Abstract
Despite significant progress in the last decade, questions still remain about the complete structural, dynamic, and thermodynamic effect of the cis-syn cyclobutane pyrimidine dimer lesion (hereafter called the thymine dimer) on double-stranded genomic DNA. We examined a 19-mer oligodeoxynucleotide duplex containing a thymine dimer lesion using several small, base-selective reactive chemical probes. These molecules probe whether the presence of the dimer causes the base pairs to be more accessible to the solution, either globally or adjacent to the dimer. Though all of the probes confirm that the overall structure of the dimer-containing duplex is conserved compared to the undamaged parent duplex, reactions with both diethylpyrocarbonate and Rh(bpy)2chrysi3+ indicate that the duplex is locally destabilized near the lesion. Reactions with potassium permanganate and DEPC hint that the dimer-containing duplex may also be globally more accessible to the solution through a subtle shift in the dsDNA ⬄ ssDNA equilibrium. To begin to distinguish between kinetic and thermodynamic effects, we determined the helix-melting thermodynamic parameters for the dimer-containing and undamaged parent duplexes by microcalorimetry and UV-melting. The presence of the thymine dimer causes this DNA duplex to be slightly less stable enthalpically but slightly less unstable entropically at 298K, causing the overall free energy of duplex melting to remain unchanged by the dimer lesion within the error of the experiment. Here we consider these results in the context of what has been learned about the thymine dimer lesion from NMR, X-ray crystallographic, and molecular biological methods.
Exposure of DNA to ultraviolet light leads to the formation of various kinds of DNA damage, including the cis-syn cyclobutane pyrimidine dimer (1). This lesion is formed via a light-promoted [2+2] cycloaddition reaction between two adjacent pyrimidine bases, often two thymines, leading them to become covalently fused by a cyclobutane ring between their respective 5 and 6 positions. The cis-syn thymine cyclobutane dimer lesion, hereafter called the thymine dimer, has traditionally been considered to be one of the more ‘bulky and destabilizing’ lesions for several reasons: it involves two nucleotides locked in a rigid, nonstandard shape; it causes anomalous migration in gels and facilitates cyclization by bending DNA; it blocks replicative DNA polymerases and RNA polymerases; and it is repaired by transcription-coupled repair and nucleotide excision repair (NER) in eukaryotes(2-8). However, more recent studies hint that the effect of the thymine dimer lesion on the double-stranded duplex may be surprisingly subtle.
The structure of the thymine dimer lesion in DNA oligonucleotide duplexes has been studied both by nuclear magnetic resonance (NMR)(9-11) and X-ray crystallographic methods(12) and compared to normal B-form DNA duplexes. In the crystal structure, the dimer-containing dodecamer is somewhat distorted, but most of the distortions are localized to the immediate vicinity of the dimer with the rest of the DNA having near-normal B-form structure (12). Despite their unusual locked configuration and loss of aromaticity, the dimer thymines are buried within a right-handed helix, stacked with their neighbors, and paired with their complementary adenines in a manner reasonably similar to normal thymines. However, the duplex is subtly strained to accommodate the constrained thymine dinucleotide: the phosphate backbone is pinched, both grooves are widened, the base pairing between the 5′ thymine and its adenine pair is significantly weakened, and the base pairs on the 5′ side of the lesion have unusual tilt and twist angles as compared to canonical B-form DNA. These changes cause the DNA duplex to be bent by ∼30° toward the major groove and unwound by about 9° in the vicinity of the lesion.
The idea that the thymine dimer lesion is relatively non-disruptive to the overall DNA helix is supported by a variety of other mechanistic studies. The thermodynamic stability of duplexes containing a thymine dimer lesion have been studied by monitoring DNA melting by NMR and UV absorbance spectroscopy (9, 10, 13). In these studies the dimer reproducibly diminished the melting temperature of short oligonucleotide duplexes, but had only a small destabilizing effect on the free energy of duplex formation. Furthermore, the nucleotide excision repair recognition factors RPA, XPA, and XPC have been shown to have very little affinity for thymine dimer lesions in double-stranded DNA, hinting that duplex DNA containing a thymine dimer lesion appears normal to cellular repair machineries (7, 14, 15). Studies of DNA-mediated electron-transfer through thymine dimer duplexes also support the idea that the thymine dimer lesion is not strongly disruptive to the surrounding duplex (16, 17). Oxidation of guanine bases from a distance is sensitive to the integrity of the base stack between the oxidant and the guanine, but the presence of an intervening thymine dimer does not prevent such oxidation by disrupting the base pair stack. Indeed the thymine dimer can itself be oxidized, further demonstrating that it is relatively well-stacked with its neighboring bases and well-paired with its complementary adenines.
Conversely, these studies do not preclude the idea that the thymine dimer lesion may make the DNA more mobile or dynamic in its vicinity, and in fact, the results from NMR studies provide some hints that the thymine dimer lesion makes the DNA duplex more dynamic. The presence of sharp peaks in the NMR spectra for the imino protons of the dimer thymines and flanking bases would indicate that the complementary DNA strands form stable hydrogen bonds throughout the duplex. Interestingly Kemmink et al. noticed broadening of these peaks at low temperatures ascribed to an ‘asymmetical melting behavior’ adjacent to the dimer (11). In the latest full solution structure, only three of the twelve exchangeable imino protons can be observed in the NMR spectrum at 25° C (well below the duplex melting temperature of 53°) because the base pairs exchange with water (9). This solution structure is generally similar to the crystal structure with some small differences in local conformation, but it predicts a much smaller bending angle along the helical axis (9). Though this difference in predicted bending angle may reflect differences in the two techniques, the authors' hypothesis that the dimer may create a flexible “hinge” in the DNA is provocative (12).
These observations have led us to rethink the question of how much thymine dimer lesions resemble normal DNA, and how they are distinct. In this work, we examine the cis-syn thymine dimer lesion within a 19-mer oligonucleotide duplex using small, reactive organic and organometallic probes that are highly selective for particular sites on the DNA bases. These probes can reveal small structural or dynamic changes in base stacking and solution accessibility caused by distortions such as the thymine dimer lesion. We then compare the differences in reactivity we measure to calorimetric and spectroscopic measurements of DNA melting thermodynamics, to disentangle the lesion's kinetic and thermodynamic effects on the duplex.
Materials and Methods
Preparation of DNA Strands
DNA oligonucleotides were commercially prepared by phosphoramidite synthesis (Integrated DNA Technologies), and were further purified by reversed phase HPLC on a Zorbax C18 column (Agilent Technologies). Oligonucleotides containing a thymine dimer were initially prepared photochemically as described previously: approximately 200 μM oligonucleotide, containing only one adjacent pair of pyrimidines, was irradiated in vacuo for ∼3 hrs in a Rayonet photochemical reactor in the presence of 25 mM acetophenone (17-19). Oligonucleotides containing the cis-syn thymine dimer were separated from unmodified oligonucleotide and other products by reversed phase HPLC on a Zorbax C18 column. In a mixed 25 mM ammonium acetate buffer-acetonitrile gradient at 30°C, the cis-syn thymine dimer strand elutes from the column approximately 2 minutes before the other products. The structure of the dimer strand was confirmed by ESI-MS (University of Massachusetts), cycloreversal assay, DNA sequencing, exonuclease assay, and reactivity with potassium permanganate. DNA was lyophilized and resuspended in buffer. However, this method did not yield enough thymine dimer strand for the calorimetry experiments, so dimer-containing DNA, prepared via solid-phase synthesis with a thymine dimer phosphoramidite, was also obtained commercially (Midland Certified Reagent Company, TX). This DNA was also HPLC-purified and the mass confirmed by ESI-MS. The results of the gel experiments were the same regardless of the source of the dimer-containing DNA.
Concentration of the DNA strands was then measured spectrophotometrically at 260 nm using the following molar extinction coefficients (ε260nm): 190,000 M-1 cm-1; strand 1TT: 173,000 M-1 cm-1; strand 2: 176,500 M-1 cm-1. DNA duplexes were annealed by mixing equimolar amounts of the complementary strands in buffer, heating to 90°C on a heat block, and cooling gradually to room temperature over ∼2 hrs.
Preparation of Radiolabeled DNA Duplexes
DNA duplexes were annealed at 8 μM duplex concentration with trace amounts of 32P-labeled DNA in one of the following buffers. DMS experiments: 10 mM sodium cacodylate, 2 mM MgCl2, 0.2 mM EDTA pH 7.6. KMnO4, DEPC, and Rh(chrysi)3+ experiments: 50 mM sodium cacodylate, 2 mM EDTA pH 7.6. Samples containing radioactively-labeled strand 1 or 1-TT (“1*” or “1TT*”) were annealed in a background of 8 μM unlabeled strands 1 and 2. Samples containing radioactively-labeled strand 2 (“2*”) were annealed in a background of 8 μM unlabeled strand 1 or 1-TT (as applicable) and 8 μM strand 2.
Probing DNA Duplexes with DMS, KMnO4, and DEPC
These reactions are variations on standard DNA sequencing reactions (20, 21). DMS was added to the DNA at 0.5% v/v and the samples were incubated on ice, at room temperature, or at 37°C for 1 to 5 minutes. Permanganate was added to 1.5% w/v and the samples were incubated on ice, at room temperature, or at 37°C for 1 to 5 minutes. DEPC was added to a concentration of 10% v/v and samples were incubated on ice, room temperature, or 37°C for 5 to 30 minutes. In all three cases, the reaction was stopped by the addition of DMS Stop Solution (1.5 M sodium acetate, 1M β-mercaptoethanol, 100 μg/mL tRNA) and ethanol precipitation. To reveal which bases had been modified, the DNA was treated with 10% aqueous piperidine for 30 minutes at 90°C and then dried in vacuo. All samples were resuspended in denaturing running dye and run in 18% denaturing polyacrylamide gels according to standard techniques (22). Gels were digitized by phosphorimagery on a Molecular Dynamics Storm 820 system (Amersham Biosciences). Band intensities were measured and analyzed in ImageQuant to decide whether differences seen by eye on the gels were indeed quantitatively distinguishable. Multiple gels were run in all cases to confirm that trends in reactivity were reproducible.
Probing DNA Duplexes with Rh(bpy)2(chrysi)3+
Samples contained 8 μM DNA duplex and varying concentrations of Rh(bpy)2(chrysi)3+ between 1 and 10 μM as detailed in the figure legends Each sample was irradiated on a near-UV light source (OAI, San Jose, CA) at room temperature for up to three hours, with aliquots removed at various time points. The aliquots were then ethanol precipitated, resuspended in denaturing running dye without piperidine treatment, and run on an 18% denaturing polyacrylamide gel.
UV Melting
For all thermodynamic experiments, DNA duplexes were annealed at 0.76 to 40 μM concentration in 100 mM sodium phosphate buffer (pH 7.4) in a heat block as described above. Samples were degassed before measurement to prevent formation of bubbles.
For UV spectrophotometric experiments, DNA melting was monitored at 260 nm using a Cary 50 UV-visible spectrophotometer with a temperature-controlled cell (Varian) and quartz cells of 0.1, 0.2, 0.5, and 1.0 cm path lengths. Absorbance measurements were made every 0.1 minutes while the cell temperature was ramped upward between 20°C and 90°C at 1°C/min. The Tm was measured as the inflection point on the melting curve, determined by the Cary software as the first derivative. Three independent sets of measurements at five concentrations were made for each duplex.
The concentrations and measured Tm values for each sample were then used to determine the ΔH and ΔS using the van't Hoff equation:
| (1) |
Ctot is the total concentration of DNA strands, or twice the duplex concentration. A plot of 1/Tm versus ln Ctot is a straight line whose slope and intercept were used to determine the enthalpy and entropy changes associated with duplex formation. A line was fit to each set of five points in Excel to give three lines, from which we determined three independent values for ΔH and ΔS. These values were then used with the Gibbs free energy expression ΔG = ΔH - TΔS to determine ΔG at 298K. Values and errors given for ΔH, ΔS, and ΔG represent the averages and standard deviations of the three sets of measurements.
Differential Scanning Calorimetry
Calorimetry experiments were performed in a differential scanning calorimeter (Microcal, Northampton, MA) with a 0.58 mL sample cell compartment. DNA samples contained 40 μM duplex in 100 mM sodium phosphate buffer (pH 7.4). Excess heat capacity (ΔCp) was measured as a function of temperature. Between 3 and 8 initial scans were performed with buffer to establish a baseline and confirm the reproducibility of the measurements before DNA sample was added. For each scan, the temperature was ramped upward from 10°C to 90°C and back to 10°C at 1°C/min; multiple scans were performed to confirm the reproducibility of the measurements. Tm's were determined from the peak of the Cp vs. T curves using Origin 7.0 software (Microcal). ΔH and ΔS were determined by measuring the integrated area under Cp vs. T and Cp/T vs. T curves, respectively (23, 24). ΔG was then calculated at 298K using the Gibbs' free energy expression. Values and errors given for ΔH, ΔS, and ΔG represent the averages and standard deviations of duplicate integrations from three sets of samples for each duplex.
Results
The 19-mer DNA oligonucleotide duplexes used in these studies are shown in Table 1. Duplexes 1&2 (the “parent duplex”) and 1TT&2 (the “dimer duplex”) are identical except for the presence of the central thymine dimer. Strand 1 contains no other adjacent pyrimidines to prevent alternative cyclobutane pyrimidine dimers from forming during photoirradiation.
Table 1.
Sequence of the 19 bp DNA oligonucleotide duplexes.
| Parent Duplex 1&2 | |
| strand 1 | 5′ G1 C2 A3 G4 T5 A6 C7 G8 T9 T10 A11G12 T13 G14 A15C16 A17C18 G19 3′ |
| strand 2 | 3′ C38G37T36C35A34T33G32C31A30A29T28C27 A26 C25 T24 G23T22 G21C20 5′ |
| Dimer Duplex 1TT&2 | |
| strand 1TT | 5′ G1 C2 A3 G4 T5 A6 C7 G8 T9□T10 A11G12 T13 G14 A15C16 A17C18G19 3′ |
| strand 2 | 3′ C38G37T36C35A34T33G32C31A30 A29T28C27 A26 C25 T24 G23T22 G21C20 5′ |
location of the cyclobutane ring between T9 and T10, forming the cis-syn thymine dimer.
Probing Guanines with Dimethyl Sulfate
We first probed both duplexes with dimethyl sulfate (DMS). DMS reacts with the N7 of guanines in the major groove, methylating them with facility in both single-stranded and double-stranded DNA. These methylated guanine products are then piperidine labile and can be cleaved and visualized on a gel (20, 21). Guanines throughout both strands of both parent and dimer duplexes are methylated by DMS (Figure. 1a, b), indicating that the N7 positions on all guanines are accessible to the solution. The dimer duplex is equally reactive to the parent duplex, showing no preferential increase in accessibility due to perturbation by the thymine dimer lesion. Not only is the absolute amount of methylation the same between the parent and dimer duplexes, but the pattern of reactivity is the same, i.e. G8, G12, and G32 that are closest to the thymine dimer show no preferential reactivity. Also, the methylation on both strands, i.e. both 1 or 1TT (Figure 1A) and complementary strand 2 (Figure 1B), is indistinguishable between parent and dimer duplex. A comparable degree and pattern of methylation is seen at room temperature and 37°C.
Figure 1.
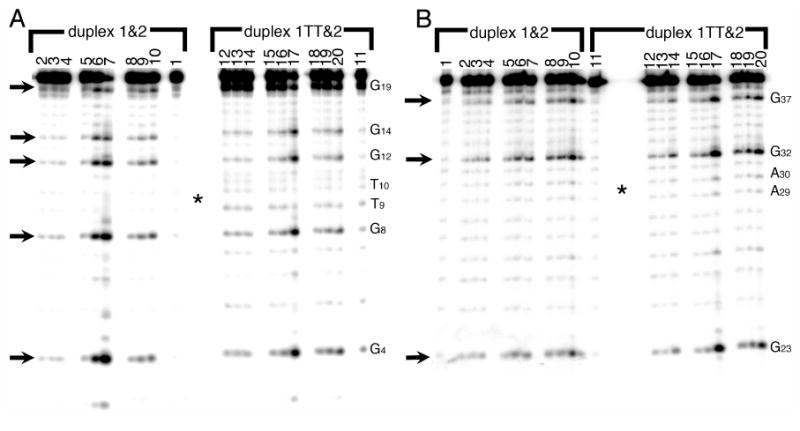
Reaction of the parent DNA duplex 1&2 (left) and dimer duplex 1TT&2 (right) with dimethyl sulfate (DMS), which methylates the N7 on guanines in the major groove (arrows). In panel A, strand 1 has the radioactive label on its 5′ end; in panel B, strand 2 bears the radioactive label. Lanes 1 and 11 : control (piperidine but no DMS). Lanes 2-4 and 12-14: reactions on ice for 1, 2, and 5 minutes. Lanes 5-7 and 15-17: reactions at room temperature for 1, 2, and 5 minutes. Lanes 8-10 and 18-20: reactions at 37°C for 1, 2, and 5 minutes. The dimer is located at T9-T10 (complementary to A29-A30) at the center of the gel, indicated by an asterisk. Note that the 5′ end of each labeled strand is at the bottom of the gel, and the 3′ end is at the top.
Probing Purines with Diethylpyrocarbonate
Diethylpyrocarbonate (DEPC) reacts predominantly with the N7 of adenine, and less so with the N7 of guanine (21, 25). The N7 position of purines lies in the major groove, but unlike methylation by DMS, reaction of the electrophilic DEPC is quite sensitive to the structure of the DNA to the point that DEPC is generally considered unreactive with duplex DNA. However, at a concentration of 10% and incubation times on the order of minutes to an hour, DEPC will react weakly with dsDNA and more strongly with ssDNA and open DNA structures (26-28).
When the parent and thymine dimer duplexes were probed with DEPC, the parent duplex is weakly alkylated by DEPC at all adenines and guanines on strand 1(Figure 2A). This reactivity is quite small even after three hours (data not shown), as we would expect for a double-stranded DNA duplex. In the thymine dimer duplex, the amount of reactivity at all adenines and guanines on strand 1TT is stronger than on strand 1 of the parent duplex at 37°C and 15-30 minute incubation times (Figure 2A). This increased reactivity on the dimer duplex indicates that the thymine dimer-containing DNA strand is unusually accessible to the solution, at least at 37°C. Purines throughout the duplex are reactive (black arrows); the adenine immediately flanking the dimer is not measurably more reactive than are purines in the rest of the duplex.
Figure 2.
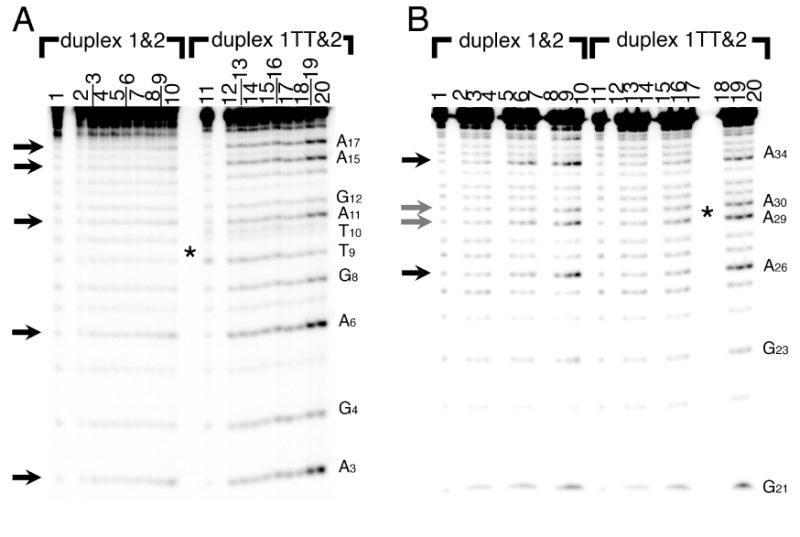
Reaction of the parent DNA duplex 1&2 (left) and dimer duplex 1TT&2 (right) with diethylpyrocarbonate (DEPC), which alkylates the N7 on adenines and guanines in the major groove (arrows). In panel A, strand 1 has the radioactive label on its 5′ end; in panel B, strand 2 bears the radioactive label. Lanes 1 and 11: control lanes (piperidine but no DEPC). Lanes 2-4: and 12-14: reactions on ice at 5, 15, and 30 minutes (note the offset numerical labels). Lanes 5-7 and 15-17: reactions at room temperature for 5, 15, and 30 minutes. Lanes 8-10 and 18-20: reactions at 37°C for 5, 15, and 30 minutes. The dimer is located at T9-T10 (complementary to A29-A30) at the center of the gel, indicated by an asterisk.
Similarly, little reactivity is seen on complementary strand 2 of both duplexes except at 37°C, but in contrast to what is seen on strand 1/1TT, the reactivity on complementary strand 2 of the duplex is quite similar between the parent and dimer duplexes (Figure 2B). The thymine dimer duplex 1TT&2 is only slightly more reactive than the parent duplex 1&2. The pattern of damage is subtly different between the two duplexes; the adenines A29 and A30 complementary to the thymine dimer are slightly more reactive than the corresponding adenines in the parent duplex (grey arrows).
Probing Thymines with Potassium Permanganate
The parent and thymine dimer duplexes were also probed with potassium permanganate (KMnO4), a common and water-stable but powerful organometallic oxidant. Permanganate oxidizes the 5,6 double bond on thymines to form thymine glycol, and also reacts to a lesser extent with the same bond on cytosines (21, 25). Though the thymine 5,6 bond is oriented along the major groove, the permanganate must access the π electrons on the face of the thymine base in order for the oxidation to occur. Thus for this bond to be accessible to the oxidant, the thymine base must be somewhat displaced from the DNA π stack at the center of the helix.
When probed with permanganate, the parent duplex 1&2 is rapidly oxidized at thymines T5, T9, and T10 on radioactively-labeled strand 1 (Figure 3A). Thymine T13 shows less reactivity, probably because of the strong cleavage at the other three thymines closer to the 5′ radioactive label. That this reactivity occurs even in the normal parent duplex, and at temperatures ranging from ice to 37°C, indicates that the thymine 5,6 bond in B-form DNA is always somewhat accessible to the permanganate. In the thymine dimer duplex 1TT&2 on the other hand, the reactivity at T5 and T13 is considerably stronger than in the parent duplex (Figure 3A, black arrows). Though comparison of T13 may be complicated by overcleavage in the parent strand, the increased reactivity at T5 in the dimer duplex is not. Note that the thymines at T9 and T10 (grey arrows) are unreactive because the target 5,6 double bonds were destroyed in forming the dimer; this cleavage is unrelated to any duplex destabilization (29).
Figure 3.
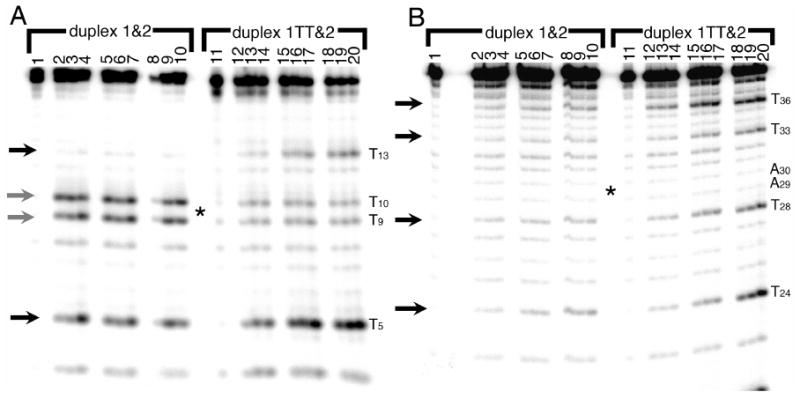
Reaction of the parent DNA duplex 1&2 (left) and dimer duplex 1TT&2 (right) with potassium permanganate (KMnO4), which oxidizes thymines at the 5 and 6 positions (arrows). In panel A, strand 1 has the radioactive label on its 5′ end; in panel B, strand 2 bears the radioactive label. Lanes 1 and 11 : control (piperidine but no KMnO4). Lanes 2-4 and 12-14: reactions on ice for 2, 4, and 6 minutes. Lanes 5-7 and 15-17: reactions at room temperature for 2, 4, and 6 minutes. Lanes 8-10 and 18-20: reactions at 37°C for 2, 4, and 6 minutes. The dimer is located at T9-T10 (complementary to A29-A30) at the center of the gel, indicated by an asterisk. Note that T22 is also reactive with the permanganate but cannot be seen on this gel, and that permanganate does not oxidize thymines involved in a thymine dimer.
All thymines (T22, T24, T28, T33, and T36) on the complementary strand 2 of the parent duplex 1&2 are weakly reactive (Figure 3B). Reactivity at all thymines on the complementary strand 2 of the dimer duplex is stronger than in the parent duplex at all temperatures but especially at 37°C, indicating that thymines in the dimer duplex are more accessible to solution than they are in the parent duplex. The thymine T28, one step away from the thymine dimer, is not selectively reactive; in fact, it is less reactive than either T24 or T36.
Probing the Duplex with Rh(bpy)2chrysi3+
The parent and dimer duplexes were also probed with Rh(bpy)2chrysi3+, an octahedral organometallic complex that has been shown to bind selectively at thermodynamically-destabilized mismatched sites in DNA (30-32). Upon irradiation with near-UV light (∼365 nm), it cleaves the DNA backbone at its binding site. In our duplexes, 5-10 μM Rh(bpy)2chrysi3+ preferentially cleaves the dimer duplex immediately 5′ to the thymine dimer lesion on strand 1TT at G8 and C7 at room temperature (Figure 4A, black arrows). To a lesser degree it also cleaves 3′ to the thymine dimer lesion at A11 and G12 on strand 1TT (grey arrows). Rh(bpy)2chrysi3+ cleaves only weakly and nonspecifically throughout the same strand of the parent duplex. In the complementary strand 2 of the dimer duplex, strong cleavage is seen at G32 (complementary to C7, one step away from the dimer) (Figure 4B, black arrow). Weaker cleavage is also observed at A29 complementary to the 5′ T of the dimer (Figure 4B, grey arrow). Though these sites are also cleaved in the parent duplex, they are cleaved to a lesser extent. Additional photocleavage is visible on the complementary strand 2 of the dimer duplex 3′ to the dimer, but not in the corresponding portion of the parent strand (black arrowheads). This cleavage pattern indicates that the rhodium complex binds at the thymine dimer site on either side of the dimer but prefers the side 5′ to the dimer (i.e. 5′ relative to strand 1TT). Though no cleavage is visible on the gel between the two dimer thymines, we cannot rule out that the complex binds and cleaves the backbone here as well since the dimerized thymines would hold the strand fragments together.
Figure 4.
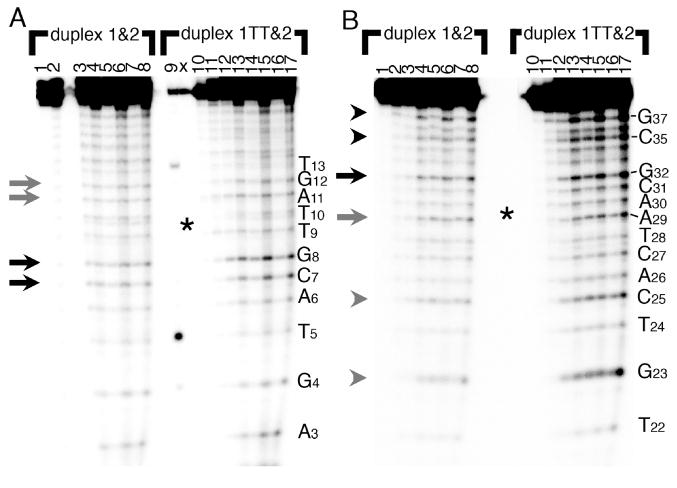
Reaction of the parent DNA duplex 1&2 (left) and dimer duplex 1TT&2 (right) with Rh(bpy)2chrysi3+, which binds to destabilized sites in dsDNA and cleaves at its binding site upon photoexcitation (arrows). In panel A, strand 1 has the radioactive label on its 5′ end; in panel B, strand 2 bears the radioactive label. Lanes 1 and 10: light control (no Rh(bpy)2chrysi3+). Lanes 2 and 11: dark control (no photoirradiation). Lane 9: KMnO4 lane (for sequencing purposes). Lanes 3-4 and 12-13: 1 μM Rh(bpy)2chrysi3+, 30 minutes and 120 minutes, respectively. Lanes 5-6 and 14-15: 5 μM Rh(bpy)2chrysi3+, 30 minutes and 120 minutes. Lanes 7-8 and 16-17: 10 μM Rh(bpy)2chrysi3+, 30 minutes and 120 minutes. The dimer is located at T9-T10 (complementary to A29-A30) at the center of the gel, indicated by an asterisk.
Thermodynamic Stability: UV Melting and Microcalorimetry
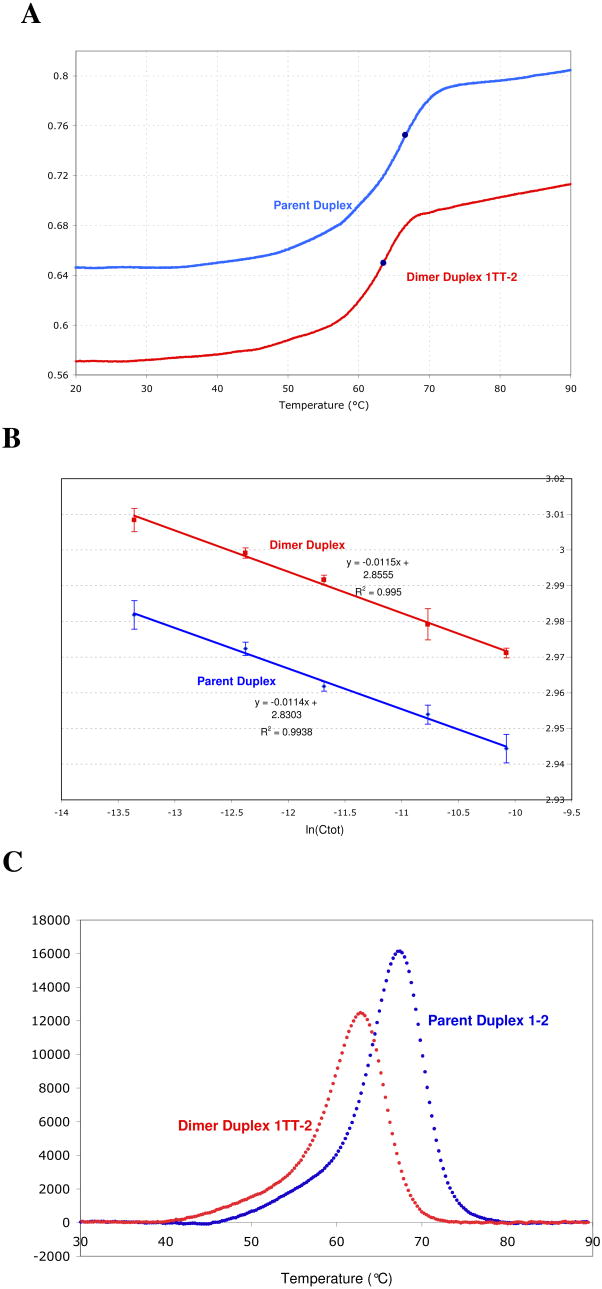
To complement the chemical probe studies we examined the dsDNA-ssDNA equilibrium for both the parent and dimer duplexes by UV melting. As a DNA duplex is heated and dissociates cooperatively to single strands, the absorbance at 260 nm increases sharply by ∼10% (Figure 5A). The melting temperature or Tm can be determined from the midpoint or inflection point of these sigmoidal melting curves. We found the Tm for parent and dimer duplexes at five duplex concentrations. We then plotted 1/Tm versus ln Ctot (Figure 5B), and used these data with the van't Hoff equation (equation 1) and the Gibbs equation to measure the enthalpy, entropy, and free energy changes associated with duplex formation. We also determined the thermodynamic parameters for the dsDNA-ssDNA equilibrium by differential scanning calorimetry (DSC), by which we measure the change in heat capacity (ΔCp) of a DNA sample as it is heated and the DNA is melted (Figure 5C). ΔH and ΔS were determined by measuring the integrated area under ΔCp vs. T and ΔCp/T vs. T curves, respectively (23).
Figure 5.
Stability of the DNA oligonucleotide duplex in the absence and presence of the thymine dimer lesion. Data for the parent duplex 1&2 are shown in blue, and for the dimer duplex 1TT&2 in red. In panel A, a representative UV melting curve at 260 nm is shown for both duplexes at a 21 μM duplex concentration. The dark blue dots indicate the inflection point on each curve, taken to be the Tm. Panel B is a van't Hoff plot showing the dependence of the melting temperature on DNA concentration for the parent and dimer duplexes. Three measurements were made for each point; error bars represent the standard deviation of the mean of each set. Equations for the best fit lines to the data are shown. Panel C shows representative DSC melting curves for the parent and dimer duplexes at 40 μM duplex concentration. The area under this curve corresponds to ΔH, and the peak of each curve is the Tm.
The thermodynamic parameters we determined by each technique are illustrated graphically in Figure 6, and exact values and errors are tabulated in Supplementary Table 1. As expected, in all cases DNA duplex formation from single strands is enthalpically favorable but entropically unfavorable at standard temperature, leading to an overall very small negative free energy.
Figure 6.
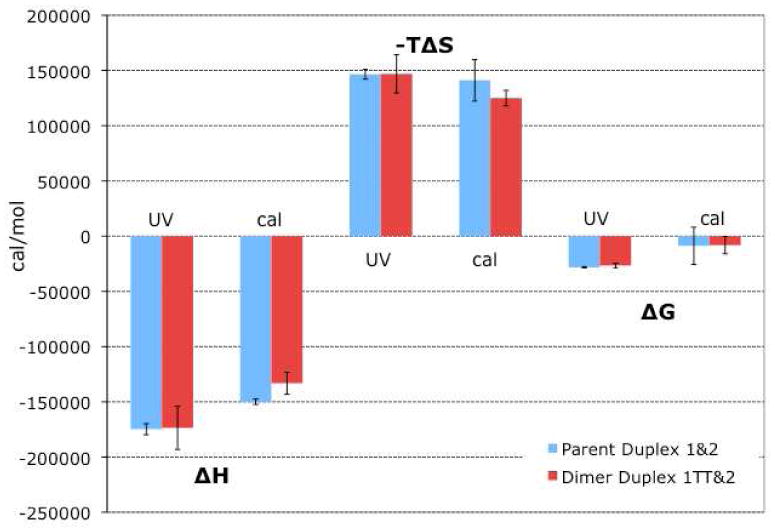
Effect of the thymine dimer lesion on the enthalpic and entropic contributions and the overall thermodynamic stability of DNA duplex formation. The source of the data is indicated as either UV melting (“UV”) or differential scanning calorimetry (“cal”). Values for the parent duplex 1&2 are shown in blue at the left of each pair; values for the dimer duplex 1TT&2 are shown in red at the right of each pair.
As determined by UV melting, the melting temperature of the dimer duplex is always approximately 3°C lower than the melting temperature of the parent strand at the same concentration. Contrary to what we might naively expect, this consistently lower melting temperature does not translate into large differences in ΔH, ΔS, and ΔG between the parent and dimer duplexes. In fact, the values determined by UV melting for ΔH, ΔS, and ΔG of dimer duplex formation are essentially the same as the values for the parent duplex within our error of measurement. A careful inspection of the plot of 1/Tm versus ln Ctot in figure 5B makes it clear that the slopes of both lines are quite similar, and though the y-intercepts are noticeably different, they are different only in the third significant digit. Thus the change in the free energy of duplex formation for the dimer duplex relative to the parent duplex is small and falls within the error of the measurement.
Though the overall trends in the thermodynamic parameters measured by DSC are not dissimilar, the differences between parent and thymine dimer duplex are more pronounced when explored by DSC. In this case we see that the thymine dimer duplex is enthalpically less stable than the parent but entropically less unstable (more disordered) than the parent. However, since ΔH and ΔS are of the same sign and thus effectively cancel each other out in the calculation of ΔG, the change in free energy of duplex formation for the dimer duplex relative to the parent duplex is again small and within the error of measurement.
Discussion
Examining the Structure of Lesion-Containing DNA with Small Reactive Probes
To examine sensitively the structure of the DNA around base lesions, we probed the DNA with small reactive molecules. Many molecules are available that have been used in the past to probe nucleic acid structure (25-28, 32-39), but they have not been exploited to examine the structure of DNA around base lesions. The ideal probes would not distort the DNA structure, dramatically change the solution pH or ionic strength, nor react preferentially with the lesion; they would ideally be stable, work in neutral buffered aqueous solution between 4 and 40°C, and generate piperidine-labile products that can be detected on a gel as strand breaks. Most importantly, our ideal chemical probes would subtly and selectively probe the base pairing and stacking, not react with the backbone. Thymine dimers and 8-oxo-guanine have been examined previously with hydroxyl radical probes (13, 40). Unfortunately, hydroxyl radicals react relatively non-specifically with the sugar-phosphate backbone on DNA, and thus, these reactions do not reveal anything about non-canonical base pairing, accessibility, opening, nor destabilization of the helical core. We determined that the reactions of the DNA duplexes with potassium permanganate (KMnO4), diethylpyrocarbonate (DEPC), and dimethylsulfate (DMS) best fit these qualifications.
The parent and thymine dimer duplexes are methylated and cleaved identically when probed with dimethyl sulfate followed by piperidine. DMS is small and relatively insensitive to the secondary structure of DNA, though its ability to methylate guanine can be blocked by the presence of another molecule such as a protein or triplex-forming strand in the major groove or by the formation of Hoogsteen base pairs. This similar reactivity reveals that, to a first approximation, the parent and dimer duplexes are structurally similar and only more sensitive probes of structure or dynamics can differentiate between them.
The dimer duplex is more reactive with KMnO4 and DEPC than is the parent duplex, indicating that the presence of the thymine dimer makes the DNA bases more accessible to the solution. To our surprise, this increased reactivity occurs all over the dimer duplex, not just immediately adjacent to the thymine dimer itself. We had originally hypothesized that a localized region of kinetic and/or thermodynamic destabilization might form in the immediate vicinity of the lesion as a “bubble” within a normal duplex, but only in the DEPC reactions on the adenines complementary to the thymine dimer is there a small hint of such preferential, localized reactivity. Instead, these chemical probes revealed a larger, duplex-wide accessibility to the solution, almost as if the entire duplex was flapping or peeling open.
Considering the implications of reactivity throughout the thymine dimer duplex, we first confirmed that the dimer duplex did not contain some reactive contaminant nor was inherently labile to the piperidine. Control reactions of dimer and parent duplexes with piperidine in the absence of organic probe (performed for each experiment and shown in the first lanes of each gel) showed almost no reactivity, so clearly the reactivity in the dimer duplex cannot be ascribed to inherent piperidine lability nor another reactive contaminant. The similarity of reactivity of both duplexes with DMS confirms these controls. A second possibility might be that the thymine dimer duplex was not properly annealed. However, the DNA melting experiments demonstrated that at the temperatures of the chemical probe experiments (ice, room temperature, and 37°C), both DNA duplexes are fully annealed (Figure 5A). Furthermore, an examination of the sequence provides no evidence for competition by a hairpin or other secondary structure.
Because both permanganate and DEPC are known to be more reactive with ssDNA than with dsDNA, another, more interesting possibility might be that there is a small amount of ssDNA in the duplex reaction mixtures. According to that hypothesis, under our conditions the probes do not react significantly with dsDNA but actually react with ssDNA. Excess single strands could occur in the solution from a dimer-induced shift in the dsDNA-ssDNA equilibrium. Even well below the melting temperature, some fraction of the strands must be single-stranded, and if the thymine dimer duplex is thermodynamically less stable than the parent, the fraction of single stranded DNA would be higher in the dimer-containing duplex solution than in the parent duplex solution. We explored this possibility through our thermodynamic experiments (vide infra).
Provocatively, we observed that throughout many repetitions of these experiments, the dimer duplex strand 1TT is always significantly more reactive with KMnO4 and DEPC than the parent strand 1, but the difference in reactivity on strand 2 between parent and dimer duplex is considerably smaller and more sensitive to experimental error. Notably, the radioactive strands 1* and 1TT* are annealed by slow cooling from 90°C a solution of 8 μM unlabeled strand 1 and 2. As a result, the dimer strand 1TT* must compete for complementary strand 2 with a high concentration of strand 1, which has a higher melting temperature. Such a mixture accentuates the subtle differences between the dimer duplex and parent duplex ssDNA-dsDNA equilibrium and increases likelihood that the dimer strand will be single-stranded in solution. Conversely, the radioactive strand 2* is annealed in a background of 8 μM unlabeled 1 and 2 or 1TT and 2 respectively, so in these experiments strand 2* must be annealed to a dimer-containing strand in the dimer duplex mix. We would predict that dimer strand 1TT* annealed in a background of dimer-containing unlabeled DNA would be less reactive with KMnO4 and DEPC, and indeed this is the case (data not shown). Similarly, we determined that the pattern of reactivity of single-stranded dimer strand 1TT* with KMnO4 is the same as the pattern of reactivity of the “double-stranded” dimer duplex 1TT*-2 with this reagent (data not shown). Thus we can conclude that the reactivity we observe between DEPC or KMnO4 and the dimer duplex is largely due to reaction between ssDNA and the chemical probes, and the reactivity of the dimer duplex is only slightly enhanced relative to the parent duplex when annealed without competition.
The temperature dependence of the reactions of the DNA duplexes with both KMnO4 and DMS supports the idea that these probes are selectively reporting a shift in the ssDNA-dsDNA equilibrium rather than changes in the structure of dsDNA. It is fundamental chemistry that the rate of the reaction with these probes is always higher at higher temperatures due to increased collisions between the two molecules in question. However, at lower temperatures where the duplex formation is thermodynamically quite favorable we had expected to see some localized reactivity around the dimer itself, albeit at longer incubation times. These duplexes are relatively long and in the UV melting and DSC experiments they show indications of melting via multiple transitions instead of a single cooperative opening event (Figure 5 and vide infra). Therefore it was reasonable to hypothesize that at low temperatures the duplex might stay annealed but form a small “bubble” around the dimer lesion, but using these probes we see little evidence that this is the case. As the temperature is increased, the pattern of reactivity shifts from little or no reactivity to duplex-wide reactivity with no conclusive evidence of stable intermediate forms.
To isolate the reactivity of single-stranded DNA from that of double-stranded DNA, we looked for a reactive probe that would mark destabilized or “opened” double-stranded DNA preferentially over single-stranded or stable duplex DNA. Chrysi complexes of rhodium have been shown to preferentially bind mismatched base pairs in duplex DNA due to their unusually large aromatic, heterocyclic chrysi ligand (31, 32). Though related rhodium complexes generally bind DNA by intercalation from the major grove, binding of Rh(bpy)2(chrysi)3+ to mismatched DNA occurs from the minor groove with extrusion of the destabilized base pair (30). We hypothesized that if the thymine dimer locally destabilizes DNA similarly to a mismatched base pair, Rh(bpy)2(chrysi)3+ should selectively bind and cleave at or immediately adjacent to the thymine dimer lesion. Moreover, the binding and cleavage should report selectively about the shape and thermodynamics of the DNA duplex (as opposed to the single strands) since rhodium intercalators generally bind double-stranded DNA more avidly than single-stranded DNA. The Rh(bpy)2chrysi3+ preferentially cleaves the dimer duplex immediately 5′ and 3′ to the thymine dimer itself, indicating that this complex binds at the thymine dimer lesion. Though it is not clear how exactly this complex binds to the thymine dimer duplex (i.e. whether by intercalation or insertion with extrusion of base pairs), this pattern of photocleavage indicates that some localized destabilization or “bubbling” occurs at the thymine dimer site in addition to the duplex-wide melting observed using the other chemical probes.
Thermodynamic Comparison of Parent and Thymine Dimer Duplexes
The thymine dimer duplexes have lower melting temperatures than the normal parent duplexes at all concentrations, as determined by both UV melting and differential scanning calorimetry. However, the data here reinforce the fact that it is important to not confuse this decrease in melting temperature with a loss in the standard free energy of duplex formation, since the two parameters are not necessarily correlated (41). The loss in free energy of duplex formation (ΔΔG) caused by the presence of the thymine dimer lesion is quite small (+0.6 to +1.6 kcal/mol). A small ΔΔG indicates that formation of a thymine dimer lesion is not destructive to duplex formation, and in fact that the effect of the thymine dimer is quite subtle. This subtle shift in free energy confirms published ΔΔG values between 1.4 kcal/mol and 2.0 kcal/mol measured for dimer-containing octamers, decamers, and dodecamers by UV melting and NMR melting (9).
Though the trends in thermodynamic values determined by UV melting and DSC experiments are similar, there is one noticeable difference: in the UV melting experiments the changes in ΔH and ΔS caused by the dimer lesion fall within the error of the measurements, whereas in the calorimetry experiments ΔH and ΔS change more significantly. These differences likely reflect an essential difference in model between the two kinds of experiments: fitting of the UV melting experiments relies on the van't Hoff model, which presumes a two-state equilibrium (ssDNA ⬄ dsDNA), but fitting of the DSC data on the other hand does not assume that the equilibrium is two-state. In fact, visual inspection of the UV melting curves reveals that the DNA melting process is almost certainly not two-state, since the curve does not have a smooth sigmoidal shape. In addition to the main DNA melting event around ∼65°C, there is a pre-melting transition for both the dimer and the parent duplexes that can be seen as a gradual increase in absorbance at lower temperatures (Figure 5A). At higher temperatures, the absorbance does not plateau as it should for fully-denatured single-stranded DNA, and is in fact still increasing when we reach the upper limit of the instrument (90°C). Therefore a van't Hoff analysis of the UV melting data would be expected to provide at best a rough approximation of the helix melting parameters due to the mismatch between the 2-state model and the real helix behavior.
The shape of individual UV melting curves can be also be used to independently determine ΔH (41), but the multiple transitions present in these data complicate such an analysis. Notably, this complex melting behavior is characteristic of not only the dimer strand but also of the parent strand; the shapes of the UV melting curves and DSC curves are similar for both in having significant “pre-melting” and “post-melting” transitions. Since an analysis of this complex behavior will most likely reveal as much or more about how long duplexes behave than it will reveal about the effect of a lesion on DNA, it will be presented elsewhere.
Analysis of the DSC melting curves confirms that the melting is not two-state. A van't Hoff two-state model was used initially to fit a symmetrical, Gaussian-shaped melting curve to the asymmetrical real data melting curve, but the two curves do not overlay (data in Figure 5C; model curve not shown). Because the shapes of these melting curves thus indicate that both 19-mer DNA duplexes open via one or various partially-melted intermediates instead of opening as a single cooperative unit, which is to be expected for a duplex of this length, ΔH and ΔS were determined by measuring the integrated area under Cp vs. T and Cp/T vs. T curves, respectively. This method does not assume that the DNA exists in only two states, double and single stranded, but can accommodate various intermediate forms. Model-free DSC results better capture the thermodynamic changes that occur during the transition from dsDNA to ssDNA by including the contributions to ΔCp made by these unspecified intermediates. We are currently working to deconvolute these DSC melting data and fit them to a more complex model that explicitly includes various intermediates.
The calorimetrically-determined ΔΔH value of +16.8 ± 10.2 kcal/mol predicts a modest disruption in base pairing and base stacking due to the thymine dimer, consistent with the crystal structure and NMR structures (9, 12). The calorimetrically-determined ΔΔS value of +54 ± 68 cal/mol•K corresponds with the dimer lesion causing an increase in the disorder of the duplex or a decrease in the order of the single strands. Given the complex role that ordered water can play in the entropic contributions to the folding of biomolecules, we should not make any strong conclusions about the effect of the lesion on the structure of the duplex. However, that the dimer causes an increase in the disorder of the duplex would be consistent with the structural and NMR studies. It is particularly interesting that the dimer-induced loss in enthalpy and increase in entropy almost entirely cancel each other out in their effect on the free energy of duplex formation. Were we to look only at ΔΔG, we might believe that the dimer had almost no effect on the thermodynamics of duplex formation; the small value of ΔΔG masks the more substantial enthalpic and entropic changes that are caused by the thymine dimer lesion.
The Effect of the Thymine Dimer Lesion on a 19-Base-Pair Oligonucleotide
Small chemical probes such as DMS, DEPC, and KMnO4 have been used successfully for decades to examine subtle static or dynamic changes in DNA (25-28, 32-39). Here they reveal that the thymine dimer lesion causes little detectable structural change to 19-mer DNA duplexes. No distortions in the thymine dimer duplex are observed using DMS, which reacts with DNA in the major groove and thus reports about the global similarity of normal and thymine dimer-containing duplexes. A slight increase in DEPC reactivity is seen at the AA immediately complementary to the dimer lesion, and mismatch-detecting Rh(bpy)2(chrysi)3+ shows some binding and cleavage at the dimer lesion site, but no selective reactivity is observed at the bases flanking the lesion. These data indicate that the destabilization around the dimer lesion is small and extremely localized to the dimer itself and its complementary bases. Dimer-induced changes detected using DEPC and KMnO4 are generally both small in magnitude and duplex-wide in scope; these small changes reflect a subtle shift in the ssDNA-dsDNA equilibrium. Our direct measurements of duplex thermodynamics demonstrate this small, dimer-induced shift in the free energy of duplex formation toward ssDNA, confirming earlier UV melting and NMR melting studies (9, 10, 13).
Conclusions
Despite conventional wisdom that the thymine dimer lesion is a “bulky and destabilizing” lesion, it appears that the DNA containing a thymine dimer lesion is surprisingly similar to native, undamaged DNA. Changes to the duplex free energy are extremely small, and consistent with both the NMR and crystal structures, base stacking and pairing near the lesion appear normal as probed directly with small reactive molecules. At the thymine dimer lesion itself, the duplex may be slightly or transiently melted, reflecting that perhaps the dimer can act as either a kink or hinge. Breslauer and colleagues have illustrated that other thermodynamically-destabilizing DNA lesions can have near-native structures, and unusual DNA lesion structures can have near-native thermodynamic parameters(42); in the case of the thymine dimer, both the lesion structure and thermodynamics are similar to native DNA.
Supplementary Material
Supplemental Table 1, “Thermodynamic Data for the ssDNA ⬄ dsDNA Equilibrium,” is available free of charge via the internet at http://www.pubs.acs.org.
Acknowledgments
We would like to thank Professor Jacqueline Barton for providing us with Rh(bpy)2chrysi3+.
Abbreviations
- DSC
differential scanning calorimetry
- DMS
dimethyl sulfate
- DEPC
diethylpyrocarbonate
- KMnO4
potassium permanganate
- Rh(bpy)2(chrysi)3+
bis(2,2′-bipyridine)(5,6-chrysenequinone diimine)rhodium(III)
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA
Footnotes
This work was supported by the Camille and Henry Dreyfus Foundation, the Clare Boothe Luce Foundation, the Radcliffe Institute for Advanced Study, and the National Institutes of Health/NIGMS (1R15GM083250-01).
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- 2.Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA. 2000;97:5681–5683. doi: 10.1073/pnas.120152397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 4.Smith CA, Baeten J, Taylor JS. The ability of a variety of polymerases to synthesize past site-specific cis-syn, trans-syn-II, (6-4), and Dewar photoproducts of thymidylyl-(3′-5′)-thymidine. J Biol Chem. 1998;273:21933–21940. doi: 10.1074/jbc.273.34.21933. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husain I, Griffith J, Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci USA. 1988;85:2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAteer K, Jing Y, Kao JY, Taylor JS, Kennedy MA. Solution-state structure of a DNA dodecamer duplex containing a cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA. J Mol Biol. 1998;282:1013–1032. doi: 10.1006/jmbi.1998.2062. [DOI] [PubMed] [Google Scholar]
- 10.Taylor JS, Garrett DS, Brockie IR, Svoboda DL, Telser J. 1H NMR assignment and melting temperature study of cis-syn and trans-syn thymine dimer containing duplexes of d(CGTATTATGC)-d(GCATAATACG) Biochemistry. 1990;29:8858–8866. doi: 10.1021/bi00489a049. [DOI] [PubMed] [Google Scholar]
- 11.Kemmink J, Boelens R, Koning T, van der Marel GA, van Boom JH, Kaptein R. 1H NMR study of the exchangeable protons of the duplex d(GCGT<>TGCG)*d(CGCAACGC) containing a thymine photodimer. Nucleic Acids Res. 1987;15:4645–4653. doi: 10.1093/nar/15.11.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H, Zang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc Natl Acad Sci USA. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barone F, Bonincontro A, Mazzei F, Minoprio A, Pedone F. Effect of thymine dimer introduction in a 21 base pair oligonucleotide. Photochem Photobiol. 1995;61:61–67. doi: 10.1111/j.1751-1097.1995.tb09243.x. [DOI] [PubMed] [Google Scholar]
- 14.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 16.Dandliker PJ, Holmlin RE, Barton JK. Oxidative thymine dimer repair in the DNA helix. Science. 1997;275:1465–1468. doi: 10.1126/science.275.5305.1465. [DOI] [PubMed] [Google Scholar]
- 17.Dandliker PJ, Núñez ME, Barton JK. Oxidative charge transfer to repair thymine dimers and damage guanine bases in DNA assemblies containing tethered metallointercalators. Biochemistry. 1998;37:6491–6502. doi: 10.1021/bi980041w. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee SK, Christensen RB, Lawrence CW, LeClerc JE. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee SK, Borden A, Christensen RB, LeClerc JE, Lawrence CW. SOS-dependent replication past a single trans-syn TT cyclobutane dimer gives a different mutation spectrum and increased error rate compared with replication past this lesion in uninduced cells. J Bacteriol. 1990;172:2105–2112. doi: 10.1128/jb.172.4.2105-2112.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth Enzymol. 1980;65:499–513. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 21.Ambrose BJB, Pless RC. DNA sequencing: chemical methods. Meth Enzymol. 1987;152:522–539. doi: 10.1016/0076-6879(87)52059-6. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- 23.Breslauer KJ, Freire E, Straume M. Calorimetry: a tool for DNA and ligand-DNA studies. Meth Enzymol. 1992;211:533–567. doi: 10.1016/0076-6879(92)11030-m. [DOI] [PubMed] [Google Scholar]
- 24.Poklar N, Pilch DS, Lippard SJ, Redding EA, Dunham SU, Breslauer KJ. Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability, and energetics of a 20-mer DNA duplex. Proc Natl Acad Sci. 1996;93:7606–7611. doi: 10.1073/pnas.93.15.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloomfield V, Crothers D, Tinoco I. Nucleic acids: structures, properties, and functions. University Science Books; Sausalito, California: 2000. [Google Scholar]
- 26.Bui CT, Rees K, Lambrinakos A, Bedir A, Cotton RGH. Site-selective reactions of imperfectly matched DNA with small chemical molecules: applications in mutation detection. Biooorg Chem. 2002;300:216–232. doi: 10.1016/s0045-2068(02)00019-6. [DOI] [PubMed] [Google Scholar]
- 27.Fox K, Grigg GW. Diethylpyrocarbonate and permanganate provide evidence for an unusual DNA conformation induced by binding of the antitumor antibiotics bleomycin and phleomycin. Nuc Acids Res. 1988;16:2063–2075. doi: 10.1093/nar/16.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohshima K, Kang S, Larson J, Wells RD. TTA*TAA triplet repeats in plasmids form a non-H bonded structure. J Biol Chem. 1996;271:16784–16791. doi: 10.1074/jbc.271.28.16784. [DOI] [PubMed] [Google Scholar]
- 29.Ramaiah D, Koch T, Orum H, Schuster GB. Detection of thymine [2+2] photodimer repair in DNA: selective reaction of KMnO4. Nuc Acids Res. 1998;26:3940–3943. doi: 10.1093/nar/26.17.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc Natl Acad Sci USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson BA, Alekseyev VY, Barton JK. A versatile mismatch recognition agent: specific cleavage of a plasmid DNA at a single mispair. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 32.Jackson BJ, Barton JK. Recognition of base mismatches in DNA by 5,6-chrysenequinone diimine complexes of rhodium(III): a proposed mechanism for preferential binding to destabilized regions of the double helix. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy J, Rich A. Detection of an unusual distortion in A-tract DNA using KMnO4: effect of temperature and distamycin on the altered conformation. Nuc Acids Res. 1991;19:3421–3429. doi: 10.1093/nar/19.12.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts E, Deeble VJ, Woods CG, Taylor GR. Potassium permanganate and tetraethylammonium chloride are a safe and effective substitute for osmium tetroxide in solid-phase fluorescent chemical cleavage of mismatch. Nuc Acids Res. 1997;25:3377–3378. doi: 10.1093/nar/25.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palacek E, Boublikova P, Jelen F, Krejcova A, Makaturova E, Nejedly K, Pecinka P, Vojtiskova M. Chemical probing of DNA polymorphic structure in vitro and in situ. In: Sarma RH, Sarma MH, editors. Structure and Methods. Adenine Press; 1990. pp. 237–253. [Google Scholar]
- 36.John DM, Weeks KM. Chemical interrogation of mismatches in DNA-DNA and DNA-RNA duplexes under nonstringent conditions by selective 2′-amine acylation. Biochemistry. 2002;41:6866–6874. doi: 10.1021/bi025611d. [DOI] [PubMed] [Google Scholar]
- 37.Tullius T. The use of chemical probes to analyze DNA and RNA structures. Curr Op Struct Biol. 1991;1:428–434. [Google Scholar]
- 38.Hanvey JC, Klysik J, Wells RD. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine*oligopyrimidine inserts in plasmids. J Biol Chem. 1988;263:7386–7396. [PubMed] [Google Scholar]
- 39.McCarthy J, Williams LD, Rich A. Chemical reactivity of potassium permanganate and diethylpyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990;29:6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- 40.Barone F, Cellai L, Giordano C, Matzei F, Pedone F. Gamma-ray footprinting and fluorescence polarization anisotropy of a 30-mer synthetic DNA fragment with one 2′-deoxy-7-hydro-8-oxoguanosine lesion. Eur Biophys J. 2000;28:621–628. doi: 10.1007/s002490050002. [DOI] [PubMed] [Google Scholar]
- 41.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry. 1992;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 42.Plum GE, Breslauer KJ. DNA lesions: a thermodynamic perspective. Ann NY Acad Sci. 1994;726:45–56. doi: 10.1111/j.1749-6632.1994.tb52796.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1, “Thermodynamic Data for the ssDNA ⬄ dsDNA Equilibrium,” is available free of charge via the internet at http://www.pubs.acs.org.