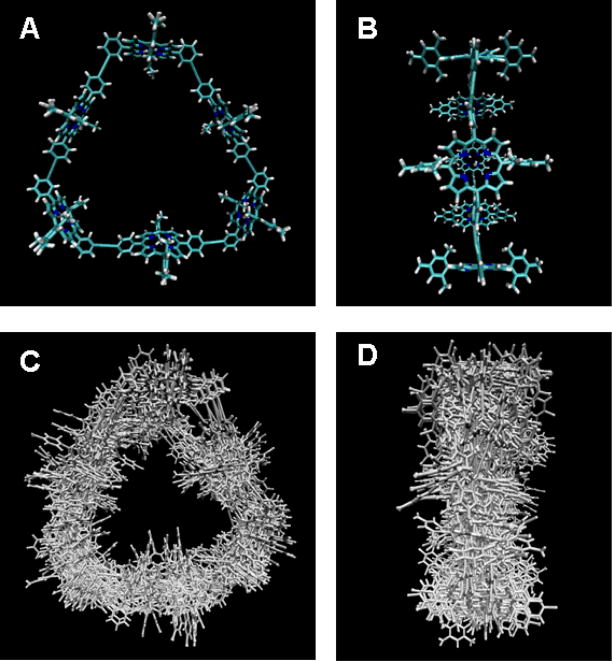
Figure 8.
Coordinate models for a porphyrin hexamer cyclic array. Parts A and B show front and side views, respectively, of the energy minimized conformer. Parts C and D show front and side views of a superposition of conformers taken from a thermally equilibrated 2 ns MD simulation of the porphyrin array. Note that the porphyrin rings tip, the overall size of the array expands and contracts, and the diphenyl-linkers exhibit large motions. This conformer motion results in the peak broadening and dampening seen in eperimental scattering data (Figure 7).
Molecular dynamics simulations were used to create ensembles of porphyrin array conformer using atomic force fields based on the CHARMM 22 force field (Mardis et al. 2009). Figure 8(C and D) shows representatives of an ensemble of 1700 conformers populated during a 2.05 ns constant pressure simulation in a bath of explicit solvent toluene molecules. The scattering profile for each member of the ensemble is shown plotted in Figure 7 using grey lines and the overall ensemble scattering is obtained as the average of the individual scattering patterns (black line). The ensemble average simulates a scattering pattern that can be experiment. Compared to scattering calculated from a single energy-minimized single conformer, the scattering pattern calculated from the MD ensemble can be seen to provide a better model for the experimental scattering data, notably by a close matching of the position of the first oscillatory peak and by the strong dampening of oscillatory features, particularly for q values above 0.5 Å−1 (Mardis et al. 2009).
The thermal fluctuations present in solution result in distance-dependent structural fluctuations that are directly detected in an x-ray scattering experiment by q-dependent broadening of x-ray scattering features. A superposition of these structural fluctuations in the simulated ensemble is shown in Figure 8. These conformations are obviously much less ordered than the energy minimized structure shown in Figure 8. Their corresponding scattering, shown as a grey line in Figure 7, form a shaded area that is the reciprocal space representation of distance-dependent atomic pair disorder simulated in solution. Careful investigation of this ensemble shows large amplitude motions that include porphyrin ring “tipping” around the porphyrin linkage axes and extended hexameric porphyrin array “breathing” motions involving torsional distortions collectively distributed along porphyrin and diphenylethyne groups. These motions result in dampening of the scattering profile.
While the overall agreement between MD and experiment is encouraging, mismatches in the magnitudes of experimental and simulated dampening, particularly in the insufficient dampening of the first two oscillatory peaks in the 0.2 Å−1 < q < 0.5 Å−1 region, show that large amplitude, hexamer array breathing-type motions are significantly under-represented in the simulated ensembles. Increases in the simulation temperature and time were found not to substantially shift the simulation ensemble. These shortfalls suggest that the force field parameters may be unrealistically stiff, or that accessible timescales for simulation do not allow the full conformational landscape to be explored (Mardis et al. 2009). Accelerated molecular dynamics (Hamelberg et al. 2007; Markwick et al. 2007; Yang et al. 2007), course graining (Marrink et al. 2004; Tozzini 2005), and multiscale (Schatz 2007) methods have been developed to provide more accurate simulation methods for modeling complex supramolecular dynamic landscapes. The scattering analysis described here provides a means to evaluate the ability of these advanced simulation methods to accurately model solution-phase ensembles compared to other experimental information which focus on a single, low-energy structure. This comparison between experimental solution-phase scattering and MD simulation provides a means not only to interpret scattering data in terms of an explicit atomic model, but more generally demonstrates the use of solution x-ray scattering as an experimental benchmark for the development of simulation methods that more accurately predict configurational dynamics of supramolecular assemblies.
Solution Structure Refinement
Numerous approaches are being developed for enhancing macromolecular structure resolution in the solution by combining x-ray scattering data with coordinate data from complementary structural techniques. For example, crystal coordinate data has been used in combination with configuration search algorithms to derive coordinate models consistent with solution scattering data (Putnam et al. 2007). Numerous methods have also been developed to exploit the complementarities of NMR and x-ray scattering data from both biomolecular and transition metal complexes. The biological NMR structural method is mainly based on the nuclear Overhauser effect (NOE) that measures hydrogen-hydrogen distances in short ranges, i.e., < 5 Å. As a result global biomolecular structure is often under-determined because the number of detectable NOE-based distance restraints is far less than the number of freedom degree of atoms in the molecules. This has been a particularly significant problem in NMR structure determination in DNA and RNA. For example, the overall conformation and details of base rise parameters for a model dodecamer DNA were found to vary significantly in different NMR structure refinements using the identical NMR dataset, demonstrating that the set of NMR distance constraints was not sufficient to uniquely determine the structure (Kuszewski et al. 2001; Zuo and Tiede 2005).
The global, all atom nature of the x-ray scattering measurement provides a useful complement to the predominately short range distance constraints measured by NMR and thus x-ray scattering data can provide additional constraints in structure refinement fitting procedures. Small-angle scattering data have been employed in the structure refinements as a global dimension restraint for NMR structure refinement for proteins and polynucleic acids (Gabel et al. 2006; Grishaev et al. 2005b; Grishaev et al. 2008; Lee et al. 2007). In general, the inclusion of scattering data as fitting constraints have been found to improve global structure and refinement convergence, and are a benefit particularly for biomolecules with elongated or flexible structures because the number of available NOE distance restraints tends to be relatively small compared to the total number of atoms. High angle X-ray scattering data have also been used in structural refinement of a model dodecamer DNA (Schwieters and Clore 2006).
Another increasingly important application of scattering data is for the resolution of global structure for multiple domain or multiple component supramolecular complexes. Frequently the structures of subunits can be determined using crystallographic or NMR approaches. However, structure determination for complex, multi-component architectures has become a major challenge for structural biology. Complex architecture determination can be described as the problem of determining relative translational positions and orientations of subunits within the complex. Approaches have been developed that use small angle X-ray scattering, SAXS, data to screen all possible arrangements of subunits within both translational and rotational spaces (Bernadó et al. 2007; Petoukhov and Svergun 2005; Svergun and Koch 2003). These approaches often are computationally expensive and result in ambiguous structures because the degeneracy in SAXS data. Similarly, NMR methods have been developed to resolve the relative positions of subunits within a complex by measuring inter-subunit NOEs. This approach requires sophisticated sample preparation and isotope-label filter/edited NOE experiments to define the interface between subunits. As a result, the inter-subunit NOE approach is costly, time-consuming, and often difficult to obtain a sufficient number of inter-subunit to unambiguously define the interface.
To relieve these limitations, another NMR technique, residual dipolar coupling (RDC), has been introduced that measures the orientations of bonds comprising of active nuclei, such as 13C-H and 15N-H in isotope-labeled biomolecules relative to the external magnetic field (Grishaev et al. 2008; Zuo et al. 2008). With knowledge of individual subunit structure, RDC measurements on an assembled complex provide a measure of the relative orientations of bonds, and therefore, the relative orientations of subunits. Use of RDC data can dramatically reduce the configurational search load for structure refinement because one set of RDC data yields only four possible discrete orientations for each subunit. The combination of SAXS and RDC data constraints simplifies the structural search and also dramatically reduces the degeneracy in both data, or even completely lifts the degeneracy in many cases.
The joint use of SAXS and RDC for determining the bimolecular complex structure has been demonstrated by using a tetraloop-receptor RNA:RNA homodimer with 86 nucleotides as an example (Zuo et al. 2008). The structure of the monomeric subunit of this dimer is highly asymmetric and elongated, determined solely using NMR data. The RDC measurements provide four possible orientations for the two subunits within the dimer, shown in Figure 9A. Each of these four subunit orientations can be grid searched with respect to the coordinates (φ,θ,r) of an inter-subunit position vector, r, that connects the center of masses for the two subunits, Figure 9A. At each rigid-body grid step, φ+iΔφ, θ+jΔθ, r+kΔr, the scattering pattern for the trial dimer complex is calculated and ranked with respect to experimental SAXS data. This search can be expedited by including Rg, steric hindrance, Dmax (largest distance within the complex obtained from experimental PDF analysis), and symmetry filters. The resulting best fit dimer complex structure is shown in green in Figure 9B. This structure has an identical dimer interface (rmsd 0.4 Å) as the one determined using NMR data containing 72 inter-subunit NOEs alone, and shown by the red structure in Figure 9B. This structure can be further refined by using a combination of SAXS data and NMR distance constraints to achieve the structure shown in cyan in Figure 9B that differs from the structure obtained by NMR data alone by a backbone rmsd of 3.2 Å. This new structure illustrates the deficiency of obtaining a solution structure determined solely from NMR data without including SAXS as global restraints (Zuo et al. 2008).
Figure 9.

Part A. Scheme for a grid search in polar coordinatesystem (φ,θ,r) and the four possible orientations (O1 - O4) of monomeric sub-unit in the tetraloop-receptor RNA:RNA complex. Part B. Structures of the tetraloop-receptor RNA:RNA homodimer: green, determined with NMR data alone including 72 inter-subunit NOE restraints; red, determined by grid search without inter-subunit NOEs; cyan, refined with both SAXS and NMR data. The interfaces of the homodimer are indicated by the black arrows.
This method can be readily modified to solve structures for multi-component complexes. We expect this methodology could become a routine method for rapid structure determination of modular architectures composed of structurally determined building blocks. The combined NMR and x-ray scattering refinement analyses gains relevance because both sets of data can be recorded using identical conditions, and they provide fully complementary data.
Conclusions
The examples cited in this review demonstrate the opportunityto gain an atomic-scale understanding of structure and structural dynamics for supramolecular assemblies in solution by combining x-ray scattering with coordinated based modeling. These approaches can be expected to have a broad range of applications for structure-function determination, particularly for a variety of structurally dynamic supramolecular assemblies in natural and artificial photosynthesis. In addition, the emerging ultrafast time resolved scattering techniques offer entirely new possibilities to resolve fundamental mechanisms for solar energy conversion by mapping out structure in light-excited reaction states.
Acknowledgments
This work was supported by the Office of Science, Basic Energy Sciences, U. S. Department of Energy under contract numbers DE-AC02-06CH11357 (D.M.T. and work at APS Sector 12), National Science Foundation IL-LSAMP grant HRD-0413000 and National Institutes of Health Grant 1SC2GM083717 (K.L.M.). The software program, solX, used for coordinated based x-ray scattering calculations is available by request to D.M.T. or X.Z.
Abbreviations
- DNA
Deoxyribonucleic acid
Pair Distribution Function
- MD
molecular dynamics
- NMR
nuclear magnetic resonance
- SAXS
small angle x-ray scattering
- WAXS
wide angle x-ray scattering
Contributor Information
David M. Tiede, Email: tiede@anl.gov.
Kristy L. Mardis, Email: kmardis@csu.edu.
Xiaobing Zuo, Email: zuox@mail.nih.gov, xiaobing.zuo@gmail.com.
References
- Bernadó P, Mylonas M, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Borgstahl GEO, Williams DR, Getzoff ED. 1.4 angstrom structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, Cupane A, Wulff M, Ihee H. Tracking the structural dynamics of proteins in solution using time-resolved wide-angle X-ray scattering (vol 5, pg 881, 2008) Nat Methods. 2008;5:988–988. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, van Gunsteren WF. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: A review. J Comput Chem. 2008;29:157–166. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- Ducruix A, Guilloteau JP, Ries-Kautt M, Tardieu A. Protein interactions as seen by solution X-ray scattring prior to crystallogenesis. J Cryst Growth. 1996;168:28–39. [Google Scholar]
- Fedorov BA, Denesyuk AI. Large-angle x-ray diffuse scattering, a new method for investigating changes in the conformation of globular proteins in solution. J Appl Cryst. 1978;11:473–477. [Google Scholar]
- Fischetti RF, Rodi DJ, Gore DB, Makowski L. Wide-angle X-ray solution scattering as a probe of ligand-induced conformational changes in proteins. Chem Biol. 2004;11:1431–1443. doi: 10.1016/j.chembiol.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Fraser RDB, MacRae TP, Suzuki E. An improved method for calculating the contribution of solvent to the x-ray diffraction pattern of biological molecules. J Applied Crystallography. 1978;11:693–694. [Google Scholar]
- Gabel F, Simon B, Sattler M. A target function for quaternary structural refinement from small angle scattering and NMR orientational restraints. Eur Biophys J. 2006;35:313–327. doi: 10.1007/s00249-005-0037-3. [DOI] [PubMed] [Google Scholar]
- Genick UK, Borgstahl GEO, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J Am Chem Soc. 2005a;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multidomain protein structures by combination of solution small-angle X-ray scattering and NMR data. J Am Chem Soc. 2005b;127:16621–8. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]
- Grishaev A, Ying J, Canny MD, Pardi A, Bax A. Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data. J Biomol NMR. 2008;42:99–109. doi: 10.1007/s10858-008-9267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinier A, Fournet G. Small Angle Scattering of X-rays. Wiley; New York: 1955. [Google Scholar]
- Gust D, Moore TA, Moore AL. Mimicking photosynthetic solar energy transduction. Acc Chem Res. 2001;34:40–48. doi: 10.1021/ar9801301. [DOI] [PubMed] [Google Scholar]
- Hamelberg D, de Oliveira CAF, McCammon JA. Sampling of slow diffusive conformational transitions with accelerated molecular dynamics. J Chem Phys. 2007;127:155102-1–155102-9. doi: 10.1063/1.2789432. [DOI] [PubMed] [Google Scholar]
- Hellemans A. X-rays find new ways to shine. Science. 1997;277:1214–1215. [Google Scholar]
- Hirai M, Koizumi M, Hayakawa T, Takahashi H, Abe S, Hirai H, Miura K, Inoue K. Hierarchical map of protein unfolding and refolding at thermal equilibrium revealed by wide-angle X-ray scattering. Biochemistry. 2004;43:9036–9049. doi: 10.1021/bi0499664. [DOI] [PubMed] [Google Scholar]
- Howard AE, Kollman PA. An analysis of current methodologies for conformational searching of complex molecules. J Med Chem. 1988;31:1669–1675. doi: 10.1021/jm00117a001. [DOI] [PubMed] [Google Scholar]
- Ihee H, Lorenc M, Kim TK, Kong QY, Cammarata M, Lee JH, Bratos S, Wulff M. Ultrafast X-ray diffraction of transient molecular structures in solution. Science. 2005;309:1223–1227. doi: 10.1126/science.1114782. [DOI] [PubMed] [Google Scholar]
- Kamikubo H, Shimizu N, Harigai M, Yamazaki Y, Imamoto Y, Kataoka M. Characterization of the solution structure of the M intermediate of photoactive yellow protein using high-angle solution x-ray scattering. Biophysical J. 2007;92:3633–3642. doi: 10.1529/biophysj.106.097287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Dumont C, Gruebele M. Simulation-based fitting of protein-protein interaction potentials to SAXS experiments. Biophysical Journal. 2008;94:4924–4931. doi: 10.1529/biophysj.107.123240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Zuo X, Tiede DM, Ihee H. Exploring fine structures of photoactive yellow protein in solution using wide-angle X-ray scattering. Bull Korean Chem Soc. 2004;25:1676–1680. [Google Scholar]
- Koch MHJ. X-ray scattering of non-crystalline biological systems using synchrotron radiation. Chem Soc Rev. 2006;35:123–133. doi: 10.1039/b500858c. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kezuka Y, Nonaka T, Hiragi Y, Watanabe T, Kimura K, Takahashi K, Yanagi S, Kihara H. SaxsMDView: a three-dimensional graphics program for displaying force vectors. J Synchrot Radiat. 2008;15:535–537. doi: 10.1107/S0909049508015951. [DOI] [PubMed] [Google Scholar]
- Kojima M, Timchenko AA, Higo J, Ito K, Kihara H, Takakhashi K. Structural refinement by restrained molecular-dynamics algorithm with small-angle X-ray scattering constraints for a biomolecule. J Appl Cryst. 2004;37:103–109. [Google Scholar]
- Kuszewski J, Schwieters C, Clore GM. Improving the accuracy of NMR structures of DNA by means of a database potential of mean force describing base-base positional interactions. J Am Chem Soc. 2001;123:3903–18. doi: 10.1021/ja010033u. [DOI] [PubMed] [Google Scholar]
- Lee D, Walsh JD, Yu P, Markus MA, Choli-Papadopoulou T, Schwieters CD, Krueger S, Draper DE, Wang YX. The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes. Journal of molecular biology. 2007;367:1007–22. doi: 10.1016/j.jmb.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Mulfort KL, O’Donnell JL, Zuo X, Goshe AJ, Nguyen ST, Hupp JT, Tiede DM. Supramolecular porphyrinic prisms: coordinative assembly and preliminary solution-phase X-ray structural characterization. Chem Commun. 2006:4581–4583. doi: 10.1039/b610025b. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Mulfort KL, Zuo X, Goshe AJ, Wesson PJ, Nguyen ST, Hupp JT, Tiede DM. Coordinative self-assembly and solution-phase X-ray structural characterization of cavity-tailored porphyrin boxes. J Am Chem Soc. 2008;130:836–838. doi: 10.1021/ja077661h. [DOI] [PubMed] [Google Scholar]
- Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu Rev Biophys Biomol Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- Makowski L, Rodi DJ, Mandava S, Devarapalli S, Fischetti RF. Characterization of Protein Fold by Wide-Angle X-ray Solution Scattering. Journal of Molecular Biology. 2008;383:731–744. doi: 10.1016/j.jmb.2008.08.038. [DOI] [PubMed] [Google Scholar]
- Mardis KL, Sutton HM, Zuo XB, Lindsey JS, Tiede DM. Solution-State Conformational Ensemble of a Hexameric Porphyrin Array Characterized Using Molecular Dynamics and X-ray Scattering. J Phys Chem A. 2009;113:2516–2523. doi: 10.1021/jp808318x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick PRL, Bouvignies G, Blackledge M. Exploring Multiple Timescale Motions in Protein GB3 Using Accelerated Molecular Dynamics and NMR Spectroscopy. J Amer Chem Soc. 2007;129:4724–4730. doi: 10.1021/ja0687668. [DOI] [PubMed] [Google Scholar]
- Marone PA, Thiyagarajan P, Wagner AM, Tiede DM. The association state of a detergent-solubilized membrane protein measured during crystal nucleation and growth by small-angle neutron scattering. J Cryst Growth. 1998;191:811–819. [Google Scholar]
- Marrink SJ, de Vries AH, Mark AE. Coarse Grained Model for Semiquantitative Lipid Simulations. J Phys Chem B. 2004;108:750–760. [Google Scholar]
- Megyes T, Balint S, Bako I, Grosz T, Palinkas G. Complete structural characterization of metallacyclic complexes in solution-phase using simultaneously X-ray diffraction and molecular dynamics simulation. J Amer Chem Soc. 2008;130:9206–9209. doi: 10.1021/ja802793g. [DOI] [PubMed] [Google Scholar]
- Megyes T, Jude H, Grosz T, Bako I, Radnai T, Tarkanyi G, Palinkas G, Stang PJ. X-ray diffraction and DOSY-NMR characterization of self-assembled supramolecular metallocyclic species in solution. J Am Chem Soc. 2005;127:10731–10738. doi: 10.1021/ja0523690. [DOI] [PubMed] [Google Scholar]
- O’Donnell JL, Zuo X, Goshe AJ, Sarkisov L, Snurr RQ, Hupp JT, Tiede DM. Solution-phase structural characterization of supramolecular assemblies by molecular diffraction. J Am Chem Soc. 2007;129:1578–1585. doi: 10.1021/ja0659065. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Eady NAJ, Brown KA, Svergun DI. Addition of missing loops and domains to protein models by X-ray solution scattering. Biophysical J. 2002;83:3113–3125. doi: 10.1016/S0006-3495(02)75315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip AF, Eisenman KT, Papadantonakis GA, Hoff WD. Functional Tuning of Photoactive Yellow Protein by Active Site Residue 46. Biochemistry. 2008;47:13800–13810. doi: 10.1021/bi801730y. [DOI] [PubMed] [Google Scholar]
- Plech A, Wulff M, Bratos S, Mirloup F, Vuilleumier R, Schotte F, Anfinrud PA. Visualizing chemical reactions in solution by picosecond X-ray diffraction. Phys Rev Let. 2004;92:125505-1–5. doi: 10.1103/PhysRevLett.92.125505. [DOI] [PubMed] [Google Scholar]
- Pontius J, Richelle J, Wodak J. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J Mol Biol. 1996;264:121–136. doi: 10.1006/jmbi.1996.0628. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Riekel C, Bosecke P, Diat O, Engstrom P. New opportunities in small-angle X-ray scattering and wide-angle X-ray scattering at a third generation synchrotron radiation source. J Molecular Structure. 1996;383:291–302. [Google Scholar]
- Schatz GC. Using theory and computation to model nanoscale properties. Proc Natl Acad Sci USA. 2007;104:6885–6892. doi: 10.1073/pnas.0702187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Clore GM. A physical picture of atomic motions within the Dickerson DNA dodecamer in solution derived from joint ensemble refinement against NMR and large-angle X-ray scattering data. Biochemistry. 2006;46:1152–1166. doi: 10.1021/bi061943x. [DOI] [PubMed] [Google Scholar]
- Seifert S, Winans RE, Tiede DM, Thiyagarajan P. Design and performance of an ASAXS instrument at the Advanced Photon Source. J Appl Cryst. 2000;33:782–784. [Google Scholar]
- Semenyuk AV, Svergun DI. GNOM- a program package for small-angle scattering data processing. J Appl Cryst. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaramakrishnan S, Spink BJ, Sim AYL, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson B, Tiede DM, Barry BA. Small-angle X-ray scattering studies of the manganese stabilizing subunit in photosystem II. J Phys Chem B. 2002;106:8485–8488. [Google Scholar]
- Svensson B, Tiede DM, Nelson DR, Barry BA. Structural studies of the manganese-stabilizing subunit in Photosystem II. Biophysical J. 2004;86:1807–1812. doi: 10.1016/S0006-3495(04)74247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL-a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66:1735–1782. [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophysical J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F, Brooks CL. SYMMETRY, FORM, AND SHAPE: Guiding Principles for Robustness in Macromolecular Machines. Ann Rev Biophys Biomol Struct. 2006;35:115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- Tiede DM, Littrell K, Marone PA, Zhang R, Thiyagarajan P. Solution structure of a biological bimolecular electron transfer complex: characterization of the photosynthetic reaction center-cytochrome c2 protein complex by small angle neutron scattering. J Applied Crystallography. 2000;33:560–564. [Google Scholar]
- Tiede DM, Zhang R, Chen LX, Yu L, Lindsey JS. Structural Characterization of Modular Supramolecular Architectures in Solution. J Am Chem Soc. 2004;126:14054–14062. doi: 10.1021/ja048209q. [DOI] [PubMed] [Google Scholar]
- Tiede DM, Zhang R, Seifert S. Protein conformations explored by difference high-angle solution X-ray scattering: oxidation state and temperature dependent changes in cytochrome c. Biochemistry. 2002;41:6605–6614. doi: 10.1021/bi015931h. [DOI] [PubMed] [Google Scholar]
- Tozzini V. Coarse-grained models for proteins. Theory and simulation/Macromolecular assemblages. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Tsuruta H, Irving TC. Experimental approaches for solution X-ray scattering and fiber diffraction. Curr Opinion Struct Biol. 2008;18:601–608. doi: 10.1016/j.sbi.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren WF, Bakowies D, Baron R, Chandrasekhar I, Christen M, Daura X, Gee P, Geerke DP, Glättli A, Hünenberger PH, Kastenholz MA, Oostenbrink C, Schenk M, Trzesniak D, van der Vegt NFA, Yu HB. Biomolecular Modeling: Goals, Problems, Perspectives. Angewandte Chemie International Edition. 2006;45:4064–4092. doi: 10.1002/anie.200502655. [DOI] [PubMed] [Google Scholar]
- Vaughan PA, Sturdivant JH, Pauling L. The determination of the structures of complex molecules and ions from X-ray diffraction by their solutions: the structures of the groups PtBr6− −, PtCl6− −, Nb6Cl12++, TaBr12++, and Ta6Cl12++ J Am Chem Soc. 1950;72:5477–5486. [Google Scholar]
- Vengadesan K, Gautham N. A new conformational search technique and its applications. Curr Sci. 2005;88:1759. [Google Scholar]
- Vigil D, Gallagher SC, Trewhella J, Garcia AE. Functional Dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophysical J. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ossowski I, Eaton JT, Czjzek M, Perkins SJ, Frandsen TP, Schulein M, Panine P, Henrissat B, Receveur-Brechot V. Protein disorder: Conformational distribution of the flexible linker in a chimeric double cellulase. Biophysical Journal. 2005;88:2823–2832. doi: 10.1529/biophysj.104.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren BE. X-Ray Diffraction. Dover Publications, Inc; New York: 1990. [Google Scholar]
- Wasielewski MR. Energy, charge, and spin transport in molecules and self-assembled nanostructures inspired by photosynthesis. Journal of Organic Chemistry. 2006;71:5051–5066. doi: 10.1021/jo060225d. [DOI] [PubMed] [Google Scholar]
- Winick H. Synchrotron radiation sources - Present capabilities and future directions. J Synchrot Radiat. 1998;5:168–175. doi: 10.1107/S0909049597018761. [DOI] [PubMed] [Google Scholar]
- Xie AH, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Formation of a new buried charge drives a large-amplitude protein quake in photoreceptor activation. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]
- Yang L, Grubb MP, Gao YQ. Application of the accelerated molecular dynamics simulations to the folding of a small protein. The Journal of chemical physics. 2007;126:125102. doi: 10.1063/1.2709639. [DOI] [PubMed] [Google Scholar]
- Yeremenko S, van Stokkum IHM, Moffat K, Hellingwerf KJ. Influence of the crystalline state on photoinduced dynamics of photoactive yellow protein studied by ultraviolet-visible transient absorption spectroscopy. Biophysical Journal. 2006;90:4224–4235. doi: 10.1529/biophysj.105.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Friesner RA. Ab initio calculation of electronic coupling in the photosynthetic reaction center. Proc Natl Acad Sci USA. 1998;95:13603–13605. doi: 10.1073/pnas.95.23.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Cui G, Mertz KM, Zhang L, Lewis FD, Tiede DM. X-ray diffraction “fingerprinting” of DNA structure in solution for quantitative evaluation of molecular dynamics simulation. Proc Natl Acad Sci USA. 2006;103:3534–3539. doi: 10.1073/pnas.0600022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Tiede DM. Resolving Conflicting Crystallographic and NMR Models for Solution-State DNA with Solution X-ray Diffraction. J Am Chem Soc. 2005;127:16–17. doi: 10.1021/ja044533+. [DOI] [PubMed] [Google Scholar]
- Zuo X, Wang J, Foster TR, Schwieters CD, Tiede DM, Butcher SE, Wang Y-X. Global Architecture and Interface: Refining a RNA:RNA Complex Structure Using Solution X-ray Scattering Data. J Am Chem Soc. 2008;130:3292–3293. doi: 10.1021/ja7114508. [DOI] [PubMed] [Google Scholar]