Abstract
Congenital diaphragmatic hernia (CDH) is a common and well-studied birth defect. The etiology of most cases remains unknown but increasing evidence points to genetic causation. The data supporting genetic etiologies which are detailed below include the association of CDH with recurring chromosome abnormalities, the existence of CDH-multiplex families, and the co-occurrence of CDH with additional congenital malformations.
Keywords: congenital diaphragmatic hernia, Bochdalek hernia, Morgagni hernia, classification, mortality, epidemiology, isolated CDH, syndromic CDH, genetic syndromes, multiplex families, chromosome abnormalities
INTRODUCTION
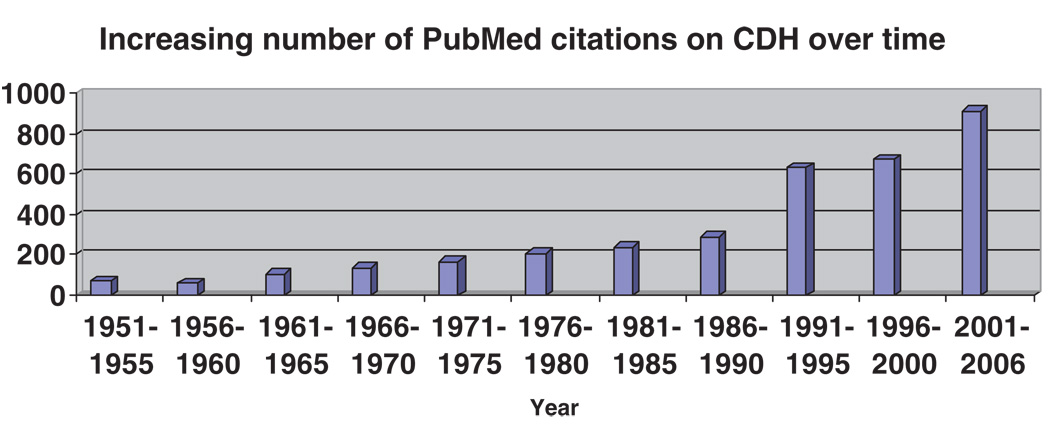
Congenital diaphragmatic hernia (CDH) is a common, dramatic, and often fatal birth defect. It is not surprising that CDH has long been the focus of scrutiny, with hand-drawn illustrations of diaphragmatic defects dating back over 500 years. There are an abundance of papers reporting on surgical techniques, management of common co-existing medical problems such as pulmonary hypoplasia and pulmonary hypertension, complications for long-term survivors, and imaging modalities for prenatal diagnosis and prognostication. The number of publications on “congenital diaphragmatic hernia” listed in PubMed has exploded in the last 15 years, in part attributable to methodological advances that permit genetic dissection of complex birth defects (Fig. 1).
Figure 1.
Increase in the number of PubMed citations containing “Congenital Diaphragmatic Hernia” (CDH) in the title from 1951 to 2006.
DEFINITION OF CDH
A congenital diaphragmatic hernia (CDH), sometimes referred to as a congenital diaphragmatic defect, is an abnormality in the integrity of the diaphragm, most often a discontinuity (e.g., “hole”), but sometimes an under-muscularization (e.g., thinning or eventration), in one or more of the diaphragmatic leaflets. CDH arises during prenatal development and the majority of cases can be detected in utero by either ultrasound examination or fetal MRI scanning.
CLASSIFICATION OF CDH
Traditionally, diaphragmatic hernias have been classified according to their presumed anatomic location. Almost 90% of diaphragmatic hernias are reported to involve the posterolateral aspect of the diaphragm; hernias in this location are referred to as Bochdalek hernias. Non-posterolateral or nonBochdalek hernias occur elsewhere, most often in the anterior portion of the diaphragm; usually anterior hernias are called Morgagni hernias. However, close scrutiny of these two supposedly distinct hernia types demonstrates considerable imprecision, since defects do not always localize to either the anterior or to the posterolateral diaphragmatic quadrant [Ackerman et al., 2006a]. A further limitation ofthe anatomic classification system is that diaphragmatic eventration and diaphragmatic hernia, previously classified as unrelated entities, have been reported to co-exist in the same patient, as well as in different affected family members [Mertins, 1952; Varpella and Lehtovaara, 1962; Thomas et al., 1976; Rodgers and Hawks, 1986].
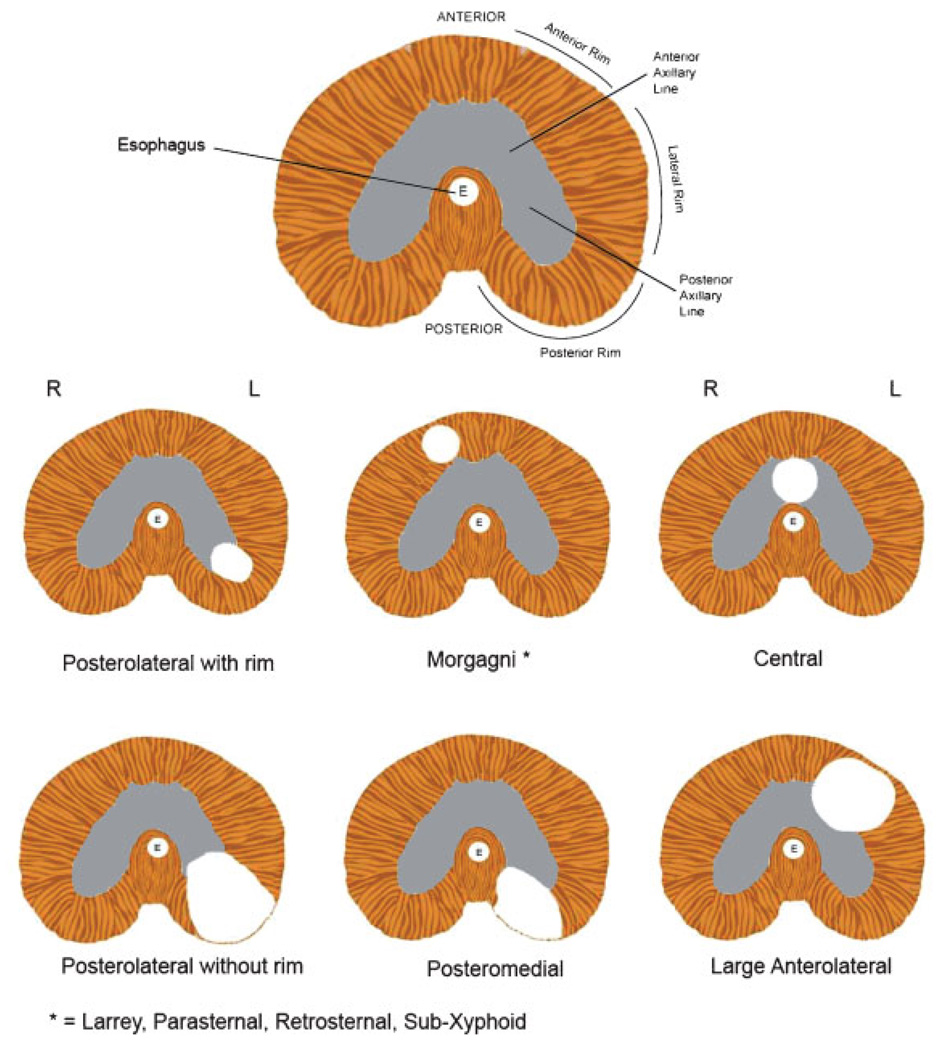
Despite the shortcomings just mentioned, an anatomic system of classification continues to be used, as follows (see Fig. 2):
Figure 2.
Stylized drawings of the diaphragm (looking up from the abdominal cavity). The first drawing shows several anatomic landmarks found in the normal diaphragm. Anterior refers to the front or ventral surface; posterior refers to the back or dorsal surface. The Anterior Rim, Lateral Rim, and Posterior Rim refer to specific portions of the peripheral rim of diaphragmatic musculature. The most commonly observed hernias are left-sided Posterolateral (e.g., Bochdalek) hernias with either an intact Posterior Rim of musculature or with an absent Posterior Rim of musculature. Morgagni hernias are also referred to as Parasternal, Retrosternal, Sub-Xyphoid, or Morgagni-Larrey hernias (the latter designation when they occur on the left). Anterolateral hernias are often associated with a hernia sac. Adapted from [Pober et al., 2006].
Posterolateral hernias (also called Bochdalek hernias) are the most common hernia type. Approximately 90–95% of all congenital diaphragmatic hernias are classified as being posterolateral, though the actual location of the defect is not confirmed in most patients reported. The majority of(presumed) Bochdalek hernias occur on the left side, while right-sided and bilateral hernias account for only 15–20% of all cases [Torfs et al., 1992; Dott et al., 2003]. Varying degrees of deficiency of the posterior rim musculature surrounding the diaphragm defect are observed; in some cases the posterior musculature is completely absent while in others the rim is well-preserved (Fig. 2). When the CDH is large involving loss of most of the hemi-diaphragm including the posterior rim of muscle it is classified as agenesis. Thus diaphragm agenesis is likely to represent a very large Bochdalek hernia.
-
Non-posterolateral (i.e., non-Bochdalek) hernias are reported to be relatively rare, and are categorized into several sub-types:
retro-sternal or parasternal hernias (also called Morgagni or Morgagni-Larrey hernias): defects in the most anterior portion ofthe diaphragm, usually accompanied by a hernia sac. Although some patients are asymptomatic with incidental discovery of their Morgagni hernia, acute or sub-acute pulmonary and GI symptoms can presage the diagnosis [Loong and Kocher, 2005].
other anterior hernias: hernias in the anterior portion of the diaphragm extending into the anterior portion ofthe central tendon, which may be due to defective development of the septum transversum. Slit3 knock-out mouse models [Liu et al., 2003; Yuan et al., 2003] demonstrate anterior hernias in this location validating their classification as a separate sub-type. Many patients with Pentalogy of Cantrell are described as having anterior diaphragmatic defects (sometimes referred to as pars sternalis defects [Milne et al., 1990]) that when combined with additional characteristic defects (e.g., congenital heart defect including ectopia cordis, midline supraumbilical abdominal wall defect, and pericardial defect) constitute this malformation complex [Cantrell et al., 1958].
and central hernias: defects primarily involving the non-muscular or central tendinous portion of the diaphragm.
Eventration refers to an abnormally thin diaphragmatic leaflet which can allow migration of abdominal contents into the chest cavity. The extent of thinning is variable, ranging from a small discrete region to virtually an entire hemi-diaphragm. Patients with eventration may present later in life with few or no clinical symptoms, and case reports describing these patients often have left the impression that eventration defects are always milder than those of traditional Bochdalek hernias [Yazici et al., 2003]. However, it is clear that eventration defects may be associated with severe pulmonary hypoplasia and death. This is substantiated by the work of Ackerman and colleagues who reviewed an autopsy series of 32 patients clinically diagnosed with CDH in which one-quarter actually had diaphragmatic eventration (personal communication, K.A.) and by cases in the literature where eventration patients have severe pulmonary disease [Rais-Bahrami et al., 1996]. Another complicating factor is that the term eventration is not used consistently. Some diaphragmatic hernias have “sacs” which are thought to represent focal thinning of the diaphragmatic musculature. Thus the terminology “sac” hernia and eventration are poorly defined, and these entities may represent a continuum.
A mixture of hernia types can occur as well, such as a typical posterolateral (Bochdalek) defect in one diaphragmatic leaflet, combined with an eventration in the contralateral leaflet or another portion of the ipsilateral leaflet [Akel and Nasr, 2001]. Although such a combination of defects is rarely seen, their true prevalence may not be known possibly due to underreporting.
Sac hernias, as noted above in the discussion of Eventration, are difficult to distinguish from eventrations and are inconsistently defined. Since a sac can co-exist with any of the CDH subtypes defined above, it should not be considered a separate hernia type on its own.
EPIDEMIOLOGY OF CDH
CDH is a common birth defect that is associated with considerable morbidity and mortality. A discussion of epidemiology of CDH is complicated by lack of uniform data collection methods as some studies are population-based, capturing all CDH cases detected early in pregnancy, while others are tertiary-care based capturing only CDH patients who survive delivery and subsequent referral to a specialty center. This discrepancy reveals the “hidden mortality” of CDH, a concept first introduced by Dr. Michael Harrison in 1978 [Skari et al., 2000]. Studies that fail to account for the impact of hidden mortality can demonstrate biased estimates of prevalence, the frequency of “isolated” versus “complex” CDH cases, and of mortality.
Prevalence
The average prevalence of CDH, derived from a meta-analysis of 16 population-based studies, was 1/4000 births [Skari et al., 2000]. Individual study prevalence estimates varied considerably, from a high of 1/1750 [Langham et al., 1996] to a low of 1/5880 [Philip et al., 1991].
Isolated versus Complex CDH
Patients with CDH can be divided into two broad phenotypic categories:
Isolated CDH
Isolated CDH is diagnosed when CDH is the only apparent major malformation. Additional abnormalities, such as pulmonary hypoplasia, intestinal malrotation, and left heart hypoplasia, may co-exist with CDH and can be considered part of a CDH sequence.
Complex CDH
Complex CDH is diagnosed when CDH occurs with additional abnormalities either as part of a recognized syndrome, a chromosome abnormality, or a non-syndromic constellation of major malformations (e.g., CDH plus ventriculoseptal defect, or CDH plus cleft lip). Averaging together data from many series complex CDH is found in 30–40% of all cases, though frequencies in individual studies range from a low of 10% to a high of 50%, in part attributable to methodologic differences in case ascertainment (as reviewed in [Skari et al., 2000]. Population-based, as well as prenatally diagnosed-based, studies have more complete ascertainment with ~40% of cases being classified as complex [Torfs et al., 1992; Skari et al., 2000; Dott et al., 2003; Colvin et al., 2005]. On the other hand, studies based at treatment centers usually reflect a biased cohort in that more seriously ill patients are less likely to be transferred for further care (i.e., so-called “hidden mortality”). Accordingly, the frequency of complex CDH cases in these series is lower, on average 23% of all cases [Skari et al., 2000].
Mortality
There continues to be considerable controversy whether medical and surgical advances have improved survival for infants with CDH. Historically, CDH that was diagnosed in the newborn period was almost uniformly fatal. Hopes for better outcomes followed the introduction of several therapies, such as delayed surgical repair, high frequency ventilation, permissive hypercapnea, extracorporeal membrane oxygenation (ECMO), and nitric oxide.
All studies demonstrate that infants with complex CDH have a higher mortality than those with isolated CDH. This is attributed to a variety of reasons including: higher frequency of spontaneous miscarriage; higher termination rate of pregnancy; more neonatal deaths ascribed to the severity of associated malformations; and the decision not to offer aggressive treatment. A meta-analysis published in 2000 found that the average mortality for cases ascertained prenatally was 75%, for cases ascertained as part of a population-based study was 48%, while for those cases transferred to a treatment center was 45% [Skari et al., 2000]. Several recent studies from tertiary care centers report excellent survival with very low mortality of 10–20% [Downard et al., 2003; Javid et al., 2004]. This remarkable survival rate reflects technical and management expertise gained from a high volume of cases combined with case selection, as would be expected in a referral center cohort. Two recent population-based studies continue to find overall mortality remains greater than 50–60%, despite the introduction of new therapies such as ECMO [Stege et al., 2003; Colvin et al., 2005]. An increasing number of elective terminations also contribute to the persistently high overall CDH mortality.
One small but particularly useful study allows comparison of population-based survival versus “immediate postnatal planned care” survival (equivalent to survival in a tertiary care center) within the same cohort [Betremieux et al., 2004]. Survival for the former was only 40% (12 of 30 cases), with the majority of fatalities occurring in cases without a prenatal diagnosis, or in complex CDH patients who either had a genetic syndrome or additional major malformations. Among those who fulfilled study criteria for receiving “immediate post-natal planned care” survival rose to 92% (11 of12 cases).
Thus, survival can be excellent in cases with isolated CDH, especially if patients are born in, or are stabilized and transferred to, a center with expertise in CDH surgery and medical management. However, survival for all cases recognized to have CDH, including those with complex CDH, does not exceed 50% on average.
EVIDENCE OF GENETIC CONTRIBUTIONS TO CDH
Most individuals diagnosed with CDH are the only member of their family to be affected. Thus, CDH typically has been viewed as a “sporadic” birth defect, which has erroneously been equated with “non-genetic”. There is growing evidence supporting genetic causation of CDH, such as genetically modified model organisms, recurring human chromosome abnormalities, and single gene mutations (Table I). A summary of the current evidence supporting genetic etiologies of CDH is presented below.
TABLE I.
Summary of Evidence Supporting Genetic Causation of CDH
| Evidence of genetic contributions to human CDH | Examples |
|---|---|
| Genetic mouse models of CDH | Fog2a, Couptf2, Wt1a, Slit3, Gata4 |
| Monogenic syndromes associated with CDH | Syndromes caused by known genes: de Lange, Simpson-Golabi- Behmel, Spondyolocostostal dysplasia |
| Syndromes caused by unknown genes: Fryns, Donnai-Barrow | |
| Multiplex human kindreds with CDH | Familial clustering consistent with AR and XL inheritance |
| Co-occurrence of CDH with other congenital malformations | CDH co-exists with CVMs, NTDs, and limb defects more often than expected by chance |
| Recurring chromosome abnormalities and CDH | ‘Large’ aneuploidy-iso12p, trisomy 18; |
| ‘Small’ aneuploidy-del 15(q26), del 8(p23) |
AR, autosomal recessive; CVMs, cardiovascular malformations; NTDs, neural tube defects; XL, X-linked.
Mutation in human homologue found in association with CDH.
“Sporadic” Birth Defects can Have Genetic Causation
Many common and seemingly sporadic birth defects are now known to be caused by mutations in one or more genes often in related pathways, or by the multiplicative effects of variations in several genes. Hirschsprung (HSCR) disease is a prime example. Prior to surgical advances that allowed improved survival, scant data demonstrated family clustering of HSCR. Now that survival is routine, many multiplex families are documented. The current understanding of HSCR genetics is considerable, with identification of mutations in the RET proto-oncogene and its ligands, as well as the endothelin B receptor and some of its ligands, among others [Lantieri et al., 2006]. CHARGE syndrome and several forms of congenital heart defects are due to mutations in CHD7 and NKX2.5, respectively [Goldmuntz et al., 2001; Elliott et al., 2003; Vissers et al., 2004]. These precedents suggest that gene defects, either in isolation or as combinatorial gene-environment or gene-gene interactions, are likely to play a significant role in diaphragmatic defect causation, even among “sporadic” cases of CDH. Discovery of a de novo mutation in FOG2 in a lethal case whose only abnormalities on autopsy were a posterior muscularization defect in the eventration-sac hernia spectrum and pulmonary hypoplasia [Ackerman et al., 2005] provides proof of principle.
Animal Models of CDH
Reports of several naturally occurring animal models of CDH are consistent with, but not confirmatory of, genetic causation. More compelling evidence derives from mouse genetic models with mutations in single genes such as COUPTFII [You et al., 2005], Fog2 [Ackerman et al., 2005], Slit3 [Liu et al., 2003; Yuan et al., 2003], Wt1 [Kreidberg et al., 1993], and Gata4 [Jay et al., 2006]. These model organisms are reviewed in greater detail in the article by Ackerman and Greer, in this monograph.
Known Monogenic Syndromes Associated with CDH
The frequency of CDH in several single gene disorders is greater than expected by chance alone, such as in Brachman-de Lange [Cunniff et al., 1993; Pankau and Janig, 1993; Marino et al., 2002], Craniofronto-nasal [Brooks et al., 2002; McGaughran et al., 2002; Vasudevan et al., 2006], Simpson-Golabi-Behmel [Chen et al., 1993], Spondylocostal dysotosis [Park et al., 1993b; Martinez-Frias et al., 1994; Lam et al., 1999; Rodriguez et al., 2004], and Donnai-Barrow syndromes [Donnai and Barrow, 1993; Gripp et al., 1997; Chassaing et al., 2003]. Accordingly, it is reasonable to hypothesize that mutations in the genes, NIPBL, EPNB1, GPC3, and DLL3, responsible for the first four disorders, respectively, interfere with normal diaphragm development or act to disrupt a downstream target.
The importance of monogenic disorders whose phenotypes include CDH can be gleaned from progress in the genetic understanding of another common birth defect, cleft lip ± cleft palate. Specifically, mutations in interferon regulatory factor 6 (IRF6) protein binding and DNA binding domains cause autosomal dominant lip pit or van der Woude syndrome [Kondo et al., 2002]. However, a common variant in IRF6 increases the risk of developing non-syndromic isolated cleft lip ± cleft palate [Zucchero et al., 2004; Scapoli et al., 2005]. It therefore follows that common variants in genes such as NIPBL or EPNB1 could be risk factors for isolated CDH.
Our group is working to identify the gene responsible for the autosomal recessive disorder, Donnai-Barrow syndrome (DBS) [Kantarci et al., 2006b]. CDH is present in more than half of reported individuals; additional phenotypic details are provided in the article in this issue by Dr. Slavotinek. To identify the locus for DBS we recruited a large consanguineous family from the United Arab Emirates and performed linkage analysis to identify regions of identity by descent. Four affected members of this kindred shared a ~17 centimorgan homozygous region that mapped to chromosome 2q23-q31. Microsatellite marker analysis narrowed this region to 17–18 million base pairs using DNA samples from members of the United Arab Emirates kindred, as well as from multiplex kindreds previously published by Donnai and Barrow and Chassaing et al. [Donnai and Barrow, 1993; Chassaing et al., 2003]. Sequencing of candidate genes within this region is underway. We hypothesize that loss of function mutations will be responsible for the multiple anomaly disorder DBS, while missense mutations or functional genetic variants may contribute to the causation of some ofthe individual anomalies found in DBS, including CDH [Kantarci et al., 2006b].
Additional examples of monogenic syndromes associated with CDH are provided in the article by Dr. Slavotinek in this monograph.
Multiplex human kindreds with CDH
There are published reports describing >50 families containing two or more relatives affected with CDH, for which a causative syndrome or chromosome abnormality has not been identified [Butler and Claireaux, 1962; Welch and Cooke, 1962; Scott and Patterson, 1966; Passarge et al., 1968; ten Kate and Anders, 1970; Feingold, 1971; Harberg et al., 1976; Thomas et al., 1976; Crane, 1979; David et al., 1979; Pollack and Hall, 1979; Arad et al., 1980; Wolff, 1980; Gencik et al., 1982; Norio et al., 1984; Czeizel and Kovacs, 1985; Lipson and Williams, 1985; Toriello et al., 1985; Bocian et al., 1986; Toriello et al., 1986; Farag et al., 1989; Hitch et al., 1989; Carmi et al., 1990; Frey et al., 1991; Sripathi and Beasley, 1992; Narayan et al., 1993; Farag et al., 1994; Gibbs et al., 1997; Mitchell et al., 1997; Kufeji and Crabbe, 1999; Manouvrier-Hanu et al., 2000]. As part of our own research over the last five years, we ascertained more than two dozen multiplex families in whom the diagnosis of a syndrome was not established. Additionally, data from The Association of Congenital Diaphragmatic Hernia Research, Advocacy, and Support (CHERUBS- a nonprofit CDH parents support group) show that 2% of the probands in their survey have an affected 1st degree relative. Thus, multiplex CDH families are not difficult to identify and collectively they suggest genetic causation. However, these multiplex kindreds constitute a heterogeneous group, both in terms of the mode of inheritance as well as whether the diaphragmatic defect is isolated or is occurring along with additional major malformations. Use of multiplex families for linkage analysis to identify genes associated with CDH has not been possible to date (aside from the work on DBS just mentioned above). This is due not only to etiologic heterogeneity, but also to the high mortality rate among those affected limiting the availability of DNA samples.
Multiplex CDH kindreds can be categorized as follows:
Consanguineous CDH families: In several families, healthy parents of two siblings affected with CDH are consanguineous [Arad et al., 1980; Norio et al., 1984; Farag et al., 1989; Farag et al., 1994]. The second affected sibling reported by Arad et. al. had additional findings making it likely that an undiagnosed syndrome was present in this sib pair. The remaining sib pairs had either isolated CDH or additional anomalies found in other family members who did not have CDH. The occurrence of CDH in inbred families makes it likely that a single mutant gene is responsible, though multifactorial inheritance and subtle chromosome abnormalities cannot be ruled out as an explanation for familial clustering.
Nonconsanguineous CDH families suggestive of autosomal recessive or X-linked inheritance: Several kindreds in the medical literature, as well as in our study cohort, contain two or more children diagnosed with isolated CDH [Welch and Cooke, 1962; Passarge et al., 1968; Pollack and Hall, 1979; Gencik et al., 1982; Norio et al., 1984; Czeizel and Kovacs, 1985; Bocian et al., 1986; Toriello et al., 1986; Hitch et al., 1989]. In some cases the affected siblings are opposite genders, while in the majority both are the same gender, most often males. Maternal male cousins with isolated CDH have also been reported [Turpin et al., 1959]. These findings are consistent with, even suggestive though not conclusive of, single gene inheritance for CDH. Additionally, isolated CDH may not be the correct diagnosis as in some instances the presence of additional anomalies may have been missed due to neonatal death or lack of a post-mortem examination.
Apparent autosomal dominant inheritance: There are no published reports of vertical transmission of isolated posterolateral or Bochdalek hernia consistent with autosomal dominant inheritance. However, we have identified two such families as part of our ongoing studies. Furthermore, the report of a de novo FOG2 mutation in a newborn with non-syndromic but lethal diaphragmatic defect and pulmonary hypoplasia [Ackerman et al., 2005], raises the possibility that other CDH cases may be due to de novo mutations; if such cases survive to reproduce then they would be at increased risk for having affected offspring. Two unrelated kindreds in which a parent with diaphragmatic eventration had offspring with typical CDH were reported in 1952 and 1969. Although these findings are suggestive of autosomal dominant inheritance, limited conclusions can be drawn from these reports, as additional abnormalities were present in several affected individuals and the diagnostic work-ups were incomplete by today’s standards [Mertins, 1952; Varpella and Lehtovaara, 1962].
Co-occurrence of CDH with Other Congenital Malformations
Approximately 30–40% of cases with CDH have additional major malformations, often involving the cardiovascular, central nervous, musculoskeletal, and/or genitourinary systems. Despite the presence of malformations in multiple organs, the diagnosis of a syndrome or chromosome abnormality cannot be established so these cases are classified as being multiply-malformed or “non-syndromic”. It is reasonable to hypothesize that a mutant gene, either acting alone or in concert with other genetic variants or environmental triggers, is responsible for all the anomalies in a given patient including the CDH. Non-syndromic associations might help identify genes responsible for CDH. Specifically, if cardiovascular malformations (CVMs) and diaphragmatic defects share a common (genetic) etiology then knowledge about genes causing CVMs may provide knowledge about genes causing CDH. As an example, if the group of cardiovascular defects referred to as laterality defects are over-represented among CDH cases, then mutations in laterality genes, such as lefty1 or 2, could receive further scrutiny as candidate genes in future studies of CDH.
CDH and Cardiovascular Malformations
Cardiovascular malformations (CVMs) are present in ~10–15% of those cases with non-syndromic CDH, as discussed in detail in the article by Lin et al. in this volume. If all cases, including those with underlying syndromes and chromosome abnormalities are included, then the prevalence of CVM’s rises to 2ndash;40% of cases [Philip et al., 1991; Robert et al., 1997; Migliazza et al., 1999a; Migliazza et al., 1999b; Dillon et al., 2000; Dott et al., 2003; Tonks et al., 2004]. The most common CVMS that co-occur with CDH reflect those found in the general population, such as ventricular septal defect, atrial septal defect, and tetralogy of Fallot. Hypo-plastic left heart syndrome also occurs but is sometimes over-diagnosed since left heart structures can be small due to abnormal hemodynamics caused by a left-sided diaphragm defect. A far more detailed discussion of the type and prevalence of CVMs found in cases with CDH is presented in the article by Lin, Pober and Adatia in this monograph.
CDH and central nervous system abnormalities
The most common CNS anomalies, present in 5–10% of non-syndromic CDH cases, are neural tube defects and hydrocephalus [David and Illingworth, 1976; Dillon et al., 2000; Dott et al., 2003]. Some authors speculate that problems of schisis-fusion or midline instability account for the cooccurrence of CDH and neural tube defects [Czeizel and Vitez, 1981].
CDH and limb abnormalities
A variety of limb defects occur in ~10% of patients with non-syndromic CDH including polydactyly, syndactyly, and reduction defects [Torfs et al., 1992; van Dooren et al., 2003]. Some authors speculate that co-existence of CDH and limb reduction defects reflects abnormal cervical neural crest migration or mutations in specific genes that are important in limb and diaphragm development though no data to support these hypotheses yet exist [McCredie and Reid, 1978; Lerone et al., 1992; Hou and Wang, 1999].
Chromosome abnormalities and CDH
Routine chromosome analysis performed on unselected patients with CDH identifies an abnormality in ~10% of cases [Thorpe-Beeston et al., 1989; Philip et al., 1991; Bollmann et al., 1995; Howe et al., 1996; Faivre et al., 1998; Garne et al., 2002; Tonks et al., 2004]. Some common abnormalities are trisomy 18, tetrasomy 12p, deletion 15q26, deletion 8p23, and + der 22 t(11;22) [Pecile et al., 1990; Howe et al., 1996; Faivre et al., 1998; Lurie, 2003; Borys and Taxy, 2004; Tonks et al., 2004; van Dooren et al., 2004]; a far more exhaustive list is nicely presented in a recent review [Lurie, 2003]. It is currently unknown why larger regions of aneuploidy (trisomy 18 and isochromosome 12p) increase the risk for CDH, and the genetic basis for these associations is not likely to be elucidated in the near future. However, smaller regions of aneuploidy are proving very powerful for gene discovery. An increasing number of subtle chromosome abnormalities are likely to be detected with use of newer technologies, such as array-based comparative genomic hybridization (aCGH).
CLINICAL ASPECTS OF SELECTED “LARGER” CHROMOSOME REARRANGEMENTS ASSOCIATED WITH CDH
Isochromosome 12p (tetrasomy 12p; Pallister-Killian Syndrome [PKS])
PKS is a unique disorder in which affected individuals are tissue mosaics for four copies of the short arm of chromosome 12. Two copies of the chromosome 12 short arm belonging to the normal chromosome 12 homologues are present, along with two additional copies of 12p that have formed a supernumerary isochromosome. The isochromosome is not present in all cell lineages. It is rarely identified in a standard blood (lymphocyte) chromosome analysis, though a recent report [Wu et al., 2006] demonstrated 7% lymphocyte isochromosome recovery compared to 90% skin fibro-blast recovery in a characteristically dysmorphic infant who did not have CDH. Among individuals with PKS, isochromosome 12p is detected in varying percentages (ranging from 5 – 100%) in amniocytes, chorionic villi, buccal cells, and skin fibroblasts [Doray et al., 2002].
Mechanisms underlying PKS vary but the isochromosome most often seems to derive from a maternal pre-meiotic or meiotic error accompanied by centromere misdivision. Several studies demonstrate that the detection frequency of the isochromosome declines over time, both in vitro with continued sub-cultures of growing cells and in vivo with advancing patient age.
In the setting of prenatally diagnosed CDH, chromosome 12p targeted-FISH can be an adjunct to routine karyotyping, and may identify PKS cases due to low-level mosaicism. Similarly, array-based comparative genomic hybridization (aCGH) may detect low level mosiacism, possibly even in lymphocytes, and has already been used to establish the diagnosis using DNA extracted from formalin fixed paraffin embedded autopsy tissue [Delahaye et al., 2006]. Systematic application of these newer molecular cytogenetic techniques will be needed before conclusions about their diagnostic utility can be drawn.
Postnatal findings of PKS include a coarse face, temporal alopecia, broad short hands, streaky cutaneous hyper-pigmentation, shortened neck, developmental delay/ mental retardation, and seizures. The frequency of CDH is low, 10–20%, among individuals postnatally diagnosed with PKS [Mathieu et al., 1997; Schaefer et al., 1997]. Among prenatally diagnosed cases, the major malformations likely to suggest the diagnosis include: polyhydramnios, CDH (found in 33%), relatively shortened limbs, CNS anomalies, cranio-facial dysmorphology (which can be appreciated on 3D ultrasound), and fetal edema [Doray et al., 2002]. Phenotypic severity is extremely broad, ranging from multiple malformations incompatible with life to relatively mild medical and developmental abnormalities, possibly due to the 12p tissue mosiacism. There may be under ascertainment of PKS as skin chromosome analysis is not routinely performed on less severely affected individuals.
PKS can be difficult to distinguish from Fryns syndrome [OMIM 229850] solely based on clinical features. CDH can occur in both disorders though occurs more frequently in Fryns syndrome. The occurrence of bilateral CDH is a rarity in either condition, but if present is more suggestive of the diagnosis of Fryns syndrome. Other features that more commonly complicate Fryns syndrome include cleft palate, distal phalangeal and/or nail hypoplasia, cardiovascular malformations, and renal malformations [Slavotinek, 2004]. Features more suggestive of PKS include a high forehead, streaky skin hyper-pigmentation, and sparseness of hair bitemporally [McPherson et al., 1993; Veldman et al., 2002]. It is especially important to perform thorough cytogenetic and molecular cytogenetic testing using the appropriate tissues to confirm or exclude the diagnosis of PKS since this is a sporadic disorder with no known risk of recurrence.
Trisomy 18
Trisomy 18 is the most common chromosome abnormality detected in all prenatal series of CDH cases. CDH (as well as omphalocele) occurs more often in male trisomy 18 fetuses than in female fetuses; this suggests that males are more likely to be multiply malformed and die in utero which, in turn, could account for the excess of female live births with trisomy 18 [Chen, 2005].
Trisomy 21
Although both Bochdalek and Morgagni hernias have been reported in trisomy 21, CDH is a relatively low frequency occurrence in trisomy 21. Morgagni hernia seems to be the most common type in trisomy 21 suggesting that three copies of one or more chromosome 21 genes predispose to abnormal development of the anterior portion of the diaphragm [Torfs et al., 1992; Parmar et al., 2001; Marin and Lopoo, 2006].
Trisomy 22 and t(11;22)
There are several reports of CDH occurring in infants found to have multiple anomalies with trisomy 22 [Kim et al., 1992; Ladonne et al., 1996]. CDH has also been observed in patients with “partial trisomy 22q” syndrome due to +der 22 t(11;22) (q23.3;q11.2) inherited from a balanced translocation carrier parent [Schinzel et al., 1981; Lin et al., 1986; Kadir et al., 1997]. Taken together, these cytogenetic abnormalities suggest that three copies of one or more genes in distal 22q confers risk for developing CDH, though neither a critical region nor candidate genes have been identified. However, patients who carry +der 22t(11;22) are also trisomic for the distal end of 11q which is further discussed below.
CLINICAL ASPECTS OF SELECTED “SMALLER” CHROMOSOME REARRANGEMENTS ASSOCIATED WITH CDH AND THE POTENTIAL FOR GENE DISCOVERY
There are several clinical reasons for routine karyotyping: a) to establish the correct diagnosis; b) to provide natural history and prognosis; c) for accurate recurrence risk counseling; and d) to guide prenatal diagnostic testing. The detection of small chromosome abnormalities can inform on these topics but also can identify “hot spots” likely contain genes contributing to CDH. “Hot spots” 15q26.1–q26.2 (DIH1, OMIM 142340), 8p23.1 (DIH2, OMIM 222400) and 8q23 (DIH3, OMIM 610187), are among the bestcharacterized to date and they, as well as selected other hot spots, will be described below.
Del (15)(q26.1-q26.2) (DIH1, OMIM %142340)
Almost two dozen unrelated cases with deletion of the 15q26.1 –15q26.2 region share a common phenotype that includes CDH, growth retardation, cardiovascular and limb malformations [Aviram-Goldring et al., 2000; Schlembach et al., 2001; Biggio et al., 2004; Hengstschlager et al., 2004; Klaassens et al., 2005; Slavotinek et al., 2005]. Most of the abnormalities, consisting of de novo deletions, unbalanced reciprocal translocation products and ring chromosomes have been detected with standard chromosome banding techniques. However, several small deletions detected only by aCGH have recently been reported [Slavotinek et al., 2005]. These findings form the basis of the hypothesis that hemizygosity for one or more genes located in 15q26.1 – 15q26.2 confer risk for CDH. Application of careful deletion mapping recently led to the delineation of a ~five megabase CDH critical region containing four candidate genes [Klaassens et al., 2005]. Subsequent analyses suggest a slightly more telomeric location of the critical region containing several candidate genes [Castiglia et al., 2005; Scott et al., 2007], with a particularly compelling candidate being COUP-TFII (Chick Ovalbumin Upstream Promotor Transcription Factor 2, also known as Nuclear Receptor Subfamily 2, Group F, Member 2), a receptor belonging to the steroid/thyroid hormone family. Converging lines of evidence that implicate COUP-TFII hemizygosity as a major risk factor for CDH are: a) recurring cytogenetic abnormalities, detailed above, that result in deletion of COUP-TFII; b) presence of a diaphragmatic hernia in the majority of conditionalCOUP-TFII mutant mice [You et al., 2005], c) the role that COUP-TFII plays in regulation of the retinol pathway, a pathway important for diaphragm development, and d) that COUP-TFII interacts with FOG2, mutations of which cause diaphragmatic abnormalities in humans and mouse models [Ackerman et al., 2005]. To date no mutations have yet been identified in COUP-TFII exons or flanking exonintron boundaries in >150 CDH patients [Slavotinek et al., 2006; Scott et al., 2007]. It remains possible that mutations contributing to CDH lie in COUPTF-II upstream regulatory elements rather than in coding regions, that COUP-TFII acts combinatorially with other genetic or environmental factors, or that it plays a minor role in human CDH causation.
Del (8)(p23.1) (DIH2, OMIM %222400)
The second most common small chromosome abnormality found in association with CDH is del (8)(p23.1) [Pecile et al., 1990; Hutchinson et al., 1992; Faivre et al., 1998; Borys and Taxy, 2004; Shimokawa et al., 2005; Slavotinek et al., 2005; Lopez et al., 2006]. Among the patients reported to date, CDH has been present in ~20% of cases. Depending on the size of the deleted segment patients also demonstrate developmental delay, cardiovascular malformations, genitourinary malformations, subtle facial dys-morphology, and minor limb anomalies. The smallest deletion characterized at the molecular level in a del 8p23 patient who had CDH is ~3.5 Mb [Shimokawa et al., 2005]. In addition to narrowing the DIH2 critical interval, the patient’s deletion breakpoints lie within two low copy repeats (LCRs) suggesting that meiotic misalignment between homologous LCRs predisposes to this deletion, and therefore is mechanistically similar to other deletion and microdeletion disorders [Lupski, 2006]. GATA4 lies within this region and recent data show that in additional to its importance in cardiac development, Gata4 also plays a role in diaphragm and pulmonary development [Ackerman et al., 2006b; Jay et al., 2006].
Chromosome 8q23 (DIH3, OMIM #610187)
Disruption of 8q22-q24 by either translocation or deletion has been reported in several patients with CDH [Harnsberger et al., 1982; Temple et al., 1994; Capellini et al., 1996; Howe et al., 1996], though exact breakpoints were not always provided. These cytogenetic abnormalities, combined with detection of a FOG2 (8q23.1) mutation in a patient with diaphragmatic defect and pulmonary hypoplasia, provide compelling evidence for the importance of this locus.
Del (4)(p16) (Wolf-Hirschhorn Syndrome, WHS, OMIM #194190)
Wolf-Hirschhorn syndrome is a clinically well-characterized chromosome deletion disorder. Although CDH is an infrequent complication, its documentation in several cases suggests that occurrence of CDH is not due to chance but rather is related to loss of a specific gene or genes in this region [Tachdjian et al., 1992; Howe et al., 1996; Sergi et al., 1998; Tapper et al., 2002; Van Buggenhout et al., 2004; van Dooren et al., 2004; Casaccia et al., 2006; Slavotinek et al., 2006]. Using standard chromosome techniques, variably sized-deletions have been noted in WHS cases, both with and without CDH. Only four cases with CDH have been characterized at the molecular level [Van Buggenhout et al., 2004; van Dooren et al., 2004; Casaccia et al., 2006; Slavotinek et al., 2006], the break point occurring in 4p16.3 in the case with the smallest deletion [Casaccia et al., 2006]. Based on current data, it is not possible to pinpoint the CDH-associated critical region.
Del (1)(q42)
Several cases with CDH, plus additional anomalies, have been reported in association with chromosome rearrangements producing disruption or deletion of chromosome band 1q41–q42.12 [Youssoufian et al., 1988; Smith et al., 1994; Rogers et al., 1995; Slavotinek et al., 2006; Kantarci et al., 2006a]. The smallest rearrangement characterized to date is ~5 Mb deletion, and this interval contains 29 candidate genes [Kantarci et al., 2006a]. Further characterization of additional patients with aneuploidy at this locus will be needed before a critical region, or a candidate gene, can be identified.
Dup (11)(q23-qter)
Trisomy for the qter region of chromosome 11, arising through inheritance of an unbalanced chromosome complement from a balanced translocation carrier parent has been reported in several CDH patients. Most commonly the parent is a carrier for t(11;22) (q23.3;q11.2), but two additional CDH patients whose parents carried t(11;12) and t(11;13) have also been reported [Park et al., 1993a; Klaassens et al., 2006]. The 11q duplication is ~19 Mb in the only patient whose aneuploidy has been characterized at the molecular level [Klaassens et al., 2006].
APPARENT “NON-GENETIC” PATTERNS IN CDH
Two observations which suggest genetic factors play only a small role in the development of CDH, particularly in non-syndromic or isolated cases, merit further discussion. “Precurrence” risk studies show a low rate of clustering of CDH among siblings [David and Illingworth, 1976; Czeizel and Kovacs, 1985; Pober et al., 2005]. Despite a low absolute sibling recurrence in non-syndromic or isolated cases, there is a 20-fold CDH excess in siblings compared to the occurrence rate in the general population. Additionally, the presence of de novo point mutations and cytogenetic aberrations raise the possibility that many cases of CDH are in fact caused by de novo events which would rarely be associated with sibling recurrence.
The second observation questioning the importance of genetic causation in CDH derives from twin studies. Monozygotic (MZ) twin pairs concordant for CDH have been reported, but several MZ pairs discordant for CDH have been published as well (reviewed in [Pober et al., 2005]). However, there are precedents demonstrating that phenotypic discordance in MZ twins does not rule out genetic mechanisms [Machin, 1996; Mansilla et al., 2005], including: de novo dominant gene mutations, deletions or duplications; autosomal recessive mutations (some with incomplete penetrance); multifactorial or 2nd hit mutations; and epigenetic modifications. Furthermore, a contributory role of non-genetic factors, such as stochastic events and environmental exposures, which may be acting synergistically with genetic risk factors is highly likely.
SUMMARY
Collectively, the evidence presented in this article strongly support genetic causation in some, if not many, cases of CDH. The evidence is most compelling for syndromic cases of CDH, e.g., patients with multiple anomalies for whom an underlying genetic or chromosome etiology has been established. However, genetic causation is also suggested in cases with isolated CDH, given reports of mouse genetic models, multiplex human families with seemingly isolated CDH, and report of a FOG2 mutation in a patient with apparent non-syndromic CDH. We anticipate that mutations in at least several additional genes, either acting singly or as part of gene-gene or gene-environmental combinations, will be identified as contributing to most cases of CDH. Such genes can then be studied to determine whether they represent different branch points within the same signaling pathways, and furthermore, whether they provide targets for potential therapies.
Acknowledgments
Grant sponsor: RO1; Grant number: HD55150-01.
Footnotes
NOTE ADDED IN PROOF
A complete review of cytogenetic abnormalities in CDH is in press by Holder AM et al. “Genetic Factors in Congenital Diaphragmatic Hernia” in Am J Hum Genet. Accepted 2/1/07.
REFERENCES
- Ackerman KG, Herron BJ, Vargas SO, Huang H, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, Greer JJ, Beier DR. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman KG, Vargas SO, Kozakewich HP. Phenotypic Variation of Congenital Diaphragmatic Defects (573). Presented at the annual meeting of The American Society of Human Genetics; October 9–13; New Orleans, LA. 2006a. Available from http://wwwashgorg/genetics/ashg05s/. [Google Scholar]
- Ackerman KG, Wang J, Luo L, Fujiwara Y, Orkin SH, Beier DR. Gata4 is Necessary for Normal Pulmonary Lobar Development. Am J Respir Cell Mol Biol. 2006b doi: 10.1165/rcmb.2006-0211RC. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akel S, Nasr W. Multiple ipsilateral congenital diaphragmatic pathologies: rarities to consider. Eur J Pediatr Surg. 2001;11:200–203. doi: 10.1055/s-2001-15492. [DOI] [PubMed] [Google Scholar]
- Arad I, Lijovetzky GC, Starinsky R, Laufer N, Cohen T. Diaphragmatic defects in children of consanguineous parents. Hum Genet. 1980;55:275–277. doi: 10.1007/BF00291778. [DOI] [PubMed] [Google Scholar]
- Aviram-Goldring A, Daniely M, Frydman M, Shneyour Y, Cohen H, Barkai G. Congenital diaphragmatic hernia in a family segregating a reciprocal translocation t(5;15)(p15.3;q24) Am J Med Genet. 2000;90:120–122. doi: 10.1002/(sici)1096-8628(20000117)90:2<120::aid-ajmg6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Betremieux P, Gaillot T, de la Pintiere A, Beuchee A, Pasquier L, Habonimana E, Le Bouar G, Branger B, Milon J, Fremond B, Wodey E, Odent S, Poulain P, Pladys P. Congenital diaphragmatic hernia: prenatal diagnosis permits immediate intensive care with high survival rate in isolated cases. A population-based study. Prenat Diagn. 2004;24:487–493. doi: 10.1002/pd.909. [DOI] [PubMed] [Google Scholar]
- Biggio JR, Jr, Descartes MD, Carroll AJ, Holt RL. Congenital diaphragmatic hernia: is 15q26.1-26.2 a candidate locus? Am J Med Genet Part A. 2004;126A:183–185. doi: 10.1002/ajmg.a.20464. [DOI] [PubMed] [Google Scholar]
- Bocian M, Spence MA, Marazita ML, Walker AP, Weissberg DL. Familial diaphragmatic defects: early prenatal diagnosis and evidence for major gene inheritance. Am J Med Genet. 1986 Suppl 2:163–176. doi: 10.1002/ajmg.1320250620. [DOI] [PubMed] [Google Scholar]
- Bollmann R, Kalache K, Mau H, Chaoui R, Tennstedt C. Associated malformations and chromosomal defects in congenital diaphragmatic hernia. Fetal Diagn Ther. 1995;10:52–59. doi: 10.1159/000264193. [DOI] [PubMed] [Google Scholar]
- Borys D, Taxy JB. Congenital diaphragmatic hernia and chromosomal anomalies: autopsy study. Pediatr Dev Pathol. 2004;7:35–38. doi: 10.1007/s10024-003-2133-7. [DOI] [PubMed] [Google Scholar]
- Brooks AS, van Dooren M, Hoogeboom J, Gischler S, Willems PJ, Tibboel D. Congenital diaphragmatic hernia in a female patient with craniofrontonasal syndrome. Clin Dysmorphol. 2002;11:151–153. doi: 10.1097/00019605-200204000-00019. [DOI] [PubMed] [Google Scholar]
- Butler N, Claireaux AE. Congenital diaphragmatic hernia as a cause of perinatal mortality. Lancet. 1962;1:659–663. doi: 10.1016/s0140-6736(62)92878-7. [DOI] [PubMed] [Google Scholar]
- Cantrell JR, Haller JA, Ravitch MM. A syndrome of congenital defects involving the abdominal wall, sternum, diaphragm, pericardium, and heart. Surg Gynecol Obstet. 1958;107:602–614. [PubMed] [Google Scholar]
- Capellini A, Sala E, Colombo D, Villa N, Mariani S. Monosomy 8q and features of Fryns syndrome. Eur J Hum Genet. 1996;4 S1:29. [Google Scholar]
- Carmi R, Meizner I, Katz M. Familial congenital diaphragmatic defect and associated midline anomalies: further evidence for an X-linked midline gene? Am J Med Genet. 1990;36:313–315. doi: 10.1002/ajmg.1320360314. [DOI] [PubMed] [Google Scholar]
- Casaccia G, Mobili L, Braguglia A, Santoro F, Bagolan P. Distal 4p microdeletion in a case of Wolf-Hirschhorn syndrome with congenital diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol. 2006;76:210–213. doi: 10.1002/bdra.20235. [DOI] [PubMed] [Google Scholar]
- Castiglia L, Fichera M, Romano C, Galesi O, Grillo L, Sturnio M, Failla P. Narrowing the candidate region for congenital diaphragmatic hernia in chromosome 15q26: contradictory results. Am J Hum Genet. 2005;77:892–894. doi: 10.1086/497082. Author reply 894–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing N, Lacombe D, Carles D, Calvas P, Saura R, Bieth E. Donnai-Barrow syndrome: four additional patients. Am J Med Genet Part A. 2003;121A:258–262. doi: 10.1002/ajmg.a.20266. [DOI] [PubMed] [Google Scholar]
- Chen CP. Omphalocele and congenital diaphragmatic hernia associated with fetal trisomy 18. Prenat Diagn. 2005;25:421–423. doi: 10.1002/pd.1140. [DOI] [PubMed] [Google Scholar]
- Chen E, Johnson JP, Cox VA, Golabi M. Simpson-Golabi-Behmel syndrome: congenital diaphragmatic hernia and radiologic findings in two patients and follow-up of a previously reported case. Am J Med Genet. 1993;46:574–578. doi: 10.1002/ajmg.1320460523. [DOI] [PubMed] [Google Scholar]
- Colvin J, Bower C, Dickinson JE, Sokol J. Outcomes of congenital diaphragmatic hernia: a population-based study in Western Australia. Pediatrics. 2005;116:e356–363. doi: 10.1542/peds.2004-2845. [DOI] [PubMed] [Google Scholar]
- Crane JP. Familial congenital diaphragmatic hernia: prenatal diagnostic approach and analysis of twelve families. Clin Genet. 1979;16:244–252. doi: 10.1111/j.1399-0004.1979.tb00996.x. [DOI] [PubMed] [Google Scholar]
- Cunniff C, Curry CJ, Carey JC, Graham JM, Jr, Williams CA, Stengel-Rutkowski S, Luttgen S, Meinecke P. Congenital diaphragmatic hernia in the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1018–1021. doi: 10.1002/ajmg.1320470716. [DOI] [PubMed] [Google Scholar]
- Czeizel A, Kovacs M. A family study of congenital diaphragmatic defects. Am J Med Genet. 1985;21:105–117. doi: 10.1002/ajmg.1320210115. [DOI] [PubMed] [Google Scholar]
- Czeizel A, Vitez M. Birth prevalence of five congenital abnormalities of medium frequency in Budapest. Acta Paediatr Acad Sci Hung. 1981;22:299–308. [PubMed] [Google Scholar]
- David TJ, Illingworth CA. Diaphragmatic hernia in the south-west of England. J Med Genet. 1976;13:253–262. doi: 10.1136/jmg.13.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David TJ, Parker VM, Illingworth CA. Anencephaly with diaphragmatic hernia in sibs. J Med Genet. 1979;16:157–159. doi: 10.1136/jmg.16.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahaye A, Pipiras E, Delorme-Vincent C, Benkhalifa M, Kasakyan S, Devisme L, Wolf JP, Benzacken B. Retrospective Diagnosis of Pallister-Killian Syndrome by CGH Array. Fetal Diagn Ther. 2006;21:485–488. doi: 10.1159/000095658. [DOI] [PubMed] [Google Scholar]
- Dillon E, Renwick M, Wright C. Congenital diaphragmatic herniation: antenatal detection and outcome. Br J Radiol. 2000;73:360–365. doi: 10.1259/bjr.73.868.10844860. [DOI] [PubMed] [Google Scholar]
- Donnai D, Barrow M. Diaphragmatil hernia, exomphalos, absent corpus callo-sum, hypertelorism, myopia, and sensorineural deafness: a newly recognized autosomal recessive disorder? Am J Med Genet. 1993;47:679–682. doi: 10.1002/ajmg.1320470518. [DOI] [PubMed] [Google Scholar]
- Doray B, Girard-Lemaire F, Gasser B, Baldauf JJ, De Geeter B, Spizzo M, Zeidan C, Flori E. Pallister-Killian syndrome: difficulties of prenatal diagnosis. Prenat Diagn. 2002;22:470–477. doi: 10.1002/pd.342. [DOI] [PubMed] [Google Scholar]
- Dott MM, Wong LY, Rasmussen SA. Population-based study of congenital diaphragmatic hernia: risk factors and survival in Metropolitan Atlanta, 1968–1999. Birth Defects Res A Clin Mol Teratol. 2003;67:261–267. doi: 10.1002/bdra.10039. [DOI] [PubMed] [Google Scholar]
- Downard CD, Jaksic T, Garza JJ, Dzakovic A, Nemes L, Jennings RW, Wilson JM. Analysis of an improved survival rate for congenital diaphragmatic hernia. J Pediatr Surg. 2003;38:729–732. doi: 10.1016/jpsu.2003.50194. [DOI] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, Feneley M, Harvey RP. Cardiac homeobox gene NKX 2–5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- Faivre L, Morichon-Delvallez N, Viot G, Narcy F, Loison S, Mandelbrot L, Aubry MC, Raclin V, Edery P, Munnich A, Vekemans M. Prenatal diagnosis of an 8p23.1 deletion in a fetus with a diaphragmatic hernia and review of the literature. Prenat Diagn. 1998;18:1055–1060. doi: 10.1002/(sici)1097-0223(1998100)18:10<1055::aid-pd405>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Farag TI, Bastaki L, Marafie M, al-Awadi SA, Krsz J. Autosomal recessive congenital diaphragmatic defects in the Arabs. Am J Med Genet. 1994;50:300–301. doi: 10.1002/ajmg.1320500316. [DOI] [PubMed] [Google Scholar]
- Farag TI, Issa MA, Mahfouz ES. Discordant, non-syndromic, congenital diaphragmatic defects in sibs. J Med Genet. 1989;26:781–782. doi: 10.1136/jmg.26.12.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold M. Aplasia of the diaphragm. Pediatrics. 1971;47:601–603. [PubMed] [Google Scholar]
- Frey P, Glanzmann R, Nars P, Herzog B. Familial congenital diaphragmatic defect: transmission from father to daughter. J Pediatr Surg. 1991;26:1396–1398. doi: 10.1016/0022-3468(91)91044-y. [DOI] [PubMed] [Google Scholar]
- Garne E, Haeusler M, Barisic I, Gjergja R, Stoll C, Clementi M. Congenital diaphragmatic hernia: evaluation of prenatal diagnosis in 20 European regions. Ultrasound Obstet Gynecol. 2002;19:329–333. doi: 10.1046/j.1469-0705.2002.00635.x. [DOI] [PubMed] [Google Scholar]
- Gencik A, Moser H, Gencikova A, Kehrer B. Familial occurrence of congenital diaphragmatic defect in three families. Helv Paediatr Acta. 1982;37:289–293. [PubMed] [Google Scholar]
- Gibbs DL, Rice HE, Farrell JA, Adzick NS, Harrison MR. Familial diaphragmatic agenesis: an autosomal-recessive syndrome with a poor prognosis. J Pediatr Surg. 1997;32:366–368. doi: 10.1016/s0022-3468(97)90212-8. [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Geiger E, Benson DW. N KX2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Donnai D, Clericuzio CL, McDonald-McGinn DM, Guttenberg M, Zackai EH. Diaphragmatic hernia-exomphalos-hypertelorism syndrome: a new case and further evidence of autosomal recessive inheritance. Am J Med Genet. 1997;68:441–444. doi: 10.1002/(sici)1096-8628(19970211)68:4<441::aid-ajmg13>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Harberg FJ, Meagher D, Wetchler S, Harris F. Congenital anomalies of the diaphragm. Personal experience with thirty-five consecutive cases. Am J Surg. 1976;132:747–748. doi: 10.1016/0002-9610(76)90449-9. [DOI] [PubMed] [Google Scholar]
- Harnsberger J, Carey JC, Morgan M, Livingston GK. Interstitial deletion of the long arm of the number 8 chromosome and the Langer-Giedion syndrome (LG) Proceedings of the 1982 Birth Defects Conference. 1982;19:175–176. [Google Scholar]
- Hengstschlager M, Mittermayer C, Repa C, Drahonsky R, Deutinger J, Bernaschek G. Association of deletions of the chromosomal region 15q24-ter and diaphragmatic hernia: a new case and discussion of the literature. Fetal Diagn Ther. 2004;19:510–512. doi: 10.1159/000080164. [DOI] [PubMed] [Google Scholar]
- Hitch DC, Carson JA, Smith EI, Sarale DC, Rennert OM. Familial congenital diaphragmatic hernia is an autosomal recessive variant. J Pediatr Surg. 1989;24:860–864. doi: 10.1016/s0022-3468(89)80582-2. [DOI] [PubMed] [Google Scholar]
- Hou JW, Wang TR. Extreme Poland anomaly associated with congenital diaphragmatic hernia. Eur J Pediatr. 1999;158:433–434. doi: 10.1007/s004310051111. [DOI] [PubMed] [Google Scholar]
- Howe DT, Kilby MD, Sirry H, Barker GM, Roberts E, Davison EV, McHugo J, Whittle MJ. Structural chromosome anomalies in congenital diaphragmatic hernia. Prenat Diagn. 1996;16:1003–1009. doi: 10.1002/(SICI)1097-0223(199611)16:11<1003::AID-PD995>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Hutchinson R, Wilson M, Voullaire L. Distal 8p deletion (8p23.1----8pter): a common deletion? J Med Genet. 1992;29:407–411. doi: 10.1136/jmg.29.6.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javid PJ, Jaksic T, Skarsgard ED, Lee S. Survival rate in congenital diaphragmatic hernia: the experience of the Canadian Neonatal Network. J Pediatr Surg. 2004;39:657–660. doi: 10.1016/j.jpedsurg.2004.01.022. [DOI] [PubMed] [Google Scholar]
- Jay PY, Bielinska M, Erlich JM, Mannisto S, Pu WT, Heikinheimo M, Wilson DB. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev Biol. 2006;301:602–614. doi: 10.1016/j.ydbio.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadir RA, Hastings R, Economides DL. Prenatal diagnosis of supernumerary chromosome derivative (22) due to maternal balanced translocation in association with diaphragmatic hernia: a case report. Prenat Diagn. 1997;17:761–764. [PubMed] [Google Scholar]
- Kantarci S, Casavant D, Prada C, Russell M, Byrne J, Haug LW, Jennings R, Manning S, Blaise F, Boyd TK, Fryns JP, Holmes LB, Donahoe PK, Lee C, Kimonis V, Pober BR. Findings from aCGH in patients with congenital diaphragmatic hernia (CDH): a possible locus for Fryns syndrome. Am J Med Genet Part A. 2006a;140A:17–23. doi: 10.1002/ajmg.a.31025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci S, Donahoe PK, Hill RS, Al-Gazali L, Lacombe D, Chassaing D, Bieth E, Black G, Donnai D, Walsh C, Pober BR. Identification of a genetic locus for Donnai-Barrow syndrome (Abstract 1472). Presented at the annual meeting of The American Society of Human Genetics; October 10, 2005; Salt Lake City, Utah. 2006b. Available from http://wwwashgorg/genetics/ashg05s/. [Google Scholar]
- Kim EH, Cohen RS, Ramachandran P, Mineta AK, Babu VR. Trisomy 22 with congenital diaphragmatic hernia and absence of corpus callosum in a liveborn premature infant. Am J Med Genet. 1992;44:437–438. doi: 10.1002/ajmg.1320440410. [DOI] [PubMed] [Google Scholar]
- Klaassens M, Scott DA, van Dooren M, Hochstenbach R, Eussen HJ, Cai WW, Galjaard RJ, Wouters C, Poot M, Laudy J, Lee B, Tibboel D, de Klein A. Congenital diaphragmatic hernia associated with duplication of 11q23-qter. Am J Med Genet Part A. 2006;140A:1580–1586. doi: 10.1002/ajmg.a.31321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassens M, van Dooren M, Eussen HJ, Douben H, den Dekker AT, Lee C, Donahoe PK, Galjaard RJ, Goemaere N, de Krijger RR, Wouters C, Wauters J, Oostra BA, Tibboel D, de Klein A. Congenital diaphragmatic hernia and chromosome 15q26: determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet. 2005;76:877–882. doi: 10.1086/429842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Kufeji DI, Crabbe DC. Familial bilateral congenital diaphragmatic hernia. Pediatr Surg Int. 1999;15:58–60. doi: 10.1007/s003830050513. [DOI] [PubMed] [Google Scholar]
- Ladonne JM, Gaillard D, Carre-Pigeon F, Gabriel R. Fryns syndrome phenotype and trisomy 22. Am J Med Genet. 1996;61:68–70. doi: 10.1002/(SICI)1096-8628(19960102)61:1<68::AID-AJMG13>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lam YH, Eik-Nes SH, Tang MH, Lee CP, Nicholls JM. Prenatal sonographic features of spondylocostal dysostosis and diaphragmatic hernia in the first trimester. Ultrasound Obstet Gynecol. 1999;13:213–215. doi: 10.1046/j.1469-0705.1999.13030213.x. [DOI] [PubMed] [Google Scholar]
- Langham MR, Jr, Kays DW, Ledbetter DJ, Frentzen B, Sanford LL, Richards DS. Congenital diaphragmatic hernia. Epidemiology and outcome. Clin Perinatol. 1996;23:671–688. [PubMed] [Google Scholar]
- Lantieri F, Griseri P, Ceccherini I. Molecular mechanisms of RET-induced Hirschsprung pathogenesis. Ann Med. 2006;38:11–19. doi: 10.1080/07853890500442758. [DOI] [PubMed] [Google Scholar]
- Lerone M, Soliani M, Corea D, Romeo G, Martucciello G, Silengo MC. Congenital diaphragmatic hernia associated with ipsilateral upper limb reduction defects: report of a case with thumb hypoplasia. Am J Med Genet. 1992;44:827–829. doi: 10.1002/ajmg.1320440623. [DOI] [PubMed] [Google Scholar]
- Lin AE, Bernar J, Chin AJ, Sparkes RS, Emanuel BS, Zackai EH. Congenital heart disease in supernumerary der(22),t(11;22) syndrome. Clin Genet. 1986;29:269–275. doi: 10.1111/j.1399-0004.1986.tb01254.x. [DOI] [PubMed] [Google Scholar]
- Lipson AH, Williams G. Congenital diaphragmatic hernia in half sibs. J Med Genet. 1985;22:145–147. doi: 10.1136/jmg.22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang L, Wang D, Shen H, Jiang M, Mei P, Hayden PS, Sedor JR, Hu H. Congenital diaphragmatic hernia, kidney agenesis and cardiac defects associated with Slit3-deficiency in mice. Mech Dev. 2003;120:1059–1070. doi: 10.1016/s0925-4773(03)00161-8. [DOI] [PubMed] [Google Scholar]
- Loong TPF, Kocher HM. Clinical presentation and operative repair of hernia of Morgagni. Postgrad Med. 2005;81:41–44. doi: 10.1136/pgmj.2004.022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez I, Bafalliu JA, Bernabe MC, Garcia F, Costa M, Guillen-Navarro E. Prenatal diagnosis of de novo deletions of8p23.1 or 15q26.1 in two fetuses with diaphragmatic hernia and congenital heart defects. Prenat Diagn. 2006;26:577–580. doi: 10.1002/pd.1468. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genome structural variation and sporadic disease traits. Nature Genetics. 2006;38:974–976. doi: 10.1038/ng0906-974. [DOI] [PubMed] [Google Scholar]
- Lurie IW. Where to look for the genes related to diaphragmatic hernia? Genet Couns. 2003;14:75–93. [PubMed] [Google Scholar]
- Machin GA. Some causes of genotypic and phenotypic discordance in monozygotic twin pairs. Am J Med Genet. 1996;61:216–228. doi: 10.1002/(SICI)1096-8628(19960122)61:3<216::AID-AJMG5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Manouvrier-Hanu S, Besson R, Cousin L, Jeanpierre C, Kacet N, Cartigny M, Devisme L, Storme L, De Martinville B, Lequien P. Sex reversal and diaphragmatic hernia in phenotypicaly female sibs with normal XY chromosomes. J Med Genet. 2000;37:315–318. doi: 10.1136/jmg.37.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla MA, Kimani J, Mitchell LE, Christensen K, Boomsma DI, Daack-Hirsch S, Nepomucena B, Wyszynski DF, Felix TM, Martin NG, Murray JC. Discordant MZ twins with cleft lip and palate: a model for identifying genes in complex traits. Twin Res Hum Genet. 2005;8:39–46. doi: 10.1375/1832427053435373. [DOI] [PubMed] [Google Scholar]
- Marin J, Lopoo J. An infant with trisomy 21 and tachypnea. Pediatr Emerg Care. 2006;22:170–172. doi: 10.1097/01.pec.0000202457.64978.3d. [DOI] [PubMed] [Google Scholar]
- Marino T, Wheeler PG, Simpson LL, Craigo SD, Bianchi DW. Fetal diaphragmatic hernia and upper limb anomalies suggest Brachmann-de Lange syndrome. Prenat Diagn. 2002;22:144–147. doi: 10.1002/pd.281. [DOI] [PubMed] [Google Scholar]
- Martinez-Frias ML, Bermejo E, Paisan L, Martin M, Egues J, Lopez JA, Martinez S, Orbea C, Cucalon F, Gairi JM, et al. Severe spondylocostal dysostosis associated with other congenital anomalies: a clinical/epi-demiologic analysis and description of ten cases from the Spanish registry. Am J Med Genet. 1994;51:203–212. doi: 10.1002/ajmg.1320510306. [DOI] [PubMed] [Google Scholar]
- Mathieu M, Piussan C, Thepot F, Gouget A, Lacombe D, Pedespan JM, Serville F, Fontan D, Ruffie M, Nivelon-Chevallier A, Amblard F, Chauveau P, Moirot H, Chabrolle JP, Croquette MF, Teyssier M, Plauchu H, Pelissier MC, Gilgenkrantz S, Turc-Carel C, Turleau C, Prieur M, Le Merrer M, Gonzales M, Journel H, et al. Collaborative study of mosaic tetrasomy 12p or Pallister-Killian syndrome (nineteen fetuses or children) Ann Genet. 1997;40:45–54. [PubMed] [Google Scholar]
- McCredie J, Reid IS. Congenital diaphragmatic hernia associated with homolateral upper limb malformation: a study of possible pathogenesis in four cases. J Pediatr. 1978;92:762–765. doi: 10.1016/s0022-3476(78)80145-0. [DOI] [PubMed] [Google Scholar]
- McGaughran J, Rees M, Battin M. Craniofrontonasal syndrome and diaphragmatic hernia. Am J Med Genet. 2002;110:391–392. doi: 10.1002/ajmg.10176. [DOI] [PubMed] [Google Scholar]
- McPherson EW, Ketterer DM, Salsburey DJ. Pallister-Killian and Fryns syndromes: nosology. Am J Med Genet. 1993;47:241–245. doi: 10.1002/ajmg.1320470219. [DOI] [PubMed] [Google Scholar]
- Mertins H. [Familial abnormality of the diaphragm.] Zentralbl Gynakol. 1952;74:951–955. [PubMed] [Google Scholar]
- Migliazza L, Otten C, Xia H, Rodriguez JI, Diez-Pardo JA, Tovar JA. Cardiovascular malformations in congenital diaphragmatic hernia: human and experimental studies. J Pediatr Surg. 1999a;34:1352–1358. doi: 10.1016/s0022-3468(99)90010-6. [DOI] [PubMed] [Google Scholar]
- Migliazza L, Xia H, Alvarez JI, Arnaiz A, Diez-Pardo JA, Alfonso LF, Tovar JA. Heart hypoplasia in experimental congenital diaphragmatic hernia. J Pediatr Surg. 1999b;34:706–710. doi: 10.1016/s0022-3468(99)90360-3. Discussion 710–711. [DOI] [PubMed] [Google Scholar]
- Milne LW, Morosin AM, Campbell JR, Harrison MW. Pars sternalis diaphragmatic hernia with omphalocele: a report of two cases. J Pediatr Surg. 1990;25:726–730. doi: 10.1016/s0022-3468(05)80006-5. [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, Cole T, Redford DH. Congenital diaphragmatic hernia with probable autosomal recessive inheritance in an extended consanguineous Pakistani pedigree. J Med Genet. 1997;34:601–603. doi: 10.1136/jmg.34.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan H, De Chazal R, Barrow M, McKeever P, Neale E. Familial congenital diaphragmatic hernia: prenatal diagnosis, management, and outcome. Prenat Diagn. 1993;13:893–901. doi: 10.1002/pd.1970131003. [DOI] [PubMed] [Google Scholar]
- Norio R, Kaariainen H, Rapola J, Herva R, Kekomaki M. Familial congenital diaphragmatic defects: aspects of etiology, prenatal diagnosis, and treatment. Am J Med Genet. 1984;17:471–483. doi: 10.1002/ajmg.1320170210. [DOI] [PubMed] [Google Scholar]
- Pankau R, Janig U. Diaphragmatic defect in Brachmann-de Lange syndrome: a further observation. Am J Med Genet. 1993;47:1024–1025. doi: 10.1002/ajmg.1320470718. [DOI] [PubMed] [Google Scholar]
- Park JP, McDermet MK, Doody AM, Marin-Padilla JM, Moeschler JB, Wurster-Hill DH. Familial t(11;13)(q21;q14) and the duplication 11q, 13q phenotype. Am J Med Genet. 1993a;45:46–48. doi: 10.1002/ajmg.1320450113. [DOI] [PubMed] [Google Scholar]
- Park Y, Gong G, Choe G, Yu E, Kim KS, Lee I. Jarcho-Levin syndrome–a report of an autopsy case with cytogenetic analysis. J Korean Med Sci. 1993b;8:471–475. doi: 10.3346/jkms.1993.8.6.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar RC, Tullu MS, Bavdekar SB, Borwankar SS. Morgagni hernia with Down syndrome: a rare association–case report and review of literature. J Postgrad Med. 2001;47:188–190. [PubMed] [Google Scholar]
- Passarge E, Halsey H, German J. Unilateral agenesis of the diaphragm. Humangenetik. 1968;5:226–230. doi: 10.1007/BF00281959. [DOI] [PubMed] [Google Scholar]
- Pecile V, Petroni MG, Fertz MC, Filippi G. Deficiency of distal 8p–report of two cases and review of the literature. Clin Genet. 1990;37:271–278. doi: 10.1111/j.1399-0004.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- Philip N, Gambarelli D, Guys JM, Camboulives J, Ayme S. Epidemiological study of congenital diaphragmatic defects with special reference to aetiology. Eur J Pediatr. 1991;150:726–729. doi: 10.1007/BF01958765. [DOI] [PubMed] [Google Scholar]
- Pober BR, Lin A, Russell M, Ackerman KG, Chakravorty S, Strauss B, Westgate MN, Wilson J, Donahoe PK, Holmes LB. Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am J Med Genet Part A. 2005;138A:81–88. doi: 10.1002/ajmg.a.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR, Russell MR, Ackerman KG. GeneReviews at GeneTests: Medical Genetics Information Resource (database online) Seattle: Copyright, University of Washington; 2006. Congenital Diaphragmatic Hernia Overview. 1997–2007. Available at http://wwwgenetestsorg. [Google Scholar]
- Pollack LD, Hall JG. Posterolateral (Bochdalek’s) diaphragmatic hernia in sisters. Am J Dis Child. 1979;133:1186–1188. doi: 10.1001/archpedi.1979.02130110094019. [DOI] [PubMed] [Google Scholar]
- Rais-Bahrami K, Gilbert JC, Hartman GE, Chandra RS, Short BL. Right diaphragmatic eventration simulating a congenital diaphragmatic hernia. Am J Perinatol. 1996;13:241–243. doi: 10.1055/s-2007-994372. [DOI] [PubMed] [Google Scholar]
- Robert E, Kallen B, Harris J. The epidemiology of diaphragmatic hernia. Eur J Epidemiol. 1997;13:665–673. doi: 10.1023/a:1007395727819. [DOI] [PubMed] [Google Scholar]
- Rodgers BM, Hawks P. Bilateral congenital eventration of the diaphragms: successful surgical management. J Pediatr Surg. 1986;21:858–864. doi: 10.1016/s0022-3468(86)80008-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez LM, Garcia-Garcia I, Correa-Rivas MS, Garcia-Fragoso L. Pulmonary hypoplasia in Jarcho-Levin syndrome. P R Health Sci. 2004;J23:65–67. [PubMed] [Google Scholar]
- Rogers JC, Harris DJ, Pasztor LM. Interstitial deletion of the long arm of chromosome 1: del(1)(pter!42.11::q42.3!q-ter) Am J Hum Genet. 1995;57 [Google Scholar]
- Scapoli L, Palmieri A, Martinelli M, Pezzetti F, Carinci P, Tognon M, Carinci F. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer GB, Jochar A, Muneer R, Sanger WG. Clinical variability of tetrasomy 12p. Clin Genet. 1997;51:102–108. doi: 10.1111/j.1399-0004.1997.tb02429.x. [DOI] [PubMed] [Google Scholar]
- Schinzel A, Schmid W, Aufder Maur P, Moser H, Degenhardt KH, Geisler M, Grubisic A. Incomplete trisomy 22. I. Familial 11/22 translocation with 3:1 meiotic disjunction. Delineation of a common clinical picture and report of nine new cases from six families. Hum Genet. 1981;56:249–262. doi: 10.1007/BF00274675. [DOI] [PubMed] [Google Scholar]
- Schlembach D, Zenker M, Trautmann U, Ulmer R, Beinder E. Deletion 15q 24-26in prenatally detected diaphragmatic hernia: increasing evidence of a candidate region for diaphragmatic development. Prenat Diagn. 2001;21:289–292. doi: 10.1002/pd.50. [DOI] [PubMed] [Google Scholar]
- Scott DA, Klaassens M, Holder AM, Lally KP, Fernandes CJ, Galjaard RJ, Tibboel D, de Klein A, Lee B. Genome-Wide Oligonucleotide-Based Array Comparative Genome Hybridization Analysis of Non-Isolated Congenital Diaphragmatic Hernia. Hum Mol Genet. 2007;16:424–430. doi: 10.1093/hmg/ddl475. [DOI] [PubMed] [Google Scholar]
- Scott JM, Patterson L. Monozygous anencephalic triplets–a case report. J Obstet Gynaecol Br Commonw. 1966;73:147–151. doi: 10.1111/j.1471-0528.1966.tb05134.x. [DOI] [PubMed] [Google Scholar]
- Sergi C, Schulze BR, Hager HD, Beedgen B, Zilow E, Linderkamp O, Otto HF, Tariverdian G. Wolf-Hirschhorn syndrome: case report and review of the chromosomal aberrations associated with diaphragmatic defects. Pathologica. 1998;90:285–293. [PubMed] [Google Scholar]
- Shimokawa O, Miyake N, Yoshimura T, Sosonkina N, Harada N, Mizuguchi T, Kondoh S, Kishino T, Ohta T, Remco V, Takashima T, Kinoshita A, Yoshiura K, Niikawa N, Matsumoto N. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am J Med Genet Part A. 2005;136A:49–51. doi: 10.1002/ajmg.a.30778. [DOI] [PubMed] [Google Scholar]
- Skari H, Bjornland K, Haugen G, Egeland T, Emblem R. Congenital diaphragmatic hernia: a meta-analysis of mortality factors. J Pediatr Surg. 2000;35:1187–1197. doi: 10.1053/jpsu.2000.8725. [DOI] [PubMed] [Google Scholar]
- Slavotinek A, Lee SS, Davis R, Shrit A, Leppig KA, Rhim J, Jasnosz K, Albertson D, Pinkel D. Fryns syndrome phenotype caused by chromosome microdeletions at 15q26.2 and 8p23.1. J Med Genet. 2005;42:730–736. doi: 10.1136/jmg.2004.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM. Fryns syndrome: a review of the phenotype and diagnostic guidelines. Am J Med Genet Part A. 2004;124A:427–433. doi: 10.1002/ajmg.a.20381. [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Moshrefi A, Davis R, Leeth E, Schaeffer GB, Burchard GE, Shaw GM, James B, Ptacek L, Pennacchio LA. Array comparative genomic hybridization in patients with congenital diaphragmatic hernia: mapping of four CDH-critical regions and sequencing of candidate genes at 15q26.1 –15q26.2. Eur J Hum Genet. 2006;14:999–1008. doi: 10.1038/sj.ejhg.5201652. [DOI] [PubMed] [Google Scholar]
- Smith SA, Martin KE, Dodd KL, Young ID. Severe microphthalmia, diaphragmatic hernia and Fallot’s tetralogy associated with a chromosome 1;15 translocation. Clin Dysmorphol. 1994;3:287–291. [PubMed] [Google Scholar]
- Sripathi V, Beasley SW. Familial occurrence of complete agenesis of the diaphragm. J Paediatr Child Health. 1992;28:190–191. doi: 10.1111/j.1440-1754.1992.tb02640.x. [DOI] [PubMed] [Google Scholar]
- Stege G, Fenton A, Jaffray B. Nihilism in the 1990s: the true mortality of congenital diaphragmatic hernia. Pediatrics. 2003;112:532–535. doi: 10.1542/peds.112.3.532. [DOI] [PubMed] [Google Scholar]
- Tachdjian G, Fondacci C, Tapia S, Huten Y, Blot P, Nessmann C. The Wolf-Hirschhorn syndrome in fetuses. Clin Genet. 1992;42:281–287. doi: 10.1111/j.1399-0004.1992.tb03257.x. [DOI] [PubMed] [Google Scholar]
- Tapper JK, Zhang S, Harirah HM, Panova NI, Merryman LS, Hawkins JC, Lockhart LH, Gei AB, Velagaleti GV. Prenatal diagnosis of a fetus with unbalanced translocation (4;13)(p16;q32) with overlapping features of Patau and Wolf-Hirschhorn syndromes. Fetal Diagn Ther. 2002;17:347–351. doi: 10.1159/000065383. [DOI] [PubMed] [Google Scholar]
- Temple IK, Barber JC, James RS, Burge D. Diaphragmatic herniae and translocations involving 8q22 in two patients. J Med Genet. 1994;31:735–737. doi: 10.1136/jmg.31.9.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Kate LP, Anders GJ. Unilateral agenesis of the diaphragm. Humangenetik. 1970;8:366–367. doi: 10.1007/BF00280341. [DOI] [PubMed] [Google Scholar]
- Thomas MP, Stern LM, Morris LL. Bilateral congenital diaphragmatic defects in two siblings. J Pediatr Surg. 1976;11:465–467. doi: 10.1016/s0022-3468(76)80206-0. [DOI] [PubMed] [Google Scholar]
- Thorpe-Beeston JG, Gosden CM, Nicolaides KH. Prenatal diagnosis of congenital diaphragmatic hernia: associated malformations and chromosomal defects. Fetal Ther. 1989;4:21–28. doi: 10.1159/000263386. [DOI] [PubMed] [Google Scholar]
- Tonks A, Wyldes M, Somerset DA, Dent K, Abhyankar A, Bagchi I, Lander A, Roberts E, Kilby MD. Congenital malformations of the diaphragm: findings of the West Midlands Congenital Anomaly Register 1995 to 2000. Prenat Diagn. 2004;24:596–604. doi: 10.1002/pd.908. [DOI] [PubMed] [Google Scholar]
- Torfs CP, Curry CJ, Bateson TF, Honore LH. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- Toriello HV, Higgins JV, Jones AS, Radecki LL. Pulmonary and diaphragmatic agenesis: report of affected sibs. Am J Med Genet. 1985;21:87–92. doi: 10.1002/ajmg.1320210113. [DOI] [PubMed] [Google Scholar]
- Toriello HV, Landenburger G, Kapur SJ, Higgins JV. Isolated diaphragmatic defect in three sibs. Am J Med Genet. 1986 Suppl2:177–181. doi: 10.1002/ajmg.1320250621. [DOI] [PubMed] [Google Scholar]
- Turpin R, Petit P, Chigot P, Lafourcade J, De Barochez Y. [Congenital diaphragmatic hernia of the embryonic type (left pleuro-peritoneal cavity): Coincidence of this isolated malformation in 2 first cousins.] Sem Hop. 1959;35:1786–1793/P. [PubMed] [Google Scholar]
- Van Buggenhout G, Melotte C, Dutta B, Froyen G, Van Hummelen P, Marynen P, Matthijs G, de Ravel T, Devriendt K, Fryns JP, Vermeesch JR. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet. 2004;41:691–698. doi: 10.1136/jmg.2003.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dooren MF, Brooks AS, Hoogeboom AJ, van den Hoonaard TL, de Klein JE, Wouters CH, Tibboel D. Early diagnosis of Wolf-Hirschhorn syndrome triggered by a life-threatening event: congenital diaphragmatic hernia. Am J Med Genet Part A. 2004;127A:194–196. doi: 10.1002/ajmg.a.20613. [DOI] [PubMed] [Google Scholar]
- van Dooren MF, Brooks AS, Tibboel D, Torfs CP. Association of congenital diaphragmatic hernia with limb-reduction defects. Birth Defects Res A Clin Mol Teratol. 2003;67:578–584. doi: 10.1002/bdra.10079. [DOI] [PubMed] [Google Scholar]
- Varpella E, Lehtovaara R. Familial occurrence of diaphragmatic abnormalities. Ann Chir Gynaecol Fenn. 1962;58:62–64. [PubMed] [Google Scholar]
- Vasudevan PC, Twigg SR, Mulliken JB, Cook JA, Quarrell OW, Wilkie AO. Expanding the phenotype of craniofrontonasal syndrome: two unrelated boys with EFNB1 mutations and congenital diaphragmatic hernia. Eur J Hum Genet. 2006;14:884–887. doi: 10.1038/sj.ejhg.5201633. [DOI] [PubMed] [Google Scholar]
- Veldman A, Schlosser R, Allendorf A, Fischer D, Heller K, Schaeff B, Fuchs S. Bilateral congenital diaphragmatic hernia: Differentiation between Pallister-Killian and Fryns syndromes. Am J Med Genet. 2002;111:86–87. doi: 10.1002/ajmg.10438. [DOI] [PubMed] [Google Scholar]
- Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Welch RG, Cooke RT. Congenital Diaphragmatic Hernia. Lancet. 1962;279:975. [Google Scholar]
- Wolff G. Familial congenital diaphragmatic defect: review and conclusions. Hum Genet. 1980;54:1–5. doi: 10.1007/BF00279041. [DOI] [PubMed] [Google Scholar]
- Wu HC, Lin LH, Tsai LP, Huang CH, Hung KL, Liao HT. Pallister-Killian syndrome: report of one case. Acta Paediatr Taiwan. 2006;47:139–141. [PubMed] [Google Scholar]
- Yazici M, Karaca I, Arikan A, Erikci V, Etensel B, Temir G, Sencan A, Ural Z, Mutaf O. Congenital eventration ofthe diaphragm in children: 25 years’ experience in three pediatric surgery centers. Eur J Pediatr Surg. 2003;13:298–301. doi: 10.1055/s-2003-43573. [DOI] [PubMed] [Google Scholar]
- You LR, Takamoto N, Yu CT, Tanaka T, Kodama T, Demayo FJ, Tsai SY, Tsai MJ. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2005;102:16351–16356. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H, Chance P, Tuck-Muller CM, Jabs EW. Association of a new chromosomal deletion [del(1)(q32q42)] with diaphragmatic hernia: assignment of a human ferritin gene. Hum Genet. 1988;78:267–270. doi: 10.1007/BF00291674. [DOI] [PubMed] [Google Scholar]
- Yuan W, Rao Y, Babiuk RP, Greer JJ, Wu JY, Ornitz DM. A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci USA. 2003;100:5217–5222. doi: 10.1073/pnas.0730709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]