Abstract
Sarcomeres, bundled into thick and thin filaments, are the units of contraction in the striated muscle. The thick filaments comprise several hundred hexameric myosin molecules, composed of 2 myosin heavy chain (MyHC) proteins, the molecular motor of contraction, and 2 regulatory and 2 essential light chains. The globular head of MyHC contains the binding domains for cardiac α-actin and adenosine triphosphate (ATP) and is attached to a hinge region, which when flexed, moves the globular head over the thin filaments. The thin filaments comprise the cardiac troponin C (cTnC), T (cTnT), and I (cTnI) complex, α-tropomyosin dimers, and cardiac α-actin, maintained in a tight 1:1:7 stoichiometry. Several additional sarcomeric proteins, such as myosin-binding protein C, titin, obscurin, and telethonin contribute to the stabilization and function of the sarcomeres.
Keywords: Editorials, heart failure, myosin isoforms, troponins, genetics
The troponin-tropomyosin complex regulates the calcium-dependent displacement of the thin filaments by the globular head of MyHC, which results in sarcomere shortening and generation of the force of muscle contraction.1–3 Depolarization of membrane potential opens the L-type calcium channels and allows Ca2+ influx, which triggers calcium-induced calcium release by opening the ryanodine receptors in the junctional sarcoplasmic reticulum (SR). The cytosolic Ca2+ binds to cTnC at the low-affinity site and induces conformational changes, which lead to removal of the inhibitory domain of cTnI away from the α-tropomyosin-actin complex (Figure).3 Consequently, MyHC binds to actin, hydrolyzes ATP to adenosine diphosphate (ADP) and inorganic phosphate, and displaces the actin filament. Each single myosin generates 3 to 4 pN force and displaces the actin by 11 nmol.4 Uptake of Ca2+by the SR reverses the process. The number and the biochemical composition of the sarcomeres in myocytes determine the magnitude of the force generated and the velocity of contraction.
Figure 1.
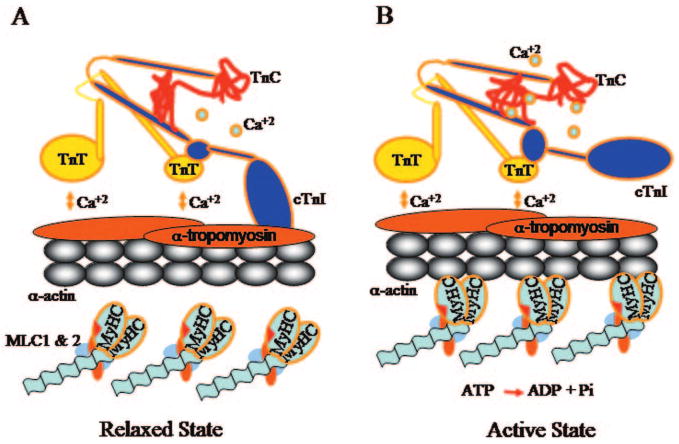
Essential role of the inhibitory arm of cardiac troponin I (cTnI) in regulating cardiac contraction and relaxation. (A) In the resting state, cytosolic Ca2+ concentration is low and troponin C (cTnC) is unbound to Ca2+. The inhibitory arm of cTnI binds to α-tropomyosin and blocks binding of the globular head of myosin to cardiac α-actin. (B) Increased cytosolic Ca2+ concentration on opening of the ryonidine channels results in the binding of Ca2+ to cTnC, which leads to removal of the inhibitory arm of cTnI from α-tropomyosin. The latter allows displacement of the thin filaments by the globular head of myosin on hydrolysis of ATP to ADP and Pi.
Two MyHC isoforms, referred to as the α-MyHC and β-MyHC, encoded by 2 distinct genes, located in tandem on chromosome 14, are expressed in the mammalian ventricular myocardium.5 The 2 MyHC isoforms share >90% sequence identify but differ significantly in ATPase activity and the rate of displacement of actin.6,7 The velocity of actin displacement is 2- to 4-fold faster, the duration of force transients is shorter, and the ATPase activity is 2- to 3-fold higher for the α-MyHC (fast) than for the β-MyHC (slow) isoform.6,7 Hence, to perform the same workload, α-MyHC uses more ATP than the β-MyHC.8 Accordingly, the relative distribution of the α-MyHC and β-MyHC isoforms in the myocardium could have significant effects on the phenotypic expression of cardiac diseases.
In this issue of Circulation, Sanbe et al and James et al report the phenotypic consequences of changes in 2 sarcomeric proteins in the heart in transgenic rabbit models of human heart failure.9,10 One model is designed to delineate the pathogenesis of human hypertrophic cardiomyopathy (HCM) caused by a missense mutation (R146G) in cardiac troponin I (cTnI),9 and the other is designed to define the functional significance of α-MyHC in human heart failure.11,12 The phenotypes entail the wide spectrum of the responses of the heart to injury, expressed as either HCM or dilated cardiomyopathy (DCM). The choice of the rabbit as the transgenic model to study human heart failure is commendable because similarities in the composition of sarcomeric proteins between humans and the selected animal model are critical for recapitulating and delineating the pathogenesis of phenotype. The mouse, the conventional transgenic model of human diseases, differs significantly from the human in sarcomeric protein composition. The differences could limit the utility of the mouse models in resolving the underlying mechanisms of human heart muscle diseases. The concern has been especially applicable to the phenotype resulting from mutations in the β-MyHC because of the major differences in the expression levels of MyHC isoforms in human and mice hearts.5 The β-MyHC is the predominant isoform in the ventricles of humans, comprising >90% of the total myofibrillar myosin.5 This is in contrast to the mouse, in which the α-MyHC isoform predominates and comprises >95% of the total myofibrillar myosin protein.5 The rabbit heart has a sarcomeric protein composition more similar to that in the human heart, with the β-MyHC comprising 80% of the myofibrillar myosin protein pool.5 Accordingly, biophysical and biochemical properties of myofibers isolated from the rabbit heart largely resemble the properties of the human heart muscle. The differences in ATPase activity and the kinetics of actomyosin cross-bridge cycling between the 2 MyHC isoforms could affect the phenotypic response of the heart to the external stimuli (eg, pressure overload) or internal stimuli (eg, genetic mutations).
The cTnI-G146 transgenic rabbits largely recapitulate the phenotype of human HCM and exhibit several notable features including cardiac hypertrophy, myocyte disarray, interstitial fibrosis, and enhanced myofibrillar Ca2+ sensitivity.9 Comparison of the phenotype between the cTnI-G146 transgenic rabbits and the previously published cTnI-G145 (146 in rabbits and humans) transgenic mice13 show similarities as well as differences. Both models showed myocyte disarray, interstitial fibrosis, and enhanced Ca2+ sensitivity of myofibrillar force generation; however, only the transgenic rabbits showed cardiac hypertrophy, albeit relatively modest.9,13 Other more subtle differences included increased left ventricular contractility (+dp/dt) in the cTnI-G145 mice, which was unchanged in the cTnI-G146 transgenic rabbits, despite an increased left ventricular ejection fraction.9 The cTnI-G145 mice also showed diastolic dysfunction, which was absent in the cTnI-G146 transgenic rabbits.9 Part of the phenotypic differences between the 2 models could reflect the difficulty in precise phenotypic quantification. They also could, however, underscore model-specific phenotypes, and hence emphasize the necessity for careful selection of the animal model in which one wishes to resolve the pathogenesis of the phenotype of interest.
A notable finding in the cTnI-G146 rabbits was enhanced Ca2+ sensitivity of myofibrillar force generation, which is also in accord with the previous reports on ATPase activity and force generation.14,15 It is noteworthy that mutations in the components of thin filaments that cause DCM are associated with decreased Ca2+ sensitivity of the myofibrillar ATPase activity, force generation, or both.16,17 It is intriguing to postulate that the impact of the thin filament mutations on Ca2+ sensitivity of myofibrillar force generation, ATPase activity, or both is the main determinant of the ensuing phenotype being DCM or HCM. Ca2+ sensitivity of myofibrils is probably determined by the impact of mutation on the affinity of the inhibitory arm of cTnI for the α-tropomyosin-actin complex. Slower dissociation of the cTnI from the complex could decrease contractility and lead to DCM. In contrast, an enhanced dissociation could facilitate actomyosin cross-bridge cycling, increase contractility, and lead to HCM.
An intriguing phenotype in the cTnI-G146 transgenic rabbits is the perinatal fatality in lines that mutant cTnI-G146 comprised >40% of the total myofibrillar cTnI.9 The finding is in apparent dichotomy with human HCM, whereby the affected individuals, having 1 mutant and 1 wild-type allele, are expected to express equal levels of the mutant and wild-type proteins. Unfortunately, limited human data are available to corroborate this notion and the available data are restricted to myofibrillar incorporation of the mutant β-MyHC and not cTnI.18,19 One possible explanation for perinatal fatality could be the inefficient incorporation of the mutant sarcomeric proteins into myofibrils, as suggested by Sanbe et al. Accordingly, a >40% incorporation of the mutant cTnI in the transgenic rabbit may not reflect human HCM, whereby a lower percentage of incorporation is expected because of inefficient incorporation. A previous report showing inefficient incorporation of mutant β-MyHC V606M and G584R in the soleus muscles of 2 patients with HCM supports this notion.19 In another study, however, inefficient myofibrillar incorporation of the mutant β-MyHC-Q403 in the soleus muscle of patients with HCM was not apparent.18 In addition, in the previously described β-MyHC-Q403 transgenic rabbit model of human HCM, the mutant comprised ≈50% of the total myofibrillar myosin, implying equal myofibrillar incorporation of the wild-type and mutant proteins.20 Nonetheless, factors that regulate sarcomere assembly and the impact of mutant sarcomeric proteins are largely unknown. It is possible that the early neonatal fatality reflected a direct effect of relatively excessive incorporation of the mutant protein into myofibrils. It is also possible the excessively expressed and unincorporated cTnI-G146 imparted fortuitous interactions with other cytosolic proteins or competition for ubiquitylation and contributed to perinatal fatality.
There is considerable interest in the potential significance of changes in the expression levels of MyHC isoforms in human heart failure. The α-MyHC protein is expressed at a low level in normal adult ventricular myocardium and is composed of ≈5% to 10% of the total myofibrillar myosin pool.11,12 In the failing ventricular myocardium, expression level of the α-MyHC is downregulated to almost undetectable levels.11,12 The α-MyHC expression level returns to normal levels on recovery of the left ventricular function.21 The concordant changes could simply reflect load-dependent effects in gene expression and not necessarily a cause and an effect. Nevertheless, given the higher ATPase activity of α-MyHC, downregulation of α-MyHC protein in human heart failure could be considered an energy-conserving mechanism.8 In contrast, because of the faster rate of actomyosin cross-bridge cycling, downregulation could have a negative inotropic effect. To address the functional significance of expression of α-MyHC in the heart, James et al has used an indirect approach and force expressed the α-MyHC isoform in the rabbit heart, which predominantly expresses the β-MyHC isoform.10 The α-MyHC is made up of ≈15% to 40% of the total myofibrillar myosin in the transgenic rabbits, and it increased actin-activated ATPase activity by ≈50%; however, no discernible phenotype was noted at the baseline. After 30 days of rapid ventricular pacing, the α-MyHC-transgenic rabbits showed a lesser degree of pacing-induced cardiomyopathy than did the control rabbits, which implies a cardioprotective role for the α-MyHC. Overall, the observed pre- and postpacing differences between the transgenic and the control rabbits were modest and subject to differences in the loading conditions, such as the blood pressure, which differed significantly between the groups. It is also noteworthy that Robbins and colleagues had previously expressed the β-MyHC in the background of α-MyHC in mice.22 Near-complete replacement of α-MyHC with the β-MyHC isoform moderately reduced cardiac contractile function but caused no other discernible phenotype at rest.22 Chronic infusion of isoproterenol in the β-MyHC transgenic mice led to augmented hypertrophy with signs of failure.22 Collectively, the findings in β-MyHC transgenic mice and α-MyHC transgenic rabbits suggest that under unstressed conditions, the MyHC isoform switch does not impart a significant effect on cardiac function; however, under severe stress, α-MyHC may be cardioprotective and β-MyHC detrimental. The conclusions, nonetheless, are indirect, based on the assumption of linearity on the effect of MyHC isoforms on cardiac function and the imperfectness of the models in representing the changes in human heart failure, wherein the expression level of α-MyHC is downregulated to almost undetectable levels.11,12
Another line of evidence suggesting a functional role for the α-MyHC in the human heart is provided by identification of rare mutations in MYH6, which encode the α-MyHC isoform.23 Unfortunately, genetic data on MYH6 mutations have been restricted to the identification of rare mutations in small families or probands with cardiomyopathies, and alone are not sufficiently robust to establish the causality. Thus, additional genetic and functional studies are necessary to delineate the functional significance of expression of α-MyHC at low levels in the human heart and its disappearance in the failing myocardium. Experiments to suppress the expression level of the α-MyHC in the background of β-MyHC through either gene targeting or siRNA experiments would be necessary and await refinement of existing gene targeting techniques.
In conclusion, the transgenic rabbit models of altered sarcomeric proteins9,10 provide considerable insight into the functional and phenotypic consequences of perturbations in sarcomeric proteins in the heart. At the same time, it is evident that there are considerable difficulties in developing and characterizing appropriate models for human heart failure and in the application of the results to the human phenotype. The findings beg for additional genetic and molecular studies to delineate molecular mechanisms that regulate sarcomere assembly and function in human heart failure with the ultimate goal of developing new interventions to prevent and treat human cardiomyopathies.
Acknowledgments
Disclosure: Dr Marian is supported by grants from the National Heart, Lung, and Blood Institute, Specialized Centers of Research (P50-HL42267), R01-HL68884, and the TexGen Fund from the Greater Houston Community Foundation.
Footnotes
The opinions expressed in this article are not necessarily those of the editors or of the American Heart Association.
References
- 1.Solaro RJ, Rarick HM. Troponin and tropomyosin: proteins that switch on and tune in the activity of cardiac myofilaments. Circ Res. 1998;83:471–480. doi: 10.1161/01.res.83.5.471. [DOI] [PubMed] [Google Scholar]
- 2.Moss RL, Razumova M, Fitzsimons DP. Myosin crossbridge activation of cardiac thin filaments: implications for myocardial function in health and disease. Circ Res. 2004;94:1290–1300. doi: 10.1161/01.RES.0000127125.61647.4F. [DOI] [PubMed] [Google Scholar]
- 3.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 4.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 5.Swynghedauw B. Developmental and functional adaptation of contractile proteins in cardiac and skeletal muscles. Physiol Rev. 1986;66:710–771. doi: 10.1152/physrev.1986.66.3.710. [DOI] [PubMed] [Google Scholar]
- 6.Lompre AM, Schwartz K, d'Albis A, Lacombe G, Van Thiem N, Swynghedauw B. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- 7.Palmiter KA, Tyska MJ, Dupuis DE, Alpert NR, Warshaw DM. Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms. J Physiol. 1999;519:669–678. doi: 10.1111/j.1469-7793.1999.0669n.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiura S, Kobayakawa N, Fujita H, Yamashita H, Momomura Si, Chaen S, Omata M, Sugi H. Comparison of unitary displacements and forces between 2 cardiac myosin isoforms by the optical trap technique: molecular basis for cardiac adaptation. Circ Res. 1998;82:1029–1034. doi: 10.1161/01.res.82.10.1029. [DOI] [PubMed] [Google Scholar]
- 9.Sanbe A, James J, Tuzcu V, Nas S, Martin L, Gulick J, Osinska H, Sakthivel S, Klevitsky R, Ginsburg KS, Bers DM, Zinman B, Lakatta EG, Robbins J. Transgenic rabbit model for human troponin I-based hypertrophic cardiomyopathy. Circulation. 2005;111:2330–2338. doi: 10.1161/01.CIR.0000164234.24957.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.James J, Martin L, Krenz M, Quatman C, Jones F, Klevitsky R, Gulick J, Robbins J. Forced expression of alpha-myosin heavy chain in the rabbit ventricle results in cardioprotection under cardiomyopathic conditions. Circulation. 2005;111:2339–2346. doi: 10.1161/01.CIR.0000164233.09448.B1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakao K, Minobe W, Roden R, Bristow MR, Leinwand LA. Myosin heavy chain gene expression in human heart failure. J Clin Invest. 1997;100:2362–2370. doi: 10.1172/JCI119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86:386–390. doi: 10.1161/01.res.86.4.386. [DOI] [PubMed] [Google Scholar]
- 13.James J, Zhang Y, Osinska H, Sanbe A, Klevitsky R, Hewett TE, Robbins J. Transgenic modeling of a cardiac troponin I mutation linked to familial hypertrophic cardiomyopathy. Circ Res. 2000;87:805–811. doi: 10.1161/01.res.87.9.805. [DOI] [PubMed] [Google Scholar]
- 14.Lang R, Gomes AV, Zhao J, Housmans PR, Miller T, Potter JD. Functional analysis of a troponin I (R145G) mutation associated with familial hypertrophic cardiomyopathy. J Biol Chem. 2002;277:11670–11678. doi: 10.1074/jbc.M108912200. [DOI] [PubMed] [Google Scholar]
- 15.Elliott K, Watkins H, Redwood CS. Altered regulatory properties of human cardiac troponin I mutants that cause hypertrophic cardiomyopathy. J Biol Chem. 2000;275:22069–22074. doi: 10.1074/jbc.M002502200. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto S, Yanaga F, Minakami R, Ohtsuki I. Ca2+-sensitizing effects of the mutations at Ile-79 and Arg-92 of troponin T in hypertrophic cardiomyopathy. Am J Physiol. 1998;275:C200–C207. doi: 10.1152/ajpcell.1998.275.1.C200. [DOI] [PubMed] [Google Scholar]
- 17.Morimoto S, Lu QW, Harada K, Takahashi-Yanaga F, Minakami R, Ohta M, Sasaguri T, Ohtsuki I. Ca(2+)-desensitizing effect of a deletion mutation Delta K210 in cardiac troponin T that causes familial dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2002;99:913–918. doi: 10.1073/pnas.022628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuda G, Fananapazir L, Zhu WS, Sellers JR, Epstein ND. Skeletal muscle expression and abnormal function of beta-myosin in hypertrophic cardiomyopathy. J Clin Invest. 1993;91:2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nier V, Schultz I, Brenner B, Forssmann WG, Raida M. Variability in the ratio of mutant to wildtype myosin heavy chain present in the soleus muscle of patients with familial hypertrophic cardiomyopathy. A new approach for the quantification of mutant to wildtype protein. FEBS Lett. 1999;461:246–252. doi: 10.1016/s0014-5793(99)01433-7. [DOI] [PubMed] [Google Scholar]
- 20.Marian AJ, Wu Y, Lim DS, McCluggage M, Youker K, Yu QT, Brugada R, DeMayo F, Quinones M, Roberts R. A transgenic rabbit model for human hypertrophic cardiomyopathy. J Clin Invest. 1999;104:1683–1692. doi: 10.1172/JCI7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, Wolfel EE, Lindenfeld J, Tsvetkova T, Robertson AD, Quaife RA, Bristow MR. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med. 2002;346:1357–1365. doi: 10.1056/NEJMoa012630. [DOI] [PubMed] [Google Scholar]
- 22.Krenz M, Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J Am Coll Cardiol. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 23.Niimura H, Patton KK, McKenna WJ, Soults J, Maron BJ, Seidman JG, Seidman CE. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]


