Abstract
Bacillus anthracis proliferates to high levels within vertebrate tissues during the pathogenesis of anthrax. This growth is facilitated by the acquisition of nutrient iron from host heme. However, heme acquisition can lead to the accumulation of toxic amounts of heme within B. anthracis. Here, we show that B. anthracis resists heme toxicity by sensing heme through the HssRS two-component system, which regulates expression of the heme-detoxifying transporter HrtAB. In addition, we demonstrate that B. anthracis exhibits elevated HssRS function compared to its evolutionary relative S. aureus. Elevated heme sensing is likely required by B. anthracis due to the significant heme sensitivity exhibited by members of the genus Bacilli. We also demonstrate that B. anthracis depends on conserved residues within the previously uncharacterized sensing domain of the histidine kinase HssS for HssS function. Finally, we show that the heme- and HssRS-regulated hrtAB promoter is activated in a murine model of anthrax. These results demonstrate the evolutionary conservation of heme sensing among multiple Gram-positive bacteria and begin to provide a mechanistic explanation for the heme resistance of B. anthracis. Further, these data suggest that heme stress is experienced by bacterial pathogens during infection.
Keywords: Heme, two-component system, Staphylococcus, Bacillus, anthrax
INTRODUCTION
Bacillus anthracis is capable of causing devastating disease upon breaching host epithelial barriers (Dixon et al., 1999; Mock and Fouet, 2001). A spore-forming Gram-positive pathogen that primarily infects livestock, B. anthracis can cause a rare zoonotic disease in humans known as anthrax (Dixon et al., 1999; Mock and Fouet, 2001). Anthrax arises when spores germinate within host tissues and can progress to systemic bacteremia followed by sudden death (Dixon et al., 1999; Mock and Fouet, 2001). Like all bacterial pathogens, B. anthracis must circumvent mechanisms of innate immunity to replicate within host tissues and cause disease.
One mechanism of innate immunity that represents a challenge to bacterial pathogens is nutritional immunity, which refers to the host-mediated sequestration of nutrients such as metal ions required for bacterial growth (Bezkorovainy, 1981; Corbin et al., 2008). In particular, free iron is maintained at a concentration within host tissues that is too low to support the growth of most bacteria, including B. anthracis (Bullen, 1999; Reniere et al., 2007). However, iron in the form of the porphyrin heme is an abundant molecule within host erythrocytes as the cofactor of hemoglobin and represents a source of nutrient iron for invading bacteria (Reniere et al., 2007). B. anthracis and related pathogens express a set of proteins known collectively as the iron-regulated surface determinant (Isd) system devoted to the acquisition and degradation of host heme for nutrient iron needs (Gat et al., 2008; Maresso et al., 2006; Maresso et al., 2008; Mazmanian et al., 2003; Skaar and Schneewind, 2004). The importance of heme acquisition to pathogenesis is highlighted by the decreased virulence observed for pathogens inactivated for members of the Isd system (Reniere and Skaar, 2008; Skaar et al., 2004; Torres et al., 2006).
Although heme is a valuable source of iron for B. anthracis during infection, heme and hemin (the free, Fe(III) form of heme found in solution) can catalyze the formation of reactive oxygen species, leading to protein and lipid damage that cause bacterial cell death (Everse and Hsia, 1997; Reniere et al., 2007; Torres et al., 2007). S. aureus, an evolutionary relative of B. anthracis, is susceptible to hemin toxicity; however, staphylococci are able to adapt to growth in the presence of hemin (Torres et al., 2007). In S. aureus, adaptation to hemin toxicity depends on expression of the heme-regulated ABC transporter (HrtAB), which is up-regulated upon exposure of staphylococci to hemin and alleviates hemin toxicity (Friedman et al., 2006; Torres et al., 2007). It has been hypothesized that HrtAB may directly export excess heme from the bacterial cytoplasm or may detoxify heme stress by exporting a toxic metabolite that accumulates in cells exposed to excess heme (Stauff and Skaar, 2008). Although the mechanism of heme detoxification by HrtAB remains unclear, the ability of S. aureus to express HrtAB upon heme exposure demonstrates that staphylococci sense and respond to heme in a way that prepares bacterial cells for growth in high heme concentrations.
In S. aureus, HrtAB expression is regulated by a two-component system (TCS) designated the heme sensor system (HssRS) (Stauff et al., 2007; Torres et al., 2007). HssS is a membrane-localized histidine kinase that senses heme through an unknown mechanism and undergoes histidine phosphorylation (Stauff et al., 2007). HssR is a cytoplasmic response regulator that recognizes phosphorylated HssS and catalyzes phosphotransfer to a conserved HssR aspartic acid residue (Stauff et al., 2007). When phosphorylated, HssR binds to a direct repeat (DR) within the hrtAB promoter, inducing expression of HrtAB (Stauff et al., 2007). Importantly, interfering with proper heme metabolism through mutation of hrtAB or hssRS alters the virulence properties of S. aureus, highlighting the importance of heme sensing to the physiology and pathogenesis of staphylococci (Stauff et al., 2008; Stauff and Skaar, 2008; Torres et al., 2007).
It is currently unclear whether heme sensing and detoxification is a general feature of Gram-positive pathogens that associate with host tissues rich in heme. It is likely that a pathogen such as B. anthracis, which is capable of replicating to a high density within the bloodstream of its host (Dixon et al., 1999), inducing hemolysis (Shannon et al., 2003), and acquiring heme (Gat et al., 2008; Maresso et al., 2006; Maresso et al., 2008), may require systems devoted to the avoidance of heme toxicity. Consistent with this supposition, Van Heyningen demonstrated in 1948 that B. anthracis is capable of proliferating on plates containing high concentrations of hemin (Heyningen, 1948). In contrast, B. subtilis (a bacterium not commonly associated with vertebrate tissues containing heme) is sensitive to hemin toxicity (Heyningen, 1948). Accordingly, B. anthracis encodes putative orthologues of S. aureus hssRS/hrtAB (Torres et al., 2007), whereas hss/hrt genes are absent from the genome of B. subtilis (Stauff and Skaar, 2008).
Here, we demonstrate that B. anthracis encodes functional HssRS and HrtAB systems required for protection from hemin toxicity. We show that differential hemin resistance within the Bacilli observed by Van Heyningen correlates with the presence of hssRS/hrtAB genes. Furthermore, B. anthracis HssRS exhibits an elevated autophosphorylation and phosphotransfer rate that correlates with the increased in vivo activity of this TCS. More specifically, we report that domains of the histidine kinase HssS are likely responsible for the elevated function of B. anthracis HssS. B. anthracis is highly sensitive to hemin toxicity in the absence of HrtAB function, and may therefore require an increased ability to mount a heme detoxification response through heme sensing by HssS and signal transduction through HssR. We also demonstrate that mutation of residues within the previously uncharacterized signal recognition domain of HssS eliminates hemin sensing by this histidine kinase. Last, we show that the hrtAB promoter is activated in a cutaneous model of anthrax, suggesting a role for heme sensing during B. anthracis infection. These results demonstrate that heme sensing by HssRS and subsequent heme detoxification by HrtAB are conserved in B. anthracis and that heme stress may be encountered by a wide range of pathogenic or saprotrophic Gram-positive bacteria.
RESULTS
Phylogenetically related Bacilli display differential hemin resistance
To test whether encoding hss/hrt correlates with resistance to hemin toxicity, we assessed the hemin resistance of four Bacilli that contain these genes (B. anthracis, B. cereus, B. thuringiensis, and B. israelensis) and two with no identifiable hss/hrt orthologues (B. licheniformis and B. subtilis) (Fig. S1). We found that each of the four Bacilli encoding putative Hss/Hrt systems are able to proliferate in the presence of hemin, while both Bacilli lacking Hss/Hrt systems succumb to hemin toxicity (Fig. S1). These data link the presence of hss/hrt genes with resistance to hemin toxicity.
B. anthracis encodes hss/hrt genes required for hemin resistance
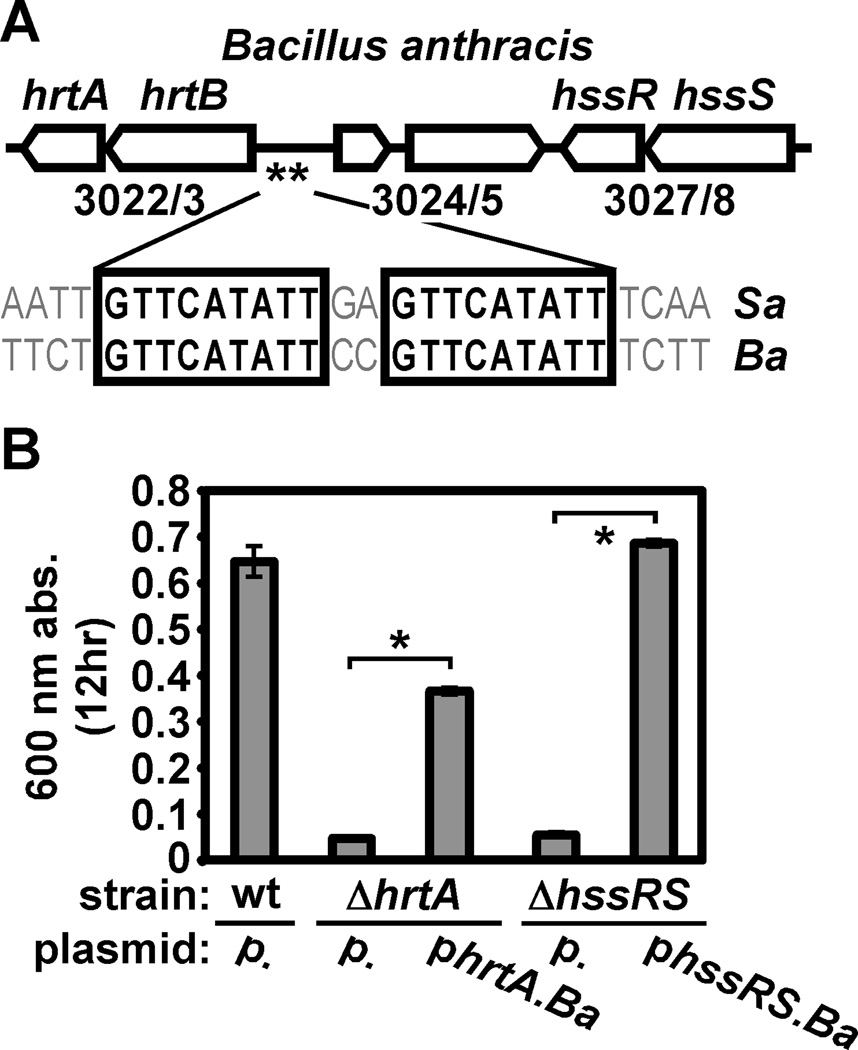
We chose to investigate Hss/Hrt function in B. anthracis based on the fact that this organism is a mammalian pathogen and one that displays an intimate association with host blood during infection. B. anthracis HrtAB and HssRS share significant sequence identity with the corresponding proteins in S. aureus (HrtA, 49.3%; HrtB, 30.9 %; HssS, 35.3%; HssR, 48.4%) (Stauff and Skaar, 2008; Torres et al., 2007). The hrtAB and hssRS operons of B. anthracis are located less than 3 kb apart on the chromosome and are separated by two open reading frames, one of which encodes an uncharacterized transcription factor (BAS3024, Fig. 1A). An HssR consensus binding site can be found within the promoter of B. anthracis hrtAB (GTTCATATT(N2)GTTCATATT) (de Been et al., 2008; Stauff et al., 2007), suggesting that HssRS may regulate HrtAB expression in B. anthracis (Fig. 1A). Furthermore, we found that B. anthracis is capable of adapting to hemin toxicity (Fig. S2), a phenotype that is likely due to expression of HrtAB in hemin-treated cells prior to exposure to toxic levels of hemin (Torres et al., 2007). These observations are consistent with the proposition that B. anthracis adapts to hemin through HssRS-dependent expression of HrtAB.
Figure 1. B. anthracis requires HrtAB and HssRS for hemin resistance.
A. Schematic of the B. anthracis hss/hrt locus. Open arrows represent open reading frames (ORF). Designations above ORFs denote predicted gene function (hrtA: ATPase; hrtB: permease; hssS: histidine kinase; hssR: response regulator). Numbers below ORFs correspond to gene numbers in the B. anthracis Sterne genome. Double asterisks signify a putative HssR binding site (consensus sequence GTTCATATT(N2)GTTCATATT) upstream of hrtAB; alignment shows the known S. aureus (Sa) and predicted B. anthracis (Ba) HssR binding sites. B. Contribution of hrt and hss to hemin resistance of B. anthracis. B. anthracis wildtype (wt) harboring plasmid pOS1 (p.), ΔhrtA with either pOS1 or pOS1 encoding hrtA (phrtA.Ba), and ΔhssRS with either pOS1 or pOS1 encoding hssRS (phssRS.Ba) were grown for 12 hours in BHI. Bacteria were sub-cultured 1:100 into BHI containing 10 µM hemin (a hemin concentration known to inhibit S. aureus hss/hrt mutants) in triplicate, grown for 12 hours at 37 °C, and culture densities were determined and averaged. Error bars correspond to one standard deviation from the mean; asterisks denote statistically significant complementation by Student’s t test (p<0.05).
Accordingly, we assessed the ability of B. anthracis mutants lacking hrtA and hssRS to resist hemin toxicity. While wildtype B. anthracis is able to proliferate in 10 µM hemin, B. anthracis ΔhrtA and ΔhssRS are unable to proliferate in this hemin concentration, phenotypes that can be at least partially complemented by providing a full-length copy of the corresponding gene in trans (Fig. 1B; see Hemin preparation and concentrations in Experimental Procedures for choice of hemin concentration here and throughout). Importantly, B. anthracis ΔhrtA and ΔhssRS display no growth defect in the absence of hemin (Fig. S3). This demonstrates that B. anthracis requires functional Hss/Hrt systems for hemin resistance.
The B. anthracis hrtAB promoter displays enhanced constitutive and hemin-induced activity
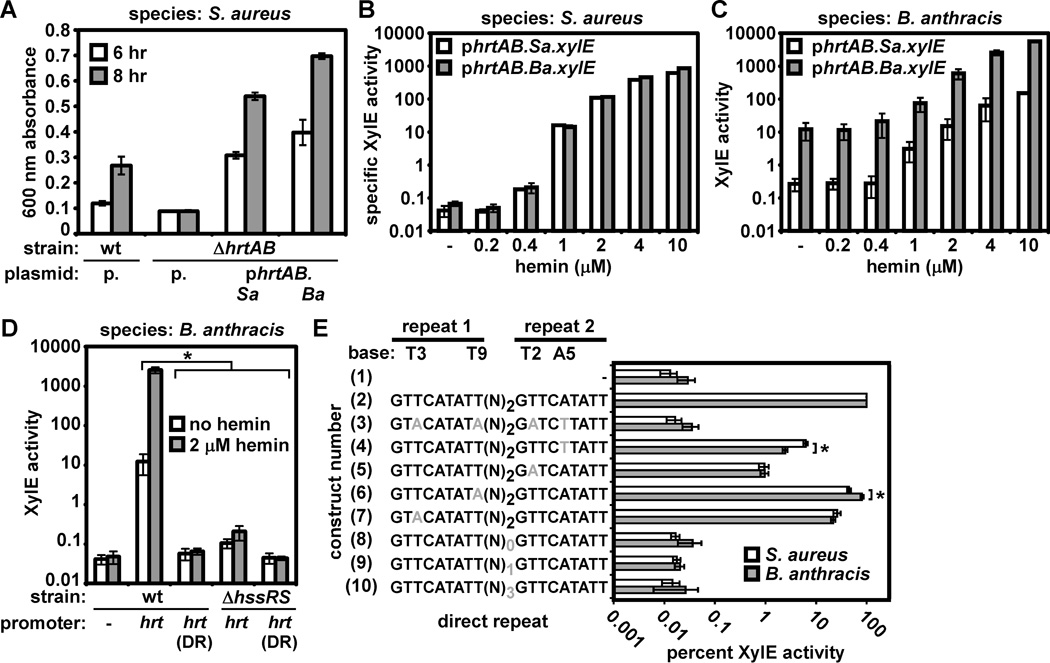
Although S. aureus and B. anthracis both require HrtAB for hemin resistance, it is unclear whether these systems display overlapping function. We therefore performed interspecies complementation experiments and found that B. anthracis or S. aureus hrtAB restore hemin resistance to S. aureus ΔhrtAB, suggesting orthology at the level of transporter function between HrtAB from these organisms (Fig. 2A). In addition, we found that in S. aureus both the S. aureus and B. anthracis hrtAB promoters respond to hemin in a similar manner (Fig. 2B). These results confirm that S. aureus HssR is capable of activating the S. aureus and B. anthracis hrtAB promoters, consistent with the presence of an HssR consensus site within these promoters (Fig. 1A).
Figure 2. Elevated activity of the B. anthracis hrtAB promoter.
A. Complementation of S. aureus ΔhrtAB with S. aureus or B. anthracis hrtAB. S. aureus wildtype (wt) transformed with pOS1 (p.), ΔhrtAB with pOS1, ΔhrtAB with pOS1 encoding S. aureus hrtAB (phrtAB.Sa)), and ΔhrtAB with pOS1 encoding B. anthracis hrtAB (phrtAB.Ba) were grown in TSB + 10 µM hemin (a hemin concentration known to inhibit growth of S. aureus hss/hrt mutants). Proliferation was tracked by growth curve analysis. Culture densities at 6 hour (white bars) and 8 hour (grey bars) time points are shown. B. Reporter assay monitoring S. aureus and B. anthracis hrtAB promoter activity in S. aureus. S. aureus harboring either an S. aureus (phrtAB.Sa.xylE, white bars) or B. anthracis (phrtAB.Ba.xylE, grey bars) hrtAB promoter XylE reporter were grown without hemin (−) or in the indicated concentration of hemin and reporter activity was measured. C. Reporter assay monitoring S. aureus and B. anthracis hrtAB promoter activity in B. anthracis. Strains were grown in LB to avoid contaminating hemin in BHI. D. The dependence on HssRS and the hrtAB promoter direct repeat (DR) for constitutive and hemin-induced hrtAB promoter activity in B. anthracis. B. anthracis wildtype (wt) or ΔhssRS harboring a plasmid containing either a promoterless xylE (−, wildtype only), xylE under the control of the B. anthracis hrtAB promoter (hrt), or xylE under the control of the hrtAB promoter mutated at four conserved DR bases (hrt(DR), shown in schematic form in E, construct #3) were grown in LB (white bars) or LB + 2 µM hemin (a hemin concentration that activates the hrtAB promoter without any detectable growth inhibition of B. anthracis ΔhssRS; grey bars) and XylE activity was measured. E. Reporter assay measuring sensitivity of S. aureus and B. anthracis HssRS to perturbations in the hrtAB promoter DR. S. aureus (white bars) or B. anthracis (grey bars) harboring a promoterless xylE (construct number 1), xylE under the control of the S. aureus hrtAB promoter (2), or xylE under the control of the S. aureus hrtAB promoter containing the indicated DR mutations (3–10; mutations are in grey) were grown in BHI or BHI + 2 µM hemin. Reporter activity was measured and is expressed as the percent of hemin-induced activity observed for the wildtype hrtAB promoter. All results are representative of at least three independent experiments. Error bars correspond to one standard deviation from the mean of triplicate samples within the same experiment; asterisks denote statistically significant differences by Student’s t test (p<0.05).
However, in B. anthracis the hrtAB promoter exhibits elevated levels of activation in response to hemin (Fig. 2C). Furthermore, the B. anthracis hrtAB promoter displays increased activity in the absence of hemin when compared with the S. aureus hrtAB promoter (Fig. 2C). In keeping with this, we tested whether the elevated constitutive activity of the B. anthracis hrtAB promoter depends on HssRS signaling and binding of HssR to the hrtAB promoter direct repeat (DR). Mutation of the DR or disruption of hssRS eliminates the hemin responsiveness and reduces the constitutive activity of the B. anthracis hrtAB promoter (Fig. 2D). This demonstrates that B. anthracis HssRS responds to hemin and activates the hrtAB promoter in a DR-dependent manner. Furthermore, these data suggest that in B. anthracis, HssRS constitutively induces HrtAB expression even in the absence of hemin, an activity not exhibited by S. aureus HssRS (Stauff et al., 2007; Torres et al., 2007).
The fact that hrtAB promoters from both species respond to hemin in a similar manner in S. aureus (Fig. 2B) but not in B. anthracis (Fig. 2C), despite similar DR sequences, indicates that bases outside of the hrtAB promoter DR may be important for promoter activity in B. anthracis. Accordingly, we found that single-base DR mutations have similar effects on the ability of S. aureus and B. anthracis HssR to activate the hrtAB promoter. Specifically, we found that mutation of four conserved DR residues, or removal or addition of spacer residues between the repeats eliminates the ability of S. aureus and B. anthracis HssR to activate the S. aureus hrtAB promoter in response to hemin (Fig. 2E, constructs 3, 8 – 10). Furthermore, mutation of T3 of the first repeat or T2 of the second repeat decreases hemin responsiveness in both species to the same extent (Fig. 2E, constructs 5 and 7). While B. anthracis HssR is more sensitive to mutation of A5 of repeat 2 and S. aureus is more sensitive to mutation of T9 of repeat 1 (Fig. 2E, constructs 4 and 6), both mutations decrease hrtAB promoter activity in response to hemin. This provides the first indication that HssR is exquisitely sensitive to alterations in the distance between each repeat and mutation of single DR residues. Overall, this suggests that HssR-hrtAB promoter interactions display similarity in S. aureus and B. anthracis, pointing to sequences outside of the DR to explain the hemin-independent, HssRS-dependent hrtAB promoter activity observed in B. anthracis.
S. aureus and B. anthracis HssRS provide differential hemin resistance
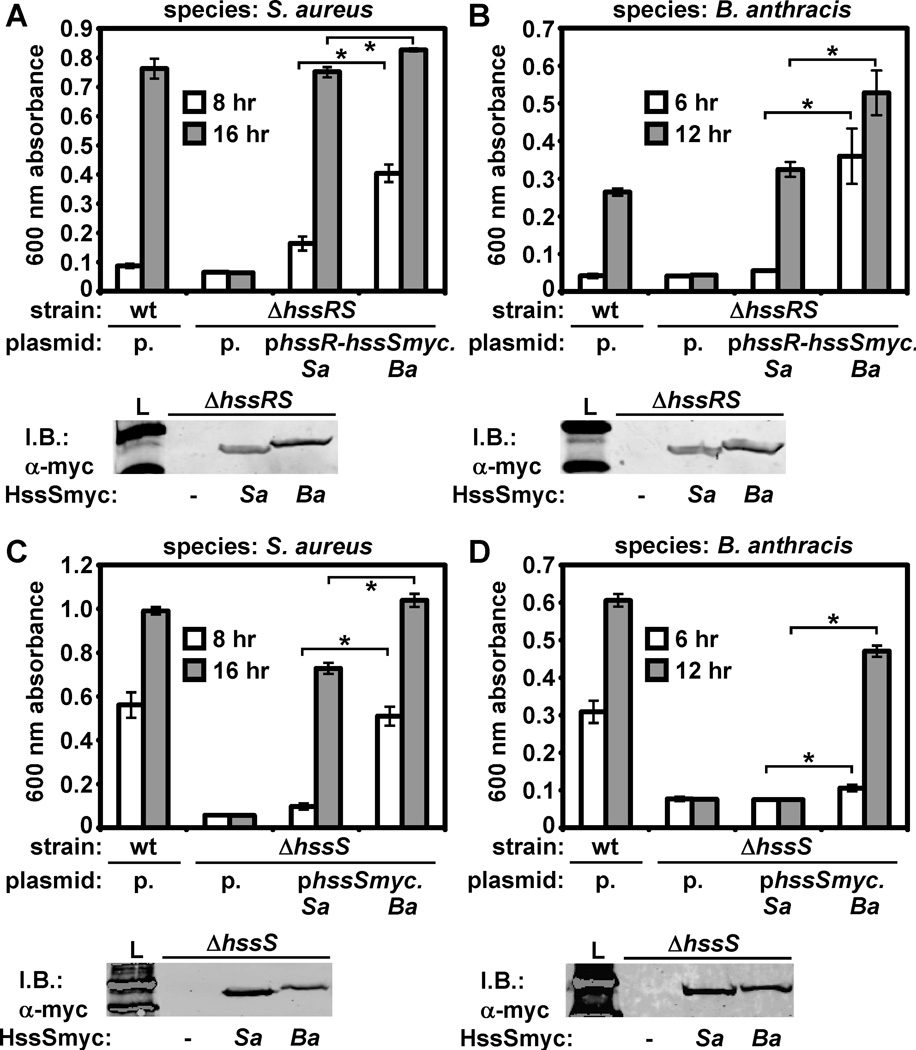
We next tested whether S. aureus and B. anthracis HssRS display differential activity in vivo by assessing the ability of HssRS from S. aureus and B. anthracis to complement S. aureus ΔhssRS. To account for potential differences in protein abundance, we tagged each hssS in the context of an hssRS-encoding plasmid, creating hssR-hssSmyc plasmids for the expression of HssR and HssSmyc from either species. We found that S. aureus and B. anthracis HssSmyc are expressed equivalently in these strains and that HssR-HssSmyc from either species is able to complement S. aureus ΔhssRS (Fig. 3A).
Figure 3. B. anthracis HssRS displays elevated hemin sensing activity in vivo.
A. Complementation of S. aureus ΔhssRS by S. aureus and B. anthracis HssRS. Top: triplicate cultures of S. aureus wildtype (wt) harboring pOS1 (p.), ΔhssRS with pOS1, ΔhssRS with pOS1 encoding S. aureus HssR-HssSmyc (phssR-hssSmyc.Sa), and ΔhssRS with pOS1 encoding B. anthracis HssR-HssSmyc (phssR-hssSmyc.Ba) were grown in TSB + 30 µM hemin (a hemin concentration chosen due to the exceptional hemin resistance of strains containing the phssR-hssSmyc.Ba plasmid). Proliferation was tracked by growth curve analysis. Average culture density at 8 hour (white bars) and 16 hour (grey bars) time points are shown. Bottom: anti-C-Myc immunoblot showing expression of S. aureus and B. anthracis HssSmyc in S. aureus ΔhssRS. Lane 1: molecular weight ladder (L). Lanes 2–4: S. aureus ΔhssRS transformed with pOS1 (−), phssR-hssSmyc.Sa (Sa), or phssR-hssSmyc.Ba (Ba). B. Top: Complementation of B. anthracis ΔhssRS by S. aureus and B. anthracis HssRS (same plasmids described in A). Experiment was carried out as described in A, except that earlier time points (6 hour (white) and 12 hour (grey)) are shown due to sporulation and autolysis observed at late time points in B. anthracis cultures. Bottom: anti-C-Myc immunoblot showing expression of S. aureus and B. anthracis HssSmyc in B. anthracis ΔhssRS. Lanes are the same as those described in A. C. Top: Complementation of S. aureus ΔhssS by S. aureus and B. anthracis HssS. Top: S. aureus wildtype harboring plasmid pOS1, ΔhssS with pOS1, ΔhssS with pOS1 encoding S. aureus HssSmyc (phssSmyc.Sa), and ΔhssS with pOS1 encoding B. anthracis HssSmyc (phssSmyc.Ba) were analyzed as described in A, except that subcultures were performed in 20 µM hemin (chosen due to lower hemin resistance of complemented ΔhssS strains compared to strains described in A). Bottom: anti-C-Myc immunoblot showing expression of S. aureus and B. anthracis HssSmyc in S. aureus ΔhssS. Lane 1: molecular weight ladder (L). Lanes 2–4: S. aureus ΔhssS transformed with empty plasmid (−), phssSmyc.Sa (Sa), or phssSmyc.Ba (Ba). D. Top: Complementation of B. anthracis ΔhssS by S. aureus and B. anthracis HssS. Experiment was performed as described in C. Bottom: anti-C-Myc immunoblot showing expression of S. aureus and B. anthracis HssSmyc in B. anthracis ΔhssS. Lanes are the same as those described in C. All results are representative of at least three independent experiments. Error bars correspond to one standard deviation from the mean of triplicate samples within the same experiment. Asterisks denote statistically significant differences in complementation by Student’s t-test (p < 0.05).
In these experiments, we noticed that B. anthracis HssR-HssSmyc provides significantly greater complementation to S. aureus ΔhssRS than HssR-HssSmyc from S. aureus (Fig. 3A). We tested whether differential complementation is conserved in B. anthracis ΔhssRS and observed a similar trend (Fig. 3B). These results indicate that the molecular function of B. anthracis HssRS may differ from that of S. aureus HssRS in a way that provides for an increased ability to sense and respond to heme.
S. aureus and B. anthracis HssS exhibit differential function in vivo
The ability of B. anthracis HssR-HssSmyc to impart elevated hemin resistance may be due to differences in HssR, HssS, or both. We reasoned that HssS differences are likely to contribute to differential activity of this TCS, as B. anthracis/S. aureus HssS share significantly less sequence identity than HssR (Stauff and Skaar, 2008). We performed complementation experiments with HssSmyc alone and found that B. anthracis HssSmyc provides significantly greater hemin resistance to S. aureus ΔhssS than HssSmyc from S. aureus (Fig. 3C). We next attempted to repeat these observations in B. anthracis. Notably, HssSmyc from B. anthracis complements B. anthracis ΔhssS, while HssSmyc from S. aureus fails to complement this strain in spite of similar expression of both proteins (Fig. 3D). These results suggest that, although S. aureus and B. anthracis HssRS are functionally orthologous TCS, B. anthracis HssRS provides elevated hemin resistance to bacteria due to differences in HssS function.
HssS chimeras reveal a complex domain interplay leading to elevated B. anthracis HssS activity
The ability of B. anthracis HssS to provide greater hemin resistance than S. aureus HssS may be due to differences in the function of any of the domains comprising HssS. A membrane-localized histidine kinase, HssS is predicted to contain four domains: an N-terminal signal recognition domain flanked by two transmembrane helices (Mascher et al., 2006), and a tripartite signaling domain consisting of a HAMP linker domain (Aravind and Ponting, 1999), a phosphate-accepting and response regulator recognition HisKA domain (Grebe and Stock, 1999), and a C-terminal ATPase domain (Grebe and Stock, 1999).
To define HssS domains required for the increased in vivo activity of B. anthracis HssS, we constructed chimeric forms of HssSmyc with either the sensing, HAMP, HisKA, or ATPase domain of S. aureus HssS in place of the respective B. anthracis HssS domain (Fig. 4; for detailed description of HssS chimeras, see Table S1). We reasoned that if a single B. anthracis HssS domain is responsible for the increased in vivo activity of this histidine kinase, replacing this domain with the corresponding S. aureus domain would decrease the function of the resulting chimera. The function of each chimera was tested in S. aureus instead of B. anthracis due to the inability of S. aureus HssS to rescue B. anthracis ΔhssS (Fig. 3D) or to signal to B. anthracis HssR (see Fig. 6F).
Figure 4. Rescue of S. aureus ΔhssS by S. aureus/B. anthracis HssS chimeras.
Left: schematic of HssS chimeras. Black: S. aureus HssS; grey: B. anthracis HssS; white: C-terminal C-Myc epitope tag. TMD1/2, predicted transmembrane domains; ECD, predicted extracytoplasmic sensing domain; HAMP, intracytoplasmic linker domain; HisKA, phosphate acceptor domain; ATPase, ATP hydrolysis domain. Chimera junctions are defined in Table S1. Right side: chimera functional data. Column 1: Expression of HssS chimeras. Triplicate cultures of S. aureus ΔhssS expressing each shown HssS variant were prepared and HssS expression was analyzed by immunoblot against the C-Myc epitope tag of HssS followed by LiCor-based quantification. Chimera levels relative to abundance of S. aureus HssS were determined and averaged. Standard deviations are shown. Data corresponding to chimeras with less than 50% expression are highlighted in grey. Columns 2 and 3: rescue of ΔhssS by HssS chimeras. Column 2: strains were inoculated into medium containing 20 µM hemin (a hemin concentration that partially inhibits wt S. aureus and completely inhibits S. aureus ΔhssS) and proliferation was tracked by growth curve analysis. The length of time required to initiate growth (OD600 = 0.2) was determined. Column 3: strains were inoculated into medium containing 10, 20, or 30 µM hemin. The highest hemin concentration each strain was able to grown in by 24 hours is indicated. Results are representative of at least three experiments.
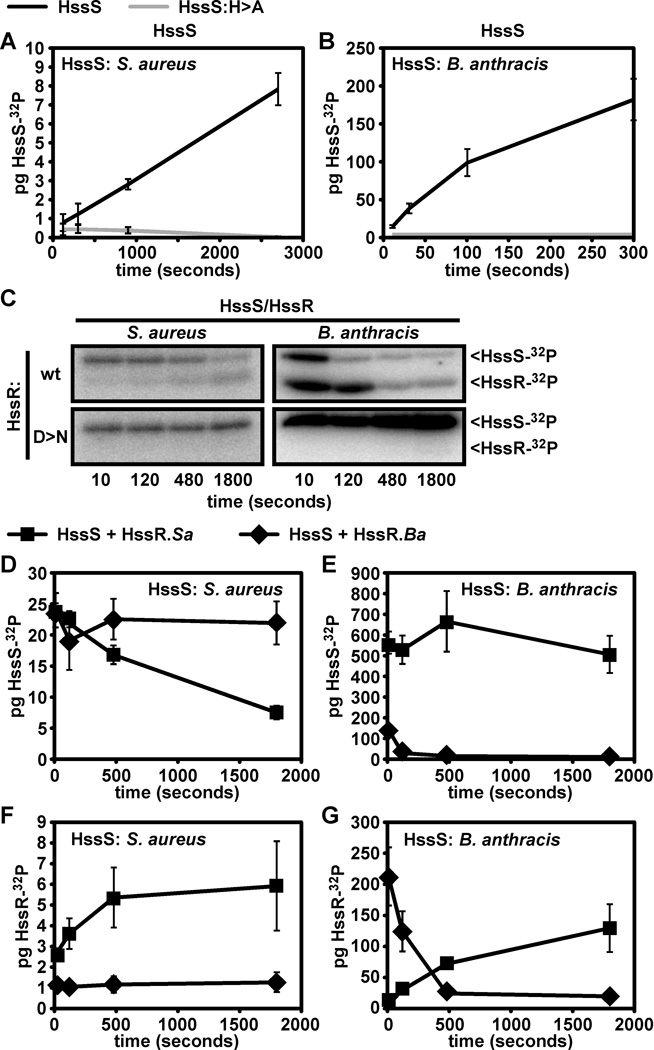
Figure 6. B. anthracis HssS–HssR exhibit enhanced autophosphorylation and phosphotransfer in vitro.
Symbols for proteins used in A and B. Black lines: wildtype HssS signaling domain (S. aureus HssS in A, B. anthracis HssS in B); grey lines: mutant HssS with alanine substituted for predicted phosphorylated histidine residue (HssS:H>A). A. Autophosphorylation of S. aureus HssS and phosphorylation site mutant. The described S. aureus HssS variants were incubated with 32P-ATP for the indicated time and samples were taken and analyzed by SDS-PAGE followed by quantification via phosphorimager analysis. B. Autophosphorylation of the described B. anthracis HssS variants. Experiments were performed as described in A. C. Intraspecies HssS-HssR phosphotransfer using S. aureus and B. anthracis HssRS. Phosphorylated S. aureus (left panels) or B. anthracis (right panels) HssS was added to wildtype HssR (wt, top panels) or HssR mutated at the predicted phosphorylated residue (D>N, bottom panels). Samples were analyzed as described in A. D through G: interspecies phosphotransfer. Symbols for reactions analyzed in D through G: Lines with squares, reactions containing HssS (S. aureus HssS in D and F, B. anthracis HssS in E and G) and wildtype S. aureus HssR (HssR.Sa); lines with diamonds, reactions containing HssS and wildtype B. anthracis HssR (HssR.Ba). D. Amount of phosphorylated HssS in phosphotransfer reactions containing S. aureus HssS. Panel shows levels of phosphorylated HssS after the addition of HssR from the indicated species. E. Amount of phosphorylated HssS in phosphotransfer reactions containing B. anthracis HssS. Experiment was performed as in D using B. anthracis HssS. F. HssR phosphorylation in phosphotransfer reactions containing S. aureus HssS. G. HssR phosphorylation in phosphotransfer reactions containing B. anthracis HssS. Quadruplicate reactions were performed and averaged for all experiments. Error bars correspond to one standard deviation from the mean of data from all four experiments.
Surprisingly, all chimeras which express in S. aureus and contain a single S. aureus domain allow this organism to proliferate to similar levels as S. aureus expressing full length B. anthracis HssSmyc (Fig. 4). In contrast, all chimeras containing two or more S. aureus HssS domains (with the exception of a chimera containing the entire S. aureus signaling domain, which is non-functional) function similar to S. aureus HssS in vivo, requiring over 8 hours to proliferate in 20 µM hemin (Fig. 4). These data indicate that B. anthracis HssS is able to withstand single domain swapping with S. aureus HssS and retain its elevated function. It is likely that a complex interplay between HssS domains is responsible for the differential activity between S. aureus and B. anthracis HssS, an interplay that is perturbed by the insertion of more than one S. aureus HssS domain into B. anthracis HssS.
B. anthracis HssS requires conserved residues in the predicted extracytoplasmic sensing domain for function in vivo
In HssS, signal recognition is predicted to occur within the putative extracytoplasmic domain, which is flanked by transmembrane helices (see Fig. 5A for schematic). However, no studies have addressed the importance of this input domain in HssS function. In this regard, we subjected the predicted sensing domain of B. anthracis HssS to mutational analysis to test whether it is required for HssS activity.
Figure 5. Residues within the predicted B. anthracis HssS sensing domain required for HssS function.
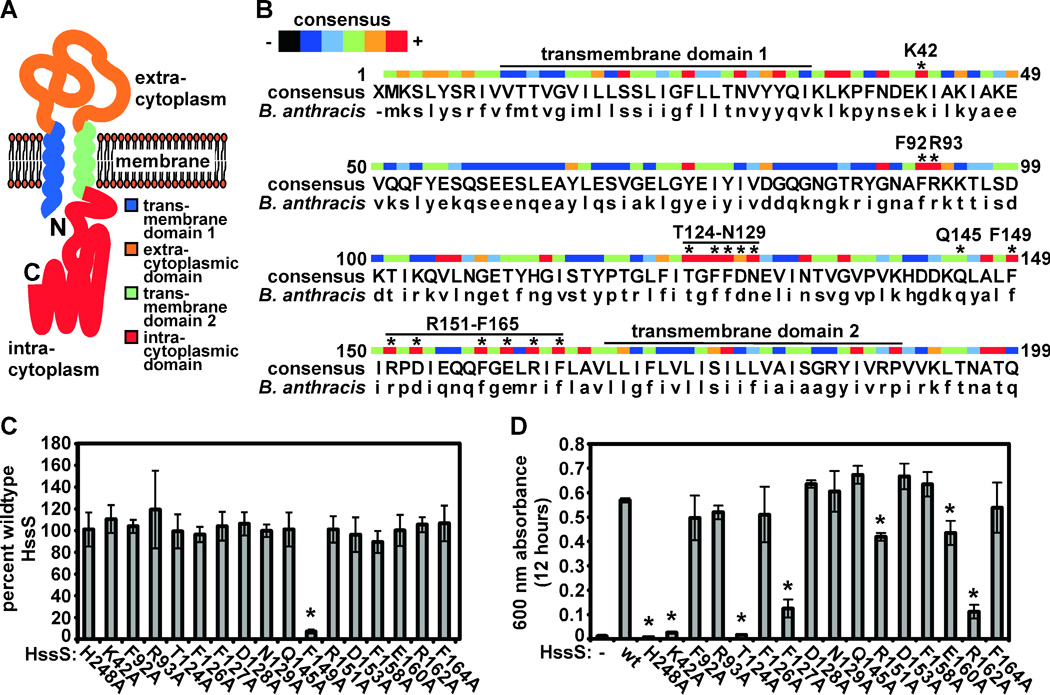
A. Schematic of B. anthracis HssS. B. anthracis HssS is predicted to consist of an N-terminal signal peptide/transmembrane domain (blue), an extracytoplasmic sensing domain (orange), a second transmembrane domain (green), and an intracytoplasmic signaling domain (red). B. Alignment of conserved HssS sensing domain residues. An HssS consensus sequence was generated from an alignment performed using data from all species with complete genome sequences that are predicted to contain hss/hrt (capital letters). Color code indicates level of conservation (blue, little conservation; red, absolute conservation). Asterisks above residues designate amino acids subjected to alanine substitution mutagenesis. The signaling domain autophosphorylated histidine residue (H248) was also selected for mutagenesis. C. Expression of B. anthracis HssSmyc point mutants. Triplicate cultures of B. anthracis ΔhssS expressing each HssSmyc point mutant were prepared and HssS expression was analyzed by immunoblot against the C-Myc epitope tag of HssSmyc followed by LiCor-based quantification. Mutant levels relative to abundance of wildtype B. anthracis HssSmyc were determined and averaged. Error bars correspond to standard deviations; asterisk denotes point mutant with expression significantly lower than wild type. D. Rescue of B. anthracis ΔhssS by HssS point mutants. Strains were inoculated into BHI containing 20 µM hemin (a hemin concentration that does not significantly inhibit wt B. anthracis in BHI but completely inhibits B. anthracis ΔhssS) and grown for 12 hours. The mean optical density of triplicate cultures was determined. Error bars correspond to standard deviations; asterisks denote strains with culture density significantly lower than those of ΔhssS expressing wildtype HssSmyc.
To identify residues within the predicted sensing domain that may be required for HssS activity, we aligned HssS sequences from Firmicutes predicted to encode hss/hrt. This analysis revealed a number of conserved residues within the predicted sensing domain (Fig. 5B). We selected 15 perfectly or highly conserved residues and a single non-conserved residue in B. anthracis HssS for alanine substitution mutational analysis, focusing on two patches of conserved sequence (T124-N129 and F149-F165, Fig. 5B). We also mutated the predicted phosphorylated histidine residue (H248) of HssS. We then expressed these point mutants in B. anthracis ΔhssS and analyzed their expression and function.
By immunoblotting against the C-terminal C-Myc epitope tag added to HssS, we found that all point mutants with the exception of HssS:F149A are expressed to levels similar to wildtype HssS (Fig. 5C). As expected, HssS:H248A is unable to rescue the hemin sensitivity of B. anthracis ΔhssS, presumably due to an inability to signal to HssR (Fig. 5D). Furthermore, HssS:Q145A, a mutant in a non-conserved HssS residue, complements ΔhssS at levels similar to wildtype HssS (Fig. 5D). However, many of the point mutants are unable to complement B. anthracis ΔhssS (Fig. 5D). Specifically, B. anthracis HssS mutants at residues K42, T124, F127, R151, E160, or R162 are unable to complement ΔhssS to varying degrees. These data suggest that conserved residues within the predicted HssS sensing domain are required for HssS function.
B. anthracis HssS-HssR undergoes rapid autophosphorylation and phosphotransfer
Because autophosphorylation is the major catalytic activity of histidine kinases (Mascher et al., 2006), we hypothesized that differences in autophosphorylation between S. aureus and B. anthracis HssS may correlate with the increased ability of B. anthracis HssS to provide hemin resistance (Fig. 3C, Fig. 4). In vitro, B. anthracis HssS undergoes autophosphorylation that is dependent on conserved histidine 248 (Fig. 6B), correlating with its lack of functionality in vivo (Fig. 5D). Significantly, we found that the autophosphorylation rate of B. anthracis HssS is 208 (+/− 50) times greater than that of S. aureus HssS (Fig. 6A, 6B). This correlates with the ability of B. anthracis HssS to provide enhanced hemin resistance to S. aureus ΔhssS (Fig. 3C, Fig. 4). Furthermore, these data also correlate with the increased in vivo activity of an HssS chimera consisting of the S. aureus HssS sensing domain fused to the B. anthracis HssS signaling domain (Fig. 4, chimera 1).
We next tested HssS-HssR phosphotransfer and found that both S. aureus and B. anthracis HssRS are capable of engaging in intraspecies phosphotransfer dependent on a conserved aspartic acid residue of HssR (Fig. 6C) (Stauff et al., 2007). We also analyzed interspecies phosphotransfer using S. aureus HssS and found that B. anthracis HssR fails to catalyze phosphotransfer from S. aureus HssS (Fig. 6D, 6F), providing a potential explanation for the inability of S. aureus HssS to complement B. anthracis ΔhssS (Fig. 3D). This indicates that evolutionary divergence has occurred between S. aureus and B. anthracis HssRS. We also tested interspecies phosphotransfer with B. anthracis HssS and found that HssR from both species are capable of catalyzing phosphotransfer from B. anthracis HssS (Fig. 6C, 6E, 6G). Notably, when phosphorylated B. anthracis HssS is incubated with S. aureus HssR, phosphorylated B. anthracis HssS levels do not decrease because B. anthracis HssS continues to undergo autophosphorylation even after HssR is added (Fig. 3E).
In these experiments, we observed differences in the rate of B. anthracis and S. aureus HssS-HssR phosphotransfer. Specifically, we found that B. anthracis HssS-HssR phosphotransfer occurs rapidly (peaking in under 10 seconds, Fig. 6G) while S. aureus HssS-HssR phosphotransfer occurs at a relatively slow rate (peaking at about 10 minutes, Fig. 6F). The rate of phosphotransfer appears to be dictated by HssR rather than HssS, as slow phosphotransfer on the part of S. aureus HssR also occurs in a phosphotransfer reaction containing B. anthracis HssS (Fig. 6G). These results indicate evolutionary divergence between S. aureus and B. anthracis HssR that favors rapid phosphorylation of B. anthracis HssR. Importantly, the increased rate of B. anthracis HssS phosphorylation and HssS-HssR phosphotransfer may in part explain the ability of this TCS to provide elevated hemin resistance to S. aureus (Fig. 3A).
B. anthracis ΔhrtA is more sensitive to hemin toxicity than S. aureus ΔhrtA
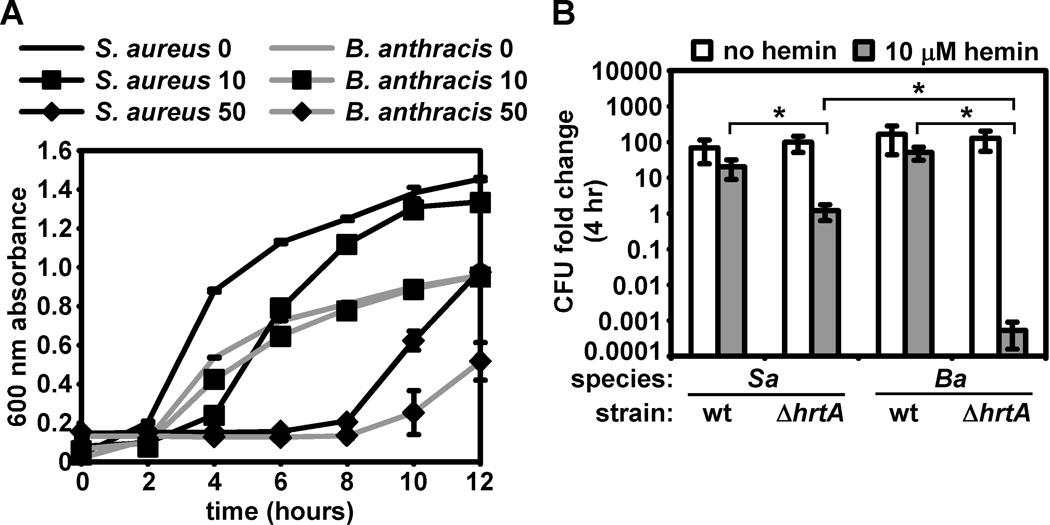
We reasoned that the enhanced function of B. anthracis HssRS may confer elevated hemin resistance to B. anthracis when compared with S. aureus. However, we found that both species display similar hemin resistance by growth curve analysis (Fig. 7A). We next reasoned that in the absence of Hss/Hrt function, B. anthracis may display enhanced sensitivity to hemin toxicity and may therefore require elevated HssRS activity to compensate. We therefore tested the relative hemin sensitivity of S. aureus ΔhrtA and B. anthracis ΔhrtA and found that after 4 hours in 10 µM hemin, S. aureus ΔhrtA exhibits a slight growth inhibition, while B. anthracis ΔhrtA exhibits a dramatic decrease in surviving bacteria (Fig. 7B). Together with previous data, these results suggest that B. anthracis may have evolved a highly active HssRS system to cope with the fact that this organism is extremely susceptible to hemin toxicity.
Figure 7. Differential hemin sensitivity of S. aureus and B. anthracis ΔhrtA.
A. Proliferation of wildtype S. aureus and B. anthracis in hemin. Growth curve analysis was performed on S. aureus (black lines) and B. anthracis (grey lines) grown in BHI without hemin (no symbols), BHI + 10 µM hemin (a hemin concentration that inhibits hss/hrt mutants; boxes), or BHI + 50 µM hemin (a hemin concentration that inhibits wt S. aureus and B. anthracis; diamonds). B. Resistance of wildtype and ΔhrtA S. aureus and B. anthracis to hemin. S. aureus (Sa) and B. anthracis (Ba) wildtype (wt) and ΔhrtA were grown overnight and subcultured into BHI or BHI + 10 µM hemin. At the time of subculture and after a four-hour incubation, serial dilutions were performed and plated and colony forming units (CFU) were enumerated. Data is presented as the fold change in CFU following incubation in the presence or absence of hemin (log scale). Asterisks denote statistically significant differences by Student’s t-test (p < 0.05). All results are representative of triplicate independent experiments. Error bars correspond to one standard deviation from the mean of triplicate samples within the same experiment; asterisks denote statistically significant differences by Student’s t test (p<0.05).
B. anthracis experiences heme stress during growth within vertebrates
Given the capacity of B. anthracis to acquire heme (Gat et al., 2008; Maresso et al., 2006; Maresso et al., 2008; Skaar et al., 2006) and to avoid hemin toxicity through HssRS-regulated expression of HrtAB (Fig. 1 and Fig. 2), we wondered whether HrtAB is expressed during systemic anthrax infection. To this end, we constructed a luminescence-based hrtAB promoter reporter by fusing the S. aureus hrtAB promoter to the lux genes of Photorabdus luminescens in the plasmid pXen-1 (Francis et al., 2000). We utilized the S. aureus hrtAB promoter instead of the B. anthracis hrtAB promoter because the latter displays significant activity in the absence of hemin (Fig. 2C), making hemin-dependent induction and constitutive promoter activity difficult to differentiate. When B. anthracis is transformed with this construct, bacilli undergo luminescence in response to hemin (Fig. 8A).
Figure 8. HrtAB expression in an animal model of cutaneous anthrax.
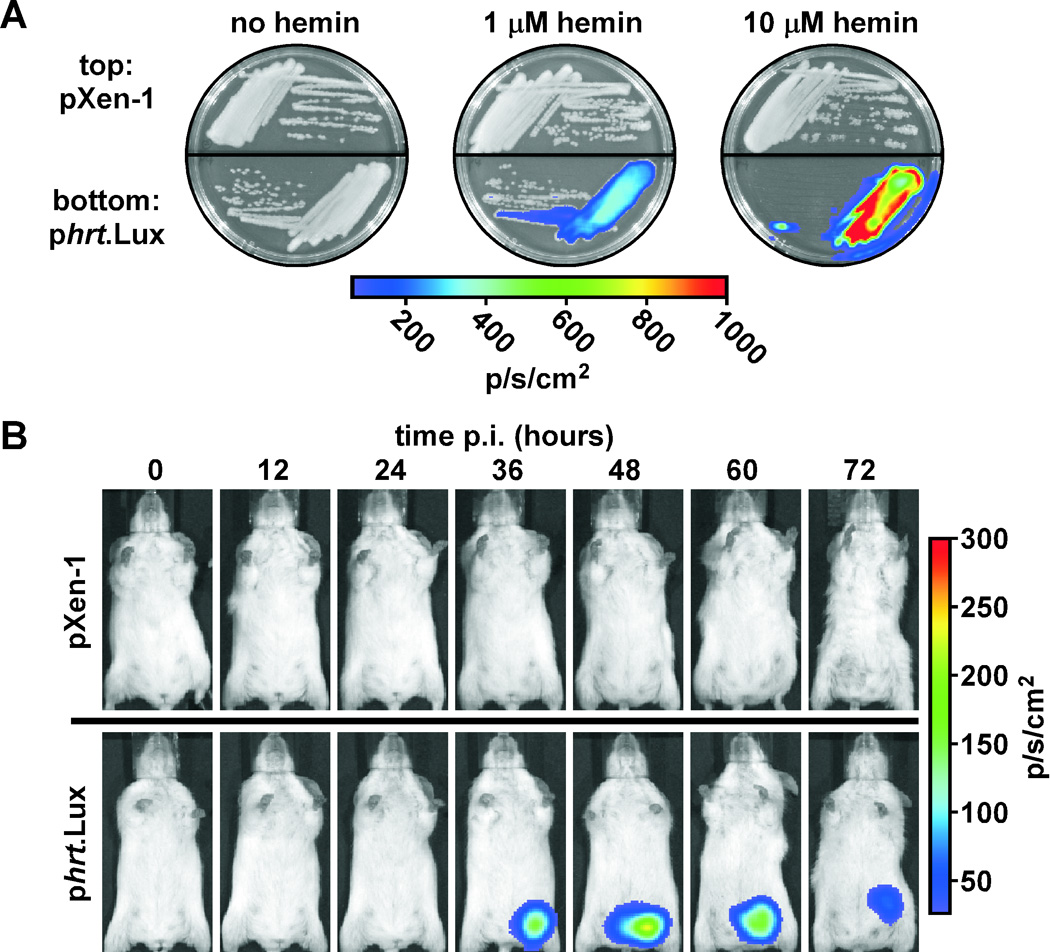
A. Hemin-dependent luminescence of B. anthracis harboring phrt.Lux. B. anthracis harboring promoterless pXen-1 or pXen-1 with the Lux genes under the control of the hrtAB promoter (phrt.Lux) were grown on BHI agar containing no hemin, BHI + 1 µM hemin, or BHI + 10 µM hemin. Plates were visualized by IVIS. B. Activation of the hrtAB promoter during anthrax infection. Spores of B. anthracis harboring pXen-1 or phrt.Lux were used to infect groups of five A/J mice via a sub-dermal injection in the left inguinal region. Luminescence of B. anthracis was tracked every 12 hours post infection (p.i.) by IVIS. Shown is one mouse representative of each group.
We next infected mice subcutaneously (Chand et al., 2009; Glomski et al., 2007; Lamanna and Jones, 1963; Watts et al., 2009) with spores prepared from B. anthracis harboring either pXen-1 or phrt.Lux and tracked bioluminescence of germinating B. anthracis cells over time. We found that B. anthracis harboring pXen-1 do not undergo detectable bioluminescence during infection (Fig. 8B, top), but that B. anthracis harboring phrt.Lux induces detectable levels of bioluminescence 24–36 hours post infection (Fig. 8B, bottom). Based on the strict hemin responsiveness of the S. aureus hrtAB promoter in B. anthracis (Fig. 2B, Fig. 8A), these data suggest that during infection, B. anthracis activates HssRS to alleviate heme toxicity.
DISCUSSION
Bacillus anthracis is capable of invading host tissues to cause disease. During infection, B. anthracis acquires host heme to satisfy nutrient iron needs (Gat et al., 2008; Maresso et al., 2006; Maresso et al., 2008; Reniere et al., 2007). It appears that for B. anthracis, amassing of exogenous heme as a nutrient must be balanced with the need to overcome the toxicity of the heme molecule.
We have demonstrated a high level of orthology between the S. aureus and B. anthracis Hss/Hrt systems. S. aureus and B. anthracis mutants lacking members of these operons display a similar hemin-sensitive phenotype (Fig. 1) (Torres et al., 2007). Also, B. anthracis hssRS and hrtAB are able to complement S. aureus mutants lacking these genes, indicating orthologous function of both systems (Fig. 2, Fig. 3). Furthermore, our data demonstrate functional overlap between the S. aureus and B. anthracis heme sensor systems at the levels of signal sensing, signal transduction, autophosphorylation by HssS, phosphotransfer to HssR, interaction between HssR and the hrtAB promoter, and activation of the hrtAB promoter by HssR (Fig. 2, Fig. 3, Fig. 6) (Stauff et al., 2007). These similarities point to the overall evolutionary conservation of Hss/Hrt function between S. aureus and B. anthracis, a further indication that these systems are important for the fitness of both bacteria at some point throughout their life cycles.
Nonetheless, we have observed significant functional differences between HssRS/HrtAB from S. aureus and B. anthracis. First, the B. anthracis hrtAB promoter displays significant activity in the absence of hemin (Fig. 2C, 2D). This activity is dependent on HssRS and the DR to which HssR binds, suggesting that B. anthracis HssRS engages in constitutive signaling that induces low-level activation of the hrtAB promoter. The fact that this occurs only in B. anthracis and that most DR mutations are tolerated to a similar extent by both species points to sequences outside of the DR that may be important for HrtAB expression. It is possible that other unidentified factors expressed only by B. anthracis (such as the putative transcriptional regulator located between hssRS and hrtAB, Fig. 1A) may interact with the hrtAB promoter outside of the DR, functioning with HssR to increase expression of HrtAB. Alternatively, another B. anthracis-specific two-component systems may utilize the hrtAB promoter DR, overlapping with HssRS to induce expression of HrtAB (de Been et al., 2008).
Another key functional difference between the S. aureus and B. anthracis heme sensor systems occurs at the levels of HssS autophosphorylation and phosphotransfer to HssR (Fig. 6). The signaling domain of B. anthracis HssS displays a significantly elevated rate of autophosphorylation in vitro compared to the corresponding S. aureus HssS domain. Although the molecular basis for the increased autophosphorylation rate of the B. anthracis HssS signaling domain is unclear, it is likely that it involves interplay between the three cytoplasmic sub-domains. This is indicated by data demonstrating that the increased in vivo activity of full-length B. anthracis HssS can be reversed by insertion of multiple HssS domains from S. aureus HssS (Fig. 4). We have also found that B. anthracis HssS-HssR phosphotransfer occurs more rapidly than S. aureus HssS-HssR phosphotransfer. These activities are likely to allow the rapid induction of HrtAB expression in B. anthracis, alleviating heme toxicity. Accordingly, S. aureus expressing B. anthracis HssRS is more resistant to hemin toxicity than wildtype S. aureus. These results suggest that a certain degree of functional divergence has occurred throughout the course of evolution between S. aureus and B. anthracis HssRS.
In the absence of the ability to respond to growth in the presence of hemin, B. anthracis is hypersensitive to hemin toxicity (Fig. 7). It is possible that B. anthracis may amass hemin more rapidly than S. aureus or that B. anthracis may be more sensitive to hemin-induced perturbation than S. aureus. It should be noted that the Isd heme iron acquisition system is not expressed in the conditions used in these experiments (data not shown), indicating that heme toxicity can occur independently of Isd-mediated heme uptake. Regardless of the reason for increased hemin susceptibility in the absence of a functional heme detoxification response, the hemin resistance of wildtype B. anthracis is similar to that of wildtype S. aureus. This is an indication that in B. anthracis, HssRS and/or HrtAB function more efficiently than the corresponding systems in S. aureus. Throughout the course of evolution, B. anthracis may have accumulated amino acid substitutions in HssRS that lead to constitutive activity of this TCS, increased autophosphorylation of HssS, and rapid HssS-HssR phosphotransfer. It is possible that these activities compensate for the hemin sensitivity of B. anthracis and that such compensatory mutations may have been necessitated by the association between B. anthracis and host tissues rich in heme.
As the first component in the HssRS/HrtAB-mediated process of heme detoxification, the histidine kinase HssS controls heme resistance by detecting exogenous heme and transducing signals through the membrane. However, the mechanism of heme sensing by this histidine kinase remains elusive (Stauff and Skaar, 2008). Here, we present data showing that mutation of any of a number of conserved residues in the predicted HssS sensing domain eliminates the ability of B. anthracis to overcome heme toxicity. These residues cluster within two conserved patches, a possible indication that these regions of the protein interface with the HssS ligand. Although these studies do not identify the signal directly recognized by HssS, they do suggest that the previously uncharacterized sensing domain of HssS is involved in detection of heme. The fact that this sensing domain is flanked by transmembrane helices and is predicted to reside in the extracytoplasmic space indicates that HssS detects a signal located either between the membrane and the cell wall, or within the membrane itself. We hypothesize that the HssS ligand may be heme or a molecule that is either modified or induced by heme exposure, and that this molecule is recognized by conserved HssS sensing domain residues shown here to be required for HssS activity.
The fact that B. anthracis activates the hrtAB promoter during infection of vertebrates (Fig. 8) and heme detoxification appears to be involved in S. aureus virulence (Torres et al., 2007) points to a role for heme sensing during the life cycles of two Gram-positive pathogens. Furthermore, a number of pathogenic and saprotrophic relatives of S. aureus and B. anthracis including B. cereus and Listeria monocytogenes are predicted to express HssRS/HrtAB (Stauff and Skaar, 2008; Torres et al., 2007). It is therefore likely that heme sensing and detoxification are processes engaged in by a number of related Gram-positive organisms. This is supported by the observation made by Van Heyningen of differential hemin resistance among phylogenetically related Bacilli (Heyningen, 1948). Van Heyningen’s method may have differentiated organisms that express functional Hss/Hrt systems from those that do not based solely upon the hemin resistance of the organism. Importantly, the fact that HssRS/HrtAB orthologues are only found in those Bacilli that exhibit pathogenic or saprotrophic lifestyles indicates that these systems are not required for the fitness of bacteria that do not encounter exogenous heme throughout their life cycles.
It has been suggested that heme sensing and detoxification evolved in dense microbial communities rich in heme-synthesizing bacteria (Miller et al., 2007; Pedersen et al., 2008). This may explain the recent discovery of a heme-regulated HrtAB orthologue required for hemin resistance in the lactic acid bacterium Lactococcus lactis (Pedersen et al., 2008). Although L. lactis is not a pathogen, it may encounter environmental heme synthesized by nearby bacteria (Tatsumi and Wachi, 2008), necessitating HrtAB function (Pedersen et al., 2008). Notably, L. lactis does not possess hssRS genes and may regulate expression of hrtAB through a mechanism other than TCS-based signal sensing (Pedersen et al., 2008). This indicates evolutionary conservation of HrtAB function but divergence of HrtAB regulation between the Lactobacillales (which include the Streptococci as well as L. lactis) and the Bacillales (including S. aureus and B. anthracis).
Throughout the course of evolution, the pathogenic or saprotrophic Firmicutes may have acquired heme detoxification systems from bacteria such as L. lactis, thereby being provided with a means to facilitate survival within host tissues rich in heme. Within the Firmicutes, selective pressure to maintain a heme sensor system and a heme detoxification system may only be exerted on bacteria that commonly associate with exogenous heme. The evolutionary origin of the bacterial heme detoxification response will become clearer as the mechanism of heme sensing and detoxification are elucidated and as more organisms are identified that are capable of sensing and responding to heme toxicity.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
S. aureus strain Newman (Duthie and Lorenz, 1952), ΔhrtAB (Friedman et al., 2006), ΔhrtA and ΔhssS (Torres et al., 2007) have been described previously. B. cereus, B. thuringiensis, B. israelensis, B. licheniformis, and B. subtilis were acquired from the Bacillus Genetic Stock Center (College of Biological Sciences, The Ohio State University, www.bgsc.org). B. anthracis Sterne was used for all experiments (Sterne, 1937). S. aureus was grown in tryptic soy broth (TSB) or brain-heart infusion broth (BHI); Bacilli were grown in BHI or Luria broth (LB). All plasmid construction was performed using Escherichia coli DH5α. Plasmid DNA to be introduced into B. anthracis was propagated in the E. coli strain K1077 (Kim et al., 2004). Plasmid selection was performed in S. aureus and B. anthracis by supplementing media with 10 µg/ml chloramphenicol (complementation and reporter plasmids) or 20 µg/ml kanamycin (knockout constructs).
Genetic manipulation of S. aureus
Genetic manipulation of S. aureus and construction of S. aureus ΔhssRS was performed as described previously (Bae and Schneewind, 2006; Stauff et al., 2007).
Genetic manipulation of B. anthracis
Electroporations were performed as previously described (Kim et al., 2004), with modifications. E. coli K1077 harboring plasmids to be introduced into B. anthracis were grown overnight in 15 ml of LB in 50 ml conical tubes. DNA was miniprepped from K1077 and dialyzed against water to remove traces of salt prior to electroporation. Electrocompetent B. anthracis cells were prepared by growing bacteria in BHI + 0.5 M D-sorbitol to OD600 = 0.2 and washing cells three times in SMG (0.5 M D-sorbitol, 0.5 M D-mannitol, 10% glycerol) without proteinase K treatment. Four µl of DNA was added to 50 µl competent cells and mixtures were incubated on ice for 10 minutes. Electroporations were carried out in 1 mm gap cuvettes with a Bio-Rad Gene Pulser using an exponential decay protocol set to 2500 V, 1 ms time constant. After electroporation, cells were recovered in BHI + 0.5 M D-sorbitol for 2 hours at 30 °C with shaking and plated onto BHI + antibiotic. For generation of B. anthracis hrtA, hssRS, and hssS knockout constructs for insertion/deletion mutagenesis, the erythromycin resistance gene ermC was inserted between 1 kilobase regions flanking hrtA, hssRS, or hssS in the thermosensitive mutagenesis plasmid pLM4 (Kern and Schneewind, 2008). Mutagenesis was performed as described previously (Kern and Schneewind, 2008). The incorporation of mutations, loss of pLM4, and maintenance of pXO1 were confirmed by PCR.
Epitope tagging of hssS
A C-terminal c-myc epitope tag was incorporated into S. aureus and B. anthracis hssS alone and hssS within the hssRS operon using a PCR-based approach as described previously (Stauff et al., 2007). Briefly, PCR reactions were assembled containing a 5’ primer annealing to the 5’ end of hssR (for amplification of hssR-hssSmyc) or hssS (for hssSmyc), dilutions of a 3’ primer annealing to the 3’ end of hssSmyc and encoding the c-myc epitope tag (EQKLISEEDL), and excess of a 3’ primer annealing to the c-myc coding region and incorporating a restriction endonuclease site. Tagged hssS is designated hssSmyc and tagged hssRS is designated hssR-hssSmyc.
Construction of complementation plasmids
All complementation and reporter plasmids were constructed in a pOS1 (Schneewind et al., 1992) or pOS1plgt (Bubeck Wardenburg et al., 2006) background. A B. anthracis hrtA complementation construct was generated by PCR-SOE (Horton et al., 1990) using a method described previously (Torres et al., 2007). This resulted in a seamless fusion between the B. anthracis hrtAB promoter and the B. anthracis hrtA open reading frame. PCR-SOE DNA was digested with EcoRI and BamHI and inserted into pOS1plgt. For generation of a complementation plasmid containing B. anthracis hssRS under the control of its native promoter, hssRS was amplified by PCR and inserted into pOS1 between the PstI and BamHI sites. For generation of complementation plasmids containing S. aureus/B. anthracis hssR-hssSmyc under the control of the S. aureus hssRS promoter, the S. aureus hssRS promoter was first inserted between the EcoRI and NdeI sites of pOS1plgt to make pOS1phssRS. Promoterless S. aureus and B. anthracis hssR-hssSmyc were then amplified by PCR and inserted between the NdeI and BamHI sites of pOS1phssRS. For generation of complementation plasmids containing S. aureus/B. anthracis hrtAB under the control of their native promoters, hrtAB was amplified by PCR and inserted into pOS1plgt between the XhoI and EcoRI sites. To construct plasmids encoding hssSmyc under the control of the S. aureus hssRS promoter, hssSmyc was PCR amplified and inserted into pOS1phssRS between the NdeI and BamHI sites. Chimeric forms of hssSmyc were constructed by cloning S. aureus and B. anthracis hssSmyc in tandem in pCR2.1 (Invitrogen) and joining HssS domains together by inverse PCR followed by blunt-end ligation (see Construction of XylE reporter plasmids below). Chimeric forms of hssSmyc were then inserted into pOS1phssRS. HssSmyc chimeras are described in Table S1.
B. anthracis HssS alignment and mutagenesis
An alignment of conserved residues in HssS was created by assembling HssS amino acid sequences from all available sequenced Firmicute genomes (with the exception of Exiguobacterium sibiricum, which clusters outside of the Staphylococcus, Listeria, and Bacillus clusters) and aligning by Clustal V method using the Lasergene 6 software package.
HssSmyc immunoblotting
For S. aureus HssSmyc immunoblots, cell lysates were prepared from bacteria grown in 5 ml TSB in 15 ml conical tubes for 15 hours at 37 °C with shaking. For B. anthracis HssSmyc immunoblots, cell lysates were prepared from bacteria grown in 5 ml BHI in 15 ml conical tubes for 12 hours at 30 °C with shaking. Bacteria were pelleted, washed in 5 ml TBS (50 mM tris, pH 7.5, 150 mM NaCl), digested in 0.5 ml TSM (100 mM tris, pH 7.0, 500 mM sucrose, 10 mM MgCl2) with 12.5 µg/ml lysostaphin (S. aureus) or 2 mg/ml lysozyme (B. anthracis), and protoplasts were sonicated in 0.5 ml TBS. Equal amounts of lysate and 2X SDS loading buffer were mixed, boiled and loaded onto 10% acrylamide gels, subjected to SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were probed with 9E10 anti-C-Myc monocolonal primary and AlexaFluor-680-conjugated anti-mouse secondary (Invitrogen) antibodies. Membranes were dried and scanned using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Construction of XylE reporter plasmids
The B. anthracis hrtAB promoter-xylE fusion was created by PCR-SOE (Horton et al., 1990) as described for the S. aureus hrtAB promoter-xylE fusion (Torres et al., 2007). Mutagenesis of the S. aureus hrtAB promoter-xylE fusion was carried out by inverse PCR-based mutagenesis. Briefly, a non-overlapping non-mutagenic primer and a primer introducing the desired mutation were designed which amplify in opposite directions on the plasmid template. Inverse PCR was carried out using S. aureus hrtAB promoter-xylE fusion as template and 5’ phosphorylated oligos. Amplified DNA was blunt end ligated, transformed into E. coli, miniprepped, and mutagenized constructs were validated by DNA sequencing.
Construction of Lux reporter plasmids
To generate a construct in which the lux genes of pXen-1 (Xenogen) (Francis et al., 2000) are under the control of the S. aureus hrtAB promoter (selected for lack of activity in the absence of hemin, Fig. 2C), primers containing a 5’ EcoRI site and a 3’ BamHI site were used to amplify the hrtAB promoter from S. aureus genomic DNA. PCR-amplified DNA was inserted into pXen-1 to create the plasmid phrt.Lux.
Construction of plasmids for expression of recombinant proteins
Construction of protein expression constructs for S. aureus HssS, HssS:H249A, and HssR has been described previously (Stauff et al., 2007). Construction of expression constructs for S. aureus HssR:D52N and B. anthracis HssR, HssS, HssS:H248A and HssR:D54N was performed as described previously (Stauff et al., 2007). Briefly, B. anthracis hssR or hssS were amplified by PCR and inserted between the NdeI and BamHI sites of pET15b (Invitrogen). Constructs were validated by sequencing and were then mutagenized by inverse PCR (see Construction of XylE reporter plasmids above). For protein expression, pET15 constructs were introduced into E. coli BL21 (DE3). Bacteria were grown overnight in terrific broth (TB) and subcultured 1:100 into fresh TB. Bacteria were incubated at 37 °C with shaking to OD600 = 0.4, and were then transferred to a 16 °C incubator with shaking. Cells were grown for another hour and then IPTG was added to a final concentration of 0.2 mM. Cells were grown overnight and hexahistidine-tagged proteins were purified as described previously (Stauff et al., 2007). To validate the integrity of recombinant HssS, S. aureus and B. anthracis HssS were analyzed by mass spectrometry (Vanderbilt University Proteomics Core Facility). Intact masses were determined for both proteins and N- and C-termini were validated by MS/MS following digestion with trypsin protease.
XylE reporter assays
XylE assays were performed as previously reported (Stauff et al., 2007) with modifications. Bacteria harboring reporter plasmids were grown overnight in BHI (S. aureus and B. anthracis when analyzing hemin induction of hrtAB promoter) or LB (B. anthracis when analyzing hemin-independent hrtAB promoter activity) and subcultured 1:50 into 2 ml fresh medium in 15 ml conical tubes. Bacteria were grown for 5 hours at 37 °C and cytoplasmic fractions were prepared and analyzed as previously reported (Stauff et al., 2007) except that for B. anthracis, lysozyme (2 mg/ml) was used instead of lysostaphin. Absolute XylE activities were determined for B. anthracis reporters due to lysozyme interference during protein quantification.
Bacterial growth curve analyses
Triplicate cultures of bacteria were grown 12 hours at 30 °C with shaking in 5 ml of medium in 15 ml tubes. Each culture was subcultured 1:50 into 150 µl fresh medium in a 96-well round-bottom plate. Plates were wrapped to avoid evaporation and were grown at 37 °C with shaking at 180 rpm. 600 nm absorbance readings were taken on a Cary 50 MPR microplate reader coupled to a Cary 50 Bio UV-visible spectrophotometer (Varian, Inc.). For all experiments, absorbance readings of the triplicate subcultures were averaged and one standard deviation from the mean was plotted as error bars.
Hemin preparation and concentrations
Hemin (Fluka) stocks were made by dissolving hemin in 0.1 M NaOH to a concentration of 10 mM. Hemin concentrations were chosen based on the following considerations. For all experiments attempting to assess hrtAB promoter activity without any detectable interference of hemin toxicity with strain growth (such as in hemin-sensitive mutants), promoter induction, or XylE synthesis, 2 µM hemin was utilized. Five µM hemin fully induces the hrtAB promoter in S. aureus with no detectable hemin toxicity and was therefore utilized to pre-adapt B. anthracis to hemin toxicity. Ten µM hemin was selected for growth curve analysis of B. anthracis hss/hrt mutants as a hemin concentration known to completely inhibit growth of S. aureus hss/hrt mutants within 12 hours of subculture, and as a hemin concentration known to partially inhibit growth of wildtype S. aureus in TSB. Twenty µM hemin is known to inhibit growth of S. aureus in BHI and was therefore used to inhibit growth of Bacilli; 20 µM hemin also completely inhibits growth of S. aureus and B. anthracis ΔhssS up to 24 hours and was therefore used in experiments assessing the hemin resistance of complemented derivatives of ΔhssS. Thirty µM hemin was used to inhibit the growth of S. aureus and B. anthracis ΔhssRS due to the significant hemin resistance of strains harboring hssRS complementation plasmids. Fifty µM hemin was chosen in experiments analyzing the native or pre-adapted hemin resistance of wildtype strains in BHI due to the inherent hemin resistance of these strains.
HssS-HssR autophosphorylation and phosphotransfer assays
To analyze autophosphorylation of S. aureus and B. anthracis HssS signaling domain, 25 µl reaction mixtures were prepared containing 288 µg/ml HssS, 50 mM tris, pH 8.0, 5 mM MgCl2, 200 mM KCl, 0.2 mM DTT, 10 % glycerol, 20 µM unlabeled ATP, 5 µCi [γ32P]ATP. 5 µl samples were mixed with 2X SDS-PAGE loading buffer at various time points after the addition of radiolabeled ATP, samples were resolved by SDS-PAGE, and gels were dried and analyzed using a PhosphorImager. To analyze transphosphorylation from HssS to HssR, B. anthracis and S. aureus HssS were labeled for 15 and 50 minutes, respectively, in reaction mixtures described above. HssR or HssR mutant was added to this reaction to a final concentration of 890 µg/ml. Samples were taken and analyzed as described above.
Bacterial hemin killing assays
Five ml cultures of S. aureus and B. anthracis wildtype and ΔhrtA were grown in 15 ml conical tubes for 12 hours at 30 °C with shaking at 180 rpm. Bacteria were subcultured 1:100 into BHI or BHI + 10 µM hemin and incubated at 37 °C for 4 hours. At the time of subculture and following incubation, bacterial colony forming units (CFU) were enumerated by performing serial dilutions in PBS and plating cells on BHI agar.
B. anthracis infections and IVIS
Spores for infection of mice were prepared as described previously (Marraffini and Schneewind, 2006) with modifications. Briefly, B. anthracis harboring pXen-1 or phrt.Lux were grown for 12 hours in 5 ml 2X SG medium (Leighton and Doi, 1971) containing 10 µg/ml chloramphenicol in 15 ml conical tubes at 30 °C with shaking. Cells were subcultured 1:100 into 250 ml flasks containing 30 ml 2X SG with 10 µg/ml chloramphenicol. Flasks were shaken at 37 °C for 4 days. Spore-containing cultures were then pelleted and washed three times with water, heated to 70 °C for 30 minutes, and then washed again with water. Purified spores were serially diluted and plated onto BHI to obtain a viable spore count. Spore preparations were stained using malachite green to microscopically confirm the presence of spores and the absence of vegetative bacilli. Spores harboring Lux plasmids were diluted to a concentration of 200 spores/µl in sterile PBS. One-hundred µl of spore dilutions (20000 spores/mouse) were injected subcutaneously into the inguinal region of 6-week-old A/J mice (two groups of 5 mice). Following spore inoculation, mice were visualized at 12-hour intervals using a Xenogen IVIS 200. Mice were monitored for signs of disease and were sacrificed when found to be moribund.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anthony Maresso and Justin Kern from the laboratory of Dr. Olaf Schneewind for technical advice and assistance with B. anthracis genetic manipulations and infections. We thank David Friedman from the Vanderbilt University Department of Biochemistry for mass spectral analyses of recombinant proteins. We are grateful for critical advice from Dr. David Wright of the Vanderbilt University Department of Chemistry. We also thank Dr. Henry C. Manning of the Vanderbilt University Institute of Imaging Science for assistance with IVIS experiments. We thank members of the Skaar laboratory and Dr. Phil Hanna for critical reading of this manuscript. We thank Xenogen for the plasmid pXen-1. We thank the Bacillus Genetic Stock Center for providing us with Bacillus strains. This work was supported by the Searle Scholars Program, and United States Public Health Service Grants 5 U54 AI057157-06 (SERCEB), AI69233 and AI073843-01A2 from the NIAID, National Institutes of Health. D.L.S was supported by NIAID, NIH Grant T32 HL069765. E.P.S. is a Burroughs Wellcome Fellow in the Pathogenesis of Infectious Disease.
REFERENCES
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bezkorovainy A. Antimicrobial properties of iron-binding proteins. Adv Exp Med Biol. 1981;135:139–154. doi: 10.1007/978-1-4615-9200-6_8. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen JJ, Griffiths E. Iron and Infection: Molecular, Physiological and Clinical Aspects. New York: John Wiley and Sons; 1999. [Google Scholar]
- Chand HS, Drysdale M, Lovchik J, Koehler TM, Lipscomb MF, Lyons CR. Discriminating virulence mechanisms among Bacillus anthracis strains by using a murine subcutaneous infection model. Infect Immun. 2009;77:429–435. doi: 10.1128/IAI.00647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- de Been M, Bart MJ, Abee T, Siezen RJ, Francke C. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ Microbiol. 2008;10:2796–2809. doi: 10.1111/j.1462-2920.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- Dixon TC, Meselson M, Guillemin J, Hanna PC. Anthrax. N Engl J Med. 1999;341:815–826. doi: 10.1056/NEJM199909093411107. [DOI] [PubMed] [Google Scholar]
- Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- Everse J, Hsia N. The toxicities of native and modified hemoglobins. Free Radic Biol Med. 1997;22:1075–1099. doi: 10.1016/s0891-5849(96)00499-6. [DOI] [PubMed] [Google Scholar]
- Francis KP, Joh D, Bellinger-Kawahara C, Hawkinson MJ, Purchio TF, Contag PR. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect Immun. 2000;68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus Redirects Central Metabolism to Increase Iron Availability. PLoS Pathog. 2006;2 doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gat O, Zaide G, Inbar I, Grosfeld H, Chitlaru T, Levy H, Shafferman A. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol Microbiol. 2008;70:983–999. doi: 10.1111/j.1365-2958.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- Glomski IJ, Corre JP, Mock M, Goossens PL. Noncapsulated toxinogenic Bacillus anthracis presents a specific growth and dissemination pattern in naive and protective antigen-immune mice. Infect Immun. 2007;75:4754–4761. doi: 10.1128/IAI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Heyningen V. Inhibition of aerobic sporing Bacilli by Haematin. Nature. 1948;162:114. doi: 10.1038/162114b0. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Kern JW, Schneewind O. BslA, a pXO1-encoded adhesin of Bacillus anthracis. Mol Microbiol. 2008;68:504–515. doi: 10.1111/j.1365-2958.2008.06169.x. [DOI] [PubMed] [Google Scholar]
- Kim HS, Sherman D, Johnson F, Aronson AI. Characterization of a major Bacillus anthracis spore coat protein and its role in spore inactivation. J Bacteriol. 2004;186:2413–2417. doi: 10.1128/JB.186.8.2413-2417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna C, Jones L. Lethality for Mice of Vegetative and Spore Forms of Bacillus Cereus and Bacillus Cereus-Like Insect Pathogens Injected Intraperitoneally and Subcutaneously. J Bacteriol. 1963;85:532–535. doi: 10.1128/jb.85.3.532-535.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton TJ, Doi RH. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971;246:3189–3195. [PubMed] [Google Scholar]
- Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Miller SI, Hoffman LR, Sanowar S. Did bacterial sensing of host environments evolve from sensing within microbial communities? Cell Host Microbe. 2007;1:85–87. doi: 10.1016/j.chom.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Pedersen MB, Garrigues C, Tuphile K, Brun C, Vido K, Bennedsen M, Mollgaard H, Gaudu P, Gruss A. Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J Bacteriol. 2008;190:4903–4911. doi: 10.1128/JB.00447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere ML, Torres VJ, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- Shannon JG, Ross CL, Koehler TM, Rest RF. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect Immun. 2003;71:3183–3189. doi: 10.1128/IAI.71.6.3183-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff DL, Torres VJ, Skaar EP. Signaling and DNA-binding Activities of the Staphylococcus aureus HssR-HssS Two-component System Required for Heme Sensing. J Biol Chem. 2007;282:26111–26121. doi: 10.1074/jbc.M703797200. [DOI] [PubMed] [Google Scholar]
- Stauff DL, Bagaley D, Torres VJ, Joyce R, Anderson KL, Kuechenmeister L, Dunman PM, Skaar EP. Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J Bacteriol. 2008;190:3588–3596. doi: 10.1128/JB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauff DL, Skaar EP. The heme sensor system (HssRS) of Staphylococcus aureus. Contributions to Microbiology. 2008 doi: 10.1159/000219376. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne M. Avirulent anthrax vaccine. Onderstepoort J Vet Sci Animal Ind. 1937;21:41–43. [PubMed] [Google Scholar]
- Tatsumi R, Wachi M. TolC-dependent exclusion of porphyrins in Escherichia coli. J Bacteriol. 2008;190:6228–6233. doi: 10.1128/JB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman P, Joyce S, Skaar EP. A Staphylococcus aureus Regulatory System that Responds to Host Heme and Modulates Virulence. Cell Host & Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts CJ, Hahn BL, Sohnle PG. Resistance of Athymic Nude Mice to Experimental Cutaneous Bacillus anthracis Infection. J Infect Dis. 2009 doi: 10.1086/596631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.