Abstract
Although post-translational modifications by the small ubiquitin-like modifiers (SUMO) are known to be important in DNA damage response, it is unclear whether they have a role in double-strand break (DSB) repair by non-homologous end joining (NHEJ). Here, we analyzed various DSB repair pathways upon inhibition of SUMO-mediated protein–protein interactions using peptides that contain the SUMO-interaction motif (SIM) and discriminate between mono- and SUMO-chain modifications. The SIM peptides specifically inhibit NHEJ as shown by in vivo repair assays and radio-sensitivity of cell lines deficient in different DSB repair pathways. Furthermore, mono-SUMO, instead of SUMO-chain, modifications appear to be involved in NHEJ. Immunoprecipitation experiments also showed that the SIM peptide interacted with SUMOylated Ku70 after radiation. This study is the first to show an important role for SUMO:SIM-mediated protein–protein interactions in NHEJ, and provides a mechanistic basis for the role of SIM peptide in sensitizing genotoxic stress of cancer cells.
Keywords: SUMO, NHEJ, SUMO-interacting motif, SBM, inhibitor, DNA repair
Introduction
DNA double-strand breaks (DSBs) are generated by ionizing radiation and replication of chromosomes that have been damaged by chemotherapeutic drugs such as doxorubicin (Dox). It has been known for more than a decade that post-translational modifications by the small ubiquitin-like modifier (SUMO) are crucial events in cellular response to radiation and a wide range of DNA-damaging agents (Seufert et al., 1995; Shayeghi et al., 1997; Andrews et al., 2005; Zhao and Blobel, 2005); however, the role of SUMOylation in the major DSB repair pathway, non-homologous end joining (NHEJ), is still unknown. DNA repair pathways are not only essential to maintain genome stability, but are also important in cancer treatment, because radiation and most chemotherapeutic drugs kill cancer cells by inducing lethal DNA damage. The DNA repair capacities of tumor cells contribute significantly to the resistance of cancer cells to radiation and chemotherapeutic drugs that cause DNA damage (Bird et al., 2002; Carson et al., 2004).
Unlike ubiquitination, SUMOylation does not target proteins for degradation, but instead mostly alters protein–protein interactions. Our previous studies have shown that SUMO mediates protein–protein interactions by binding to a SUMO-binding motif or a SUMO-interacting motif (SBM or SIM) on receptor proteins (Song et al., 2004, 2005). Unlike the SUMO modification consensus sequence, the SIM functions as a receptor for the SUMO moiety of SUMOylated proteins during the downstream effects of SUMOylation. In this way, the SUMO:SIM-mediated protein–protein interactions can be quickly controlled through SUMO conjugation and deconjugation, and can thereby rapidly regulate cellular functions in response to extracellular stimulation (Song et al., 2004). Structural and biochemical studies of the SUMO:SIM interaction have shown that the core consensus sequence of a SIM is sufficiently short to design peptidomimetics to inhibit the SUMO:SIM interaction (Song et al., 2005).
In this study, we show that inhibition of SUMO: SIM-mediated protein–protein interactions by a SIM peptide significantly inhibits DNA repair and increases cancer cell sensitivity to radiation as well as to Dox. Analysis of the various DNA repair pathways showed that inhibition of the SUMO-dependent protein–protein interactions significantly inhibits NHEJ. In addition, mono-SUMO, instead of SUMO-chain, modifications are likely involved in NHEJ. This study is the first to show that SUMO:SIM-mediated protein–protein interactions are important in NHEJ. In addition, this study is proof-of-principle that SIM mimetics can be developed as sensitizers to enhance radiation and chemotherapy.
Results
The SIM-containing peptides recognize SUMOylated proteins in cells
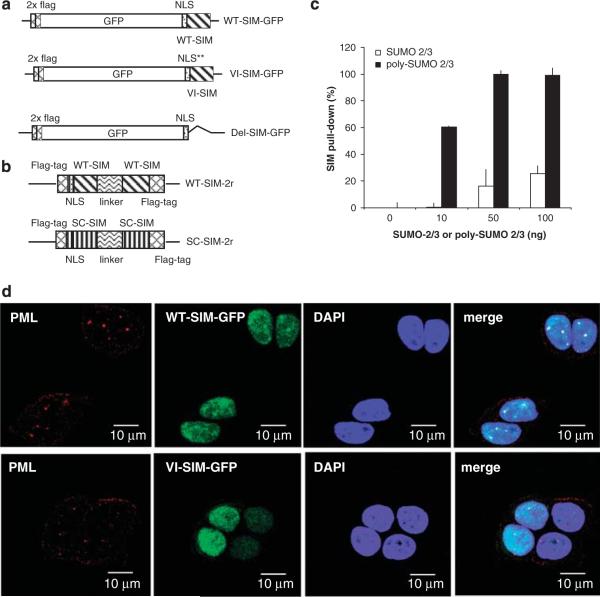
A SIM peptide should compete with the SIM sites in receptor proteins for SUMOylated targets, and thus inhibit SUMO-dependent protein–protein interactions. For our studies, we chose the SIM sequence (KVDVIDLTIE) from the protein PIASx, which binds to SUMO-1, SUMO-2 or SUMO-3 with a Kd of approximately 4–6 μM (Song et al., 2005). We designed two approaches to deliver a SIM peptide into cells. In one approach, we synthesized cellular transduction peptides that fused the SIM to the HIV-1 TAT nuclear localization signal (GYGRKKRRQRRRG), which leads to cellular uptake and nuclear localization of these peptides (Krebber and Silver, 2000). A control peptide was also synthesized that contained the scrambled (SC) sequence of the SIM fused to the same HIV-1 TAT nuclear localization signal. In a second approach, to circumvent the lack of peptide stability, we expressed the SIM peptide and its controls in cells as fusion proteins using green fluorescent protein (GFP) (Figure 1a). The SV40 nuclear localization sequence was used for nuclear targeting of the peptide and for separating GFP from the SIM to eliminate any potential hindrance of SUMO binding by the large GFP molecule. The two control plasmids included one with two mutations, V4A/I5A, in the SIM to eliminate SUMO binding (referred to as VI-SIM-GFP in subsequent discussions), and one in which the SIM was deleted (referred to as SIM-Del in subsequent discussions). The FLAG tag was included in all plasmids.
Figure 1.
The design of the SIM peptides and their interaction with SUMOylated proteins in cells. (a) A schematic representation of the cDNA fragments inserted into pcDNA3.1 for production of WT (WT-SIM-GFP), SUMO binding-deficient (VI-SIM-GFP) and a deletion mutant (Del-SIM-GFP) of SIM. The peptides are fused with two FLAG-tag repeats, GFP and a nuclear localization sequence. (b) A schematic representation of the construction of the double-SIM peptide (WT-SIM-2r) and SC -SIM control (SC-SIM-2r). The SIM or SC-SIM sequences are separated by the linker (RSPSPPVETSISSTN) and fused with a FLAG tag and a nuclear localization signal. (c) Preferential binding of the double-SIM peptide to poly-SUMO-2/3 chains. GST-fusion WT-SIM-2r was used to pull down poly-SUMO-2/3 (■) or mono-SUMO2/3 (□) at the indicated concentration. The measurement was normalized as 100% (poly-SUMO2/3 at 50 and 100 ng/ml) and control (0 ng/ml) defined as 0%. (d) Localization of WT-SIM-GFP and VI-SIM-GFP (green) in MCF7 cells. PML (red) was visualized by immunocytochemistry and nuclei (blue) were stained by DAPI. DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; GST, glutathione-S-transferase; PML, SC, scrambled; SIM, SUMO-interaction motif; SUMO, small ubiquitin-like modifiers; WT, wild type.
It is known that SUMO-2 or SUMO-3 carry internal SUMOylation sites for forming polymers (Tatham et al., 2001). This type of polymers is recognized by SUMO-targeted ubiquitin ligases that are important in DNA damage response (Sun et al., 2007; Prudden et al., 2007; Xie et al., 2007; Uzunova et al., 2007). To target poly-SUMO-chain-mediated protein–protein interactions more effectively, a construct containing two SIMs separated by the sequence between the SIMs from the protein Rfp1 was designed (Sun et al., 2007; Prudden et al., 2007; Figure 1b). To verify that the double-SIM peptide binds to the known type of poly-SUMO chains with higher affinity than to mono-SUMO, we synthesized such poly-SUMO-2/3 chains in vitro and performed binding assays. In this experiment, Escherichia coli-expressed GST-fusion double-SIM peptide was used to pull down the poly-SUMO-2/3 chains or mono-SUMO2/3 (Figure 1c). The double-SIM peptide showed higher affinity for the poly-SUMO chains. However, we do not rule out the possibility that unknown types of poly-SUMO chains cannot be blocked by the double-SIM peptide. In addition, a control was designed that contained two copies of the SC sequence of the SIM separated by the same linker (Figure 1b).
Localization of the single SIM and control fusion proteins was evaluated by fluorescence microscopy (Figure 1d). Wild-type (WT) SIM specifically localized to nuclear foci, as indicated by the formation of focused green puncta, but the control (VI-SIM-GFP) did not. Most of the WT SIM puncta colocalized with the PML nuclear bodies (red puncta, stained with an anti-PML antibody). This suggested that the expressed SIM bound to SUMO-modified proteins, because many SUMO-modified proteins, including the PML protein, are localized in the PML nuclear bodies. The SIM peptide did not disrupt the apparent morphology of the PML bodies, despite the fact that formation of PML bodies strictly depends on the SIM sequence of the PML protein and SUMO modification of PML (Muller et al., 1998). A similar observation was made with cells transfected with the double-SIM construct (data not shown).
The SIM peptide sensitizes cancer cells to genotoxic stress
We tested whether the SIM peptide affects DNA damage response, because SUMO modifications are known to be involved in these processes (Kerscher et al., 2006). To test the effect of the SIM peptide in DNA damage response, the HIV-TAT-SIM or the corresponding control peptide (20 μM) was pre-transduced into the breast cancer cell line MCF-7 for 2 h. The cells were then treated with increasing concentrations of the chemotherapeutic drug Dox, which induces DNA DSBs (Minotti et al., 2004). After 48 h of Dox treatment, cell viability was monitored by MTS assay. The SIM peptide, but not the control, increased the sensitivity of cancer cells to Dox treatment (Supplementary Figure S1A). In another experiment, we also tested the transiently expressed SIM and control peptides in either breast cancer MCF-7 cells or prostate cancer PC3 cells. Cells expressing the WT SIM were more sensitive to Dox than the controls (Supplementary Figure S1B and C).
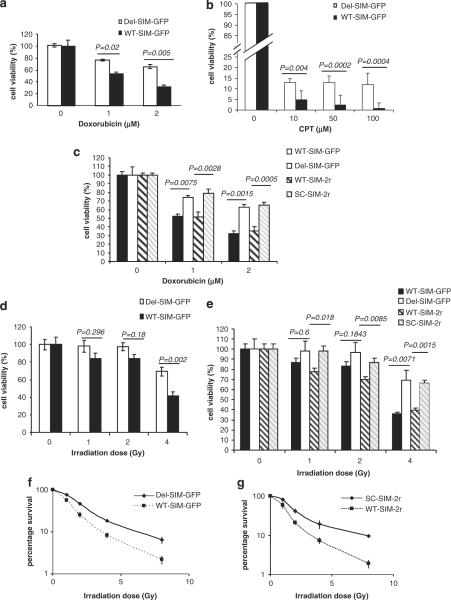
To further investigate the mechanism of the SIM peptides in DNA damage response, we established stably transformed MCF-7 cell lines that expressed either WT SIM-GFP (Figure 1a) or the double-SIM peptides (Figure 1b) by using pcDNA-based stable transformation and G418 selection. In addition, we established controls with MCF-7 cell lines that stably expressed the control plasmids: Del-SIM-GFP, VI-SIM-GFP (Figure 1a) or the double-repeats of SC SIM sequence (Figure 1b). Successful establishment cell lines showing stable SIM expression provided further evidence that neither the single-SIM nor the double-SIM peptides are toxic to cells at unstressed stage (same viability without Dox or radiation). The stably expressing WT and control SIM cell lines were then treated with increasing concentrations of the chemotherapeutic drug Dox. After 48 h of treatment, cell viability was monitored by MTS assay. Similar to experiments using membrane transduction peptides or transient transfection, cells stably expressing the WT SIM were more sensitive to Dox relative to the controls (Figure 2a). The SIM peptide also effectively sensitized camptothecin (CPT) treatment (Figure 2b). Unlike Dox, CPT inhibits topoisomerase-II that produces covalent DNA damage. However, the SIM peptide does not sensitize ICRF-193 treatment (Supplementary Figure S1D). This is consistent with the literature that ICRF-193 inhibits topoisomerase-II without generating covalent DNA adducts. In addition, the double-SIM construct did not induce more significant sensitivity to Dox than the WT-SIM-GFP construct (Figure 2c). These results suggest that the effect of the SIM peptide in sensitizing the cancer cell lines is due to its effect in DNA damage response.
Figure 2.
The SIM peptide sensitizes MCF-7 cells to the chemotherapeutic drugs Dox and CPT, and to radiation. (a, b) Viability of MCF-7 cells that stably expressed WT-SIM-GFP and Del-SIM-GFP upon Dox (a) and CPT (b) treatment. The cells were treated with the indicated Dox and CPT concentrations and cell viability, measured by MTS assay, was detected 48 h after treatment. (c) Comparison of the effect of a single-SIM and double-SIM constructs on MCF-7 cell sensitivity to Dox. Cell viability was measured by MTS assay. (d) Cell viability of MCF-7 cell lines stably expressing WT-SIM-GFP and Del-SIM-GFP upon irradiation at the indicated dosages. Cell viability, measured by MTS assay, was detected 48 h after radiation. (e) Comparison of the effect of a single-SIM and double-SIM constructs on MCF-7 cell sensitivity to radiation. Cell viability was measured by MTS assay. Clonogenic assays of cells stably expressing WT-SIM-GFP or the deletion control, Del-SIM-GFP (f), and WT-SIM-2r or the control, SC-SIM-2r (g), after irradiation at the indicated doses. Cell colonies were visualized by crystal violet and counted on day 14 after irradiation. The error bars represent the standard error of the mean of triplicate experiments. P-values are indicated. CPT, camptothecin; Dox, doxorubicin; GFP, green fluorescent protein; SC, scrambled; SIM, SUMO-interaction motif; WT, wild type.
To determine whether SUMO-mediated protein–protein interactions are involved in cellular response to another form of DNA damage, radiation, MCF-7 cells that stably express WT or control peptide were γ-irradiated at increasing dosages (1–4 Gy). Cell viability was measured 48 h after irradiation. We found that WT SIM significantly enhanced the sensitivity of MCF-7 cells to irradiation (Figure 2d) and the double-SIM construct induced similar sensitivity to irradiation (Figure 2e). Next, we used a clonogenic assay (Franken et al., 2006) to determine the long-term cell survival rates after DNA damage caused by radiation (1–8 Gy). Cell colonies were visualized by crystal violet staining and counted 14 days after irradiation. Both single-SIM and double-SIM peptides caused significant sensitivity to radiation (Figures 2f and g), and the double-SIM peptide did not induce significantly greater sensitivity to radiation than the single-SIM peptide. To determine the effect of the SIM peptide on normal cells, we performed normal human cell controls in parallel using the mammary epithelial cell line MCF-10A, which is a relevant control for the breast cancer cell line MCF-7. The SIM peptides do not sensitize MCF-10A to chemotherapy and radiation (Supplementary Figure S2A and B).
The SIM peptide inhibits DNA repair
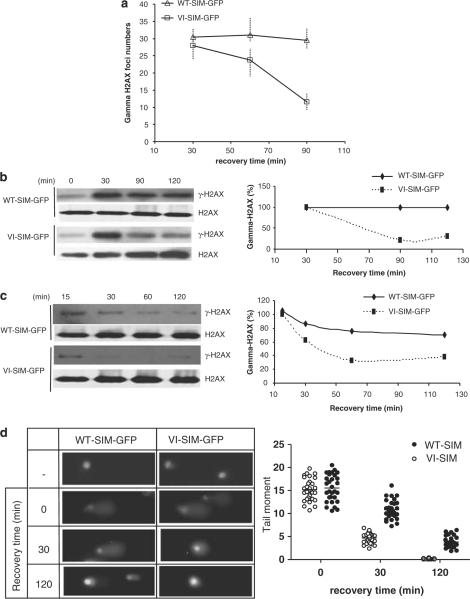
To better understand the mechanism by which the SIM peptide sensitizes cancer cells to radiation and chemotherapeutic drugs, we investigated whether it inhibits DNA repair. One of the markers of DNA DSBs in eukaryotes is serine-139 phosphorylation of the histone variant H2AX (γ-H2AX) (Rogakou et al., 1998), which recruits a number of proteins to the damaged sites, such as MDC1, ATM, BRCA1, the p53-binding protein 53BP1, the MRN complex (Mre–Nbs1–Rad50) and DNA repair enzymes (Stucki and Jackson, 2006). These proteins form large complexes (γ-H2AX foci) at the sites of DNA damage to coordinate DNA repair and transduce signals to prevent cell-cycle progression. These complexes can be visualized by confocal microscopy, and the number of γ-H2AX foci correlates with the number of DSB sites (Rogakou et al., 1999). After 4-Gy irradiation, we quantified the number of γ-H2AX foci by confocal microscopy and γ-H2AX levels by western blotting. MCF-7 cells that expressed either the SIM (WT-SIM-GFP) or the control (VI-SIM-GFP) showed similar levels of γ-H2AX and numbers of γ-H2AX foci at 30 min after radiation (Figures 3a and b). After 1.5–2 h, as repair progressed the γ-H2AX level and the number of foci decreased significantly in cells expressing the control peptides, indicating that the DNA damage had been repaired. However, the γ-H2AX level and the number of γ-H2AX foci remained high in cells expressing the WT SIM peptide, suggesting delayed DNA repair (Figures 3a and b). These data suggested that the SIM peptide inhibited DNA repair after treatment with ionizing radiation.
Figure 3.
The SIM peptide inhibits DNA repair. (a) γ-H2AX foci formation after irradiation. The graph shows the number of γ-H2AX foci in cells expressing WT-SIM-GFP or VI-SIM-GFP after irradiation (4 Gy) and a 30- to 90-min recovery period. The γ-H2AX foci numbers represent those counted from an average of 40 cells. (b, c) SIM peptide delays γ-H2AX de-phosphorylation in MCF-7 cells after γ-irradiation (4 Gy) (b) or 2 μM Dox treatment (c). γ-H2AX levels were measured at the indicated time points after treatment. γ-H2AX was detected and normalized with H2AX by western blotting and quantified. The western blot images are shown on the left and the quantifications are shown on the right. (d) Comet assays were used to measure the amount of unrepaired DNA damage in cells expressing WT-SIM-GFP and VI-SIM-GFP after γ-radiation (4 Gy) and recovery for 0, 30 or 120 min. Non-irradiated cells were used as controls (−). Representative images are shown on the left. Tail moments (% DNA in the tail × tail length), measured using Cometscore (TriTek), are shown on the left. Every spot represents a single cell, and 30 comet images were measured for each treatment. Dox, doxorubicin; GFP, green fluorescent protein; SIM, SUMO-interaction motif; WT, wild type.
We also examined γ-H2AX levels after treatment with Dox. MCF-7 cells expressing either WT-SIM-GFP or VI-SIM-GFP were treated with Dox and western blotting was used to measure the levels of γ-H2AX at various time points (15–120 min) after Dox treatment. γ-H2AX levels decreased as recovery time increased in cells expressing the control peptide, but remained high in cells expressing the WT SIM peptide (Figure 3c). This result, again, indicates that the SIM peptide delays DNA repair induced by Dox.
To further confirm the inhibitory effect of the SIM peptide on DNA repair, we used the comet assay to measure the amount of DNA damage in cells expressing WT-SIM-GFP and VI-SIM-GFP after 4 Gy γ-radiation and recovery for 30 or 120 min. In the comet assay, damaged DNA fragments migrate out of the cell nucleus as a streak similar to the tail of a comet, and the quantified tail moments are directly proportional to the amount of DNA damage. Immediately after irradiation, cells expressing either the SIM peptide or the control peptide produced comet tails of similar lengths and intensities (Figures 3d and e). However, after 30 and 120 min, the tail moments were significantly larger in cells expressing the SIM peptide than in cells expressing the control (Figures 3d and e). These data were consistent with the γH2AX data described above, indicating that repair of the damaged DNA was delayed by expression of the SIM peptide.
The SIM peptide suppresses the NHEJ DNA repair pathway
Because radiation, Dox and CPT all induce DNA DSBs, we investigated how the SIM peptide affects the DSB repair pathways. Vertebrate cells repair DSBs by two main pathways: NHEJ (Pastwa and Blasiak, 2003) and homologous recombination (HR) (Pastink et al., 2001). The HR pathways are further separated into single-strand annealing and gene conversion. The various pathways are controlled by different protein factors (Sancar et al., 2004; Sonoda et al., 2006).
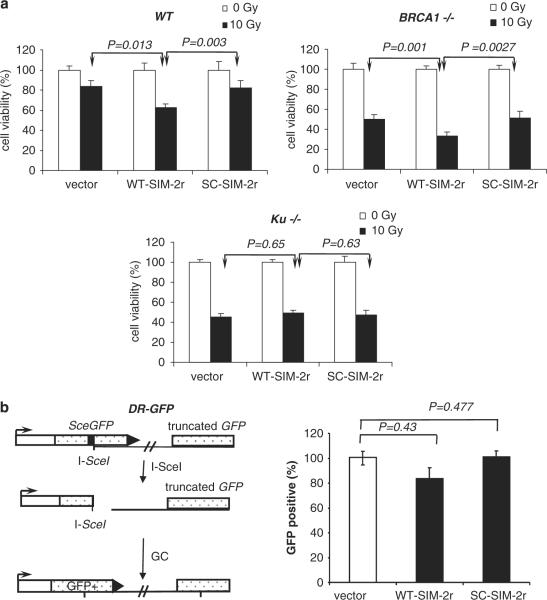
Mutant cell lines deficient in either the NHEJ or the HR pathways were used to investigate the role of SUMOylation in the DSB repair pathways. Because the Ku70/80 heterodimer is essential for NHEJ, we used a Ku70-knockout mouse embryonic stem (ES) cell line (Ku70−/−) to address NHEJ and HR. We measured the cell viability (using MTS assay) of WT and Ku70−/− cells after irradiation. WT SIM significantly reduced cell viability in the parental mouse ES cell line but not in the Ku70-knockout line (Figure 4a). Because DSB repair should occur primarily through the HR pathways in the Ku70−/− cell line, these data indicate that the HR pathways are not significantly affected by the SIM peptide, and thus, the NHEJ pathway is likely affected by the SIM peptide. BRCA1-deficient (BRCA1−/−) mouse ES cell lines, which are deficient in HR, were used to address the role of SUMOylation in NHEJ. Because the HR DNA repair pathway is inhibited, the NHEJ DNA repair pathway would be primarily responsible for DNA repair in BRCA1-deficient cells. If SIM inhibits NHEJ, the SIM-expressing BRCA1−/− cells should show enhanced sensitivity to DNA damage. As expected, the BRCA1−/− cells expressing WT SIM showed lower cell viability than cells expressing the control 72 h after irradiation. These results suggest that the SIM peptide preferentially inhibits the Ku-dependent NHEJ DNA repair pathway. The sensitization effect of the SIM peptide was not dependent on the cell-cycle stage. Cells synchronized in G1 or M-phase were similarly sensitized by the SIM peptide (Supplementary Figure S3). This observation is consistent with the role of SIM:SUMO interaction in NHEJ.
Figure 4.
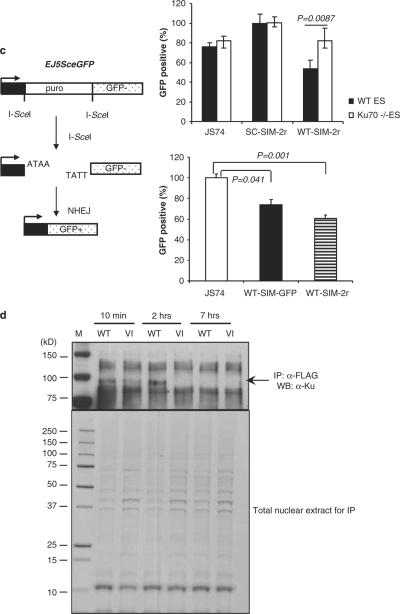
The SIM peptide inhibits NHEJ. (a) The SIM peptide sensitizes WT and BRCA1-deficient (BRCA1−/−) mouse ES cell lines, but not a Ku-deficient (Ku70−/−) cell line, to radiation. Cells were transfected with WT-SIM-2r, SC-SIM-2r or a vector cDNA, and irradiated (10 Gy) or not irradiated. Cell viability was detected 72 h after treatment and normalized to cells transfected with an empty expression vector. (b, c) Schematic representations of fluorescence-based assays for measuring HR or NHEJ DSB repair. (b) After DSBs are generated by I-SceI in cells carrying the DR-GFP reporter, HR uses the downstream GFP sequence (truncated GFP) as a template, which restores the coding sequence of functional GFP. The panel on the right shows the results of repair of DR-GFP in GFP-positive cells from mouse ES cells co-transfected with an I-Sce I expression vector and one of the following: WT-SIM-2R, SC-SIM-2r or expression vector (JS74). (c) EJ5-GFP contains a promoter that is separated from a GFP coding region by a puro gene that is separated by two I-SceI sites. Once DSBs are generated by I-SceI and the puro gene is excised by NHEJ repair, the promoter is joined to the rest of the expression region, leading to restoration of functional GFP. The graph on the upper right panel shows the frequency of repair of EJ5-GFP (resulting in GFP-positive cells) in WT and Ku70−/− ES cells co-transfected with an I-SceI expression vector and one of the following: WT-SIM-2r, SC-SIM-2r or vector control. P-values are indicated. The graph on the lower right panel shows the comparison of the effect of a single-SIM and double-SIM constructs on DNA DSB repair by NHEJ using a GFP-based chromosomal reporter in WT ES cells. (d) The WT-SIM-GFP (WT) or the VI-SIM-GFP (VI) mutant control peptide was immunoprecipitated by an anti-FLAG antibody, followed by western blotting with an anti-Ku70 antibody. A band corresponding to the molecular weight of SUMOylated Ku70 was repeatedly identified. The lower panel shows 10% of the input for immunoprecipitation, stained with Coomassie blue. DSB, double-strand break; ES, embryonic stem; GFP, green fluorescent protein; HR, homologous recombination; NHEJ, non-homologous end joining; SC, scrambled; SIM, SUMO-interaction motif; SUMO, small ubiquitin-like modifiers; WT, wild type.
To further confirm the effect of the SIM peptide in NHEJ, we measured the repair of endonuclease I-SceI generated DSBs using two fluorescence-based assays in vivo (Weinstock and Jasin, 2006; Weinstock et al., 2006; Bennardo et al., 2008). Briefly, to detect the gene conversion pathway of HR, we used the DR-GFP reporter. This reporter contained an upstream GFP gene repeat (SceGFP) that was non-functional because of insertion of a site for a rare-cutting endonuclease, I-SceI (Figure 4b). Thus, DSBs were generated in the middle of the SceGFP coding region by transfecting cells with an endonuclease I-SceI expression vector. The gene conversion repair pathway of HR uses the downstream GFP sequence (truncated GFP) as a template, which restores the coding sequence to generate a functional GFP gene, and cells that are repaired by gene conversion can be detected by assessing green fluorescence using flow cytometry. Cells expressing the SIM peptide showed only slight reduction of GFP production relative to cells expressing the control plasmids (Figure 4b). Furthermore, the double-SIM peptide did not show any additional effect than the single-SIM peptide (data not shown). These data suggest that the SIM peptide does not significantly inhibit the HR pathway, which is consistent with data on DNA damage sensitivity from the mutant cell lines (Figure 4a).
Next, we measured DSB repair by NHEJ using another GFP-based chromosomal reporter (Bennardo et al., 2008) (EJ5SceGFP) in mouse ES cells (Figure 4c). This reporter contains a promoter that is separated from a GFP coding cassette by a puro gene that is flanked by two I-SceI sites that are in the same orientation (Figure 4c). Once the puro gene is excised by NHEJ repair of the two I-SceI-induced DSBs, the promoter is joined to the rest of the expression cassette, leading to restoration of the GFP gene. Upon co-transfection of the I-SceI expression plasmid, the frequency of GFP-positive cells was significantly decreased in the cells expressing the WT SIM but not in cells expressing the SC control peptide, indicating that the WT SIM inhibits NHEJ (Figure 4c, upper right panel). Moreover, the single-SIM peptide was as inhibitory as the double-SIM peptides (Figure 4c, lower right panel), indicating that a multi-SUMO-chain-mediated protein–protein interaction may not be specifically involved in NHEJ. As control, the same assay was also performed in Ku70-knockout mouse ES cells, and the SIM and the control peptide had similar effect on the repair. Again, these data are consistent with all data described above, suggesting that the SIM peptide significantly inhibits the Ku-dependent NHEJ pathway.
To identify the target of the SIM peptide, the SIM peptide was immunoprecipitated by an anti-FLAG antibody. The immunoprecipitates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane and immunoblotted using various antibodies against proteins that are known to be important in DNA DSB repair, including Rad52, PCNA, Ku70, Ku80 or Mre11. In these immunoprecipitation experiments, only the anti-Ku70 antibody could consistently detect a band that was preferentially pulled down by the SIM relative to the control, and the band was visible up to 2 h after radiation, but not detectable at 7 h after radiation (Figure 4d). This band corresponds to a molecular weight of approximately 90 kDa, which is consistent with the size of Ku70 modified by a single SUMO protein. in vitro, recombinant Ku70 protein was promiscuously modified by SUMO at multiple sites (data not shown) (Yurchenko et al., 2008), and thus it was difficult to knock out the SUMOylation sites in Ku70. Although it was found previously that XRCC4 could be modified by SUMO (Yurchenko et al., 2006), the SIM peptide could not pull down SUMOylated XRCC4 or DNA-PKcs.
Discussion
This is the first study to uncover an important role of SUMO:SIM-mediated protein–protein interactions in the major DSB repair pathway, NHEJ. Two NHEJ factors, XRCC4 and Ku70, have previously been reported to be SUMOylated, although these previous studies did not elucidate whether SUMOylation of these NHEJ factors has a direct role in NHEJ. The SIM peptide could not pull down SUMOylated XRCC4, consistent with the previous finding that SUMOylation of XRCC4 was required for its nuclear localization but was removed in the nucleus (Yurchenko et al., 2006). Immunoprecipitation experiments indicated that SUMOylated Ku70 may be the target of the SIM peptide to inhibit SUMO:SIM-mediated protein–protein interactions involved in NHEJ. Overexpression of SUMO can stabilize the cellular levels of Ku70, but this effect does not appear to be due to SUMO modifications of Ku70, although inhomogeneous, multi-SUMO-modified Ku70 was observed upon SUMO overexpression (Yurchenko et al., 2008). The SIM peptide does not significantly inhibit SUMOylation in vitro or in vivo (data not shown), because it does not interfere with the binding of SUMO to the enzymes that catalyze the modifications. The mechanism by which the SIM peptide inhibits NHEJ remains to be elucidated. The SIM peptide does not appear to affect the localization of Ku to DNA damage sites, as determined by experiments using laser microbeam irradiation to induce DNA damage in live cells (Supplementary Figure S4A and B). In addition, the SIM peptide does not affect DNA-PKcs phosphorylation at serine-2056 after exposure to ionizing radiation (data not shown). The SIM peptide may inhibit NHEJ by affecting the normal dynamics of loading and uploading of proteins required for repair. Recent reports have shown that Ku70 is not only required for recognition of DSB ends, but also directly recruits other factors involved in DNA repair by NHEJ (Yano and Chen, 2008; Yano et al., 2008). Some of these protein–protein interactions may be SUMO:SIM-mediated and triggered by SUMO modifications. It is also possible that SUMO:SIM interactions are needed for removal of the Ku70/Ku80 heterodimer from DNA, as it was shown previously that the Ku proteins do not bind rigidly to DNA ends, but instead there is constant dynamic exchange between DNA-bound Ku proteins and free, unbound Ku proteins (Mari et al., 2006). As the SIM peptide does not significantly inhibit HR, our results are consistent with the previous finding of other roles for SUMOylation in HR, such as antagonizing other post-translational modifications (Sacher et al., 2006) or altering enzymatic functions (Hardeland et al., 2002). The lack of a notable effect on HR also suggests that the SIM peptides are likely to directly interfere with DNA repair, instead of having a general effect on cell proliferation or cell death.
The similar effects of the single-SIM and double-SIM peptides suggest that poly-SUMO-chain-mediated SUMO:SIM interactions are not directly involved in NHEJ. The double-SIM construct used in this study was designed on the basis of the SUMO-recognition region of the protein rfp1 (Sun et al., 2007; Prudden et al., 2007), which carries SIMs separated by the same linker sequence as in the double-SIM construct here. The SIM used in this study binds to all three SUMO paralogues with similar affinities (Kd of 4–6 μM) (Song et al., 2004). The double-SIM construct should bind to poly-SUMO-2 or poly-SUMO-3 chains with 104–106 higher affinity than to single SUMO on the basis of thermodynamic principles, whereas the single-SIM peptide does not discriminate single or poly-SUMO modifications. This is supported by the pull-down results (Figure 1c). The single-SIM and double-SIM peptides have similar effect on cellular sensitivity to genotoxic stress in MCF-7 cells, and similar inhibitory effect on NHEJ in mouse ES cells. This finding suggests that poly-SUMOylated proteins are not directly involved in NHEJ. This is consistent with pull down of Ku70 that is not modified by heterogeneous poly-SUMO chains. Another implication of the result is that SUMO-1, instead of SUMO-2 and SUMO-3, is involved in NHEJ. SUMO-2 and SUMO-3 readily form poly-SUMO chains, but SUMO-1 does not. Thus SUMO-1, instead of SUMO-2 or SUMO-3, is more likely to be important in NHEJ.
This study has shown that the SIM peptide significantly increases genotoxic stress in cell lines deficient in HR but not in cell lines deficient in NHEJ. It is important to note that many tumor-associated mutations, such as ATM, BRCA1 and BRCA2, and chk2 and p53, have been found to be clustered in the HR pathway. Therefore, inhibiting NHEJ in human tumors should be a highly useful strategy to combat cancer by enhancing the effect of radiation or DSB-inducing chemotherapy. This may be of particular importance to breast cancer therapy, because a significant percentage of hereditary breast cancers carry BRCA1 or BRCA2 mutations and thus are deficient in HR. Therefore, SIM mimetics are promising for developing sensitizers for treatment of BRCA-deficient breast cancers by DNA-damaging chemotherapeutic drugs and radiation. This study serves as proof-of-principle of targeting SUMO-dependent functions in the development of novel therapeutics, as well as in uncovering the role of SUMO modifications in various cellular functions.
Materials and methods
For Materials and methods, see Supplementary information.
Acknowledgements
This work is funded by NIH Grants R01GM074748 and R01GM086171 to Y Chen, RO1CA120954 to JM Stark, R37CA050519-20 to DJ Chen and R01DE14183 to DK Ann. Y-J Li is a recipient of the NIH National Research Service Award (F32CA134180).
Footnotes
Conflict of interest The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Carson JP, Zhang N, Frampton GM, Gerry NP, Lenburg ME, Christman MF. Pharmacogenomic identification of targets for adjuvant therapy with the topoisomerase poison camptothecin. Cancer Res. 2004;64:2096–2104. doi: 10.1158/0008-5472.can-03-2029. [DOI] [PubMed] [Google Scholar]
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Hardeland U, Steinacher R, Jiricny J, Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Krebber H, Silver PA. Directing proteins to nucleus by fusion to nuclear localization signal tags. Methods Enzymol. 2000;327:283–296. doi: 10.1016/s0076-6879(00)27284-4. [DOI] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim Pol. 2003;50:891–908. [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shayeghi M, Doe CL, Tavassoli M, Watts FZ. Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res. 1997;25:1162–1169. doi: 10.1093/nar/25.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting C, Naismith JH, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;12:12. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Jasin M. Alternative pathways for the repair of RAG-induced DNA breaks. Mol Cell Biol. 2006;26:131–139. doi: 10.1128/MCB.26.1.131-139.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, Hochstrasser M. The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem. 2007;282:34176–34184. doi: 10.1074/jbc.M706025200. [DOI] [PubMed] [Google Scholar]
- Yano K, Chen DJ. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle. 2008;7:1321–1325. doi: 10.4161/cc.7.10.5898. [DOI] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, et al. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Gama V, Matsuyama S, Sadofsky MJ. Ku70 is stabilized by increased cellular SUMO. Biochem Biophys Res Commun. 2008;366:263–268. doi: 10.1016/j.bbrc.2007.11.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Sadofsky MJ. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol. 2006;26:1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;29:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]