Abstract
The radically distinct morphologies of arthropod and tetrapod legs argue that these appendages do not share a common evolutionary origin. Yet, despite dramatic differences in morphology, it has been known for some time that transcription factors encoded by the Distalless (Dll)/Dlx gene family play a critical role in the development of both structures. Here we show that a second transcription factor family encoded by the Sp8 gene family, previously implicated in vertebrate limb development, also plays an early and fundamental role in arthropod leg development. By simultaneously removing the function of two Sp8 orthologs, buttonhead (btd) and Sp1, during Drosophila embryogenesis, we find that adult leg development is completely abolished. Remarkably, in the absence of these factors, transformations from ventral to dorsal appendage identities are observed, suggesting that adult dorsal fates become derepressed when ventral fates are eliminated. Further, we show that Sp1 plays a much more important role in ventral appendage specification than btd and that Sp1 lies genetically upstream of Dll. In addition to these selector-like gene functions, Sp1 and btd are also required during larval stages for the growth of the leg. Vertebrate Sp8 can rescue many of the functions of the Drosophila genes, arguing that these activities have been conserved, despite more than 500 million years of independent evolution. These observations suggest that an ancient Sp8/Dlx gene cassette was used in an early metazoan for primitive limb-like outgrowths and that this cassette was co-opted multiple times for appendage formation in multiple animal phyla.
Author Summary
The development of vertebrate and invertebrate appendages differs in many respects. Yet, despite these differences, genes related to the Distalless (Dll) gene of Drosophila (vertebrate Dlx genes) are important for the development of appendages in multiple animal phyla. Such findings raise the question of whether disparate animal appendages have a common evolutionary origin. In vertebrates, a second gene family, related to Drosophila Sp1, also plays a fundamental role in appendage development. Although there was some evidence to suggest that Sp1 family members may play a role in Drosophila appendage development, definitive data were lacking. Using a new deficiency that removes both Drosophila sister genes, Sp1 and buttonhead (btd), we unambiguously assess their role in Drosophila development. We find that Sp1, but not btd, is critical for specifying leg (ventral) development, and that neither gene is required for wing (dorsal) development. We also show that Sp1 lies genetically upstream of Dll. The fact that both Sp1 and Dlx gene families are used for appendage development in vertebrates and invertebrates provides striking evidence that the Sp1–Dlx relationship represents an ancient gene network that was used in a common ancestor for appendage-like outgrowths.
Introduction
During Drosophila embryogenesis, the cells that will give rise to both the dorsal (wing and haltere) and ventral (leg) appendages are allocated from a ventral region of each thoracic hemisegment [1], [2]. About a quarter of the way through embryogenesis (stage 11), these cells can be recognized by the expression of the homeobox gene Distalless (Dll) [3]. Initially, ∼30 ventral cells activate Dll in response to receiving positive input from Wingless (Wg) and negative inputs from the Decapentaplegic (Dpp) and Epidermal Growth Factor (EGF) pathways [1], [4],[5]. At this early stage, these Dll-expressing cells can contribute to any part of the leg or to any part of the wing and haltere; the distinction between ventral and dorsal appendage identities has not yet occurred [2], [6]. A few hours later (by stage 14), the cells that will give rise to the wing and haltere no longer express Dll, and the Dll-expressing cells will only contribute to the portion of the leg that is distal to the coxa, the telopodite [2], [7]. Thus, within a few hours, the Dll-expressing cells in the thorax have dramatically changed their presumptive fates. This refinement of developmental potential mirrors a change in the cis-regulatory elements used to control Dll expression. At stage 11, when the Dll-expressing cells are multi-potent, Dll is activated by the Dll-304 enhancer [2], [8]. At stage 14, when the fate of Dll-expressing cells is limited to the telopodite, Dll-304 is no longer active and a different regulatory element, Dll-LT, is used [2].
Loss-of-function experiments demonstrate that in the absence of Dll the leg telopodite fails to develop [1], [9]. Conversely, mis-expression of Dll in dorsal appendages can lead to the ectopic development of distal leg segments, most typically, tarsal segments [10]. The necessity and in some contexts sufficiency of Dll to generate leg fates has been interpreted to suggest that Dll is a selector gene for the ventral appendage. However, although Dll is transiently expressed in cells that will give rise to the coxa and dorsal appendages, it is not required to generate these fates, nor is it required to generate any other ventral structure besides the telopodite [1], [9], [10]. Most strikingly, when transplanted to wild type hosts, ventral tissue dissected from Dll null embryos retains the capacity to generate proximal leg fates, demonstrating that ventral appendage specification and, in particular, proximal leg fates, form independently of Dll [1]. The limited requirement of Dll in leg development raises the question of what gene(s) may be required to initially specify the ventral appendage primordia and proximal leg fates.
A pair of genes that could fulfill a more general role in ventral appendage specification is buttonhead (btd) and Sp1, which encode highly related transcription factors with three C2H2 Zn-fingers and a conserved ‘Btd box’ [11], [12]. Both genes share a similar expression pattern throughout Drosophila development and appear to have partially redundant functions during mechanosensory organ development [13], [14]. btd and Sp1 are also both expressed in the thoracic appendage primordia (see Figure S1) [14]. Although previous work in Drosophila suggested that one or both of these genes may play a role in ventral appendage specification, these conclusions have significant limitations [14]. Embryos homozygous for a large deficiency that removes both btd and Sp1 do not express Dll in the ventral primordia [14]. In fact, these embryos appear to have no ventral appendage primordia because expression of escargot (esg), a general marker for imaginal disc fates, is absent in the ventral thoracic segments of these embryos [14]. However, the large size of the deficiency used in these experiments (Df(1)C52), which removes >50 genes in addition to btd and Sp1, leaves open the question of whether these phenotypes are due to the loss of btd, Sp1, and/or one of the other deleted genes. Second, mis-expression experiments suggest that Btd has the ability to induce ectopic leg development and the expression of leg marker genes in dorsal tissues, such as the wing. Sp1's activity was not tested in this ectopic expression test [14]. Third, because Df(1)C52 is too large to be used for clonal analysis, loss-of-function btd and Sp1 phenotypes in the adult were analyzed by RNA interference (RNAi) [14]. Counter to the idea that these genes are essential for leg specification, RNAi knockdown of btd and Sp1 did not prevent leg development, but instead only resulted in a reduction of leg growth. These phenotypes are highly reminiscent to those observed when btd and Sp1 orthologs were knocked-down in the beetle Tribolium castaneum (Sp8) or milkweed bug Oncopeltus fasciatus (Sp8 and Sp9) [15], [16]. In sum, because previous experiments depended on a large deficiency, ectopic expression, and RNAi knockdown approaches, they do not resolve whether btd and/or Sp1 are required for the initial establishment of the ventral appendage primordia and/or for ventral appendage growth.
In vertebrates, there are two genes that are closely related to btd and Sp1, called Sp8 and Sp9. Both Sp8 and Sp9 are initially expressed in the ectoderm of the developing limb bud but are later restricted to the Apical Ectodermal Ridge (AER) [17], [18]. Interestingly, Sp8 mutant mice have truncated limbs, due to the loss of expression of several genes essential for limb formation, including those encoding FGFs, Shh and BMPs [17]–[19]. Thus, although double mutant Sp8 Sp9 mice have not been studied, the Sp8 single mutant suggests a critical role for these genes in vertebrate limb development.
Here, we use a new deletion of Sp1 and btd that allows us to unambiguously analyze the function of these genes in Drosophila. Most strikingly, the complete absence of Sp1 and btd, but not btd alone, results in the loss of all leg structures and can lead to a dramatic transformation of leg into wing and notum fates, representing a complete ventral to dorsal fate change. These phenotypes are rescued by resupplying Sp1, but not btd, suggesting that Sp1 plays an essential and early role in leg specification. Consistent with these severe phenotypes, early loss of both genes leads to the loss of expression of the telopodite genes Dll and dachshund (dac). However, in contrast to previous findings [14], appendage primordia, as assessed by esg expression, still form in the absence of btd and Sp1. We also find that, like Btd, ectopic expression of Sp1 and vertebrate Sp8 can induce ectopic leg development in dorsal appendages, suggesting that these functions have been conserved. If, however, btd and Sp1 are removed after Dll expression is initiated, ventral appendage fates still form, but leg growth is severely compromised. Together, these results suggest that Sp1 functions upstream of Dll and that it plays an early and essential selector-like function in the specification of ventral appendage and body wall fates. Later in development, both btd and Sp1 work in parallel with Dll to control the growth and morphology of the legs.
Results
Generation of a btd Sp1 deficiency
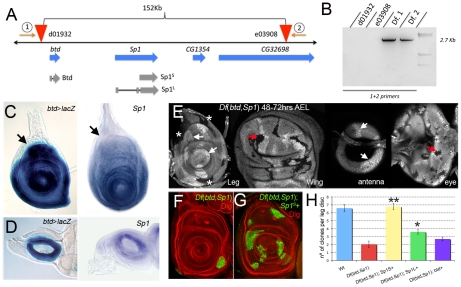
Although btd null alleles are available, there were no mutations that eliminated both btd and Sp1 that could be used for clonal analysis, thus precluding a definitive assessment of the role these genes play in adult development. The distance between btd and Sp1 is approximately 32 kilobases (kb), with no known intervening genes (Figure 1A). To generate a deficiency that removes both btd and Sp1 we used the FRT-directed recombination technique [20]. Using this method, we were able to generate a deficiency that deletes btd, Sp1, and two adjacent genes with unknown function (CG1354 and CG32698) (Figure 1A; see Materials and Methods). The generation of this deficiency, hereafter referred to as Df(btd,Sp1), was confirmed by the polymerase chain reaction (PCR) using primers that flank the FRT-containing P elements (Figure 1B).
Figure 1. Generating a deficiency for btd and Sp1.
(A) Genomic organization of the btd and Sp1 genomic region. While btd only encodes for one isoform, Sp1 encodes for two, a small one (Sp1-PB (Flybase) or Sp1S,) and a larger one (Sp1-PD (Flybase) or Sp1L). Two FRT-containing P elements, d01932 and e03908, are situated 5′ and 3′ of btd and Sp1, respectively. The PCR primers used to molecularly confirm the deficiency are indicated (1 and 2). (B) PCR confirmation of Df(btd,Sp1). Using the primers shown in (A), no product is observed in the original P element stocks, while a 2.7 kb product is observed in two independently generated Df(btd,Sp1) stocks. (C,D) btd and Sp1 expression patterns visualized by btd-Gal4>UAS-lacZ and Sp1 in situ hybridization, respectively, in third instar leg (C) and antennal (D) imaginal discs. Note the absence of btd or Sp1 expression in the presumptive body wall of the leg (arrows). (E) Df(btd,Sp1) mutant clones (absence of signal) are difficult to recover in the btd/Sp1 expression domain when they generated before the third instar (<72 hrs AEL). Twin spots (white arrows) and clones in proximal regions (asterisks) can be observed, as can clones in the wing or eye discs (red arrows). Only twin spots are recovered in the medial antenna (white arrows). The images of the antenna and eye discs represent two different confocal planes of the same disc. (F) Df(btd,Sp1) clones generated 48–72 hrs AEL positively marked by ß -Gal staining (green) in the leg survive poorly and tend to segregate from the surrounding tissue. The disc is co-stained for Discs large (Dlg) which labels all cell membranes (red). (G) MARCM Df(btd,Sp1); Sp1S+ mutant clones generated in parallel to those in (F) are recovered more frequently than Df(btd,Sp1) mutant clones, indicating rescue. (H) Quantification of rescue. Sp1 rescued the number of clones in the leg disc (only telopodite clones were scored). Note that Sp1S rescued better then Sp1L. Clones were induced 48–72 hrs AEL. The rescue experiments with Sp1S or Sp1L, but not with btd+ (p>0.05), show a statistically significant difference from the control experiment (Df(btd,Sp1); * p<0.05 and ** p<0.001 with Student's t-test).
In third instar imaginal discs, btd and Sp1 have indistinguishable expression patterns as assessed by in situ hybridization and by the expression of an enhancer trap inserted close to the transcription start of btd (Figure 1C). Both genes were expressed in the entire leg imaginal disc except for the most proximal ring of cells (Figure 1C). Based on its relationship to other markers in the leg, these genes appear to be expressed in the entire leg (coxa through tarsus), but not in the body wall (see Figure S2). Thus, unlike all previously described genes, the btd and Sp1 expression domain marks the tissue that will become leg as opposed to body wall. In the antennal disc, both genes were expressed in a medial ring of cells along the PD axis (Figure 1D). No expression was observed in wing, haltere, or eye discs. Consistent with these expression patterns, mitotic clones of Df(btd,Sp1) initiated during the second instar or before survived poorly in most of the leg disc (except from the most proximal domain, which gives rise to the body wall), but were readily recovered in wing, haltere, and eye imaginal discs (Figure 1E and 1F). When we examined the cell death marker Caspase 3 (Cas3), we found that clones in the dorsal imaginal discs (eye, wing, and haltere) had no Cas3 staining, while the few clones that survive in the ventral discs (antenna and leg) express Cas3, suggesting that cell death is occurring in these clones (see Figure S3). Consistent with this observation, when Df(btd,Sp1) clones also expressed the baculovirus cell death inhibitor p35, their growth was partially rescued (see Figure S4).
Because Df(btd,Sp1) removes two additional genes in addition to btd and Sp1, we used RNAi knockdown and rescue experiments to address which genes were responsible for the poor survival of Df(btd,Sp1) clones. Knocking down the expression of the other two genes deleted in Df(btd,Sp1) using RNAi produced no phenotype in the legs or antennae, suggesting that their absence does not contribute to the poor survival of these clones (data not shown). Using a rescue approach, we tested two different isoforms of Sp1 (Sp1S and Sp1L; Figure 1A) and btd. The recovery of Df(btd,Sp1) clones in the leg disc was rescued to wild type by Sp1S and, to a lesser degree, by Sp1L (Figure 1G and 1H). Importantly, the weak rescue provided by btd was not statistically significant (Figure 1H). Together with the data described below, these results suggest that the poor survival of Df(btd,Sp1) clones is largely due to the loss of Sp1.
To gain additional insights into the compromised growth of Df(btd,Sp1) clones, we tested which growth-promoting pathways might be able to rescue this phenotype. Co-expressing string (cdc25) and cyclinE, which promote the cell cycle by promoting the G2 to M transition [21], failed to provide any rescue of Df(btd,Sp1) clones (see Figure S4). In contrast, expressing the transcriptional co-activator Yorkie (Yki), a downstream component of the Hippo tumor suppressor pathway [22], [23], was able to rescue the growth of Df(btd,Sp1) clones, both in the leg imaginal disc and the adult leg (see Figure S4). These data suggest that Yki, which is known to activate genes required for proliferation and cell survival [22], [23], functions downstream of Sp1 to activate growth-promoting target genes.
These results show that Df(btd,Sp1) is a valuable tool to analyze the role of btd and Sp1 during Drosophila development. They further suggest that Sp1 plays a more critical role in leg development than btd, a conclusion that we further support below.
Removing btd and Sp1 functions during larval development results in leg growth defects
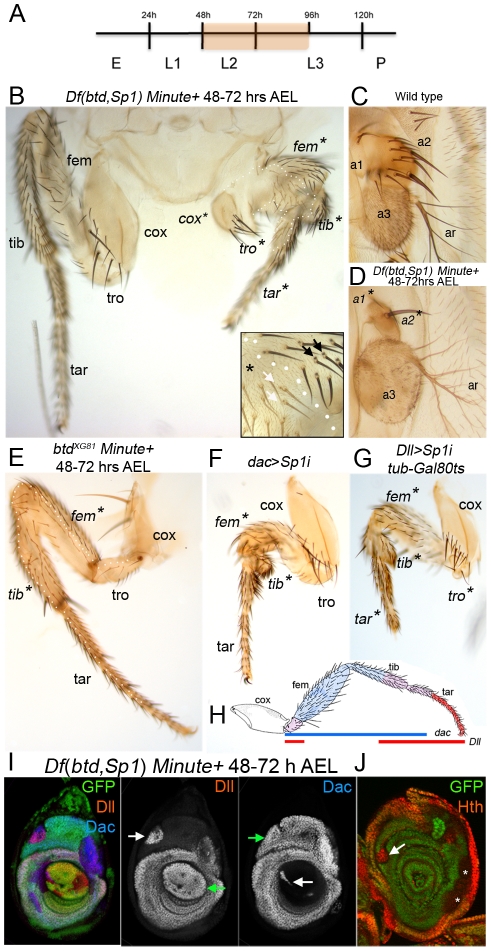
To assess the role these genes play at different times during development, we first analyzed the behavior of Df(btd,Sp1) clones in the adult that were induced in the second instar stage (48 to 72 hrs after egg laying (AEL)), long after the imaginal discs have been allocated and Dll expression has been initiated. To give these clones a growth and survival advantage, we used the Minute (M) technique, which allows the generation of tissue comprised of entirely, or almost entirely, homozygous mutant cells [24]. No phenotypes were observed in the dorsal appendages (wing or haltere) or dorsal body (see below). In contrast, legs containing Df(btd,Sp1) M+ clones were dramatically reduced in size (Figure 2B). Growth defects were observed throughout the entire leg, from the coxa to the tip of the tarsus. However, leg identity, assessed by the presence of bracted bristles, was still maintained in Df(btd,Sp1) tissue (Figure 2B, inset). When recovered in the antenna, the sizes of the a1 and a2 segments were also severely reduced, while the a3 and arista segments were unaffected (Figure 2C and 2D), consistent with the expression of btd and Sp1 in a medial ring in the antennal imaginal disc (Figure 1D). These findings suggest that, by 48 hrs of development, neither btd nor Sp1 are required to maintain leg identities, but that one or both of these genes is required for proper leg growth and morphology.
Figure 2. btd and Sp1 control leg growth.
(A) Time line showing when removing the function of btd and Sp1 affects leg growth (orange shadow). (B) A large Df(btd,Sp1) M+ clone in a T1 leg induced 48–72 hrs AEL and marked by yellow (y) bristles (the clone boundary is indicated by the white dotted line in the left leg). For comparison, the wild type right T1 leg is included in this image. The mutant tissue still maintains leg identity scored by the presence of bracted bristles (arrows, inset). Asterisks mark the segments affected by the clone. The same leg segment nomenclature has been used for all the figures: coxa (cox), trochanter (tro), femur (fem), tibia (tib) and tarsus (tar). (C) Wild type antenna with 1st antennal segment (a1), 2nd antennal segment (a2), 3rd antennal segment (a3) and arista (ar). (D) Df(btd,Sp1) M+ clone induced 48–72 hrs AEL results a strong reduction in size of the a1 and a2 antennal segments, while a3 and the ar are normal. The clone is marked by y. (E) A large btdXG81 M+ clone in a T2 leg induced 48–72 hrs AEL and marked by y (clone is outlined by white dots) results in a small growth defect in the femur (fe) and tibia (tib), which are also partially fused (arrow). The tarsus (tar), trochanter (tro) and coxa (cox) are unaffected. (F, G, H) The downregulation of Sp1 beginning at the second instar using RNAi affects the growth of the entire leg. Two different Gal4 drivers were used to examine different regions of the leg. (F) The medial part of the leg is strongly reduced in size in dac-Gal4; UAS-Sp1i flies. (G) The distal part of the leg is strongly reduced in size in Dll-Gal4; UAS-Sp1i flies. In this experiment we blocked Gal4 activity prior to the second instar using tub-Gal80ts. (H) Shows a schematic representation of the expression patterns of the two Gal4 drivers used to downregulate Sp1 function (dac in blue and Dll in red). (I, J) Df(btd,Sp1) M+ clones induced 48–72 hrs AEL and examined in 3rd instar leg discs. (I) A subset of Df(btd,Sp1) M+ clones (marked by the absence of GFP) show de-repression of Dll in the Dac domain and de-repression of dac in the Dll domain (white arrows). Note that these clones do not affect the expression of Dll and dac in their normal expression domains (green arrows). (J) A subset of Df(btd,Sp1) M+ clones (marked by the absence of GFP) show derepression of hth (arrow). Clones that do not derepress hth are indicated with asterisks.
To determine which of these two genes is required for leg growth at this stage we examined the effects of eliminating or knocking down btd and Sp1 individually. Large btdXG81 M+ clones made between 48 to 72 hrs AEL only generated weak phenotypes in the femur and tibia, which were partially fused (Figure 2E). These results support the idea that btd plays only a minor role in leg development. Because a Sp1 null allele is not available, we improved upon earlier RNAi knockdown experiments [14] to assess the role of Sp1 (see Materials and Methods). In contrast to the weak phenotypes observed in btdXG81 clones, reducing Sp1 activity by RNAi resulted in growth defects that were similar to those observed in large Df(btd,Sp1) clones (Figure 2F–2H). Analogous results were observed in the antenna: btdXG81 clones had no effect, while knockdown of Sp1 phenocopied the loss of the a1 and a2 segments seen in Df(btd,Sp1) mutant clones (see Figure S5). Taken together, these results suggest that Sp1 is playing a more important role than btd in leg and antennal growth after 48 hrs AEL.
We next examined the behavior of Df(btd,Sp1) M+ clones in the leg imaginal discs. For these experiments, we analyzed the expression of the three primary genes expressed along the proximo-distal (PD) axis, Dll, dachshund (dac), and homothorax (hth). In wild type third instar leg discs, these genes are expressed in overlapping domains along the PD axis to create five unique combinations [25]. From distal to proximal, these combinations are: 1) Dll only, 2) Dll + dac, 3) dac only, 4) Dll + dac + hth, and 5) hth only [26], [27]. In 86% (n = 23) of the Df(btd,Sp1) M+ clones recovered in the dac only domain Dll was derepressed, without any effect on dac expression (Figure 2I). In a smaller number of clones (34%; n = 32), we observed the de-repression of hth in the dac-only domain (Figure 2J). Similarly, 44% (n = 36) of the Df(btd,Sp1) M+ clones recovered in the Dll only domain de-repressed dac, without affecting Dll expression (Figure 2I). No effect on hth expression was observed in clones present in the most proximal domain of the leg disc (data not shown). Importantly, the expression patterns of Dll, dac, and hth were unaffected in btdXG81 M+ clones (data not shown). These data demonstrate that Sp1 plays an important role in generating the unique domains of gene expression that comprise the leg's PD axis.
Removing btd and Sp1 function during embryogenesis results in severe leg truncations
Previous work using a much larger deficiency (Df(1)C52) that removes btd, Sp1, and >50 other genes suggested that btd and Sp1 are required for the embryonic expression of Dll [14]. Using Df(btd,Sp1), we find that Dll expression is barely detectable in the leg primordia of stage 15 embryos (see Figure S6). In addition, in contrast to what was previously suggested based on the larger deficiency, formation of the leg primordia, as monitored by escargot (esg) expression, does not require btd and Sp1, because esg expression was still observed, although reduced, in Df(btd,Sp1) homozygous embryos (see Figure S6). As will be described below, the weak residual Dll protein that is observed in older Df(btd,Sp1) embryos is likely due to the activity of the early Dll-304 enhancer, which does not require btd or Sp1 inputs.
The near absence of Dll expression in older embryos contrasts with the relatively subtle effects on Dll expression when btd and Sp1 activities are removed 48 hrs AEL or later (Figure 2I and data not shown). One possible scenario to reconcile this difference is that btd and Sp1 have two temporally distinct functions during leg development: early, during embryogenesis, they would be required to maintain or perhaps establish ventral appendage fates, in part by activating Dll. Later in development, during larval stages, they would only be required for the proper growth of the leg.
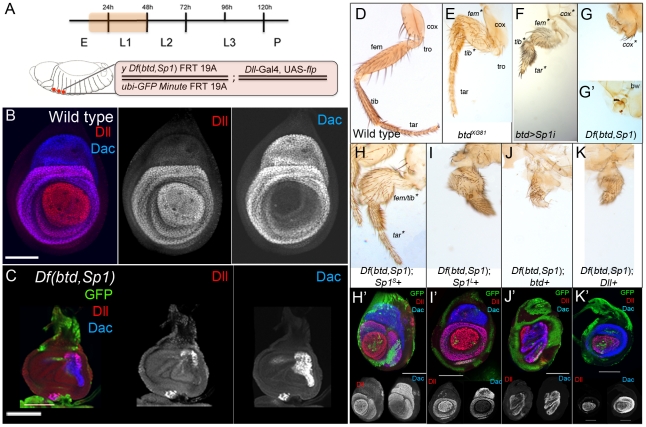
To test this idea, we analyzed the behavior of Df(btd,Sp1) M+ clones generated during embryogenesis. Using Dll-Gal4; UAS-flp to induce mitotic recombination (see Materials and Methods), 100% of the adults had severely aberrant legs. In 90% of the adults, the legs were completely absent or consisted of only a small patch of residual leg tissue (Figure 3G). When leg tissue was observed, it was invariably associated with non-mutant tissue, suggesting that these clones were generated slightly later than those samples in which no leg tissue remained. Generating btdXG81 clones at this early time produced relatively minor fusions of the femur and tibia, but left the tarsal segments largely unaffected (Figure 3E). These phenotypes are similar to those observed in later-induced btd clones (compare with Figure 2E). In contrast, reducing Sp1 activity by RNAi resulted in severe defects throughout the entire leg (Figure 3F). These results suggest that Sp1 is playing a more important role than btd, a conclusion that is supported by rescue experiments. When btd+ was resupplied in the Df(btd,Sp1) M+ legs, the resulting appendages were still highly abnormal, indicating poor rescue (Figure 3J). In contrast, when Sp1S or, to a lesser degree, Sp1L, were resupplied in Df(btd,Sp1) M+ legs, significant rescue of leg development was observed (Figure 3H and 3I).
Figure 3. Sp1 is required for leg development.
(A) Top: time line showing when the functions of btd and Sp1 (orange shadow) were removed. Bottom: In these experiments we initiated clone induction during embryogenesis using Dll-Gal4; UAS-flp to generate Df(btd,Sp1) M+ or btd M+ clones. This method results in excellent survival of animals that have all six legs completely, or nearly completely, mutant. For the rescue experiments, an additional UAS transgene (e.g. UAS-btd) was included. (B) Wild type third instar leg imaginal disc showing the expression patterns of Dll and dac. (C) Third instar Df(btd,Sp1) mutant leg generated using the genotype schematized in (A); mutant tissue is marked by the absence of GFP. These discs are much smaller than wild type and show a nearly complete loss of Dll and dac expression. White bar is 75 µm. (D) Wild type T1 adult leg with the segments indicated. (E) T1 adult leg entirely mutant for btd (marked by y) generated as shown in (A). Only the size of the femur and the tibia are affected and are partially fused together. (F) btd-Gal4; UAS-Sp1i reduces the size of the entire leg, from the coxa to the distal tip. Shown here is a T1 leg. (G,G') T1 adult legs entirely (G') or nearly entirely (G) mutant for btd and Sp1, generated as described in (A). Mutant tissue is marked by y. In (G), only a small patched of mutant tissue is visible and is associated with some non-mutant (y+) coxa tissue. In (G'), no leg tissue is observed. (H–K) Rescue of Df(btd,Sp1) T1 mutant legs (marked by y) generated as described in (A) where (H) Sp1S, (I) Sp1L, (J) btd and (K) Dll were expressed under the control of Dll-Gal4. (H'-K') show examples of leg imaginal discs of the same genotypes. White bars represents 75 µm. (H) Sp1S is able to rescue the Df(btd,Sp1) mutant phenoype and restore the Dll and dac expression domains. (I) Sp1L is able to partially rescue the adult Df(btd,Sp1) mutant phenoype and completely restore the Dll and dac expression domains. (J) btd is unable to rescue the adult Df(btd,Sp1) mutant phenoype and partially rescues the Dll and dac expression domains. (K) Dll is unable to rescue the adult Df(btd,Sp1) mutant phenoype but partially rescues Dll and dac expression domains.
Similar conclusions come from the analysis of leg imaginal discs containing Df(btd,Sp1) M+ clones generated during embryogenesis. Because these clones were generated early, entirely mutant leg discs could be obtained. In most cases, the discs were greatly reduced in size, with no or little Dll expression, and only a small patch of residual dac expression (Figure 3C). Strikingly, the normal expression domains of Dll and dac could be fully rescued by resupplying Sp1S or Sp1L (Figure 3H' and 3I'). In contrast, resupplying btd to Df(btd,Sp1) M+ discs provided a very weak rescue of these PD expression domains (Figure 3J'). Together, these experiments suggest that Sp1 is required during embryogenesis to generate leg fates, while btd plays a much more restricted role in leg development. Similarly, as noted above, Sp1 plays a much more important role in antennal development than btd (see Figure S5).
Ventral to dorsal transformations in the absence of btd and Sp1
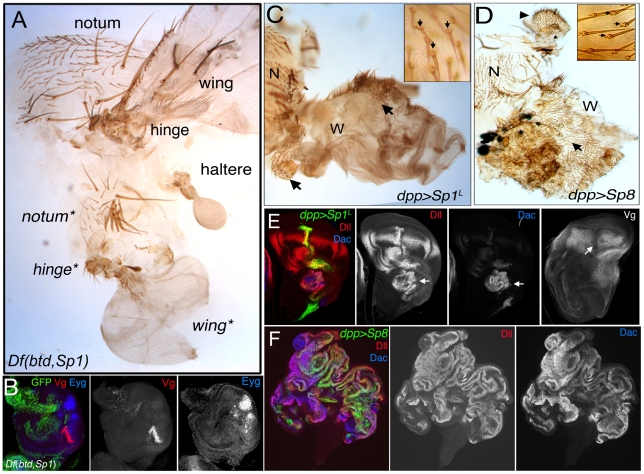
In addition to observing severely truncated or no legs, in about 10% of the adults with Dll>flp induced Df(btd,Sp1) M+ clones one or two of the legs were transformed towards a dorsal thoracic fate, including elements of the wing or haltere, and notum (Figure 4A and Table S1). In some examples we observed the triple row of bristles characteristic of the dorso-ventral border of the wing blade (data not shown). These ventral to dorsal homeotic transformations were confirmed by the presence of wing and notum molecular markers in mutant Df(btd,Sp1) leg discs, including vestigial (vg) and eyegone (eyg) (Figure 4B) [28], [29]. In addition to observing wing tissue, we also observed haltere tissue in place of the third thoracic legs in some of these animals (Figure S7 and Table S1). Curiously, these ventral to dorsal transformations did not always respect the normal thoracic identities because wing tissue, which normally develops in the second thoracic (T2) segment, was frequently observed in the T1, T2, and, to a lesser extent, the T3 segments (see Table S1). Nevertheless, these dramatic transformations indicate that btd and Sp1 are required for establishing adult ventral fates and that they inhibit the establishment of dorsal fates. Ventral to dorsal transformations were never observed when only btd function was removed at this early time, arguing that Sp1 is sufficient for executing these selector-like gene functions.
Figure 4. Dorsal to ventral transformations resulting from the loss of btd and Sp1.
(A) A T2 adult segment comprised mostly of Df(btd,Sp1) y tissue. These animals are generated via the genotype shown in Figure 3A. Dorsal is up. The normal notum, wing, and hinge are at the top; the bottom half of the tissue shows a transformation of ventral fates towards dorsal fates, including an ectopic wing, hinge, and notum (indicated by asterisks). Note that the normal notum, wing, and hinge are mutant (marked by y) but appear wild type. (B) Third instar leg imaginal disc of the same genotype as in (A) stained for GFP (absence marks the mutant tissue), Vg (red) and Eygone (Eyg; blue), which are markers for wing and notum fates, respectively. (C,D) Ectopic expression of Sp1L (C) or mouse Sp8 (D), under the control of dpp-Gal4 result in the transformation of wing towards leg in the adult. Arrows indicate leg tissue. Remaining notum (N) and wing (W) tissue are indicated. Insets show a high magnification of the leg tissue, with bracts (small arrows). Note the appearance of leg structures also in the pronotum in (E) (arrowhead). (E,F) Ectopic expression of Sp1L (E) or mouse Sp8 (F), under the control of dpp-Gal4 results in the induction of leg fates in the wing imaginal disc. These discs were stained for dpp-Gal4 expression (green), Dll (red), Dac (blue), or Vg (E, right-most panel). dpp>Sp8 also results in dramatic overgrowths that are not observed in the dpp>Sp1L wing discs.
Ectopic expression experiments also support the idea that Sp1 behaves as a ventral appendage selector gene. Using either flip-out Gal4 (not shown) or dpp-Gal4 (Figure 4), Sp1L was able to activate both Dll and dac and inhibit vg expression in the wing imaginal disc and produce wing to leg transformations in the adult appendage (Figure 4C and 4E). Ectopic expression of Sp1L was also able to induce another proximal leg gene, teashirt (tsh), in the wing, in a pattern that was reminiscent of that seen in wild type leg discs (see Figure S8). Moreover, this property has been evolutionarily conserved in this gene family because mouse Sp8 can also activate Dll and dac and induce dramatic dorsal to ventral homeotic transformations (Figure 4D and 4F).
Sp1 is required for Dll-LT, but not Dll-304, activity
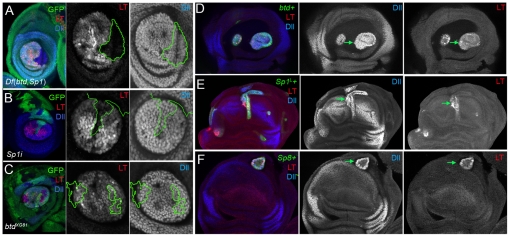
To examine the connection between btd, Sp1, and Dll at higher resolution, we analyzed the dependencies of two Dll enhancers, Dll-304 and Dll-LT, on btd and Sp1 activities. Dll-LT is directly activated by Wg and Dpp inputs [30]. Consequently, a LT-lacZ reporter gene is expressed in the center of the leg imaginal disc, in cells that receive strong input from both of these signaling pathways [30], [31]. One question that stems from these previous studies is why Dll-LT activity is specific to the ventral appendages and is not activated in other tissues where Wg and Dpp activities intersect such as the wing disc. The data described above suggest that btd and/or Sp1 may be the answer.
To test this idea, we generated Df(btd,Sp1) M+ clones and analyzed the effects on the LT-lacZ reporter gene in leg imaginal discs. Strikingly, LT-lacZ expression was absent in these clones (Figure 5A). This appears to be a consequence of the loss of Sp1 and not btd because btdXG81 clones had no effect on LT-lacZ expression (Figure 5C). Further, the down regulation of Sp1 by RNAi was sufficient to strongly reduce, but not eliminate, LT-lacZ expression (Figure 5B). Note that, consistent with our earlier studies [30], no effect on Dll expression was observed in these clones because the maintenance of Dll expression in larval stages is independent of Sp1 and btd (Figure 5A–5C). Ectopic expression of Sp1 also induced the expression of LT-lacZ and Dll in the wing disc (Figure 5E). LT-lacZ and Dll can also be activated by mouse Sp8 and by btd (Figure 5D and 5F). Thus, although btd is not required for LT activity, it has the capacity to induce its activity when ectopically expressed. This gain-of-function property of btd is consistent with previous observations that btd is sufficient to induce leg development when ectopically expressed in the wing [14].
Figure 5. Sp1, not btd, is required for Dll-LT activity.
(A) Df(btd,Sp1) M+ clone (outlined in green) generated 72–96 hrs AEL shows the absence of LT-lacZ expression, but no affect on Dll. (B) Clones expressing Sp1i strongly reduced LT-lacZ expression. (C) btdXG81 mutant clones generated 72–96 hrs AEL do not affect LT-lacZ expression. (D–F) Ectopic expression of btd (D), Sp1L (E), or mouse Sp8 (F) in the wing disc activates Dll and LT-lacZ (green arrows). These flip-out clones were generated 48–72 hrs AEL.
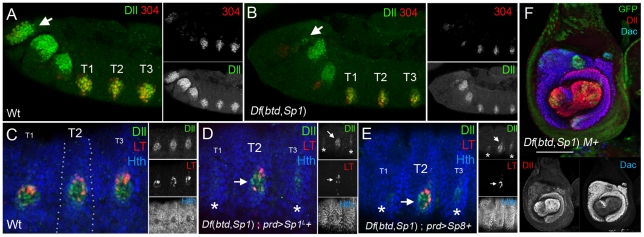
LT-lacZ is first activated in stage 14 embryos [2]. Consistent with the above findings, Df(btd,Sp1) embryos failed to express LT-lacZ (Figure 6D). In contrast, LT-lacZ is expressed in btd embryos, although at reduced levels [2]. The lack of LT-lacZ expression in Df(btd,Sp1) embryos could be rescued by resupplying only Sp1 (Figure 6D). Remarkably, mouse Sp8 was also able to rescue Dll expression and LT activity in Df(btd,Sp1) embryos (Figure 6E) and Dll and dac expression in Df(btd,Sp1) mutant leg imaginal discs (Figure 6F). In contrast to Dll-LT, the earlier-acting Dll enhancer, Dll-304, did not require btd or Sp1 because a 304-lacZ reporter gene was expressed in Df(btd,Sp1) embryos (Figure 6B). The independence of Dll-304, but dependence of Dll-LT, on Sp1 activity accounts for the observation that Df(btd,Sp1) stage 14 embryos show very weak, residual Dll protein in the leg primordia (Figure 6D and 6E).
Figure 6. Different dependencies on Sp1 for early and late Dll enhancer activities.
(A,B) Stage 11 wild type (A) and Df(btd,Sp1) (B) embryos stained for Dll (green) and Dll304-lacZ (red). Dll304-lacZ remains active in the absence of both factors. Expression of Dll in the antennal primordia, however, is nearly absent in Df(btd,Sp1) embryos (arrows). T1, T2, and T3 mark the three thoracic segments. (C) Wild type stage 14 embryo stained for Dll (green), Hth (blue), and Dll-LT-lacZ (red). The white dots mark the prd-Gal4 expression domain. (D, E) Df(btd,Sp1); prd-Gal4; UAS-Sp1L (D) or UAS-Sp8 (E) stage 14 embryos. In T1 and T3, where prd-Gal4 is not expressed, Dll expression is greatly reduced (asterisks) and LT activity is completely absent. The remaining Dll expression is likely derived from the Dll304 early enhancer. In T2, where prd>Sp1L (D) or prd>Sp8 (E) both Dll and LT-lacZ expression are rescued. btd and Sp1S can also rescue embryonic Dll and LT-lacZ expression (not shown). (F) Df(btd,Sp1) M+ mutant leg disc generated using the scheme shown in Figure 3A, rescued with UAS-Sp8. Significant rescue of the Dll (red) and dac (blue) expression domains is observed. Mutant tissue is marked by the absence of GFP (green).
btd and Sp1 function upstream of Dll
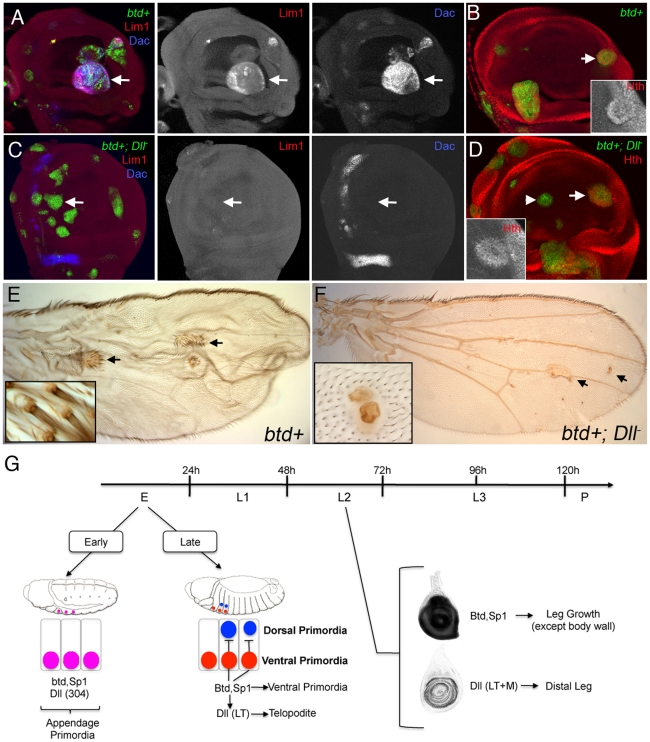
As described above, btd and Sp1 both have the ability to induce ectopic leg development when expressed in the dorsal imaginal discs, and both have the ability to induce Dll expression. Given our observation that the initiation of LT activity is also dependent on btd and Sp1, we reasoned that the ability of these factors to induce leg development, especially distal leg fates, might depend on Dll. To test this, we used the MARCM method [32] to generate clones that ectopically express btd or Sp1L and at the same time were mutant for Dll (tub>btd; Dll– or tub>Sp1L; Dll–). In control tub>btd clones (wild type for Dll), ectopic leg tissue was observed in the wing, and markers for leg development (Lim1, dac, and hth) were activated in the wing imaginal disc (Figure 7A and 7B). In contrast, when these clones were also mutant for Dll, the activation of Lim1 and dac, which are markers for the distal leg, was not observed (Figure 7C). However, tub>btd; Dll– clones close to the wing hinge were still able to activate hth, a marker for proximal fates (Figure 7D). Similar observations were obtained in tub>Sp1L; Dll– clones (see Figure S9). From these data, we conclude that btd requires Dll to generate the Lim1+, dac+ telopodite, but that btd can induce proximal, hth+, leg fates in the absence of Dll. This conclusion is further supported by the behavior of tub>btd+; Dll– clones that arise in the adult wing. Although these clones cannot generate distal leg fates, they are able to produce what appears to be proximal leg tissue (Figure 7F). In contrast, tub>btd clones have the ability to induce both proximal and distal leg structures in the adult wing (Figure 7E). Finally, the epistatic relationship between btd, Sp1, and Dll was further supported by analyzing the consequences of resupplying Dll+ in Df(btd,Sp1) M+ legs and leg discs. The resulting legs were still severely truncated, indicating poor rescue, while in the imaginal discs Dll and dac expression was only partially rescued (Figure 3K). These phenotypes likely reflect the later requirement of Sp1 and, to a lesser extent, btd, in leg growth and cell survival (see above).
Figure 7. btd and Sp1 require Dll to induce distal and medial leg development.
(A,B) btd+ MARCM clones in the wing disc activated the expression of Lim1, dac (A) and hth (B) (arrows). Clones were generated 48–72 hrs AEL and are marked by GFP+ (green). (C,D) Dll-; btd+ MARCM clones in the wing disc were unable to induce Lim1 or dac (C; arrows), but can still able activate hth close to its own domain (arrow) but not in the center of the wing pouch (D; arrowhead). Clones were generated 48–72 hrs AEL and are marked by GFP (green). (E) btd+ MARCM clones in the adult wing blade induced the formation of leg-like tissue, including distal leg identities (arrows and inset). (F) Dll-; btd+ MARCM clones failed to induce distal leg-like tissue, although they generate tissue that might correspond to proximal leg tissue (arrows and inset). (G) Schematic representation of the differential requirements for btd and Sp1 during leg development. At embryonic 11 stage, Dll (via the 304 enhancer), btd, and Sp1 are all activated independently in the appendage primordia. A few hours later, Dll expression is restricted to the telopodite precursors cells of the leg (via the LT enhancer) and depends on Sp1 activity. At this stage, Sp1 is required to promote the formation of the ventral appendage primordia (legs) and inhibit the formation of the dorsal primordia (wing and haltere). Dll is required for the entire telopodite domain. During larval second instar stage (L2), Dll expression no longer requires btd and Sp1. Dll is required only for distal leg development while btd and Sp1 are required for the growth of the entire leg but have no function in the body wall.
In summary, btd and Sp1 have the capacity to induce telopodite fates, which depend on Dll, as well as more proximal coxapodite fates, which do not require Dll.
Discussion
Prior to this study, our understanding of the roles that btd and Sp1 play in ventral appendage development in Drosophila was largely derived from ectopic expression experiments showing that btd could induce ectopic leg development when expressed in dorsal imaginal discs [14]. In addition, based on a large deficiency that removes >50 genes, it was suggested that these genes may function upstream of Dll in ventral appendage specification. What was lacking in this previous study was the ability to specifically analyze the functions of these genes, both in embryogenesis and during adult development, using loss-of-function null alleles. Here, using a newly derived deficiency, together with rescue experiments, we show for the first time that these Zn-finger transcription factors play non-redundant roles in ventral appendage development. Moreover, for all of the readouts examined here – leg allocation, leg growth, proliferation, and PD axis formation – btd plays a much more minor or no role compared to Sp1. Early, Sp1, but not btd, is required to define the group of cells that will give rise to the legs and perhaps additional ventral body structures as well. Thus, Sp1 is a selector-like gene for the entire ventral appendage. Later in development, both genes are required for the proper growth of the leg, although to very different degrees. We also show that vertebrate Sp8 retains both the selector and growth-promoting functions, suggesting that there has been a remarkable amount of functional conservation between the vertebrate and fly genes. Below we discuss both functions, and summarize how these findings contribute to our overall understanding of ventral appendage development in Drosophila.
Growth-promoting functions of btd and Sp1 during larval stages
During larval development, we find that Sp1 is required for the proper growth of the entire leg, from the coxa through the tarsus. In contrast, btd plays a much more limited role in the tibia and femur. At this stage, neither gene is required for leg identity, nor are they required for the development of ventral body structures that arise from the most proximal cells in the leg imaginal disc. These ‘late’ phenotypes are consistent with the expression patterns of these genes in the third instar leg imaginal discs, where they appear to mark the entire presumptive leg, but not more proximal cells. This is interesting, because prior to these observations there were no markers that distinguished between the hth-expressing cells that give rise to the coxa from the hth-expressing cells that give rise to the ventral body wall. Dll, for example, is expressed in the cells that give rise to the distal tibia and tarsus, and lineage tracing with the Dll-LT element marks the entire telopodite (trochanter, femur, tibia, and tarsus) [2]. The addition of the btd and Sp1 expression patterns and mutant phenotypes to previously characterized PD genes therefore adds an important demarcation that distinguishes leg from body fates.
Our analysis also reveals dramatic differences in the post-embryonic functions of btd and Sp1. Specifically, most of the growth phenotypes observed when both genes are removed can be phenocopied by knocking down only Sp1. In contrast, btdXG81 clones (or btdXA clones, see Materials and Methods) have no phenotypes in the antenna, and, in the leg, result in only partial fusions between the femur and tibia. Thus, Sp1, not btd, plays an important and non-redundant function in ventral appendage development at this stage.
Selector-like functions of Sp1
Selector and selector-like genes have the property that they specify an entire organ or body part [33]. The classic example is engrailed (en) which ‘selects’ posterior compartment identities in Drosophila [34]. Another example is eyeless (ey), which is both necessary and sufficient for eye development in Drosophila [35]. In the leg, previous work highlighted the role of Dll in ventral appendage specification. In the absence of Dll, the distal portion of the leg fails to develop, while dorsal appendages remain wild type [1]. Moreover, ectopic expression of Dll can induce distal legs to develop in dorsal positions [10]. Taken together, these observations suggested that Dll is a selector-like gene for the distal leg.
Despite the requirement for Dll in leg development, it has been known for sometime that the ventral appendage primordia form in the absence of Dll [1], [9]. Moreover, homeotic transformations are not observed in the absence of Dll. Thus, Dll cannot be considered a selector-like gene for the entire ventral appendage. These observations raise the question of what factor or factors initially specify the cells that will give rise to the ventral appendage. We propose that Sp1 fulfills this selector-like role.
The suggestion that Sp1 is a selector-like gene for the entire ventral appendage stems in part from the observation that when the function of this gene is removed early in development, ∼10% of the animals have dramatic transformations of ventral structures to dorsal structures. In many of these cases, we observe both wing and notum tissue developing in ventral positions. Molecularly, Dll and dac expression is lost in transformed leg discs, and ectopic expression of vg and eyg, two markers for the dorsal appendages, are observed instead. The expression of Dll-304, which is traditionally been considered a marker for the ventral appendage, in Df(btd,Sp1) embryos may seem at odds with the idea that Sp1 is required for the initial specification of leg fates. However, fate-mapping studies show that Dll-304-expressing cells give rise to both the ventral (leg) and dorsal (wing and haltere) appendages [2]. Thus, Dll-304 cannot be considered a ventral marker, and its activity in Df(btd,Sp1) embryos only confirms the establishment of appendage primordia without ventral or dorsal identity.
In sum, the striking transformations of fate seen in Df(btd,Sp1) animals suggest that Sp1 promotes ventral fates, both the entire leg and ventral body wall, and that in the absence of this gene, dorsal fates are de-repressed. This change in developmental fate is analogous to other classical homeotic transformations, for example, when the leg is transformed to antenna in the absence of Antennapedia (Antp) [36]. Note that btd null clones made at the same early time in development only result in mild growth defects, but legs are still generated. Thus, btd is not required for this function. However, because an Sp1 null allele (btd+) is not currently available, we cannot at this time be completely certain that btd plays no role in this process.
Because wing development is normally limited to T2, it was unexpected to observe leg to wing transformations in the T1 and, to a lesser extent, T3 segments. One potential explanation for this violation of antero-posterior identity is due to the timing of clone induction. Although the Hox genes are responsible for determining the segmental identities of the dorsal appendages [37], [38], it may be that they are deployed at different times in the ventral and dorsal primordia in the different thoracic segments. If this is the case, then the resulting transformations may be very sensitive to the time they were generated and to their segmental origins. It is also worth noting that the wing primordia and T2 identity can be generated in the absence of Hox input [39], [40]. Thus, wing fates, as opposed to haltere or humeral (dorsal T1) fates, represent a Hox-free default state, which may predominate in these aberrant developmental situations.
Gradual refinement of ventral fates
Together with previous studies, these findings allow us to present a more complete view of ventral appendage specification, which we breakdown into three main phases (Figure 7G). In the first phase, Sp1, btd, and Dll (via it’s early Dll-304 enhancer) are initially activated in parallel in a ventral domain in each thoracic hemisegment of stage 11 embryos. The activation of all three genes is dependent on Wg signaling [1], [14]. This early, Dll-304-driven expression of Dll does not require either btd or Sp1. This initial group of cells is fated to give rise to both the entire ventral and dorsal thoracic imaginal discs, in other words, the entire adult thorax. In the second phase, which begins at stage 14, Dll-304 is no longer active and Dll is controlled by late-acting enhancers such as Dll-LT, which is activated by Wg and Dpp signaling [2], [30]. Interestingly, as shown here, these late-acting Dll enhancers also require Sp1, but not btd [2], thus placing Sp1 genetically upstream of Dll. At this stage, the Dll+ cells will only give rise to the leg telopodite. Sp1 is also required for telopodite formation but is carrying out at least two additional functions. One is that, unlike Dll, Sp1 is required to specify more proximal leg segments (the coxapodite). Second, the ventral to dorsal homeotic transformations described above suggest that Sp1 is also required to repress dorsal fates. Finally, in the third phase, Dll begins to autoactivate it’s expression and no longer depends on Wg and Dpp inputs [30], [41]. At this stage, Dll also no longer requires Sp1 to be expressed. Instead of working through Dll, btd and Sp1 continue to play a critical role in leg development but now work in parallel to Dll to promote the growth of the entire leg. Thus, the specification of the ventral primordia depends on a feed-forward logic in which Sp1 activates late embryonic Dll expression followed by a phase in which both btd and Sp1 contribute to appendage growth in parallel to Dll (Figure 7G).
Appendage development: a case of “deep homology”
Besides having a PD axis, arthropod and vertebrate appendage morphologies have little in common. Moreover, the developmental logic of limb formation in Drosophila is very different from that of vertebrate limb development. In flies, Hedgehog signaling induces two antagonistic secondary signals, Dpp and Wg, which in turn establish the PD axis by activating genes such as Dll and dac [41], [42]. In vertebrate limb development, Sonic hedgehog induces the activity of fibroblast growth factor-like molecules such as FGF8 in the ectoderm, which drives the proliferation of the underlying mesenchyme and the nested expression of Hox genes to create a PD axis [43], [44]. Despite these differences, it is striking that multiple vertebrate orthologs of both Sp1 and Dll are expressed during vertebrate limb development. In addition, orthologs of both hth and exd (Meis and pbx, respectively) are expressed in the proximal domain of the developing mouse limb [45], [46]. Although the existence of multiple Dll and Sp1 orthologs (Dlx1/Dlx2/Dlx5/Dlx6 and Sp8/Sp9, respectively) makes it much more challenging to assess their functions in detail, the available data demonstrate that, as in flies, both sets of genes are critical for vertebrate limb development [17]–[19], [47], [48]. Our results, illustrating that vertebrate Sp8 can rescue many of the Sp1 and btd loss of function phenotypes in Drosophila, support the idea that appendage development in these two phyla represents a case of ‘deep homology’ [49], [50]. Interestingly, that orthologs of both Sp1 and Dll gene families are used in both phyla argue that, for appendage development, the functions of these transcription factors have been much more conserved than those of the signaling pathways used in limb development. The same conclusion holds for eye development where the transcription factors, more than the deployment of specific signaling pathways, have been conserved over vast evolutionary distances [49], [51]. These observations imply that, once established, transcription factor networks may be very stable, while the organization of signaling pathway networks may be much more plastic and easily modified to accommodate radically distinct morphologies.
Materials and Methods
Generation of the Df(btd,Sp1)
To generate Df(btd,Sp1) we used the FRT-directed recombination technique using two FRT-containing P elements (PBac{XP}d01932 and PBac{RB}CG32698e03908). Recombinants lose the miniwhite gene, providing a positive identification for the recombination event. Two independent deletions were generated and confirmed by PCR using primers flanking the genomic region or within the P elements. Besides btd and Sp1 this deletion also removes CG1354 (molecular function: GTP binding) and partially deletes CG32698 (molecular function: carbonate dehydratase activity) (DrosDel FDD-0029282, http://www.drosdel.org.uk).
Generation of UAS-Sp1L and mouse UAS-Sp8
Sp1L and Sp1S are called Sp1-RD and Sp1-RB, respectively, by FlyBase (http://flybase.org). For the UAS-Sp1L construct we isolated RNA from leg imaginal discs to generate cDNA (SuperScript III First Strand Synthesis System for RT-PCR, Invitrogen). This served as a template to amplify the Sp1L isoform by PCR, which was sequenced and cloned into the pUAST attB vector. For mouse UAS-Sp8 we cloned the mouse Sp8 cDNA (gift from A. Mansouri) into a 3XHA-tagged pUAST attB vector.
Fly stocks
Two btd mutations were studied, the strong btdXG81 mutation and the amorph btdXA [52]. We found that btdXG81 phenotypes are stronger than btdXA, in agreement with Cohen and Jurgens [52]. To knock down Sp1 function, we combined two UAS-RNAi hairpin transgenes, one described by Estella et al. [14] and one from the Vienna Drosophila Resource Center (VDRC; line #4097). The Vienna RNAi stock is reported to have no off-target affects. As confirmation of this, we only observed phenotypes in tissues where Sp1 is expressed. Both transgenes, which target both Sp1 isoforms, were used in conjunction with UAS-dicer to enhance the RNAi, and thus generated much stronger phenotypes than were previously reported [14]. Dll SA1 [8], UAS-Dll [10], UAS-btd [13], Dll-Gal4 line 212; [10], btd-Gal4 [14], dac-Gal4 [53], and prd-Gal4 [54] have been described. The dpp-Gal4; UAS-GFP, tub-Gal80ts and UAS-flp were from the Bloomington Stock Center. The UAS-Sp1 was from [13] was renamed UAS-Sp1S because it encodes the short Sp1 isoform. The two Dll elements, Dll-304-lacZ [8] and LT-LacZ [14] were described. ubi-GFP FRT19A; hs-flp and ubi-GFP M(1)osp FRT19A were gifts from G. Struhl. UAS-yki was from D.J. Pan [55] and UAS-p35, UAS-string, and UAS-cycE were from L. Johnston.
Clonal analysis
To generate these genotypes we used a duplication on the Y chromosome that covers the btd and Sp1 genes (Dp(1;Y)lz+) [13].
-btd clones
yw btdXG81 or btdXA FRT19A/ubi-GFP M(1)osp FRT19A; Dll-Gal4, UAS-flp or hs-flp.
-Df(btd,Sp1) loss of function clones
yw Df(btd,Sp1) FRT19A/ubi-GFP M(1)osp FRT19A; Dll-Gal4, UAS-flp or hs-flp.
yw Df(btd,Sp1) FRT19A/yw ubi-GFP FRT19A; hs-flp
Because Minute/+ flies are developmentally delayed by approximately 1 day we adjusted the time of the heat-shock to induce clones at the correct developmental stage. Larvae were heat shocked for 1 hour at 37°C.
btd, Sp1S, Sp1L, and Sp8 gain of function clones.
yw hs-flp; act>y+>Gal4 UAS-GFP. The larvae were heat shocked for 10 minutes at 37°C.
Dll-; UAS-btd or UAS-Sp1L MARCM clones.
yw hs-flp, UAS-GFP; FRT42D y+ tubG80/DllSa1 FRT 42D; tub-Gal4
Df(btd,Sp1); UAS-btd, UAS-Sp1S or UAS-Sp1L MARCM clones.
tubGal80 FRT19A/Df(btd,Sp1) FRT19A; tub-Gal4, UAS-lacZ
Df(btd,Sp1); UAS-btd, UAS-Sp1S or UAS-Sp1L, or UAS-Sp8 rescue experiments.
yw Df(btd,Sp1) FRT19A/ubi-GFP M(1)osp FRT19A; Dll-Gal4, UAS-flp
Immunofluorescence methods
Imaginal discs and embryos were prepared and stained using standard procedures. RNA in situ hybridizations were carried out with digoxigenin-labeled RNA probes against btd and Sp1 [14]. For the Sp1 probe, the first and second exons of Sp1L were cloned in pBSK and transcribed to generate the anti-sense probe. These exons partially overlap a non-coding exon of Sp1S, so is likely to hybridize to both Sp1 transcripts. The primary antibodies used were: rabbit and mouse anti-ßGal (Capell and Promega), rabbit anti-GFP (Invitrogen), mouse anti-Dachsund and mouse anti-Dlg (Developmental Studies Hybridoma Bank (DSHB)), rabbit anti-caspase-3 (Upstate biotechnologies), guinea pig anti-Distalles, rabbit anti-Homothorax, rat anti-Lim1 (gift from Gerard Campbell), guinea pig anti-Vestigial (gift from M. Zecca) and rabbit anti-Vestigial (gift from Sean Carroll) and guinea pig anti-Eyegone (gift from N. Azpiazu).
Supporting Information
btd and Sp1 are expressed in the leg primordia. Embryos are oriented anterior to the left and dorsal up. Sp1 (A) and btd (B) RNA in situ hybridization in stage 13 embryos reveals the expression of these genes in the leg primordia (arrows). The inset at the right show a higher magnification image of the thoracic segments.
(0.89 MB TIF)
btd-Gal4 is expressed in the coxa but not in the body wall. (A) Third instar imaginal disc stained for Dll (blue), Hth (red) and GFP (btd-Gal4; UAS-GFP). Three different views of the same imaginal disc are shown with the most proximal domains marked. Note that at this stage btd is expressed at low level is the trochanter (tro), strongly in the coxa (cox) but is not expressed in the body wall (bw). (B) Schematic representation of the imaginal disc shown in (A). Note that btd is expressed in the entire leg at different levels but is not expressed in the body wall. (C) Everting pupal leg disc stained as in (A). The double-headed arrow indicates the PD axis of the leg. Note that btd expression subdivides the hth expression domain into presumptive coxa (btd+ hth+) and body wall (btd- hth+).
(2.53 MB TIF)
btd and Sp1 mutant clones activate cell death. Df(btd,Sp1) positively marked (ß-Gal, green) mutant clones generated 48–72hrs are readily recovered in the wing (A) or eye (C) discs, while in the leg discs (B) or second segment of the antenna disc (D) are rarely recovered and tend to segregate from the surrounding tissue. When recovered, these clones activate the apoptotic program as indicated by the expression of the cell death marker Cas 3 (red). These discs were stained with Dlg (blue) to identify cell membranes. The small panels in (A) and (B) show optical cross-sections of the Df(btd,Sp1) clones in the wing and leg discs, respectively. The eye-antenna imaginal disc shown in (C) and (D) is the same disc imaged in different confocal planes.
(3.80 MB TIF)
yki rescue of Df(btd,Sp1) mutant clones. (A) MARCM Df(btd,Sp1) clones generated 48-72 hrs AEL positively marked by b-Gal staining (red) in the leg imaginal disc survive poorly. The disc is co-stained for Hth which labels the proximal domain of the leg (green). (B) MARCM Df(btd,Sp1); yki+ mutant clones generated in parallel to those in (A) are recovered more frequently than Df(btd,Sp1) mutant clones, indicating rescue. (C) Adult leg resulting from the same experiment as in (A). Note the nearly absence of Df(btd,Sp1) mutant tissue marked by yellow (y). The arrow points to one clone that has sorted out form the main epithelium. (D) Adult leg resulting from the same experiment as in (B). Note that providing Yki in Df(btd,Sp1) mutant clones can rescue the appearance of mutant clones (arrows, marked by y). The inset shows a mutant clone that has sorted out form the main tissue but maintains a leg identity. (E) Quantification of rescue. yki, and to a lesser extent p35, rescued the number of clones in the leg disc (only telopodite clones were scored). Note that stg + cyclin-E do not rescue. Clones were induced 48-72 hrs AEL. Each column shows the mean and standard error of the mean. All three independent experiments ((Df (btd,Sp1) plus p35, yki or stg and cyclin-E) are different from (Df (btd,Sp1) mutant clones (* p<0.05,** p<0.001 with Student's t-test).
(5.54 MB TIF)
Sp1, but not btd, is required for antennal growth. All antennae are labeled with: 1st segment (a1), 2nd segment (a2), 3rd (a3) and arista (ar). (A) Wild type antenna. (B) Completely btdXG81 mutant antenna marked by y of the geneotype: yw btdXG81 FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp. No phenotype is observed in the mutant antenna. (C) btd-Gal4; UAS-Sp1i reduces the size of the a1 and a2 antennal segments. Compare to (A). (D) Antenna of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp where the a1 and a2 segments are greatly reduced.
(1.34 MB TIF)
btd and Sp1 mutant embryos fail to maintain Dll expression. Thoracic regions of stage 14 embryos stained for ß-Gal (esg-LacZ, green) and Dll (red). Anterior is left and dorsal is up. (A) Wt embryo showing the thoracic appendage primordia (legs, wing and haltere primordia). (B) Df(btd,Sp1) mutant embryo that fails to maintain Dll expression, compare it to (A).
(0.71 MB TIF)
Ventral to dorsal transformation in the absence of btd and Sp1. Hemi-third thoracic segment of a fly of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp where the third leg is transformed to an haltere (asterisks). Dorsal is to the left and ventral is to the right.
(0.62 MB TIF)
Ectopic expression of Sp1 induces leg development in the wing disc. (A) dpp-Gal4; UAS-Sp1L induces the ectopic expression of the leg PD genes Dll (green), dac (blue), and tsh (red) in the wing imaginal disc. Two planes of focus are shown. Note that the tissue where Dll, dac and tsh are ectopically induced (white square) is organized as a wild type leg imaginal disc. (B) A wild type leg imaginal disc shown for comparison.
(0.90 MB TIF)
Sp1 requires Dll to induce leg development. (A) Sp1L ectopic expression clones in the wing disc induce the expression of the leg genes dac and tsh (arrows). Clones are generated 48-72 hrs AEL. Note that Sp1 is better able to induce dac expression in the notum that in the wing pouch. (B) Dll-; Sp1L+ MARCM clones fail to induce dac expression (arrows). However, these clones retain the ability to activate tsh. Clones are generated 48-72 hrs AEL.
(1.76 MB TIF)
Summary of ventral to dorsal transformations. We scored the number of animals of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp that had a leg transformation to a dorsal appendage (wing or haltere) in any of the three thoracic segments. The ambiguous category includes those animals that had dorsal transformations but could not be unambiguously scored as wing-like or haltere-like. Total number of animal counted = 41.
(0.08 MB TIF)
Acknowledgments
We thank N. Azpiazu, G. Campbell, S. Carroll, L. Johnston, A. Mansouri, D. McKay, G. Morata, D. J. Pan, G. Struhl, and M. Zecca for fly stocks and reagents and W. Zhang for technical support. We thank Oliver Hobert and Roumen Voutev for comments on the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by NIH grant GM058575 to RSM. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cohen B, Simcox AA, Cohen SM. Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development. 1993;117:597–608. doi: 10.1242/dev.117.2.597. [DOI] [PubMed] [Google Scholar]
- 2.McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen SM. Specification of limb development in the Drosophila embryo by positional cues from segmentation genes. Nature. 1990;343:173–177. doi: 10.1038/343173a0. [DOI] [PubMed] [Google Scholar]
- 4.Goto S, Hayashi S. Specification of the embryonic limb primordium by graded activity of Decapentaplegic. Development. 1997;124:125–132. doi: 10.1242/dev.124.1.125. [DOI] [PubMed] [Google Scholar]
- 5.Kubota K, Goto S, Eto K, Hayashi S. EGF receptor attenuates Dpp signaling and helps to distinguish the wing and leg cell fates in Drosophila. Development. 2000;127:3769–3776. doi: 10.1242/dev.127.17.3769. [DOI] [PubMed] [Google Scholar]
- 6.Wieschaus E, Gehring W. Clonal analysis of primordial disc cells in the early embryo of Drosophila melanogaster. Dev Biol. 1976;50:249–263. doi: 10.1016/0012-1606(76)90150-0. [DOI] [PubMed] [Google Scholar]
- 7.Snodgrass R. New York: McGraw-Hill; 1935. Principles of Insect Morphology. pp. 83–99. [Google Scholar]
- 8.Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, et al. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- 9.Campbell G, Tomlinson A. The roles of the homeobox genes aristaless and Distal-less in patterning the legs and wings of Drosophila. Development. 1998;125:4483–4493. doi: 10.1242/dev.125.22.4483. [DOI] [PubMed] [Google Scholar]
- 10.Gorfinkiel N, Morata G, Guerrero I. The homeobox gene Distal-less induces ventral appendage development in Drosophila. Genes Dev. 1997;11:2259–2271. doi: 10.1101/gad.11.17.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wimmer EA, Jackle H, Pfeifle C, Cohen SM. A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature. 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 12.Wimmer EA, Frommer G, Purnell BA, Jackle H. buttonhead and D-Sp1: a novel Drosophila gene pair. Mech Dev. 1996;59:53–62. doi: 10.1016/0925-4773(96)00575-8. [DOI] [PubMed] [Google Scholar]
- 13.Schock F, Purnell BA, Wimmer EA, Jackle H. Common and diverged functions of the Drosophila gene pair D-Sp1 and buttonhead. Mech Dev. 1999;89:125–132. doi: 10.1016/s0925-4773(99)00215-4. [DOI] [PubMed] [Google Scholar]
- 14.Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–5941. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- 15.Beermann A, Aranda M, Schroder R. The Sp8 zinc-finger transcription factor is involved in allometric growth of the limbs in the beetle Tribolium castaneum. Development. 2004;131:733–742. doi: 10.1242/dev.00974. [DOI] [PubMed] [Google Scholar]
- 16.Schaeper ND, Prpic NM, Wimmer EA. A conserved function of the zinc finger transcription factor Sp8/9 in allometric appendage growth in the milkweed bug Oncopeltus fasciatus. Dev Genes Evol. 2009. [DOI] [PMC free article] [PubMed]
- 17.Kawakami Y, Esteban CR, Matsui T, Rodriguez-Leon J, Kato S, et al. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development. 2004;131:4763–4774. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- 18.Treichel D, Schock F, Jackle H, Gruss P, Mansouri A. mBtd is required to maintain signaling during murine limb development. Genes Dev. 2003;17:2630–2635. doi: 10.1101/gad.274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell SM, Schreiner CM, Waclaw RR, Campbell K, Potter SS, et al. Sp8 is crucial for limb outgrowth and neuropore closure. Proc Natl Acad Sci U S A. 2003;100:12195–12200. doi: 10.1073/pnas.2134310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 21.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 22.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Yue T, Jiang J. Hippo signaling pathway and organ size control. Fly (Austin) 2009;3:68–73. doi: 10.4161/fly.3.1.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morata G, Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 25.Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- 26.Morata G. How Drosophila appendages develop. Nat Rev Mol Cell Biol. 2001;2:89–97. doi: 10.1038/35052047. [DOI] [PubMed] [Google Scholar]
- 27.Kojima T. The mechanism of Drosophila leg development along the proximodistal axis. Dev Growth Differ. 2004;46:115–129. doi: 10.1111/j.1440-169X.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- 29.Aldaz S, Morata G, Azpiazu N. The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development. 2003;130:4473–4482. doi: 10.1242/dev.00643. [DOI] [PubMed] [Google Scholar]
- 30.Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estella C, Mann RS. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–636. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 33.Mann RS, Morata G. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu Rev Cell Dev Biol. 2000;16:243–271. doi: 10.1146/annurev.cellbio.16.1.243. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Bellido A, Santamaria P. Developmental analysis of the wing disc in the mutant engrailed of Drosophila melanogaster. Genetics. 1972;72:87–104. doi: 10.1093/genetics/72.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 36.Struhl G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature. 1981;292:635–638. doi: 10.1038/292635a0. [DOI] [PubMed] [Google Scholar]
- 37.Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 38.Morata G, Sanchez-Herrero E. Patterning mechanisms in the body trunk and the appendages of Drosophila. Development. 1999;126:2823–2828. doi: 10.1242/dev.126.13.2823. [DOI] [PubMed] [Google Scholar]
- 39.Carroll SB, Weatherbee SD, Langeland JA. Homeotic genes and the regulation and evolution of insect wing number. Nature. 1995;375:58–61. doi: 10.1038/375058a0. [DOI] [PubMed] [Google Scholar]
- 40.Struhl G. Genes controlling segmental specification in the Drosophila thorax. Proc Natl Acad Sci U S A. 1982;79:7380–7384. doi: 10.1073/pnas.79.23.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lecuit T, Cohen SM. Proximal-distal axis formation in the Drosophila leg. Nature. 1997;388:139–145. doi: 10.1038/40563. [DOI] [PubMed] [Google Scholar]
- 42.Diaz-Benjumea FJ, Cohen B, Cohen SM. Cell interaction between compartments establishes the proximal-distal axis of Drosophila legs. Nature. 1994;372:175–179. doi: 10.1038/372175a0. [DOI] [PubMed] [Google Scholar]
- 43.Tabin C, Wolpert L. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev. 2007;21:1433–1442. doi: 10.1101/gad.1547407. [DOI] [PubMed] [Google Scholar]
- 44.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- 46.Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 47.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 48.Kraus P, Lufkin T. Dlx homeobox gene control of mammalian limb and craniofacial development. Am J Med Genet A. 2006;140:1366–1374. doi: 10.1002/ajmg.a.31252. [DOI] [PubMed] [Google Scholar]
- 49.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 50.Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- 51.Vopalensky P, Kozmik Z. Eye evolution: common use and independent recruitment of genetic components. Philos Trans R Soc Lond B Biol Sci. 2009;364:2819–2832. doi: 10.1098/rstb.2009.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen SM, Jurgens G. Mediation of Drosophila head development by gap-like segmentation genes. Nature. 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 53.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
btd and Sp1 are expressed in the leg primordia. Embryos are oriented anterior to the left and dorsal up. Sp1 (A) and btd (B) RNA in situ hybridization in stage 13 embryos reveals the expression of these genes in the leg primordia (arrows). The inset at the right show a higher magnification image of the thoracic segments.
(0.89 MB TIF)
btd-Gal4 is expressed in the coxa but not in the body wall. (A) Third instar imaginal disc stained for Dll (blue), Hth (red) and GFP (btd-Gal4; UAS-GFP). Three different views of the same imaginal disc are shown with the most proximal domains marked. Note that at this stage btd is expressed at low level is the trochanter (tro), strongly in the coxa (cox) but is not expressed in the body wall (bw). (B) Schematic representation of the imaginal disc shown in (A). Note that btd is expressed in the entire leg at different levels but is not expressed in the body wall. (C) Everting pupal leg disc stained as in (A). The double-headed arrow indicates the PD axis of the leg. Note that btd expression subdivides the hth expression domain into presumptive coxa (btd+ hth+) and body wall (btd- hth+).
(2.53 MB TIF)
btd and Sp1 mutant clones activate cell death. Df(btd,Sp1) positively marked (ß-Gal, green) mutant clones generated 48–72hrs are readily recovered in the wing (A) or eye (C) discs, while in the leg discs (B) or second segment of the antenna disc (D) are rarely recovered and tend to segregate from the surrounding tissue. When recovered, these clones activate the apoptotic program as indicated by the expression of the cell death marker Cas 3 (red). These discs were stained with Dlg (blue) to identify cell membranes. The small panels in (A) and (B) show optical cross-sections of the Df(btd,Sp1) clones in the wing and leg discs, respectively. The eye-antenna imaginal disc shown in (C) and (D) is the same disc imaged in different confocal planes.
(3.80 MB TIF)
yki rescue of Df(btd,Sp1) mutant clones. (A) MARCM Df(btd,Sp1) clones generated 48-72 hrs AEL positively marked by b-Gal staining (red) in the leg imaginal disc survive poorly. The disc is co-stained for Hth which labels the proximal domain of the leg (green). (B) MARCM Df(btd,Sp1); yki+ mutant clones generated in parallel to those in (A) are recovered more frequently than Df(btd,Sp1) mutant clones, indicating rescue. (C) Adult leg resulting from the same experiment as in (A). Note the nearly absence of Df(btd,Sp1) mutant tissue marked by yellow (y). The arrow points to one clone that has sorted out form the main epithelium. (D) Adult leg resulting from the same experiment as in (B). Note that providing Yki in Df(btd,Sp1) mutant clones can rescue the appearance of mutant clones (arrows, marked by y). The inset shows a mutant clone that has sorted out form the main tissue but maintains a leg identity. (E) Quantification of rescue. yki, and to a lesser extent p35, rescued the number of clones in the leg disc (only telopodite clones were scored). Note that stg + cyclin-E do not rescue. Clones were induced 48-72 hrs AEL. Each column shows the mean and standard error of the mean. All three independent experiments ((Df (btd,Sp1) plus p35, yki or stg and cyclin-E) are different from (Df (btd,Sp1) mutant clones (* p<0.05,** p<0.001 with Student's t-test).
(5.54 MB TIF)
Sp1, but not btd, is required for antennal growth. All antennae are labeled with: 1st segment (a1), 2nd segment (a2), 3rd (a3) and arista (ar). (A) Wild type antenna. (B) Completely btdXG81 mutant antenna marked by y of the geneotype: yw btdXG81 FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp. No phenotype is observed in the mutant antenna. (C) btd-Gal4; UAS-Sp1i reduces the size of the a1 and a2 antennal segments. Compare to (A). (D) Antenna of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp where the a1 and a2 segments are greatly reduced.
(1.34 MB TIF)
btd and Sp1 mutant embryos fail to maintain Dll expression. Thoracic regions of stage 14 embryos stained for ß-Gal (esg-LacZ, green) and Dll (red). Anterior is left and dorsal is up. (A) Wt embryo showing the thoracic appendage primordia (legs, wing and haltere primordia). (B) Df(btd,Sp1) mutant embryo that fails to maintain Dll expression, compare it to (A).
(0.71 MB TIF)
Ventral to dorsal transformation in the absence of btd and Sp1. Hemi-third thoracic segment of a fly of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp where the third leg is transformed to an haltere (asterisks). Dorsal is to the left and ventral is to the right.
(0.62 MB TIF)
Ectopic expression of Sp1 induces leg development in the wing disc. (A) dpp-Gal4; UAS-Sp1L induces the ectopic expression of the leg PD genes Dll (green), dac (blue), and tsh (red) in the wing imaginal disc. Two planes of focus are shown. Note that the tissue where Dll, dac and tsh are ectopically induced (white square) is organized as a wild type leg imaginal disc. (B) A wild type leg imaginal disc shown for comparison.
(0.90 MB TIF)
Sp1 requires Dll to induce leg development. (A) Sp1L ectopic expression clones in the wing disc induce the expression of the leg genes dac and tsh (arrows). Clones are generated 48-72 hrs AEL. Note that Sp1 is better able to induce dac expression in the notum that in the wing pouch. (B) Dll-; Sp1L+ MARCM clones fail to induce dac expression (arrows). However, these clones retain the ability to activate tsh. Clones are generated 48-72 hrs AEL.
(1.76 MB TIF)
Summary of ventral to dorsal transformations. We scored the number of animals of the genotype yw Df(btd,Sp1) FRT19A/ubi-GFP M FRT19A; Dll-Gal4, UAS-flp that had a leg transformation to a dorsal appendage (wing or haltere) in any of the three thoracic segments. The ambiguous category includes those animals that had dorsal transformations but could not be unambiguously scored as wing-like or haltere-like. Total number of animal counted = 41.
(0.08 MB TIF)