Abstract
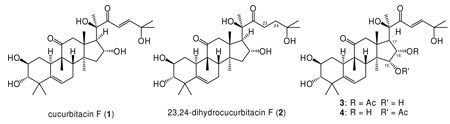
Bioassay-guided fractionation of Physocarpus capitatus yielded two new cucurbitacins (3 and 4) along with the known cucurbitacin F (1) and dihydrocucurbitacin F (2). Preliminary mechanism of action studies indicate that the cucurbitacins cause actin aggregates and inhibit cell division.
Natural products have provided the vast majority of small molecule probes for studying the cell cycle and the cytoskeleton1,2 as illustrated by the tubulin-interactive agents Taxol®, vincristine, and halichondrin B.3 Natural product probes of actin include the cytochalasins and latrunculin B, which inhibit actin polymerization, and jasplakinolide, which induces actin polymerization. These natural products have greatly facilitated our understanding of the role of actin in several fundamental processes. In a classic example from the 1970s, the actin binder cytochalasin was used to show that actin plays a key role in cytokinesis, the final step of cell division. Researchers observed that cytochalasin inhibited multiple processes such as cell migration, cell ruffling and division and they reasoned that a single protein, which turned out to be actin, was involved in these processes. Treatment of cells with cytochalasin resulted in inhibition of cellular division (cytokinesis), but not nuclear division (mitosis), providing the first evidence that the two processes can be decoupled from each other. Researchers then went on to show that the target of cytochalasin was actin and that it was a key protein in cytokinesis.4 Given the demonstrated value of natural products in studying the cytoskeleton, the discovery of additional small molecules probes should prove useful.
In an effort to find inhibitors of cytokinesis, approximately 51,000 small molecules - belonging to libraries of “drug-like” compounds, combinatorial libraries, and crude natural product extract libraries provided by the NCI - were screened in a high-throughput imaging assay.5 In this screen, Drosophila Kc167 cells were exposed to small molecules, allowed to undergo a complete cell cycle, fixed, and stained with tetramethylrhodamine-NHS ester (to visualize the cytoplasm) and Hoechst dye (to visualize the DNA). Stained cells were imaged using automated fluorescence microscopy and cells with multiple nuclei (that is, cells capable of undergoing mitosis but incapable of cytokinesis) were scored by a combination of automated image analysis and visual inspection. This cell-based approach allowed for the possibility of discovering unknown targets, and molecules that hit should also have an improved chance of providing drug leads, since they have already demonstrated cell penetration and effectiveness in vivo.6 Interestingly, although natural products represented only 5% of the total compounds screened, they accounted for 25% of the hits.
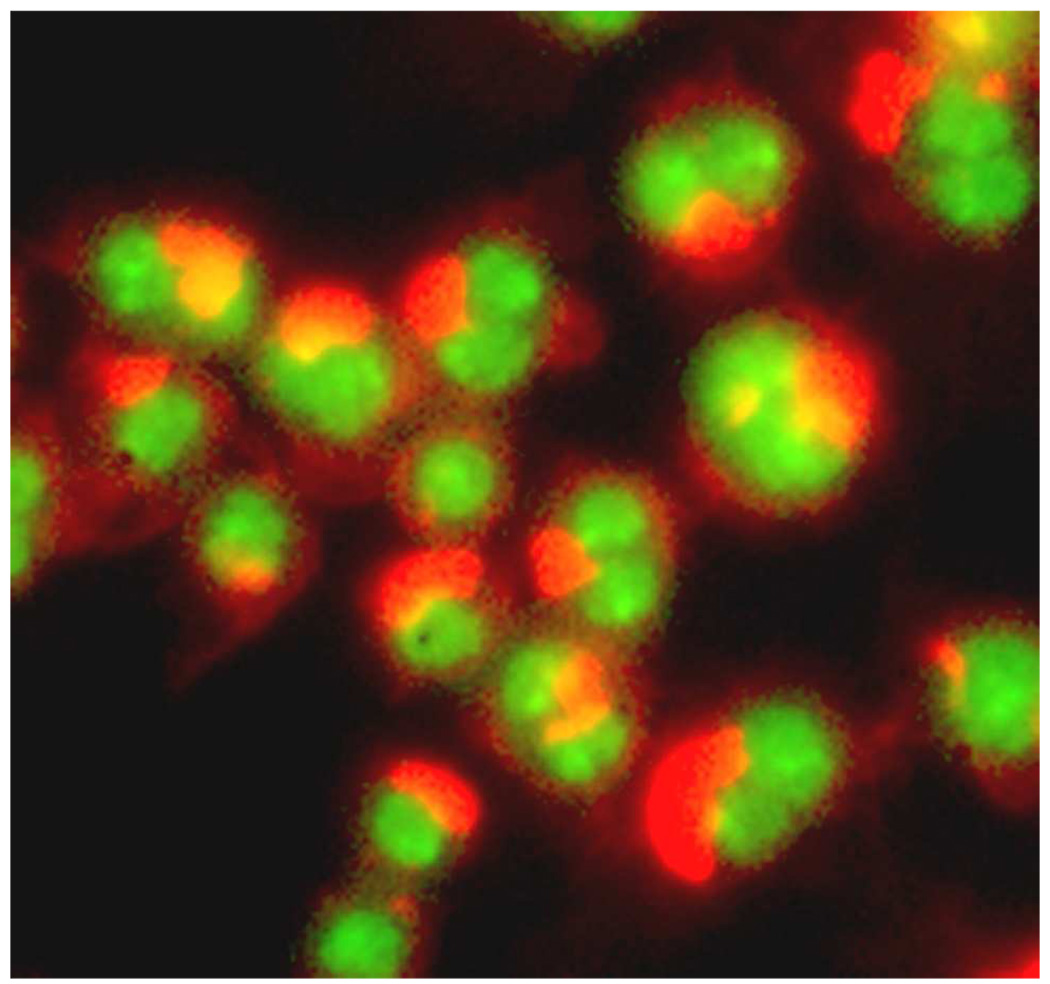
A crude extract of Physocarpus capitatus (Pursh) Kuntze (Rosaceae) (Pacific Ninebark), a shrub native to the western United States, aggregated actin in cells and induced the formation of binucleated cells, a sign of strong cytokinesis-inhibitory activity (Figure 1). The active P. capitatus extract was fractionated by several rounds of reverse-phase and silica column chromatography, followed by reverse-phase HPLC separation. Nineteen active fractions were examined by NMR and found to contain assorted cucurbitacins. Comparison of chemical shifts to literature values revealed cucurbitacin F (1)7,8 as the most potent cucurbitacin from P. capitatus, and the known cucurbitacin 23,24-dihydrocucurbitacin F (2)7,9 was also observed.
Figure 1.
Actin aggregates and cytokinesis failure resulting from cucurbitacin treatment. Drosophila Kc167 cells were treated with an extract of P. capitatus, allowed to undergo a full cell cycle, fixed, and stained with TRITC-phalloidin to visualize actin (red) and Hoechst dye to visualize DNA (green).
Closer inspection of the double quantum-filtered COSY spectra of the less active fractions revealed two new cucurbitacins (3 and 4). A spin system connecting methine carbons C-15 (δ 3.98 in 3, 4.66 in 4), C-16 (δ 5.33 in 3, 4.47 in 4), and C-17 (δ 2.65 in 3, 2.49 in 4) revealed a site of oxidation at carbon C-15. HMBC correlations from protons H-16 of 3 (δ 5.33) and H-15 of 4 (δ 4.66) to acetate carbonyls at δ 170.8 and δ 171.2, respectively, suggested that the two cucurbitacins differ only in the site of acetylation. A NOESY correlation between protons H-15 (δ 3.98 in 3; 4.66 in 4) and H-16 (δ 5.33 in 3, δ 4.47 in 4) indicates that both hydroxyls are on the same face. Remaining NMR chemical shifts indicated that the structures are in all other ways identical to cucurbitacin F (Table 1).
Table 1.
NMR chemical shifts of cucurbitacins (1–4) in CD3OD (1H 600 MHz; 13C 100 MHz)
| 1 | 2 | 3 | 4 | |||||
|---|---|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | δC | δH | |
| 1 | 34.4 | 1.02, 1.80 | 34.4* | 1.02*, 1.80* | 33.4 | 1.02, 1.78 | 33.4* | 1.02*, 1.78* |
| 2 | 71.5 | 3.54 | 71.5* | 3.54* | 70.3 | 3.55 | 70.3* | 3.55* |
| 3 | 81.9 | 2.85 | 81.9* | 2.85* | 80.7 | 2.85 | 80.7* | 2.85* |
| 4 | 43.0 | - | 43.0* | - | 42.0 | - | 42.0* | - |
| 5 | 142.5 | - | 142.5* | - | 140.9 | - | 140.9* | - |
| 6 | 119.7 | 5.75 | 119.7* | 5.74 | 118.9 | 5.72 | 118.9* | 5.72* |
| 7 | 24.6 | 1.97, 2.40 | 24.6* | 1.97*, 2.40* | 22.1 | 2.32, 2.71 | 22.1* | 2.32*, 2.71* |
| 8 | 44.2 | 1.93 | 44.2* | 1.93* | 43.1 | 2.11 | 43.2* | 2.11* |
| 9 | 49.6 | - | 49.6* | - | 48.7 | - | 48.7* | - |
| 10 | 34.9 | 2.48 | 34.9* | 2.48* | 33.8 | 2.49 | 33.8* | 2.49* |
| 11 | 215.8 | - | 215.8* | - | 214.0 | - | 214.0* | - |
| 12 | 49.7 | 2.55, 3.31 | 49.7* | 2.55*, 3.31* | 48.4 | 2.57, 3.42 | 48.4* | 2.58*, 3.42* |
| 13 | 48.8 | - | 48.9 | - | 47.5 | - | 47.5* | - |
| 14 | 51.8 | - | 51.7 | - | 49.5 | - | 48.4 | - |
| 15 | 46.4 | 1.39, 1.84 | 46.4 | 1.38, 1.82 | 75.0 | 3.98 | 79.7 | 4.66 |
| 16 | 71.8 | 4.46 | 71.2 | 4.41 | 72.8 | 5.33 | 68.5 | 4.47 |
| 17 | 59.3 | 2.60 | 59.0 | 2.57 | 53.2 | 2.65 | 56.9 | 2.49 |
| 18 | 20.4 | 0.90 | 20.1 | 0.90 | 19.4 | 0.97 | 19.4* | 0.96 |
| 19 | 20.3 | 1.07 | 20.3* | 1.07* | 18.9 | 1.09 | 18.9* | 1.09* |
| 20 | 79.6 | - | 80.5 | - | 78.5 | - | 78.5* | - |
| 21 | 25.2 | 1.38 | 25.3 | 1.38 | 23.3 | 1.38 | 23.3 | 1.38 |
| 22 | 204.7 | - | 216.9 | - | 202.8 | - | 203.3 | - |
| 23 | 120.9 | 6.82 | 33.0 | 2.72, 2.85 | 119.4 | 6.80 | 119.2 | 6.80 |
| 24 | 155.0 | 6.97 | 37.9 | 1.71, 1.72 | 155.2 | 7.04 | 154.4 | 6.98 |
| 25 | 71.3 | - | 70.7 | - | 70.5 | - | 70.5 | - |
| 26 | 29.1 | 1.32 | 29.1 | 1.19 | 27.9 | 1.32 | 27.9* | 1.32* |
| 27 | 29.1 | 1.33 | 29.1 | 1.19 | 28.1 | 1.34 | 28.1* | 1.34* |
| 28 | 22.1 | 0.96 | 22.1* | 0.96* | 20.9 | 0.97 | 20.9* | 0.97* |
| 29 | 25.2 | 1.17 | 25.2* | 1.17* | 24.0 | 1.16 | 24.0* | 1.16* |
| 30 | 28.9 | 1.30 | 28.9 | 1.30 | 11.6 | 1.20 | 12.8 | 1.31 |
| 31 | 170.8 | - | 171.2 | - | ||||
| 32 | 19.8 | 1.90 | 19.8 | 2.10 | ||||
Overlapped signals.
Effects of cucurbitacins on actin have been reported in the literature,10–13 and a recent study suggests that they may interact with actin directly.14 Studies are ongoing to investigate the mechanism of action for this interesting family of compounds.
Experimental Section
General Experimental Procedures
1H NMR and 13C NMR spectra were obtained on a Varian INOVA 600 MHz spectrometer with standard pulse sequences. CD3OD was used as solvent. Offline processing was conducted using Mestre-C NMR Software (Mestrelab Research, A Coruña, Spain; www.mestrec.com). 1H and 13C chemical shifts were referenced with the methanol solvent peaks at δ 3.31 and δ 49.05, respectively. LCMS data were obtained using a Micromass Platform LC-Z spectrometer, equipped with a Waters 2690 LC system and Waters 2690 photodiode array detector, and processed using MassLynx software (Waters Corporation, Milford, MA; www.waters.com). HPLC was carried out on an Agilent 1100 system equipped with diode array detector and operated with Chemstation software (Agilent Technologies). All HPLC separations were done using a Discovery HS-C18 column (Supelco, 250 × 10.0 mm, 5mm particle size) with a 4 mL/min flow rate. Column chromatography was carried out on silica gel 60 (FisherChemicals, 230–400 mesh) and octadecyl-functionalized silica gel (Aldrich).
Plant Material
Approximately 18 g of an organic extract of Physocarpus capitatus (NPID N102479) was provided by Gordon Cragg in the Natural Products Branch of the National Cancer Institute. The plant material was collected in August 1997 in the Umpqua National Forest, Jackson County, Oregon (Longitude: 123 005.88W; Latitude: 42 40.26N). The plant material was identified by W. Hess, and a voucher specimen is on deposit at the Smithsonian Institute, U.S. National Herbarium in Washington, DC (specimen no. 0GDK0989). Extraction was performed according to the standard NCI protocol. (In brief, plant material was extracted with 1:1 dichloromethane:methanol followed by a wash with 100% methanol. The extract and methanol wash were combined prior to evaporation.)
Isolation
The crude NCI extract (18 g) was dissolved in aqueous MeOH (90%) and partitioned against hexanes (700 mL×3), then adjusted to 60% MeOH and extracted with CH2Cl2 (1000 mL×3). The CH2Cl2 layer (4.5 g) was chromatographed on C18 silica gel and five fractions eluted with a MeOH /H2O step gradient. Fraction 2 (0.59 g) from this step was again chromatographed on C18 silica gel, this time eluting with 50% aqueous MeOH to give four fractions. Fractions 1 (412 mg) and 2 (231 mg) were combined and chromatographed on silica gel using a CH2Cl2/MeOH step gradient to give seven fractions. Fractions 4 (70 mg) and 5 (160 mg) were combined and subjected to RP-HPLC with an acetonitrile/ H2O gradient, yielding nine fractions. Finally, fraction 4 (53 mg) was separated into eleven more fractions by RP-HPLC with a MeOH/H2O gradient. Fraction 4 (16.9 mg) from this last column contained a mixture of cucurbitacins 3 and 4. Fraction 9 (2.4 mg) contained a mixture of 23,24-dihydrocucurbitacin F (2), cucurbitacin F (1), and a biphenyl, and fraction 10 (3.8 mg) contained pure cucurbitacin F (1).
Cucurbitacin F (1)
Physical constants were identical to those reported in the literature;7 13C and 1H NMR spectroscopic data: see Table 1.
23,24-Dihydrocucurbitacin F (2)
(characterized as a mixture with 1) Physical constants were identical to those reported in the literature;7 13C and 1H NMR spectroscopic data: see Table 1.
Compound 3
(characterized as a 6:1 mixture with 4) white solid; 13C NMR (CD3OD, δ): 214.0 (C-11), 202.8 (C-22), 170.8 (C-31), 155.2 (C-24), 140.9 (C-5), 119.4 (C-23), 118.9 (C-6), 80.7 (C-3), 78.5 (C-20), 75.0 (C-15), 72.8 (C-16), 70.5 (C-25), 70.3 (C-2), 53.2 (C-17), 49.5 (C-14), 48.7 (C-9), 48.4 (C-12), 47.5 (C-13), 43.1 (C-8), 42.0 (C-4), 33.8 (C-10), 33.4 (C-1), 28.1 (C-27), 27.9 (C-26), 24.0 (C-29), 23.3 (C-21), 22.1 (C-7), 20.9 (C-28), 19.8 (C-32), 19.4 (C-18), 18.9 (C-19), 11.6 (C-30); 1H NMR (CD3OD, δ): 7.04 (1H, d, J = 15.4 Hz, H-24), 6.80 (1H, d, J = 15.4 Hz, H-23), 5.72 (1H, dt, J = 6.2 Hz, J = 2.1 Hz, H-6), 5.33 (t1H,, J = 7.9 Hz, H-16), 3.98 (1H, d, J = 8.1 Hz, H-15), 3.55 (1H, ddd, J = 11.6 Hz, J = 9.4 Hz, J = 4.2 Hz, H-2), 3.42 (1H, d, J = 14.2 Hz, H-12a), 2.85 (1H, d, J = 9.4 Hz, H-3), 2.71 (1H, ddd, J = 19.1 Hz, J = 6.2 Hz, J = 1.7 Hz, H-7a), 2.65 (1H, d, J = 7.6 Hz, H-17), 2.57 (1H, d, J = 14.2 Hz, H-12b), 2.49 (1H, m, H-10), 2.32 (1H, m, H-7b), 2.11 (1H, m, H-8), 1.90 (3H, s, H-32), 1.78 (1H, dt, J = 12.3 Hz, J = 4.2 Hz, H-1a), 1.38 (3H, s, H-21), 1.34 (3H, s, H-27), 1.32 (3H, s, H-26), 1.20 (3H, s, H-30), 1.16 (3H, s, H-29), 1.09 (3H, s, H-19), 1.02 (1H, dd, J = 12.3 Hz, J = 11.6 Hz, H-1b), 0.97 (3H, s, H-18), 0.97 (3H, s, H-28). HRESIMS (m/z): [M + Na]+ calcd for C32H48O9 + Na, 599.3196; found, 599.3202.
Compound 4
(characterized as a 1:6 mixture with 3) white solid; 13C NMR (CD3OD, δ): 214.0 (C-11), 203.3 (C-22), 171.2 (C-31), 154.4 (C-24), 140.9 (C-5), 119.2 (C-23), 118.9 (C-6), 80.7 (C-3), 79.7 (C-15), 78.5 (C-20), 70.5 (C-25), 70.3 (C-2), 68.5 (C-16), 56.9 (C-17), 48.7 (C-9), 48.4 (C-14), 48.4 (C-12), 47.5 (C-13), 43.2 (C-8), 42.0 (C-4), 33.8 (C-10), 33.4 (C-1), 28.1 (C-27), 27.9 (C-26), 24.0 (C-29), 23.3 (C-21), 22.1 (C-7), 20.9 (C-28), 19.8 (C-32), 19.4 (C-18), 18.9 (C-19), 12.8 (C-30); 1H NMR (CD3OD, δ): 6.98 (1H, d, J = 15.4 Hz, H-24), 6.80 (1H, d, J = 15.4 Hz, H-23), 5.72 (1H, dt, J= 6.2 Hz, J = 2.1 Hz, H-6), 4.66 (1H, d, J = 7.9 Hz, H-15), 4.47 (1H, t, J = 7.7 Hz, H-16), 3.55 (1H, ddd, J = 11.6 Hz, J = 9.4 Hz, J = 4.2 Hz, H-2), 3.42 (1H, d, J = 14.2 Hz, H-12a), 2.85 (1H, d, J = 9.4 Hz, H-3), 2.71 (1H, ddd, J = 19.1 Hz, J = 6.2 Hz, J = 1.7 Hz, H-7a), 2.58 (1H, d, J = 14.6 Hz, H-12b), 2.49 (1H, d, J = 7.6 Hz, H-17), 2.49 (1H, m, H-10), 2.32 (1H, m, H-7b), 2.11 (1H, m, H-8), 2.10 (3H, s, H-32), 1.78 (1H, dt, J = 12.3 Hz, J = 4.2 Hz, H-1a), 1.38 (3H, s, H-21), 1.34 (3H, s, H-27), 1.32 (3H, s, H-26), 1.31 (3H, s, H-30), 1.16 (3H, s, H-29), 1.09 (3H, s, H-19), 1.02 (1H, dd, J = 12.3 Hz, J = 11.6 Hz, H-1b), 0.97 (3H, s, H-28), 0.96 (3H, s, H-18). HRESIMS (m/z): [M + Na]+ calcd for C32H48O9 + Na, 599.3196; found, 599.3202.
Supplementary Material
Acknowledgment
This research was supported NIH grant CA24487 (J.C.). K.N.M. was supported by an NSF Graduate Research Fellowship. HRESIMS was performed by Steve Mullen of the Mass Spectrometry Facility at the University of Illinois at Urbana Champaign. U.S.E. was a Merck-sponsored fellow of the Helen Hay Whitney Foundation. U.S.E., C.M.F. and T.J.M. were supported by NIH grant R01 GM023928–25 awarded to T.J.M. The organic extract of P. capitatus (NPID N102479) was provided by Gordon Cragg in the Natural Products Branch of the National Cancer Institute.
Footnotes
Supporting Information Available: NMR (1D and 2D) spectra of compounds 1–5. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Hung DT, Jamison TF, Schreiber SL. Chem. Biol. 1996;3:623–639. doi: 10.1016/s1074-5521(96)90129-5. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM, Holbeck S, Sausville EA. Curr. Cancer Drug Targets. 2002;2:279–308. doi: 10.2174/1568009023333791. [DOI] [PubMed] [Google Scholar]
- 3.Cragg GM, Boyd MR, Christini MA, Kneller R, Mays TD, Mazan KD, Newman DJ, Sausville EA. In: Phytochemical diversity: A source of new industrial products. Wrigley SK, Hays MA, Chrystal EJ, Thomas R, editors. Cambridge: Royal Society of Chemistry; 1997. pp. 1–29. [Google Scholar]
- 4.For a review of the history of small molecules that target the cytoskeleton, see: Peterson JR, JR, Mitchison TJ. Chem. Biol. 2002;9:1275–1285. doi: 10.1016/s1074-5521(02)00284-3.
- 5.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertzberg RP. Curr. Opin. Biotechnol. 1993;4:80–84. doi: 10.1016/0958-1669(93)90037-w. [DOI] [PubMed] [Google Scholar]
- 7.Fang X, Phoebe CH, Pezzuto JM, Fong HHS, Farnsworth NR. J. Nat. Prod. 1984;47:988–993. doi: 10.1021/np50036a013. [DOI] [PubMed] [Google Scholar]
- 8.Rehm S, Enslin PA, J MAD, H WJ. J. Sci. Food Agric. 1957;8:679–686. [Google Scholar]
- 9.Bittner M, Poyser KA, Poyser JP, Silva M. Phytochemistry. 1973;12:1427–1431. [Google Scholar]
- 10.Musza LL, Speight P, McElhiney S, Barrow CJ, Gillum AM, Cooper R, Killar LM. J. Nat. Prod. 1994;57:1498–1502. doi: 10.1021/np50113a004. [DOI] [PubMed] [Google Scholar]
- 11.Duncan KL, Duncan MD, Alley MC, Sausville EA. Biochem. Pharmacol. 1996;52:1553–1560. doi: 10.1016/s0006-2952(96)00557-6. [DOI] [PubMed] [Google Scholar]
- 12.Duncan MD, Duncan KL. J. Surg. Res. 1997;69:55–60. doi: 10.1006/jsre.1997.5028. [DOI] [PubMed] [Google Scholar]
- 13.Graness A, Poli V, Goppelt-Struebe M. Biochem. Pharmacol. 2006;72:32–41. doi: 10.1016/j.bcp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Momma K, Masuzawa Y, Nakai N, Chujo M, Murakami A, Kioka N, Kiyama Y, Akita T, Nagao M. Cytotechnology. 2008;56:33–39. doi: 10.1007/s10616-007-9100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.