Abstract
γ-Aminobutyric acid (GABA)-ergic transmission is critical for normal cortical function and is likely abnormal in a variety of neuropsychiatric disorders. We tested the in vivo effects of variations in two genes implicated in GABA function on GABA concentrations in prefrontal cortex of living subjects: glutamic acid decarboxylase 1 (GAD1), which encodes GAD67, and catechol-o-methyltransferase (COMT), which regulates synaptic dopamine in the cortex. We studied six single nucleotide polymorphisms (SNPs) in GAD1 previously associated with risk for schizophrenia or cognitive dysfunction and the val158met polymorphism in COMT in 116 healthy volunteers using proton magnetic resonance spectroscopy. Two of the GAD1 SNPs (rs1978340 (p=0.005) and rs769390 (p=0.004)) showed effects on GABA levels as did COMT val158met (p=0.04). We then tested three SNPs in GAD1 (rs1978340, rs11542313, and rs769390) for interaction with COMT val158met based on previous clinical results. In this model, rs11542313 and COMT val158met showed significant main effects (p=0.001 and 0.003, respectively) and a trend toward a significant interaction (p=0.05). Interestingly, GAD1 risk alleles for schizophrenia were associated with higher GABA/Cre, and Val-Val homozygotes had high GABA/Cre levels when on a GAD1 risk genotype background (N=6). These results support the importance of genetic variation in GAD1 and COMT in regulating prefrontal cortical GABA function. The directionality of the effects, however, is inconsistent with earlier evidence of decreased GABA activity in schizophrenia.
Keywords: dopamine, healthy volunteers, genes, schizophrenia, prefrontal function, single nucleotide polymorphisms
INTRODUCTION
γ-Aminobutyric acid (GABA)-ergic function is critical for numerous aspects of cognition (Gao and Goldman-Rakic, 2003a; Goldman-Rakic, 1990; Menzies et al, 2007; Seamans and Yang, 2004), and its alterations have been implicated in a number of neuropsychiatric disorders, including epilepsy (Wong et al, 2003), schizophrenia (Lewis and Hashimoto, 2007; Lewis et al, 2005), and depression (Sanacora et al, 1999). Studies of GABA activity in vivo in humans, however, have been very limited and consequently characterizing the basic physiology of the system in vivo has been an elusive goal.
Genes that contribute to the metabolism of GABA and the activity of GABA neurons are critical in GABA function, as they affect neurodevelopment and abnormalities in their function are likely to predispose to neuropsychiatric disorders. One of the synthetic enzymes for GABA, glutamic acid decarboxylase 67 (GAD67, encoded by GAD1 on 2q31, a 46 kb gene consisting of 17 exons), is particularly relevant in this context because single nucleotide polymorphisms (SNPs) in this gene have been associated with GAD1 mRNA levels (Mellios et al, 2009; Straub et al, 2007), with risk for schizophrenia (Addington et al, 2005; Du et al, 2008; Straub et al, 2007; Zhao et al, 2007), with individual measures of neuroticism (Hettema et al, 2006) as well as with measures of prefrontal cortical function in normal subjects (Straub et al, 2007). Moreover, decreases in the expression of this gene have been found in several post-mortem studies of schizophrenia (eg, Hashimoto et al, 2008; Huang and Akbarian, 2007a; Straub et al, 2007, reviewed in Akbarian and Huang, 2006). GAD1 expression and related GABA levels have also been found to have a specific role in sculpting interneuron axon growth and synapse formation during development (Chattopadhyaya et al, 2007). Another gene with potential implications for GABA function and risk for schizophrenia is catechol-o-methyltransferase (COMT), an enzyme that regulates cortical DA levels (Gogos et al, 1998; Karoum et al, 1994) and likely GABA neuronal excitability (Seamans et al, 2001), perhaps particularly during development (Tseng et al, 2007), and would thus be expected to modulate GABA function.
In this study, we tested whether several SNPs in GAD1 previously associated with risk for schizophrenia alter GABA levels in the anterior cingulate cortex (ACC) measured in vivo. We further investigated the effects of the functional variant val158met in COMT. The study by Straub et al (2007) found evidence of statistical epistasis between GAD1 and COMT in increasing risk for schizophrenia. We thus tested whether this specific interaction would be recapitulated in terms of GABA levels in healthy individuals.
Given the neuropathology and genetic studies of schizophrenia mentioned above, we hypothesized that GABA levels would be reduced in carriers of GAD1 alleles that confer risk for schizophrenia and that there would be a significant interaction with COMT, where the combination of Val alleles (relatively disadvantageous to prefrontal functioning: Blasi et al, 2005; Egan et al, 2001; Tan et al, 2007; Winterer et al, 2006) and risk alleles in GAD1 would lead to further reductions in GABA. This is the first study that addresses the impact of genetic variation in any enzyme on in vivo GABA measurements.
MATERIALS AND METHODS
Participants
We recruited 116 adult healthy volunteers (59 men, 57 women) who had participated in the ‘CBDB sibling study of the genetics of schizophrenia' (NCT00001486). Subjects were all Caucasian (mean age=32.8±10.2 years, mean±SD, range 18–55). Exclusion criteria included any current or history of psychiatric (DSM-IV axis I or II by a modified Structured Clinical Interview) or medical illness affecting the brain, pregnancy, and current psychotropic medication use. All subjects had no first- or second-degree relatives with a psychotic disorder according to an assessment performed with the Family Interview for Genetic Studies (http://zork.wustl.edu/nimh/digs/figs/). Participants were informed of the purpose of the study and written consent (NIMH protocols 95-M-0150 and 91-M-0124) was obtained. This selected population was chosen to avoid potential concomitant effects of medications, illness duration, and phase of illness that are problematic in any patient-based research. Only three subjects were smokers.
Neuroimaging
Participants were scanned on three 3 T scanners (GE, Waukesha, Wisconsin), two of which were equipped with a 14m4 software platform (n=79 and 22), whereas one ran on LX software (n=15). A quadrature transmit–receive head coil was used on all scanners (IGC-Medical Advances, Milwaukee, WI). A T1-weighted 3D SPGR sequence (TR/TE, 9.2/3.3 ms, flip angle 17°, FOV 24 mm, matrix 256 × 192, 76 slices, 2 mm thick) was used for voxel placement and for image segmentation with SPM5 (http://www.fil.ion.ucl.ac.uk/spm). This segmentation was used to obtain relative proportions of gray, white matter, and cerebrospinal fluid (CSF) present in the voxel by an automated program written in IDL (ITT Visual Information Solutions, Boulder, CO, USA) by ASB.
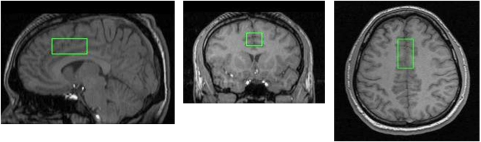
For magnetic resonance spectroscopy (MRS), a single voxel of interest (2 × 2 × 4.5 cm: 18 cm3) was placed in the medial prefrontal cortex (PFC) to maximize coverage of the gray matter in the dorsal ACC (Figure 1). We chose this location because of the ease of measurement (a large amount of gray relative to white matter can be investigated with excellent shimming despite the high magnetic field) and because this region has been shown to function abnormally in schizophrenia, with several approaches (eg, (Adams and David, 2007; Camchong et al, 2008; Carter et al, 2001; Deicken et al, 1997; Ende et al, 2000; Ford et al, 2004; Fornito et al, 2008; Ko et al, 2009; Manoach et al, 2007; Segall et al, 2009), including post-mortem studies of GAD67 mRNA transcript (Woo et al, 2004). Moreover, this region has been shown to be important in conflict and error detection and conflict monitoring (eg, Botvinick et al, 1999; Carter et al, 1999) and GABA circuitry is likely to be key to its proper functioning. It might have been preferable to study the dorsolateral prefrontal cortex (DLPFC), rather than the medial PFC, however several technical difficulties discouraged us from doing so. Measurement of the DLPFC with the same signal to noise resulted in much broader line widths in pilot studies and ended up including about 20% of gray matter in the voxel vs the current 75% for the ACC.
Figure 1.
Magnetic resonance spectroscopy (MRS) voxel position. Sagittal, coronal, and axial images showing the position of the anterior cingulate voxel of interest, placed in the medial prefrontal cortex while trying to maximize gray matter.
An interleaved PRESS-based J-editing method (Hasler et al, 2007; Sailasuta et al, 2001) was used for measurement of metabolites (TR/TE 1500/68 ms, NEX=2, 768 averages, 2048 acquisition points, sampling frequency=5000 Hz, 20 min scanning time). This method is perhaps more sensitive to the larger intracellular pool of GABA produced by GAD67 (Asada et al, 1997; Tian et al, 1999) as GAD2, which encodes for GAD65, likely produces a much smaller pool of GABA used for vesicular release (review in Soghomonian and Martin, 1998). A fully automated nonlinear fitting program written in IDL by JWvdV was used to calculate metabolite levels. The amplitudes of residual water, N-acetyl aspartate (NAA), choline (Cho), and creatine (Cre) were fitted in the unedited spectrum. Subsequently, the unedited spectrum was subtracted from the edited spectrum, resulting in a difference spectrum where the co-edited Glx (glutamate+glutamine) and GABA signals were separated from overlapping resonances. After subtraction of the water baseline, GABA and Glx were fitted automatically, using the linewidth and positions fit results from the fit of Cho, Cre, and NAA in the unedited spectrum. The entire process yielded estimates for absolute signal intensities and ratios of the metabolites to Cre and to water (see Figure 2 for an example of the output). The fitting procedure also rejected pairs of acquisitions where the differences in the absolute value of the residual water signals of a pair were more than 10%, indicating possible movement during the scan (van der Veen et al, 2007). Two further quality control procedures were put in place: (1) at the time of scanning, the voxel was shimmed to a linewidth for water below 10 Hz in all cases (the average linewidth being 7±0.96 Hz); (2) our expert physicist (JWvdV) reviewed all spectra masked to genotype to ensure adequate fitting and line shapes. A separate estimate of Cre was obtained by processing the unedited portion of the spectrum with LCmodel (Provencher, 2001), a commonly used technique to obtain absolute quantitative estimates (derived using water as a reference). Cre values were further corrected for the amount of CSF in the voxel by dividing them by the proportion of tissue in the voxel (percent gray+percent white matter).
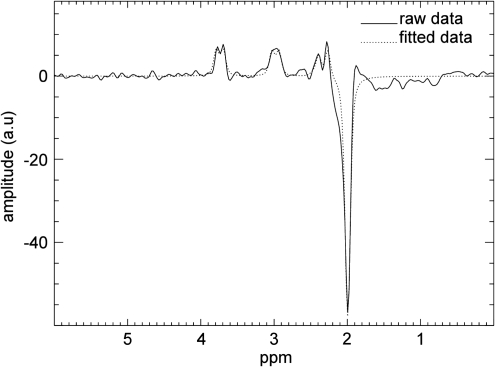
Figure 2.
Spectroscopy output. A typical difference spectrum is displayed, showing the GABA resonance at 3.0 p.p.m. and co-edited Glx at 3.8 p.p.m. The N-acetyl-aspartate (NAA) signal at 2 p.p.m. appears inverted due to the editing that suppresses frequencies around this area of the spectrum. The subtraction of the unedited from the edited spectrum will thus result in a negative peak for NAA. The dotted line represents the automated fit to the raw data (continuous line).
Genotyping
Ten SNPs were genotyped in GAD1 (rs10432420, rs7557793, rs1978340, rs872123, rs3762555, rs3749034, rs2241165, rs11542313 (formerly known as rs16823181), rs769390, and rs3791850) based on previous associations of these SNPs with risk for schizophrenia, GAD1 mRNA expression, gray matter loss, and/or cognitive performance (Addington et al, 2005; Straub et al, 2007; see Table 1), rs7557793, rs872123, rs3762555, and rs2241165 were in strong linkage disequilibrium (LD) with rs3749034 (all r2>0.8), so they were dropped from further analysis, leaving six SNPs to investigate further. rs3749034 was kept for analysis because previous evidence from our post-mortem sample indicated that this SNP was associated with GAD1 mRNA expression (Straub et al, 2007). Comparing single SNP and haplotype results with both clinical and cognitive phenotypes, we have found that when SNPs are in such strong LD (here all r2 values are >0.80) and the genotyping call rate is high, with our sample size, there is usually little advantage to including the highly redundant genotypes. Haplotype analysis would have added to the number of tests, and interpretation of results from rare haplotypes is difficult. Detailed information about the SNPs is presented in Table 1 and additional information regarding the LD structure is shown in Figure 3. Of the remaining SNPs, rs1978340, rs11542313, and rs769390 had been shown to increase risk for schizophrenia when the risk allele was present on a COMT Val homozygote background (Straub et al, 2007; Table 1). Thus, we explored specifically the potential interaction of these SNPs with COMT on GABA levels. COMT Val158Met (rs4680) was also genotyped. All genotyping was performed using the TaqMan allelic discrimination assay, as previously described (Straub et al, 2007). All genotypes were in Hardy–Weinberg equilibrium. Genotype reproducibility was assessed by comparing genotypes obtained from plates used for this study to plates genotyped for Straub et al (2007). Depending on the SNP in question, we found that somewhere between 153 and 236 individuals had duplicate genotypes. Discordance rate was below 2.3%, and discordant genotypes were removed before analysis.
Table 1. Location and Association with Function of the 10 Genotyped SNPs.
| SNPa | NCBI dbSNP rs no. | Coding strand allelesb | CBDB control MAF | Location | Chromosome 2 positionc | Intermarker distanced | Distance from M01 |
Evidence for |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Function (mRNA expression) | Clinical association | Association with cognition | Association with gray matter loss | Association with fMRI activation | ||||||||
| M01 | rs10432420 | G/A | 0.31 | 5′ Flanking | 171 485 819 | 0 | 0 | scz (Straub et al, 2007) | (Straub et al, 2007); (Addington et al, 2005) | |||
| M02 | rs7557793 | T/C | 0.27 | 5′ Flanking | 171 490 832 | 5013 | 5013 | (Straub et al, 2007) | (Addington et al, 2005) | (Straub et al, 2007) | ||
| M03 | rs1978340 | G/A | 0.29 | 5′ Flanking | 171 495 628 | 4796 | 9809 | scz (Straub et al, 2007); bpd (Lundorf et al, 2005); etoh dep (Kuo et al, 2009) | ||||
| M04 | rs872123 | T/C | 0.25 | 5′ Flanking | 171 496 673 | 1045 | 10854 | (Huang et al, 2007b) | bpd (Lundorf et al, 2005) | (Addington et al, 2005) | ||
| M07 | rs3762555 | G/C | 0.25 | 5′ Flanking | 171 497 902 | 317 | 12083 | (Straub et al, 2007) | ||||
| Start of transcript NM_000817: | 171 498 707 | 12888 | ||||||||||
| M09 | rs3749034 | G/A | 0.24 | Exon 1, 5′ UTR | 171 498 982 | 695 | 13163 | (Straub et al, 2007) | scz (Straub et al, 2007); cos (Addington et al, 2005); etoh dep (Loh el et al, 2006) | (Straub et al, 2007) | ||
| Start Codon (Met) of transcript NM_000817: | 171 500 609–11 | 14790-92 | ||||||||||
| M12 | rs2241165 | A/G | 0.25 | Intron 2 | 171 503 886 | 3683 | 18067 | (Huang et al, 2007b) | cos (Addington et al, 2005); ped bpd (Geller et al, 2008); etoh dep (Kuo et al, 2009) | (Straub et al, 2007) | (Addington et al, 2005) | |
| M13 | rs11542313e | T/C | 0.41 | Exon 3, His37His | 171 504 132 | 246 | 18313 | scz (Straub et al, 2007) | (Straub et al, 2007) | |||
| M16 | rs769390 | A/C | 0.27 | Intron 6 | 171 518 962 | 4862 | 33143 | scz (Straub et al, 2007); (Loh el et al, 2006) | ||||
| M18 | rs3791850 | G/A | 0.22 | Intron 12 | 171 533 607 | 5620 | 47788 | scz (Straub et al, 2007); cos (Addington et al, 2005); neuroticism (Hettema et al, 2006); etoh dep (Kuo et al, 2009) | (Straub et al, 2007) | |||
Abbreviations: MAF, minor allele frequency; scz, schizophrenia; cos, childhood onset schizophrenia; bpd, bipolar disorder; ped bpd, pediatric bipolar disorder; etoh dep, alcohol dependence.
Numbering refers to Straub et al (2007).
Major/minor.
From UCSC May 2004.
In base pairs.
This was formerly rs16823181.
Figure 3.
Linkage disequilibrium (r2) between the 10 genotyped single nucleotide polymorphisms (SNPs). Figure obtained with Haploview (http://www.broadinstitute.org/mpg/haploview).
Statistics
Before assessing the effects of genotype on GABA/Cre (our main dependent variable), we addressed the effects of age, sex, scanner, and percent gray matter contribution to the voxel in the whole cohort. This was carried out by testing ANCOVA models with GABA/Cre as the dependent variable, scanner and gender as factors, and age and percent gray matter as continuous variables in a general linear model. We tested the full factorial model and also reduced models with the four main effects without any interactions. This allowed us to infer which variables should be retained in the model testing for genotype. When the effect of scanner was significant in the full factorial model or the reduced model with main effects only, the values of the dependent variable were transformed to a Z-score (the mean value for the individual scanner was subtracted from the dependent variable and divided by the standard deviation), thus removing scanner effects before further analyses. The other covariates were then retested with the Z-scores for the dependent variable. If the covariates were not significant in the initial analyses, they were dropped from further consideration. If effects of genotype were significant, the r2 of the model was used to estimate the proportion of total variance accounted for by each variable. Separate analyses were repeated for the six GAD1 SNPs and COMT rs4680. In addition, the separate hypothesis was tested that COMT rs4680 would interact with rs1978340, rs11542313, and rs769390 in GAD1 based on the clinical results reported by Straub et al (2007). For these analyses, to obtain more than two individuals per cell, GAD1 genotypes were collapsed to two groups: major allele homozygotes and minor allele carriers. As a further assurance against the effects of using different scanners to collect the data, we also tested whether the different genotypes were equally distributed across scanners with a χ2-statistic.
The p-values for the analysis of individual SNPs are reported without correction, but the threshold is p=0.007 for a Bonferroni correction for seven multiple comparisons and p=0.017 for the three multiple comparisons performed to test interactions of GAD1 and COMT val158met. Because each of the hypotheses tested in this report is based on prior evidence of association with clinical diagnosis and on an extensive basic science literature about the roles of GAD67 and dopamine on GABA function, and the current sample is independent of the earlier results, these hypothesis-driven analyses would arguably not be appropriate for agnostic statistical correction.
Because significant effects of GABA/Cre levels could depend on changes in GABA, Cre, or both, we also tested Cre obtained through LCModel in the same manner as described above. A lack of a significant genotype effect for Cre strengthens the case for GABA being the main contributor to the genotype effect. Finally, we tried to rule out other potential covariates such as intelligence quotient (IQ) and years of education by running one-way ANOVAs between genotype groups.
RESULTS
For GABA/Cre, no covariate was significant for the full factorial model, but the model that included only main effects showed significance for age and scanner (Supplementary Table S1). After transforming the GABA/Cre values to Z-scores, only age remained significant in the main effects model. Thus, all further analyses of genotype were conducted with z-transformed values and age as a covariate.
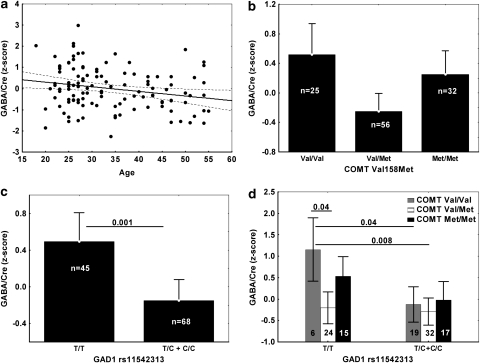
Two of the six GAD1 SNPs showed a significant effect of genotype on GABA/Cre: rs1978340 (F(2,111)=5.4, p=0.006) and rs769390 (F(2,111)=5.75, p=0.004). Both of these associations passed the criteria set by Bonferroni correction for the seven SNP multiple comparisons. COMT Val158Met was also nominally significant in this model (F(2,109)=3.4, p=0.04) (but see below for other model results). In all cases, interaction effects with age were not significant and were removed from the model. Supplementary Table S2 shows the complete results. Figure 4 shows the direction of the genotype effects, with the homozygotes for the major allele always to the left of the plot. Age accounted for about 5% and individual GAD1 SNPs contributed about 8% (Supplementary Table S3) of the total variation in GABA/Cre Z-scores. COMT Val158Met accounted for about 7% of the total variance.
Figure 4.
Directionality of main effects of GAD1 single nucleotide polymorphisms (SNPs) on GABA/Cre. The alleles associated with increased risk for schizophrenia according to Straub et al (2007) were A for rs11978340 and C for rs769390. Error bars represent 95% confidence limits. The p-values for post hoc comparisons (Tukey's ‘honest significant difference' correction for multiple comparisons) are also shown, with lines connecting the bars that showed significant differences. All results were adjusted for age in an ANCOVA model.
Interactions between the three SNPs in GAD1 (rs1978340, rs11542313, and rs769390) that had shown interaction with COMT in the clinical sample were investigated next, after collapsing the genotype groups to have more than two individuals per cell. The interaction effect for the model including rs11542313, COMT, and age (main effect of rs11542313: F(1,106)=10.6, p<0.0015 and COMT: F(2,106)=6.09, p<0.003) showed a trend toward significance (F(2,106)=3.03, p=0.05: Supplementary Table S2, corrections for multiple comparisons would require at least a p-value of 0.017). The effects of genotype are shown in Figure 5. R2 analysis indicated that the effect of rs11542313 (C allele carriers were combined in one group for this analysis) and of the interaction term each accounted for 4% of the variance in GABA/Cre (Supplementary Table S3). The full model r2 was 0.21.
Figure 5.
Effects of age (a), COMT Val158Met (b), GAD1 rs11542313 (c), and the interaction of the two genotypes (d). Error bars represent 95% confidence intervals, as does the dotted line in (a). The significant post hoc comparisons are also shown, as in Figure 4. The number of individuals in each cell is specified in (c) and (d).
Scanner and age were significant covariates in predicting Cre values determined with LCmodel (Supplementary Table S1). After correcting for scanner effects, only age was a significant covariate, thus all significant genotype effects were tested with age as a covariate. No significant main effect of genotype or GAD1–COMT interactions was found for Cre Z-scores (Supplementary Table S2). No significant differences for IQ, years of education, or percent gray matter in the spectroscopic voxel were present across GAD1 or COMT genotypes. Finally, genotypes were equally distributed across different scanners (1.78< χ2<5.5, 0.78>p>0.24 for GAD1 and COMT, respectively).
DISCUSSION
We report a set of positive associations of SNPs in the genes for GAD1 and for COMT and weak evidence of an interaction of these genes on levels of GABA measured in the living human cingulate cortex. These genes are involved in GABA synthesis and GABA neuronal activity, and thus are hypothesis driven and biologically lawful predictors of variation in GABA concentrations. Moreover, the SNPs and the interactions studied here were previously shown to increase risk for the diagnosis of schizophrenia in an independent clinical genetic association data set. Although we have performed a number of statistical tests in this report, and only a few of the results would pass agnostic statistical correction, each test was predicted strongly by the basic science data about GABA and by previous independent clinical association with schizophrenia.
Effects of Genetic Variation in GAD1 on GABA
On the basis of previous evidence of association of SNPs in GAD1 with schizophrenia and related brain phenotypes, we found significant effects of rs1978340 and rs769390 on cingulate cortex GABA concentrations in vivo (Figure 4). On the basis of post hoc tests of significance shown in Figure 4, we found that increased GABA/Cre levels appeared only in homozygotes for the minor allele, suggesting a recessive effect. These results passed the criteria set by Bonferroni correction for the seven SNPs tested. rs11542313 was significant only after collapsing the minor allele carriers, consistent with a recessive effect, and after using a model including COMT. These changes were independent of aging (see Figure 5a). In all cases, homozygosity for the GAD1 alleles thought to confer risk for schizophrenia on the background of COMT Val-Val homozygosity in our larger clinical sample (A in rs1978340, C in rs769390, and T for rs11542313; Straub et al, 2007) was associated with relatively increased GABA/Cre. The three SNPs identified as having an effect on GABA measures were in negligible LD (r2=0.2–0.3) with each other.
Various mechanisms may account for these effects: changes in the efficiency of production of the protein by alteration of the mRNA secondary structure (Nackley et al, 2006) in the case of rs11542313 (a synonymous coding SNP); being part of a regulatory element in the 5′ flanking region in the case of rs1978340 (Veyrieras et al, 2008), or in an intron for rs769390; monitoring other rare causative SNPs by LD; or modification of interaction with DNA methyltransferases or with histones thought to regulate epigenetic control of GAD1 mRNA expression (Kundakovic et al, 2007, 2009) and to be relevant for schizophrenia pathology (Huang and Akbarian, 2007a). It should be noted that neither one of these SNPs was associated with alteration in the amount of mRNA for GAD1 measured in post-mortem brain (Straub et al, 2007), however this prior finding was in a sample of mainly African-American ancestry where genetic variation is likely to be associated with different causative haplotypes. Clearly, our study results await replication in an independent sample and a biological explanation for their effect.
For all significant SNPs in GAD1, the ‘risk' allele was associated with an unexpected increase in GABA/Cre. We would have expected the opposite directionality given the post-mortem evidence for reductions in GAD1 mRNA expression (see Akbarian and Huang, 2006; Lewis et al, 2005 for reviews) and protein levels (Guidotti et al, 2000) in schizophrenia (but see Dracheva et al, 2004; Gluck et al, 2002; Hakak et al, 2001, for results in the opposite direction in elderly patients). It is unclear what mechanism might account for this phenomenon. We speculate that GABA production by GAD1 is actually reduced in risk allele carriers and that a compensatory change such as excessive production by GAD2 or decreased catabolism by GABA transaminase may be causing this unpredicted increase in GABA/Cre levels. The compensation could also be occurring in certain cortical layers, whereas others may be decreased but beyond the spatial resolution of our technique; or be present intracellularly, but correspond to a relative deficit of GABA synaptic transmission; or be limited to calretinin-positive cells that are thought to constitute the majority of GABA-containing cells (Conde et al, 1994) and therefore may provide the bulk of the signal for this technique. Alternatively, it is entirely possible that the same genetic variations may show opposite or no effects in patients with schizophrenia. We will be exploring this in future studies. Given the reported association of GAD1 SNPs with neuroticism (Hettema et al, 2006), we also checked if our study results may help elucidate the biology of the associations reported by Hettema et al (2006), but no significant SNP overlapped between the two studies.
It is also relevant to point out that GAD67 can be regulated by post-transcriptional mechanisms (Martin and Rimvall, 1993; Rimvall and Martin, 1994), and that this may occur in the absence of a change in mRNA levels, a possible mechanisms for a dissociation between GABA and GAD1 mRNA levels in schizophrenia. If our study results were to be replicated in an independent sample, detailed post-mortem studies of the effects of GAD1 genetic variation on various markers of GABA interneurons will be necessary to understand the physiology of this phenomenon.
Effects of Genetic Variation in COMT on GABA
COMT Val158Met was nominally significant on its own, but became significant beyond our threshold for multiple comparisons when tested in combination with rs11542313, indicating that the two SNPs determine GABA concentrations independently of each other and share separate portions of the variance in GABA values, likely a demonstration of independent biological mechanisms affected by these two genes. COMT val158met genotype also showed a trend toward an interaction with GAD1 rs11542313, but this trend did not survive stringent multiple comparisons correction. Nevertheless, the association of genetic variation in COMT and GABA levels and the interaction of GAD1 and COMT genotypes on GABA levels were strikingly consistent with basic science data that cortical DA regulates GABA neuronal activity. The effects of COMT rs4680 contributed 6% of the total variation in GABA/Cre, compared with 8% for rs769390 and rs1978340 in GAD1, consistent with a modulatory effect. However, the pattern of genotype associations accounting for the main effect of COMT was unexpected, with Val/Met heterozygotes showing the lowest GABA/Cre ratios. We had expected Val-Val homozygotes (having the lowest cortical synaptic DA levels) to show the lowest levels of GABA. Although the higher levels of GABA in Met homozygotes might have been expected based on higher dopamine being related to a net increase in inhibitory effect in non-fast spiking interneurons (mainly situated in the superficial layers of the cortex and containing calretinin: Gao et al, 2003b), the higher GABA level in Val homozygotes is difficult to explain. The interaction effect between COMT and GAD1 rs11542313 was mainly promoted by the six T/T homozygotes (the GAD1 ‘risk' genotype) who were also Val-Val homozygotes (Figure 5d). Given this limitation, we are inclined to await further replication or a substantially larger sample size before interpreting the finding. Although one possible explanation for these counterintuitive findings may lie in the distribution of D1 and D5 receptor subtypes on different subtypes of GABAergic neurons (Glausier et al, 2008), or in a homeostatic mechanism to maintain optimal levels of noise or tuning of the system (Rolls et al, 2008), not all aspects of our findings can be explained by these models of DA and GABA receptor modulation of working memory.
Methodological Strengths and Limitations
This study had several methodological strengths. First, our spectroscopic technique allowed for highly accurate separation of the GABA peak from other signals. This same methodology has also been shown to be sensitive to the effects of disease and has shown expected reductions of GABA levels in depression (Hasler et al, 2007). Moreover, preliminary data (Geramita et al, unpublished observations) show excellent reproducibility in repeated sessions (around 6–8% coefficient of variation) for GABA/Cre. Second, this sample was exclusively composed of Caucasian volunteers, limiting genetic heterogeneity and improving our ability to discern effects due to allelic variation. Finally, special care was put into removing individuals who had any history of psychiatric disorders or substance abuse.
Limitations of this study include that it was conducted on multiple scanners, due to forced changes in platforms during the collection of the sample. It should be kept in mind that the genotypes of the significant SNPs were equally distributed across scanners. Moreover, we cannot account for differences in macromolecules (contributing up to 56% of the GABA peak: Kegeles et al, 2001; van der Veen et al, 2007) between genotypes, but given that the macromolecule signal is formed by many different lipoprotein species, it is highly unlikely that a systematic genetic effect would be found.
Difficulties in interpretation also arise when considering ratios to Cre, however there were no effects of genotype when considering absolute values of Cre obtained by analyzing the unedited portion of the spectrum with LCmodel (Provencher, 2001), supporting the contention that this effect is due mostly to GABA. Conversely, the effect of age on GABA/Cre levels appears to be mainly explained by rising Cre values, a finding consistent with some reports (Angelie et al, 2001; Maudsley et al, 2009; Pfefferbaum et al, 1999). The contribution of partial voluming of gray and white matter in the voxel was also addressed by the use of GABA/Cre ratios as there was no statistical effect of percent gray matter in the voxel in this large sample of controls. Moreover, there were no significant differences in the percent of gray matter present in the voxel between genotypes.
Conclusion
In conclusion, we showed for the first time that genetic variation in GAD1 and in COMT was associated with alterations in GABA/Cre ratios in vivo. The directionality of the genetic relationship was opposite to that predicted. It is likely that the association detected here is determined early in development, because none of our analyses showed a significant interaction of age and genotype, therefore GABA/Cre levels are likely to represent a relatively stable trait. Our study results might be the first in vivo evidence supporting a role of GAD67 in the development of GABA circuitry (Chattopadhyaya et al, 2007) and subsequent determination of GABA levels.
Acknowledgments
This work was presented in part at the Society for Neuroscience 15–18 2008 November Meeting in Washington, DC. We thank Christine Rebsch for help in data acquisition and Iona Machado for helping with data compilation and analysis.
This work was funded entirely by the NIMH IRP. The authors declare that, except for income received by the primary employer (the NIH), no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional services and there are no personal financial holdings that could be perceived as constituting a potential conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Adams R, David AS. Patterns of anterior cingulate activation in schizophrenia: a selective review. Neuropsychiatr Dis Treat. 2007;3:87–101. doi: 10.2147/nedt.2007.3.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addington AM, Gornick M, Duckworth J, Sporn A, Gogtay N, Bobb A, et al. GAD1 (2q31.1), which encodes glutamic acid decarboxylase (GAD67), is associated with childhood-onset schizophrenia and cortical gray matter volume loss. Mol Psychiatry. 2005;10:581–588. doi: 10.1038/sj.mp.4001599. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Angelie E, Bonmartin A, Boudraa A, Gonnaud PM, Mallet JJ, Sappey-Marinier D. Regional differences and metabolic changes in normal aging of the human brain: proton MR spectroscopic imaging study. AJNR Am J Neuroradiol. 2001;22:119–127. [PMC free article] [PubMed] [Google Scholar]
- Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, et al. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi G, Mattay VS, Bertolino A, Elvevag B, Callicott JH, Das S, et al. Effect of catechol-O-methyltransferase val158met genotype on attentional control. J Neurosci. 2005;25:5038–5045. doi: 10.1523/JNEUROSCI.0476-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinick M, Nystrom LE, Fissell K, Carter CS, Cohen JD. Conflict monitoring versus selection-for-action in anterior cingulate cortex. Nature. 1999;402:179–181. doi: 10.1038/46035. [DOI] [PubMed] [Google Scholar]
- Camchong J, Dyckman KA, Austin BP, Clementz BA, McDowell JE. Common neural circuitry supporting volitional saccades and its disruption in schizophrenia patients and relatives. Biol Psychiatry. 2008;64:1042–1050. doi: 10.1016/j.biopsych.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CS, Botvinick MM, Cohen JD. The contribution of the anterior cingulate cortex to executive processes in cognition. Rev Neurosci. 1999;10:49–57. doi: 10.1515/revneuro.1999.10.1.49. [DOI] [PubMed] [Google Scholar]
- Carter CS, MacDonald AW, III, Ross LL, Stenger VA. Anterior cingulate cortex activity and impaired self-monitoring of performance in patients with schizophrenia: an event-related fMRI study. Am J Psychiatry. 2001;158:1423–1428. doi: 10.1176/appi.ajp.158.9.1423. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Wu CZ, Knott G, Kuhlman S, Fu Y, et al. GAD67-mediated GABA synthesis and signaling regulate inhibitory synaptic innervation in the visual cortex. Neuron. 2007;54:889–903. doi: 10.1016/j.neuron.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Lund JS, Jacobowitz DM, Baimbridge KG, Lewis DA. Local circuit neurons immunoreactive for calretinin, calbindin D-28k or parvalbumin in monkey prefrontal cortex: distribution and morphology. J Comp Neurol. 1994;341:95–116. doi: 10.1002/cne.903410109. [DOI] [PubMed] [Google Scholar]
- Deicken RF, Zhou L, Schuff N, Weiner MW. Proton magnetic resonance spectroscopy of the anterior cingulate region in schizophrenia. Schizophr Res. 1997;27:65–71. doi: 10.1016/S0920-9964(97)00082-0. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Elhakem SL, McGurk SR, Davis KL, Haroutunian V. GAD67 and GAD65 mRNA and protein expression in cerebrocortical regions of elderly patients with schizophrenia. J Neurosci Res. 2004;76:581–592. doi: 10.1002/jnr.20122. [DOI] [PubMed] [Google Scholar]
- Du J, Duan S, Wang H, Chen W, Zhao X, Zhang A, et al. Comprehensive analysis of polymorphisms throughout GAD1 gene: a family-based association study in schizophrenia. J Neural Transm. 2008;115:513–519. doi: 10.1007/s00702-007-0844-z. [DOI] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ende G, Braus DF, Walter S, Weber-Fahr W, Soher B, Maudsley AA, et al. Effects of age, medication, and illness duration on the N-acetyl aspartate signal of the anterior cingulate region in schizophrenia. Schizophr Res. 2000;41:389–395. doi: 10.1016/s0920-9964(99)00089-4. [DOI] [PubMed] [Google Scholar]
- Ford JM, Gray M, Whitfield SL, Turken AU, Glover G, Faustman WO, et al. Acquiring and inhibiting prepotent responses in schizophrenia: event-related brain potentials and functional magnetic resonance imaging. Arch Gen Psychiatry. 2004;61:119–129. doi: 10.1001/archpsyc.61.2.119. [DOI] [PubMed] [Google Scholar]
- Fornito A, Yung AR, Wood SJ, Phillips LJ, Nelson B, Cotton S, et al. Anatomic abnormalities of the anterior cingulate cortex before psychosis onset: an MRI study of ultra-high-risk individuals. Biol Psychiatry. 2008;64:758–765. doi: 10.1016/j.biopsych.2008.05.032. [DOI] [PubMed] [Google Scholar]
- Gao WJ, Goldman-Rakic PS. Selective modulation of excitatory and inhibitory microcircuits by dopamine. Proc Natl Acad Sci USA. 2003a;100:2836–2841. doi: 10.1073/pnas.262796399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao WJ, Wang Y, Goldman-Rakic PS. Dopamine modulation of perisomatic and peridendritic inhibition in prefrontal cortex. J Neurosci. 2003b;23:1622–1630. doi: 10.1523/JNEUROSCI.23-05-01622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller B, Tillman R, Bolhofner K, Hennessy K, Cook EH., Jr GAD1 single nucleotide polymorphism is in linkage disequilibrium with a child bipolar I disorder phenotype. J Child Adolesc Psychopharmacol. 2008;18:25–29. doi: 10.1089/cap.2007.0056. [DOI] [PubMed] [Google Scholar]
- Glausier JR, Khan ZU, Muly EC. Dopamine D1 and D5 receptors are localized to discrete populations of interneurons in primate prefrontal cortex. Cereb Cortex. 2008;19:1820–1834. doi: 10.1093/cercor/bhn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck MR, Thomas RG, Davis KL, Haroutunian V. Implications for altered glutamate and GABA metabolism in the dorsolateral prefrontal cortex of aged schizophrenic patients. Am J Psychiatry. 2002;159:1165–1173. doi: 10.1176/appi.ajp.159.7.1165. [DOI] [PubMed] [Google Scholar]
- Gogos JA, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, et al. Catechol-O-methyltransferase-deficient mice exhibit sexually dimorphic changes in catecholamine levels and behavior. Proc Natl Acad Sci USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Rakic PS.1990Cellular and circuit basis of working memory in prefrontal cortex of nonhuman primates Prog Brain Res 85325–335.discussion 335–326. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Arion D, Unger T, Maldonado-Aviles JG, Morris HM, Volk DW, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler G, van der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced prefrontal glutamate/glutamine and gamma-aminobutyric acid levels in major depression determined using proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2007;64:193–200. doi: 10.1001/archpsyc.64.2.193. [DOI] [PubMed] [Google Scholar]
- Hettema JM, An SS, Neale MC, Bukszar J, van den Oord EJ, Kendler KS, et al. Association between glutamic acid decarboxylase genes and anxiety disorders, major depression, and neuroticism. Mol Psychiatry. 2006;11:752–762. doi: 10.1038/sj.mp.4001845. [DOI] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS ONE. 2007a;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007b;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoum F, Chrapusta SJ, Egan MF. 3-Methoxytyramine is the major metabolite of released dopamine in the rat frontal cortex: reassessment of the effects of antipsychotics on the dynamics of dopamine release and metabolism in the frontal cortex, nucleus accumbens, and striatum by a simple two pool model. J Neurochem. 1994;63:972–979. doi: 10.1046/j.1471-4159.1994.63030972.x. [DOI] [PubMed] [Google Scholar]
- Kegeles LS, Mao X, Gonsalez R, Shungu DC. Evaluation of anatomic variation in macromolecule contribution to the GABA signal using metabolite nulling and the J-editing technique at 3.0 T. Proc Intl Soc Magn Reson Med. 2001;15:1391. [Google Scholar]
- Ko JH, Ptito A, Monchi O, Cho SS, Van Eimeren T, Pellecchia G, et al. Increased dopamine release in the right anterior cingulate cortex during the performance of a sorting task: a [11C]FLB 457 PET study. Neuroimage. 2009;46:516–521. doi: 10.1016/j.neuroimage.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Mol Pharmacol. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Guidotti A, Grayson DR. The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes. Mol Pharmacol. 2009;75:342–354. doi: 10.1124/mol.108.051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, Alexander J, et al. Associations of glutamate decarboxylase genes with initial sensitivity and age-at-onset of alcohol dependence in the Irish Affected Sib Pair Study of Alcohol Dependence. Drug Alcohol Depend. 2009;101:80–87. doi: 10.1016/j.drugalcdep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T. Deciphering the disease process of schizophrenia: the contribution of cortical GABA neurons. Int Rev Neurobiol. 2007;78:109–131. doi: 10.1016/S0074-7742(06)78004-7. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Loh el W, Lane HY, Chen CH, Chang PS, Ku LW, Wang KH, et al. Glutamate decarboxylase genes and alcoholism in Han Taiwanese men. Alcohol Clin Exp Res. 2006;30:1817–1823. doi: 10.1111/j.1530-0277.2006.00218.x. [DOI] [PubMed] [Google Scholar]
- Lundorf MD, Buttenschon HN, Foldager L, Blackwood DH, Muir WJ, Murray V, et al. Mutational screening and association study of glutamate decarboxylase 1 as a candidate susceptibility gene for bipolar affective disorder and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2005;135B:94–101. doi: 10.1002/ajmg.b.30137. [DOI] [PubMed] [Google Scholar]
- Manoach DS, Ketwaroo GA, Polli FE, Thakkar KN, Barton JJ, Goff DC, et al. Reduced microstructural integrity of the white matter underlying anterior cingulate cortex is associated with increased saccadic latency in schizophrenia. Neuroimage. 2007;37:599–610. doi: 10.1016/j.neuroimage.2007.04.062. [DOI] [PubMed] [Google Scholar]
- Martin DL, Rimvall K. Regulation of gamma-aminobutyric acid synthesis in the brain. J Neurochem. 1993;60:395–407. doi: 10.1111/j.1471-4159.1993.tb03165.x. [DOI] [PubMed] [Google Scholar]
- Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, et al. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI) Magn Reson Med. 2009;61:548–559. doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Menzies L, Ooi C, Kamath S, Suckling J, McKenna P, Fletcher P, et al. Effects of gamma-aminobutyric acid-modulating drugs on working memory and brain function in patients with schizophrenia. Arch Gen Psychiatry. 2007;64:156–167. doi: 10.1001/archpsyc.64.2.156. [DOI] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. In vivo spectroscopic quantification of the N-acetyl moiety, creatine, and choline from large volumes of brain gray and white matter: effects of normal aging. Magn Reson Med. 1999;41:276–284. doi: 10.1002/(sici)1522-2594(199902)41:2<276::aid-mrm10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- Rimvall K, Martin DL. The level of GAD67 protein is highly sensitive to small increases in intraneuronal gamma-aminobutyric acid levels. J Neurochem. 1994;62:1375–1381. doi: 10.1046/j.1471-4159.1994.62041375.x. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Loh M, Deco G, Winterer G. Computational models of schizophrenia and dopamine modulation in the prefrontal cortex. Nat Rev Neurosci. 2008;9:696–709. doi: 10.1038/nrn2462. [DOI] [PubMed] [Google Scholar]
- Sailasuta N, LeRoux P, Hurd R, Wang P, Sachs N, Ketter T.(eds). (2001Detection of cerebral gamma-aminobutyric acid (GABA) in bipolar disorder patients and healthy volunteers at 3T Proc Intl Soc Magn Reson Med 91011 [Google Scholar]
- Sanacora G, Mason GF, Rothman DL, Behar KL, Hyder F, Petroff OA, et al. Reduced cortical gamma-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 1999;56:1043–1047. doi: 10.1001/archpsyc.56.11.1043. [DOI] [PubMed] [Google Scholar]
- Seamans JK, Gorelova N, Durstewitz D, Yang CR. Bidirectional dopamine modulation of GABAergic inhibition in prefrontal cortical pyramidal neurons. J Neurosci. 2001;21:3628–3638. doi: 10.1523/JNEUROSCI.21-10-03628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamans JK, Yang CR. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol. 2004;74:1–58. doi: 10.1016/j.pneurobio.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Segall JM, Turner JA, van Erp TG, White T, Bockholt HJ, Gollub RL, et al. Voxel-based morphometric multisite collaborative study on schizophrenia. Schizophr Bull. 2009;35:82–95. doi: 10.1093/schbul/sbn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soghomonian JJ, Martin DL. Two isoforms of glutamate decarboxylase: why. Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, et al. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Tan HY, Chen Q, Goldberg TE, Mattay VS, Meyer-Lindenberg A, Weinberger DR, et al. Catechol-O-methyltransferase Val158Met modulation of prefrontal–parietal–striatal brain systems during arithmetic and temporal transformations in working memory. J Neurosci. 2007;27:13393–13401. doi: 10.1523/JNEUROSCI.4041-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian N, Petersen C, Kash S, Baekkeskov S, Copenhagen D, Nicoll R. The role of the synthetic enzyme GAD65 in the control of neuronal gamma-aminobutyric acid release. Proc Natl Acad Sci USA. 1999;96:12911–12916. doi: 10.1073/pnas.96.22.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng KY, Lewis BL, Lipska BK, O'Donnell P. Post-pubertal disruption of medial prefrontal cortical dopamine-glutamate interactions in a developmental animal model of schizophrenia. Biol Psychiatry. 2007;62:730–738. doi: 10.1016/j.biopsych.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen JW, Hattori N, Shen J. Quantification of co-edited macromolecules in GABA J-editing. Proc Intl Soc Magn Reson Med. 2007;15:1399. [Google Scholar]
- Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression—QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer G, Musso F, Vucurevic G, Stoeter P, Konrad A, Seker B, et al. COMT genotype predicts BOLD signal and noise characteristics in prefrontal circuits. Neuroimage. 2006;32:1722–1732. doi: 10.1016/j.neuroimage.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Wong CG, Bottiglieri T, Snead OC., 3rd GABA, gamma-hydroxybutyric acid, and neurological disease. Ann Neurol. 2003;54 (Suppl 6:S3–S12. doi: 10.1002/ana.10696. [DOI] [PubMed] [Google Scholar]
- Woo TU, Walsh JP, Benes FM. Density of glutamic acid decarboxylase 67 messenger RNA-containing neurons that express the N-methyl--aspartate receptor subunit NR2A in the anterior cingulate cortex in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2004;61:649–657. doi: 10.1001/archpsyc.61.7.649. [DOI] [PubMed] [Google Scholar]
- Zhao X, Qin S, Shi Y, Zhang A, Zhang J, Bian L, et al. Systematic study of association of four GABAergic genes: glutamic acid decarboxylase 1 gene, glutamic acid decarboxylase 2 gene, GABA(B) receptor 1 gene and GABA(A) receptor subunit beta2 gene, with schizophrenia using a universal DNA microarray. Schizophr Res. 2007;93:374–384. doi: 10.1016/j.schres.2007.02.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.