The majority of genes are inherited in 2 copies, 1 from each parent (the exceptions being sex-linked genes in males and mitochondrial genes). Whereas most such genes have identical functions, imprinted genes usually function only when inherited from either the mother or the father. Imprinted genes are marked in the male and female germline and retain molecular memory of their parental origin, resulting in allelic expression differences during development. Over 35 imprinted genes have been identified to date in the combined human and mouse genomes, representing perhaps 5–20% of those predicted to be imprinted (1, 2). Abnormalities in imprinted inheritance occur in several well-known developmental and neurobehavioral disorders, including Albright’s hereditary osteodystrophy and Angelman, Beckwith-Wiedemann, and Prader-Willi syndromes. Each of these diseases involves complex genetic loci and gene regulation and will be compared in this article. Recent studies have also demonstrated directly a role for imprinted genes in determining brain development and for 2 paternally expressed genes in regulating maternal behavior in mice. Here I will review evidence that imprinted mammalian genes influence complex neurobehavioral phenotypes, including psychiatric disorders. Such observations have significant implications for genetic approaches to identifying the etiological genes involved in these and other similar complex traits.
Prader-Willi and Angelman syndromes.
Clinical features of Prader-Willi syndrome (PWS) include postnatal failure to thrive with childhood onset of hyperphagia and severe obesity, and short stature with neurosecretory growth hormone deficiency (3). Other neurobehavioral attributes of PWS include developmental delay, mild to moderate mental retardation, learning disabilities (with strengths in reading, speech, and long-term memory and weaknesses in arithmetic and short-term memory), obsessive-compulsive disorder, temper tantrums and stubbornness, poor social interactions, abnormal sleep, hypogonadotropic hypogonadism, temperature instability, and, in 5 to 10% of patients, psychosis. In contrast, the Angelman syndrome (AS) clinical phenotype is characterized by severe neurological features, including developmental delay, ataxia, severe mental retardation, lack of speech, paroxysms of laughter, seizures, hyperactivity, attention deficit (with poor social interactions), sleeping difficulty, aggressive behavior, and tongue protrusion.
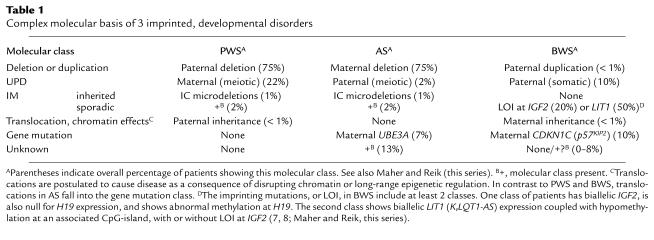
The genetic aberrations in PWS and AS include a common 4-Mb 15q11-q13 deletion of paternal or maternal origin, respectively, or maternal uniparental disomy (UPD) in PWS and paternal UPD in AS (Table 1) (4, 5). These 2 genetic classes indicate that the PWS gene(s) are normally expressed only from the paternal allele and are silent on the maternal chromosome, with the opposite pattern and a maternally expressed gene causing AS. The latter has been identified as the UBE3A gene, because specific mutations occur in familial and sporadic patients (Table 1) (5). Intriguingly, UBE3A typifies a growing number of mammalian imprinted genes, in that it only displays imprinting in a tissue-restricted manner. Thus, maternal-only expression of UBE3A occurs only in certain regions of the human and mouse brain, such as cerebellar Purkinje’s cells and hippocampal neurons (5), with biparental expression in all other tissues. These observations explain the neurological-specific phenotype of AS.
Table 1.
Complex molecular basis of 3 imprinted, developmental disorders
In contrast to AS patients, individuals with PWS of the classic type always have chromosomal abnormalities — large deletions, UPD, or an imprinting mutation (the latter discussed below) — affecting multiple genes. As seen in Table 1, no patient has inheritance consistent with a single gene mutation, suggesting that PWS requires the loss of function of 2 or more paternally expressed genes (3, 4). Indeed, multiple genes have been identified across a 1.5–2-Mb region of human chromosome 15q11-q13 that are expressed only from the paternally inherited allele (Figure 1a) (3, 4). The deletion interval associated with PWS also includes the maternally expressed UBE3A gene and several nonimprinted genes (Figure 1a). The finding of many paternally expressed genes presents a major obstacle to identifying the critical imprinted genes involved in PWS, because each of these, or others not yet identified, may play some phenotypic role in PWS. As the 15q11-q13 genes are identified from the human genome sequence and allele-specific expression assays, it becomes possible to test their contribution to the PWS phenotype, either by generating mouse models or by studying people with specific components of PWS for evidence of mutations in the gene of interest. Of the PWS candidate genes, one intriguing locus (SNURF-SNRPN) is polycistronic and encodes 2 functional proteins (3). This complex locus is of considerable interest because it specifically shows microdeletions (7–200 kb) in patients with mutations in the imprinting process, indicating an involvement in the regulation of imprinting in 15q11-q13 (see below).
Figure 1.

Complex structures of imprinted gene loci. (a) Genetic map of the 2-Mb imprinted domain in chromosome 15q11-q13 associated with PWS and AS. An IC is associated with the 5′ end of the bicistronic SNURF-SNRPN locus. Blue or pink circles, imprinted genes showing expression of only the paternal or maternal allele, respectively; open squares, nonimprinted genes; small arrows, overlapping and antisense transcription; large arrows, regional imprint control through an IC; zigzag lines, common breakpoint (BP) regions for deletions. (b) Genetic map of the 1-Mb imprinted domain in chromosome 11p15 associated with BWS. Symbols are the same as in a; colored squares represent genes imprinted in some tissues in mice, but not yet shown in humans. Three loci have been specifically implicated in the pathogenesis of BWS. The 5′ H19 region appears to act as a local IC regulating imprinting at IGF2, whereas a 5′ CpG-island for LIT1 (KvLQT1-AS) has been proposed to perhaps (denoted by ?) act as an IC for either CDKN1C, IGF2, and/or other genes in the 11p15 domain. (c) Genetic map of the 75-kb GNAS1 locus in chromosome 20q13. Three alternative promoters of transcription all lead to splicing to a common set of 12 downstream exons. Each exon 1 region has a different pattern of parental origin–specific allelic methylation and transcription, and each leads to a different encoded protein product.
Although a few balanced translocations occur in PWS (Table 1), the molecular basis is not understood (3). Two patients with classical PWS have a translocation breaking within the bicistronic SNURF-SNRPN gene and express all tested paternally expressed genes in 15q11-q13, implicating the disrupted locus in PWS. However, the translocations in 2 patients with atypical “PWS-like” phenotypes lay distal of SNURF-SNRPN but proximal to the IPW gene (Figure 1a), which may indicate a role for genes at or on the distal side of the translocation. A unifying hypothesis is that the translocations affect neuronal chromatin structure, disrupting the function of multiple genes within 15q11-q13 through some form of position effect. Whereas it is also possible that the germline imprinting process (see below) is affected, the imprint may already have been set before the chromosome rearrangement, and this hypothesis is not fully compatible with the expression and methylation data.
Several mouse models have recently been developed for PWS or AS. These include animals with UPD and strains that carry a 4-Mb deletion affecting the region of the mouse genome that corresponds to human 15q11-q13. Interestingly, the deletion model segregates mouse phenotypes that mimic both PWS and AS, depending on the inheritance of the mutation. In addition, an imprinting mutation, discussed below, causes features of PWS, and targeted mutations of the murine Ube3a gene have been used to study AS (reviewed in ref. 6). The AS models display a mild neurobehavioral phenotype, observable only on careful testing, whereas the PWS mouse models all fail to thrive and often die shortly after birth. Failure to thrive is also seen in infants with PWS, and recent mouse studies involving gene-specific knockouts suggest that this phenotype involves the additive contribution of several genes (6), including the NDN gene, SNURF-SNRPN, and/or an unknown gene between the latter locus and IPW (see Figure 1a) (6). By restoring these genes individually into PWS mouse models it may be possible to identify their contributions to the various phenotypic components of PWS. Ultimately, therapeutic intervention in PWS or AS will require a fuller understanding of the biochemistry and physiological roles of the products of these genes.
Developmental disorders involving imprinted genes.
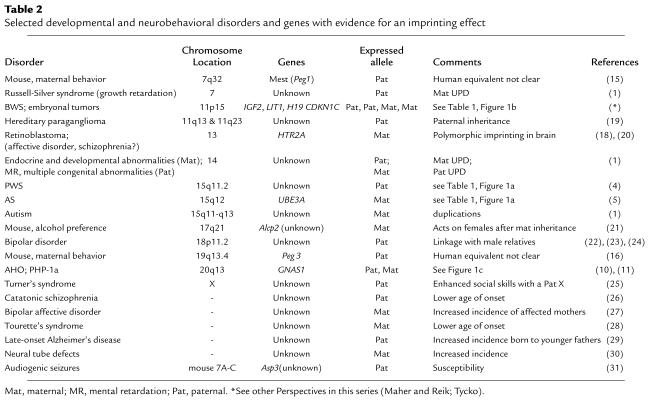
Molecular analysis of 2 disorders, Beckwith-Wiedemann syndrome (BWS) and Albright hereditary osteodystrophy (AHO), provides the clearest examples of imprinted genes that regulate growth and development, although Table 2 indicates other developmental disorders that show similar, but less well-understood, effects. As for PWS and AS, the molecular basis of BWS is extraordinarily complex. The genetic mechanisms underlying BWS include paternally derived duplications, UPD, and loss of imprinting (LOI) at IGF2, implicating a role for a paternally expressed gene, and maternally derived translocations and specific CDKN1C (p57KIP2) mutations, implicating a role for a maternally expressed gene (Table 1) (7, 8; Maher and Reik in this Perspective series). Mouse model studies suggest that in some tissues, where both are expressed, IGF2 and CDKN1C antagonize each other’s effects on cell cycle progression (9). Nevertheless, these 2 oppositely imprinted candidate genes are located over 500 kb apart and may be located in independently regulated imprinted domains (Figure 1b). Detailed mutational studies in the mouse have clearly shown that the 5′ H19 region is able to regulate the imprinting of H19, Igf2, and Ins2, but that this domain is independent of the other imprinted genes in chromosome 11p15 (Figure 1b) (2). It is unclear whether, or to what extent, the imprinting of these other genes is coordinately regulated. Evidence for long-range chromatin effects is provided by the occurrence of translocations in BWS patients within KvLQT1 (Table 1), which, though imprinted in most tissues, is clearly not the target gene, because recessive and dominant cardiac diseases occur as a consequence of the lack of imprinting in this tissue. Furthermore, recent studies have identified a paternally expressed antisense transcript (KvLQT1-AS or LIT1) associated with a differentially methylated CpG-island and shown that LOI at this locus is the most common abnormal epigenetic alteration in BWS (Table 1 and Figure 1b) (7, 8; Maher and Reik, this series). In some, but not all, of these translocation and LIT1 LOI cases, there is also LOI at IGF2. Hence, it is not clear if this genetic element acts as a general imprint regulator (Figure 1b) or if these BWS epimutations specifically interfere with gene regulation of CDKN1C (7, 8).
Table 2.
Selected developmental and neurobehavioral disorders and genes with evidence for an imprinting effect
AHO is associated with short stature and skeletal defects and arises due to dominant inactivating mutations in the GNAS1 gene encoding the α-subunit of the heterotrimeric G protein Gs, as does a more severe disorder with AHO and the hormone-resistant syndrome termed pseudohypoparathyroidism type 1a (PHP-1a; Table 2) (10). Isolated parathyroid hormone resistance (PHP-1b) is also linked to the same locus, although Gsα levels are normal. PHP-1a and PHP-1b arise from exclusive maternal inheritance, whereas AHO alone occurs within PHP-1a pedigrees after paternal inheritance. Oppositely imprinted endocrine, behavioral, and morphological phenotypes also arise from UPD at this locus or from the targeted disruption of exon 2 of the mouse Gnas gene (see ref. 11 for review). The disease complexity at this locus may be explained by its remarkable genetic structure (Figure 1c) (10, 11). Although Gsα is encoded by the nonimprinted exons 1-13 of GNAS1, 2 upstream alternative promoters are found in this locus, and, as seen in Figure 1c, these sequences are oppositely imprinted. Thus, the first exon under control of each of these promoters is spliced with exons 2–13 of GNAS1 to generate the maternally expressed NESP55 or the paternally expressed XLαs. The latter 2 proteins appear to be coexpressed, and both are involved in formation of neuroendocrine secretory granules (10). The inheritance patterns produced by mutations at this locus in the mouse or in human PHP-1a could be explained by interference with transcriptional or splicing regulation at GNAS1 (10, 11) or by antagonistic roles of the 3 proteins translated from this bizarre locus. AHO may arise solely from 50% reduction in Gsα, with or without an effect on XLαs, whereas PHP-1b mutations may occur in NESP55. Further mutational studies in both species will be needed to clarify these issues.
Molecular basis of genomic imprinting.
It is clear from the preceding discussion that imprinted genetic disease is associated with complex inheritance patterns and elaborate gene loci often comprising large clusters of imprinted genes (Figure 1). Other than allelic transcription differences, the epigenetic modification most clearly demonstrated for imprinted genes is the differential patterns of allele-specific DNA methylation in somatic tissues, dependent on the parent of origin (2, 12). Differential and specific DNA methylation imprints have been identified within several imprinted genes in sperm and oocytes, and in several cases, experimental studies in the mouse have shown that these represent the gametic imprints (molecular memory of parental origin) transmitted to the next generation. However, recent studies have implicated a wide range of gene-specific and chromatin-domain features in the regulation of imprinted gene expression in somatic cells (2, 7, 8, 12, 13). These include differential histone H4 and H3 acetylation, nuclease sensitivity, and nuclear matrix association, as well as the presence of G-rich direct-repeat sequences in or near CpG-islands, oppositely imprinted antisense RNA transcripts (see above), and asynchronous DNA replication and homologous chromosome association. Nevertheless, whereas many of these factors may help maintain imprinted domains in somatic cells or be secondary events consequent to differential packaging of chromatin on the 2 alleles, the respective roles in establishing the imprint in germ cells or after maternal and paternal pronuclei fusion after fertilization remain unknown.
Imprinting is a reversible process requiring the previous parental imprint to switch in the germline of progeny of the opposite sex. Mutations in this process occur in approximately 3% of AS and PWS patients, and particular insight into the molecular mechanism has come from study of those patients with heritable microdeletions in 15q11-q13 (Table 1) (4). Although the genotype of such imprinting mutation (IM) patients appears normal, their pattern of DNA methylation and gene expression — that is, their epigenotype — is abnormal, with maternal-only imprints in PWS and paternal-only imprints in AS.
Familial transmission of such epigenetic defects results from specific microdeletions in this region, but many IM patients are sporadic and have no detectable mutation; stochastic events during parental gametogenesis probably account for this class of imprinting defects (Table 1) (4). Inherited microdeletions in familial PWS and AS IM patients occur at the 5′ end of the SNURF-SNRPN gene, defining a region termed the imprinting center (IC; Figure 1a). The IC is thought to regulate initiation of the parental imprint switch in the male and female germline (4, 13), leading to the establishment of heterochromatic-like DNA at the IC in oocytes and euchromatic-like DNA at the IC during spermatogenesis. Subsequently, these chromatin states may propagate bidirectionally and consequently regulate imprint switching for all genes over the 2-Mb imprinted domain (Figure 1a) (13). Both the familial and sporadic forms of IM effects are remarkable among human genetic diseases in that the IM acts, not in the affected patient, but rather in the parental germline, where it blocks the switch of the grandparental imprint. Clinical manifestations occur only because the offspring has inherited the incorrect grandparental imprint.
An understanding of the molecular events underlying these cases may also lead to insight into the basis of imprinting mutations at the H19, IGF2, and LIT1 (KvLQT1-AS) genes in chromosome 11p15 in BWS and in human embryonal tumors (Table 1) (7, 8; Maher and Reik, this series; Tycko, this series). In these diseases, the epigenetic changes usually occur sporadically and hence are probably somatic in origin. At present, the primary genetic or epigenetic events causing these changes are unknown, and this is paralleled by our ignorance of the primary events causing sporadic IM in PWS and AS. Identification of these events will have a significant impact on our understanding of how imprinting occurs and whether it may one day be feasible to manipulate the epigenetic state of an normally silent imprinted allele as a therapeutic approach in imprinted disorders.
Imprinted genes play important roles in brain function and behavior.
Imprinted genes have been indirectly implicated in brain function and behavior as a consequence of the genetic defects in well-studied syndromes such as PWS, AS, and AHO, although the roles of such genes in growth processes have received greater attention. However, recent experimental studies in the mouse have more directly indicated an important role for imprinting in regulating brain development and behavior. The first evidence for imprinting in mice came from nuclear transfer experiments that generated parthenogenetic and androgenetic embryos, the former containing a diploid maternal genome and the latter containing a diploid paternal genome. This work established that the 2 parental genomes play complementary developmental roles, and subsequent experiments revealed that tissues are differentially affected by uniparental development. Using chimeras between normal and uniparental cells to overcome the lethality seen in parthenogenetic and androgenetic embryos, it has been found that androgenetic cells contribute to, or survive in, specific tissues, including placenta, muscle, and certain brain structures, but that the maternal genome contributes to other brain structures and to a greater extent to brain growth (2, 14). A high proportion of parthenogenetic cells in the brain was also associated with male aggression. The paternal genome contributed primarily to regions important for primary motivated behavior, such as the hypothalamus, but androgenetic cells were excluded from the neocortex and striatum, areas that showed selective accumulation of parthenogenetic cells (14).
Two recent studies are of particular relevance to understanding the roles imprinted genes have in growth and behavior. Mutations were targeted into the mouse Mest and Peg3 genes, resulting in both instances in a moderate postnatal growth retardation up until weaning at 4 weeks of age and an intriguing effect on maternal behavior following grandpaternal transmission of the mutation (15, 16). Thus, most offspring of such females do not survive, because the mother fails to retrieve, feed, or warm the pups; these females also lack such typical maternal behaviors as nest building and consuming the placenta after pups are born. In the case of Peg3 mutations, a maternal lactation defect with a reduced number of oxytocin-positive hypothalamic neurons may contribute to this phenotype (16). Both paternally expressed genes therefore appear to play roles in controlling postnatal growth and nurturing behavior in mothers.
Evolution of imprinting in growth and behavioral pathways.
Perhaps the best supported theory for evolution of imprinting is that a parent–offspring “conflict” results in enhancers of prenatal and postnatal growth being of paternal origin, whereas those of maternal origin should be growth suppressors (2, 17). Parent–offspring conflict arises from the dependence of mammalian offspring on maternal resources from conception through weaning and the potential involvement of multiple paternity. This model fits well with the proposed antagonistic roles of paternal IGF2 and maternal CDKN1C expression in promoting and inhibiting growth, respectively, and for several other imprinted genes (2). Similarly, we have proposed that selection for imprinting in the PWS region arose for a postnatal growth advantage to a paternally derived gene, given the failure-to-thrive phenotype of PWS neonates and PWS mouse model pups (3). However, this selection pressure need only operate on a single gene within a large, coordinately regulated imprinted domain, and most genes within the domain may display imprinting simply as a consequence of recent evolutionary acquisition by the domain or as a consequence of the spreading effect of the imprinting mechanism.
The parent–offspring conflict hypothesis has been expanded to explain the grandpaternal role of Mest and Peg3 in controlling maternal behavior for the survival and care of offspring to ensure growth enhancement postnatally (15, 16). More generally, this hypothesis can also be related to evolutionary changes in mammalian brain growth, in which the expansion of the neocortex and striatum associated with social behavior can explain the role of the maternal genome in contributing to these structures (14). Similarly, the predominant contribution of the paternal genome to the hypothalamus and other structures involved in reproduction, feeding, and growth, may account for the relative contraction of these regions in the brain during the evolution of primates and humans. These changes provide a genetic explanation for complex neural development and behavior.
Psychiatric disorders and behavioral phenotypes that may involve imprinted genes.
Recent studies provide additional genetic clues implicating imprinted genes in complex neurobehavioral phenotypes, including susceptibility genes involved in alcohol preference and audiogenic seizures (Table 2). In addition, evidence for linkage of candidate loci to bipolar affective disorder is, in some cases, sensitive to the parent of origin of the putative disease allele. Similarly, a lower age of onset of one subtype of schizophrenia after paternal inheritance and of Tourette’s syndrome after maternal inheritance, or an increased incidence of familial neural tube defects maternally transmitted and late onset Alzheimer’s disease paternally transmitted, may indicate underlying imprinted genes (Table 2). In one intriguing example, social cognitive skills were better for Turner’s syndrome patients inheriting the paternal, rather than the maternal, X chromosome (Table 2). Until the relevant imprinted genes associated with these disorders are isolated it will not be possible to prove the imprinting hypothesis in such cases, and the occurrence of tissue-specific and mRNA isoform-specific imprinting (discussed above) or polymorphic imprinting (18) will potentially complicate the identification of these genes. However, the insight from the cited studies that imprinting allows susceptibility alleles to be transmit silently from parents of one sex within a pedigree may account for some of the difficulties in psychiatric genetics that are otherwise ascribed to environmental modification of the phenotype, variable expressivity, incomplete penetrance, or genetic modifiers.
The work discussed here clearly indicates the complementary requirement for normal development and neurobehavioral function of inheritance deriving from the mother and the father. These studies also highlight the potential for significant advancement in multidisciplinary fields ranging from genetics and epigenetic regulation to behavioral sciences, development, and pathology, which will further aid development of therapeutic approaches to developmental and neurobehavioral disorders.
Acknowledgments
I thank Ben Tycko and John Ashkenas for comments on the manuscript and all students and colleagues who have contributed to our work in imprinting. In many instances recent reviews have been cited because of space restraints, and I apologize to all whose important primary work was, consequently, not cited. This work is supported by the National Institutes of Health (HD-31491, HD-36079, and HD-36436) and March of Dimes Birth Defects Foundation (6-FY99-390).
References
- 1.Morison IM, Reeve AE. A catalogue of imprinted genes and parent-of-origin effects in humans and animals. Hum Mol Genet. 1998;7:1599–1609. doi: 10.1093/hmg/7.10.1599. [DOI] [PubMed] [Google Scholar]
- 2.Tilghman SM. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls RD, Ohta T, Gray TA. Genetic abnormalities in Prader-Willi syndrome and lessons from mouse models. Acta Paediatr. 1999;88(Suppl.):99–104. doi: 10.1111/j.1651-2227.1999.tb14414.x. [DOI] [PubMed] [Google Scholar]
- 4.Nicholls RD, Saitoh S, Horsthemke B. Imprinting in Prader-Willi and Angelman syndromes. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 5.Jiang Y-H, et al. Genetics of Angelman syndrome. Am J Hum Genet. 1999;65:1–6. doi: 10.1086/302473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholls RD. Incriminating gene suspects, Prader-Willi style. Nat Genet. 1999;23:132–134. doi: 10.1038/13758. [DOI] [PubMed] [Google Scholar]
- 7.Lee MP, et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KvLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smilinich NJ, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caspary T, et al. Oppositely imprinted genes p57Kip2 and Igf2 interact in a mouse model for Beckwith-Wiedmann syndrome. Genes Dev. 1999;13:3115–3124. doi: 10.1101/gad.13.23.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward BE, Moran V, Starin L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci USA. 1998;95:15475–15480. doi: 10.1073/pnas.95.26.15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters J, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci USA. 1999;96:3830–3835. doi: 10.1073/pnas.96.7.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constância M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 13.Greally JM, et al. Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center. Proc Natl Acad Sci USA. 1999;96:14430–14435. doi: 10.1073/pnas.96.25.14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keverne EB. Genomic imprinting in the brain. Curr Opin Neurobiol. 1997;7:463–468. doi: 10.1016/s0959-4388(97)80023-2. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre L, et al. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- 16.Li L-L, et al. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 17.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 18.Bunzel R, et al. Polymorphic imprinting of the serotonin-2A (5-HT2A) receptor gene in human adult brain. Brain Res Mol Brain Res. 1998;59:90–92. doi: 10.1016/s0169-328x(98)00146-6. [DOI] [PubMed] [Google Scholar]
- 19.Struycken PM, Cremers CW, Mariman EC, Joosten FB, Bleker RJ. Glomus tumours and genomic imprinting: influence of inheritance along the paternal or maternal line. Clin Otolaryngol. 1997;22:71–76. doi: 10.1046/j.1365-2273.1997.00884.x. [DOI] [PubMed] [Google Scholar]
- 20.Kato MV, Ikawa Y, Hayashizaki Y, Shibata H. Paternal imprinting of mouse serotonin receptor 2A gene Htr2 in embryonic eye: a conserved imprinting regulation on the RB/Rb locus. Genomics. 1998;47:146–148. doi: 10.1006/geno.1997.5089. [DOI] [PubMed] [Google Scholar]
- 21.Melo JA, Shendure J, Pociask K, Silver LM. Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/6 mice. Nat Genet. 1996;13:147–153. doi: 10.1038/ng0696-147. [DOI] [PubMed] [Google Scholar]
- 22.Stine OC, et al. Evidence for linkage of bipolar disorder to chromosome 18 with a parent-of-origin effect. Am J Hum Genet. 1995;57:1384–1394. [PMC free article] [PubMed] [Google Scholar]
- 23.Gershon ES, Badner JA, Detera-Wadleigh SD, Ferraro TN, Berrettini WH. Maternal inheritance and chromosome 18 allele sharing in unilineal bipolar illness pedigrees. Am J Med Genet (Neuropsych Genet) 1996;67:202–207. doi: 10.1002/(SICI)1096-8628(19960409)67:2<202::AID-AJMG11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Nöthen MM, et al. Evaluation of linkage of bipolar affective disorder to chromosome 18 in a sample of 57 German families. Mol Psych. 1999;4:76–84. doi: 10.1038/sj.mp.4000454. [DOI] [PubMed] [Google Scholar]
- 25.Skuse DH, et al. Evidence from Turner’s syndrome of an imprinted X-linked locus affecting cognitive function. Nature. 1997;387:705–708. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- 26.Stöber G, Haubitz I, Franzek E, Beckmann H. Parent-of-origin effect and evidence for differential transmission in periodic catatonia. Psychiatr Genet. 1998;8:213–219. doi: 10.1097/00041444-199808040-00003. [DOI] [PubMed] [Google Scholar]
- 27.McMahon FJ, Stine OC, Meyers DA, Simpson SG, DePaulo JR. Patterns of maternal transmission in bipolar affective disorder. Am J Hum Genet. 1995;56:1277–1286. [PMC free article] [PubMed] [Google Scholar]
- 28.Eapen V, O’Neill J, Gurling HMD, Robertson MM. Sex of parent transmission effect in Tourette’s syndrome. Neurology. 1997;48:934–937. doi: 10.1212/wnl.48.4.934. [DOI] [PubMed] [Google Scholar]
- 29.Farrer LA, Cupples A, Connor L, Wolf PA, Growdon JH. Association of decreased paternal age and late-onset Alzheimer’s disease. An example of genetic imprinting? Arch Neurol. 1991;48:599–604. doi: 10.1001/archneur.1991.00530180051017. [DOI] [PubMed] [Google Scholar]
- 30.Byrne J, et al. Multigeneration maternal transmission in Italian families with neural tube defects. Am. J. Med. Genet. 66:303–310.31.Banko, B.L., Allen, K.M., Dolina, S., Neumann, P.E., and Seyfried, T.N. 1997. Genomic imprinting and audiogenic seizures in mice. Behav Genet. 1996;27:465–475. doi: 10.1023/a:1025626501148. [DOI] [PubMed] [Google Scholar]