Abstract
In the past decade, the potential of harnessing the ability of nuclear magnetic resonance (NMR) spectroscopy to monitor intermolecular interactions as a tool for drug discovery has been increasingly appreciated in academia and industry. In this Perspective, we highlight some of the major applications of NMR in drug discovery, focusing on hit and lead generation, and provide a critical analysis of its current and potential utility.
Nuclear magnetic resonance (NMR) spectroscopy is often valued for its ability to shed light on molecular structure, but its greatest potential in drug discovery probably lies in the information that it can reveal about molecular interactions at the atomic level1. A simple parameter, the NMR chemical shift, is highly sensitive to the exact environment of the atom and therefore yields information about whether a small molecule binds to a target protein or nucleic acid, what parts of the small molecule are interacting and to which part of the macromolecular target the small molecule is bound. Other NMR experiments, such as the saturation transfer difference2,3, or 1H NMR simple relaxation measurements, such as the T1ρ experiment4, are sensitive to the overall molecular motion of test compounds, which is very different for free versus bound ligands. Thus, these simple approaches can be used to validate ligand binding and/or to identify potential ligands in mixtures of test compounds.
The variety of readily measurable NMR parameters allows several applications of NMR in drug discovery, including assessing target druggability, pharmacophore identification, hit validation, hit optimization and potentially structure-based drug design (FIG. 1). Perhaps most notably, in the past decade, NMR has demonstrated its utility in fragment-based drug design (FBDD)5–7 (BOX 1), a novel and increasingly popular strategy for lead discovery that provides an alternative to conventional high-throughput screening, or to support hit-to-lead optimization for a particular drug target. NMR can also be used to determine low-resolution structures of target–ligand complexes for natively unstructured proteins or membrane proteins that are not amenable to crystallographic approaches.
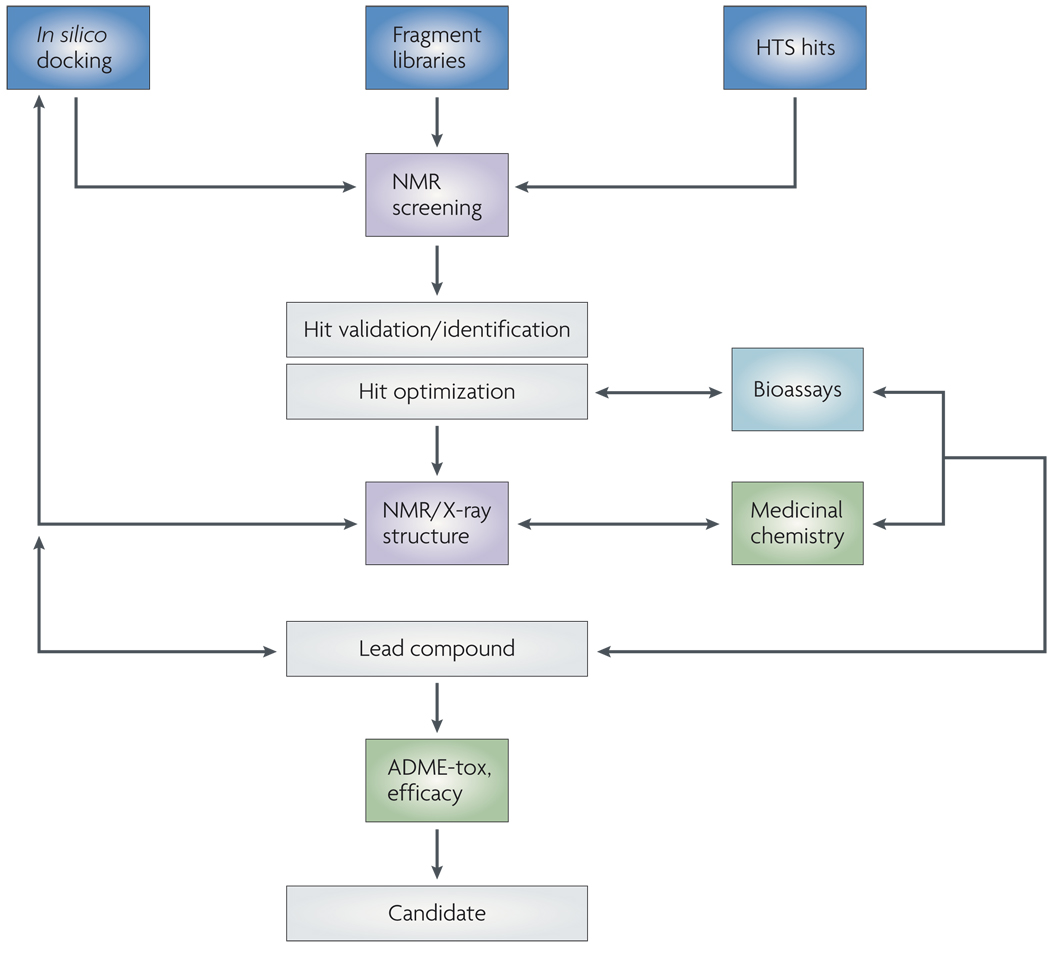
Figure 1. Overview of applications of NMR in drug discovery.
NMR spectroscopy can provide critical information at early stages of hit validation and identification. NMR measurements for binding studies can represent a key step to eliminate false positives from high-throughput (HTS) campaigns, to validate putative hits from in silico screens or to identify novel scaffolds in fragment-based programmes. NMR and X-ray crystallography can also provide unique information to subsequently guide hit-to-lead optimization. ADME-tox, absorption, distribution, metabolism, excretion and toxicity.
Box 1 Fragment-based lead discovery.
It has been estimated that the number of potential drug molecules is of the order of 1010−1050 (REF. 52). However, for a given target system, it is difficult to imagine high-throughput screening (HTS) performed with much more than 106 compounds, especially considering that such endeavours would be very expensive and subject to a sizeable number of false positives and false negatives. The traditional approach of testing variations of known drugs is not going to explore this potential pool very deeply either, but at least it has the advantage of exploring compound space based on knowledge, so the search will be made more effectively. Unfortunately, the chances of encountering cross-resistance, for example, are enhanced if searches are limited to compounds similar to those already in clinical use.
The realization that poor ADME-tox (absorption, distribution, metabolism, excretion and toxicity) of drug candidates in vivo has been a common and expensive limitation in drug development has stimulated the development of tools to identify compounds or classes of compounds that may overcome these problems. One recent school of thought has been to filter the databases to be screened so that criteria largely comply with Lipinski’s “Rule of Five”, an empirical list of desirable properties based on clinically successful drugs with good bioavailability53,54. However, as Lipinski reports, there are numerous examples of drugs that do not abide by these rules, so it is not a rule to be followed blindly; for example, the rules of Veber et al. are also well suited and provide an alternative way to look at potential drugs55. Nevertheless, ligands that are larger than ∼500 daltons and with poor solubility tend to become problematic at the developmental stages56.
These issues are exacerbated when dealing with targets that have macromolecular interaction surfaces, for which HTS campaigns tend not to provide therapeutically viable leads. However, it is currently unclear whether HTS screening campaigns are not producing valuable leads against protein–protein interactions because of the alleged ‘undruggable’ nature of the binding surfaces or simply because the compound libraries are populated by compounds that were not originally derived to complement a protein surface. Although the most likely the answer is both, it seems sensible to assume that a more rational chemical design approach, in which a compound is ‘built’ stepwise within the binding cavity of the target, probably represents a more suitable strategy to tackle targets that are otherwise very challenging for HTS.
These considerations represent the fundamental impetus for the development of the so-called fragment-based drug discovery (FBDD) approaches5,6, which have the intrinsic advantage of exploring in principle a much larger accessible chemical space than conventional HTS campaigns. The strategy consists of building up a lead compound (typically smaller than 500 daltons) from screening a database of typically 1,000–15,000 smaller molecules (fragments) that are smaller than 300 daltons and have good aqueous solubility. The most common FBDD approaches include tethering5,6,57–70, X-ray diffraction65,71–76 or NMR spectroscopy1,5–7,69,77–82, as methods for fragment screening and to guide iterative optimizations.
When introducing and developing novel drug discovery technologies in general, the response of the scientific and business communities has often been similar (C. Lipinski, personal communication): after a period of sceptical resistance, which varies depending on the current development stage of other competing technologies, considerable and often exaggerated enthusiasm is generated around the approach. As time goes by, set-backs and the overoptimistic nature of the original ‘hype’ results in the enthusiasm turning to distrust. Eventually, for those technologies that survive, their real value emerges and the realistic impact on the drug discovery process can be critically assessed and justified. This process takes on average about 5–10 years. We believe that many applications of NMR spectroscopy in drug discovery are now mature enough for such assessments. With this in mind, in this article, which is based on discussions at a recent meeting on NMR in drug discovery (see acknowledgements), we provide our collective evaluation of the past, present and future of the applications of NMR spectroscopy in hit and lead generation.
NMR-based strategies in drug discovery
Several NMR-based strategies have been developed (TABLE 1, TABLE 2) that are particularly useful for FBDD applications (BOX 1). These range from the more traditional chemical-shift mapping, to ligand-based techniques that monitor changes in ligand nuclear spin relaxation properties upon binding, to measurements of diffusion. Some of these approaches are better suited to screening methods and/or to validate hits coming from high-throughput screening (HTS) campaigns8–10 (TABLE 1), whereas others are better suited to guide hit optimization into more potent, selective and drug-like compounds9 (TABLE 2). An overview of the merits and pitfalls of some of these approaches is provided below. The detailed description and technical details of these methods fall outside the scope of this article and can be found in the citations within TABLE 1, 2, as well as in recent review articles cited in this manuscript.
Table 1.
NMR methods for compound screening and hit validation
| Approach | Observation | Use | Description and references to recent applications |
|---|---|---|---|
| Chemical-shift perturbation1 |
Target (protein or nucleic acid) resonances |
Primary screening Hit validation Site of binding |
Identifies compounds that bind by means of chemical-shift perturbation of resonances of the target11,77,83–86 |
| STD NMR2 | Ligand | Primary screening Hit validation |
Identifies compounds that bind weakly; build-up curve identifies interacting functional groups3,13,30,86–89 |
| WaterLOGSY90 | Ligand | Primary screening | Identifies compounds that bind by using water-mediated NOEs10,91 |
| SLAPSTIC (Using spin- labelled protein)92 |
Ligand | Primary screening | Highly sensitive detection of fragments that bind5,92 |
| TINS93 | Ligand | Primary screening Hit validation |
Identifies compounds that bind by screening libraries against immobilized protein targets93 |
| T1ρ and T2 relaxation; line broadening4 |
Ligand | Primary screening Hit validation |
Binding enhances relaxation; enables affinity estimates; build-up curve identifies interacting functional groups94 |
| Transferred NOEs95 |
Ligand | Hit validation Conformation of flexible ligands |
Gives information about the interaction of binders with the target96,97; determines bioactive conformation of flexible ligands such as peptides14 |
| FABS16,17 | Substrate or cofactor |
Primary screening Hit validation |
Uses reference substrates or cofactors to monitor enzymatic reactions12,98–104 |
| FAXS105,106 | Reference ligand |
Primary screening Hit validation |
Measures the displacement of a fluorinated ‘spy’ molecule104,107 |
| Diffusion measurements108,109 |
Ligand | Primary screening Hit validation |
Measures the difference in diffusion rates for ligands in the bound versus free state110 |
FABS, fluorine atoms for biochemical screening; FAXS, fluorine chemical shift anisotropy and exchange for screening; NOE, nuclear Overhauser effect; SLAPSTIC, spin labels attached to protein side chains as a tool to identify interacting compounds; STD, saturation transfer difference; TINS, target immobilized NMR screening; T1ρ, rotating frame nuclear spin longitudinal relaxation time; T2, transverse nuclear spin relaxation time; waterLOGSY, water-ligand observed via gradient spectroscopy.
Table 2.
NMR methods for hit/lead optimization
| Approach | Observation | Use | Description and references to recent applications |
|---|---|---|---|
| SAR by NMR69,111 | Ligand Target |
Structural information FBDD screening Compound optimization |
Design bi-dentate compounds20,112 |
| SLAPSTIC with first-site spin- labelled compound21 |
Ligand | FBDD screening Compound optimization |
Highly sensitive detection of fragments and weakly interacting second-site compounds113 |
| SAR by lLOEs19,114 | Ligand-to- ligand |
FBDD screening Compound optimization |
Detects protein mediated ligand–ligand interactions (compounds occupying adjacent sites)22 |
| Pharmacophore by lLOEs115 |
Ligand-to- ligand |
FBDD screening Compound optimization |
Detects protein-mediated ligand–ligand interactions and uses information for pharmacophore -based search of bi-dentate compounds115 |
| H2O/D2O exchange-rate measurements |
Target | Compound characterization |
Identifies binding epitope116 |
| INPHARMA117 | Ligand-to- ligand |
Compound characterization |
Detects protein mediated ligand–ligand interactions (competition for the same binding site) |
FBDD, fragment-based drug design; ILOE, interligand nuclear Overhauser effect; INPHARMA, interligand NOEs for pharmacophore mapping; SAR, structure–activity relationship; SLAPSTIC, spin labels attached to protein side chains as a tool to identify interacting compounds.
NMR screening and hit validation
A simple approach for ligand binding studies, and probably the most utilized approach for hit identification and validation by NMR, exploits differences in chemical shift between free and bound protein/nucleic acid targets in 15N/1H and/or 13C/1H two-dimensional correlation spectra of the target upon titration of a ligand or a mixture of ligands. When the resonance assignments are known (usually attainable for proteins smaller than 30–40 kDa), this approach, also called chemical-shift mapping, can also provide crude but meaningful structural information on the site of binding. The method can be extended to larger macromolecular targets in which an amino-acid type has been selectively labelled to reduce spectral complexity, thus extending its applicability to targets larger than 100 kDa11.
The use of chemical-shift mapping studies to monitor ligand binding has several advantages, the most obvious being that compounds that bind to a given protein can be found and characterized without the need to develop a specific assay or having knowledge of the function of the protein — a concept that is generally true for most NMR-based techniques. Second, as mentioned, when combined with resonance assignments, this approach can rapidly provide information on the site of binding. Third, when the structure of the target has been previously determined by NMR, in some instances it should be possible to rapidly derive ligand–protein distances via nuclear Overhauser effect (NOE)-type experiments that allow more precise determination of the ligand binding mode. Last but not least, the chemical shift approach is arguably one of the most robust, reliable and reproducible ligand binding assays currently available. Although X-ray crystallography could, assuming suitable crystallization conditions, provide more precise information on the binding mode, this approach does not provide information on the dissociation constant of the complex, nor can it easily be used to monitor ligand binding. However, like X-ray crystallography, the amount of protein that is needed for a single NMR experiment is relatively high for the technique to be used efficiently to test large libraries of compounds. Hence, these assays have found more widespread use in hit validation12 and in FBDD (BOX 1), in which smaller libraries are screened.
In detecting ligand binding, rather than observing the NMR spectra of the target upon complex formation, the so-called ligand-based experiments focus on the observation of the perturbations induced by a substoichiometric amount of target on the NMR spectra of the ligand. Examples of these approaches are the saturation transfer difference (STD) or simple T1ρ measurements (TABLE 1). In a typical STD experiment, simple one-dimensional 1H NMR experiments are recorded for a ligand in the presence of a small amount of target, usually at a ligand:protein ratio of about 100:1, with and without selective irradiation of the protein resonances. When the longitudinal relaxation rates for the hydrogens of the small molecule are longer than the dissociation rate constant (koff) of the complex (typically true for ligands with micromolar to millimolar dissociation constants), there will be an accumulation of saturated ligand even if the target is present in a substoichiometric concentration. The selective saturation of protein resonances can be obtained by irradiating regions of the 1H NMR spectra (typically, the aliphatic region of the spectrum, between −1 and 2 ppm) that are usually well populated by methyl groups of the protein but are not occupied by resonances from small-molecule, organic ligands. Subsequently, a difference spectrum is generated from two spectra that are recorded with and without pre-irradiation of protein resonances. Similarly, the T1ρ experiment exploits the fact that the relaxation properties of ligand nuclei differ depending on whether the molecule is in the free versus the bound state, even if the binding is transient, in the presence of a substoichiometric amount of target. These ‘transferred’ ligand-based methods are less informative than chemical-shift mapping and are used primarily for screening and/or to validate ligand binding. However, they could also be used for the determination of the binding epitope13 and the bioactive conformation of larger ligands, such as carbohydrates or peptides; this information could be very useful in pharmacophore-based design14,15.
Finally, chemical reactions that are catalysed by enzyme targets can also be easily monitored by NMR16–18, which is particularly useful not only in screening but also to validate hits that were previously identified by using spectroscopic techniques. The major advantages of the NMR-based approaches is that they require a relatively small amount of protein per sample (the typical target concentration needed for one experiment is 1–5 µM for binding studies), that their range of applicability is not limited to proteins amenable to NMR spectroscopy (as only the resonance lines of ligands are observed) and that measurements are relatively fast (1–10 minutes for binding assays) and readily automatable. Similar considerations can be made for other approaches listed in TABLE 1. These are mostly used for hit validation or for screening small libraries, of the order of hundreds up to thousands of compounds, which are usually tested in mixtures of 10–50 compounds per sample.
An obvious strategy is to combine these NMR screening approaches with in silico docking of a compound database to predict those that should bind to a target protein structure (usually static). Computational hits from this virtual screening will need to be tested experimentally, but a relatively small number of top-ranking hits can be selected (for example, 1%) for NMR screening. This approach could be less expensive and faster than HTS if the time to develop an experimental assay is considered, as this is not needed for NMR.
In summary, various NMR-based approaches can provide valuable information for hit identification and validation, and ultimately guide medicinal chemistry. The development of these applications now seems to have reached a plateau, and it is unclear whether further room for expansion remains. Rather, it is likely that applications of the approaches described above to ‘non-traditional’ targets, such as macromolecule–macromolecule interactions or membrane proteins, could be of most value.
NMR-based hit/lead optimization
A powerful application of chemical-shift mapping in hit/lead optimization is the ‘SAR (structure–activity relationship) by NMR’ strategy, which is arguably the archetypal FBDD approach (BOX 1). In this approach, a chemical-shift mapping-based screen for a second compound that binds to the target is performed in the presence of an initial weak hit compound. The latter may come from a previous NMR screen or it could be a known ligand that was discovered from other approaches such as HTS. Compounds that induce chemical-shift changes for nuclei in a region on the protein’s surface that is adjacent to the site of binding of the first ligand are considered. The structural characterization of the ternary complex by NMR allows the design of potential chemical linkers of the two compounds to give a higher-affinity ligand. The binding affinity of the resulting bi-dentate compound is, in principle, higher than that of the individual compounds because of a larger number of interactions (enthalpy factor) and because of a reduced loss in translational and rotational entropy upon binding. The approach has been demonstrated to yield bi-dentate compounds with dramatically increased affinity compared with the individual fragments and, as anticipated, the method has resulted in the successful design of compounds against protein–protein interactions or more complex macromolecular targets19,20. Alternative approaches to the design of high-affinity bi-dentate compounds include those that rely on the use of a paramagnetically labelled first ligand21 or on the detection of protein-mediated ligand–ligand magnetization transfers19,22 as screening methods (TABLE 2).
Because of their simple chemical structure and smaller size, the initial fragments, also referred to as privileged binding scaffolds23, might bind to the target with KD values ranging from low micromolar to millimolar. However, rather than the absolute potency of the binding fragments, a key parameter to follow in a FBDD campaign is the ligand efficiency (or binding efficiency index)24–26, which is defined as the free energy of binding per non-hydrogen atom. The ligand efficiency is a simple and intuitive method of normalizing molecular weight and potency of a given molecule, providing a meaningful rank ordering of hit compounds25,26. Because in successful optimizations, potency and molecular weight increase linearly (with a rate of approximately 0.3 kcal per mol per atom), it is obvious that considering initial hit fragments with the most optimal ligand efficiency is crucial for the development of a potent lead molecule with an acceptable size27.
These approaches have proven very useful in deriving high-affinity ligands for challenging targets for which other approaches have failed to produce viable leads. The current view is that NMR has found a more useful role in these approaches for drug discovery than as a structure determination tool in the hit-to-lead stage. For structure determination, X-ray crystallography can provide higher resolution structures much faster and, as a result, it is far more widely used. The only exceptions are in companies that have been set up based on NMR expertise, certain large pharmaceutical companies with the necessary expertise in which NMR has consistently been shown to be valuable in this respect, or in academic research groups that focus on these applications of NMR.
So, are there other areas of drug discovery in which NMR information is clearly superior or for which there are no alternatives? We believe that this will be the case for an increasing number of interesting targets that are classified as ‘undruggable’ after nonproductive HTS campaigns. The identification of new possible targets or of possible druggable sites on known targets can also begin with NMR-based screens of fragment libraries. In fact, the simple observation that some targets yield more free energy of binding per atom for the initial binding fragments is itself a good indicator for assessing the target’s druggability and to identify potential hot spots on the target’s surface28. We therefore predict that there will be an increase in the application of NMR-based approaches in FBDD aimed towards challenging drug targets such as protein tyrosine phosphatases29 or those involved in macromolecular recognition, including protein–protein20, protein–membrane22 and protein–nucleic-acid interactions30. It should also be possible to extend some of the NMR-FBDD approaches to in-cell NMR experiments to provide, for example, mapping information from chemical-shift perturbations for serially expressed protein systems31–33. Furthermore, it can be envisioned that these approaches may also be extended to target membrane proteins, assuming that issues related to production of these proteins, such as poor yields, lack of stability and poor solubility, can be solved.
NMR in structural characterization of drug targets
The use of macromolecular NMR as a structural tool to complement X-ray crystallography is not fully exploited in industry for various reasons, but most notably because it is not rapid. It might still be viable in academic laboratories, where the choice of protein targets is less constrained by immediate commercial therapeutic relevance. It is possible that a structural focus could re-emerge with the growing interest in integral membrane proteins, particularly with the advances in solid-state NMR techniques, and NMR-based structure determination of protein–ligand complexes could also attract more attention in the future. However, NMR-based structure determination must become faster, and should take advantage of synergies with X-ray crystallography and computational tools34,35. In particular, to be viable in the fast-paced industrial setting, structure refinement by NMR must be streamlined. There is also a lack of suitable NMR structure determination software for such industrial purposes. Establishing a consortium to develop new, compatible, easy to use software, as done by and for the X-ray community in the past, could help in this respect.
Another structure-related application that might contribute to drug discovery is in computational modelling with limited NMR structural data. Our view is that the combination of molecular docking supported by limited NMR experimental constraints (either internuclear distances information11,36 or chemical-shift mapping37,38) could represent an efficient way to rapidly gather information on ligand–target complexes without full structure determination.
Importantly, an advantage of NMR in structure-based drug design projects is that it can quickly deliver information about ligand binding properties even if the receptor cannot be characterized at high resolution. In essence, whereas X-ray crystallography is arguably a superior technique in providing a detailed picture of the binding interactions at the atomic level, NMR can provide a crude but meaningful picture of the bound ligand(s), even if the receptor cannot be characterized11,36,39. In addition, the spatial relations between two binding fragments can also be readily obtained by, for example, protein-mediated ligand–ligand NOEs22 or relaxation measurements with paramagnetic, labelled reference compounds21. Although these approaches could clearly benefit from the availability of the three-dimensional structure of the target or, better, of the ternary complex, such information is not absolutely necessary for the design of high-affinity bi-dentate compounds21,22.
However, many multidomain proteins show considerable flexibility in the organization of their components during interactions with multiple ligands, and allosteric modulation of activities is of considerable significance in their activity. In contrast to a structure determined in a crystal, in which the interdomain interactions accommodate the need for the lowest crystal packing energy, structures that are determined in solution reflect a more physiological milieu and can be used to characterize the available dynamic interconversions40. NMR methods to characterize these interactions, using relaxation properties and special isotopic labelling, can be applied to complex systems in which the plasticity of interaction with ligands (or known drugs) is evident, such as protein tyrosine kinases, which are widely recognized as significant drug targets41. Magic-angle-spinning solid-state NMR has emerged in the last five years as a potential alternative method to determine protein structure, particularly for samples which could not be easily analysed before, such as native membranes, fibrils and cytoskeletal complexes. Recently, models of a potassium-channel–toxin complex, various fibrils and other receptor–agonist complexes have been published, illustrating the advances in the field39,42–49. Projects aiming to provide well-determined structures of membrane proteins are underway in several laboratories.
Concluding remarks
Macromolecular NMR works best in drug discovery when the data can be quickly integrated with that attained from other analytical techniques, and it has to be comprehensible, portable and available on a timescale that is compatible with medicinal chemistry. It seems that some years ago, researchers in modelling and bioinformatics eschewed solution NMR structures (as opposed to crystal structures) because a series of structures could be obtained, thus calling into question the accuracy and meaning of the data. This underscores the importance of integrating NMR analysis with the views of other disciplines that are major driving forces in drug discovery.
Indeed, fragment-based approaches such as the SAR by NMR strategy require excellent integration with medicinal chemistry and possibly biology. Effective use therefore implies some degree of centralized organization and a specialization of labour. In an academic setting, this must come from collaboration. We envision that a possible solution would be to engage in collaborative programmes that would bring together the state-of-the-art design of new drugs using NMR and other technologies to optimize the speed and quality of lead optimization. In particular, there is a major need for synthetic chemistry groups to collaborate in such efforts, as well as research groups to perform biological and functional testing of intermediate new compounds to combine binding studies with functional assays more efficiently. In the United States, there are several screening centres that may provide such support (see Further information).
A further important issue is that the training of researchers able to translate basic discoveries to new drugs is not well established in academia. For example, in the United States, medicinal chemistry is predominantly learned in industry. Only recently, several schools of pharmacy have either instituted or increased their investment in educating research scientists in such disciplines (see Further information). With regard to NMR, there is some controversy over how much spectroscopy is required for its effective use, but with few exceptions, there is little educational effort in NMR relative to other means of lead generation and optimization. In screening and FBDD, medicinal chemists rather than physicists, biologists or organic chemists would be the likely users of the techniques, but for NMR, the current educational focus is strongly on chemical physics and structure-determination, which appeals more to biologists or biophysicists than medicinal chemists. However, many of these students are interested in entering drug discovery in industry, which raises the question of what kind of jobs might be available for them. There is also less interest in mastering the underlying theory of a given biophysical technique, and much more on fast downloads to rapidly summarize the results of multiple techniques. This is natural, given the urgency and competition in drug discovery, but what does this imply in terms of designing an appropriate curriculum for such students? Perhaps a curriculum that includes multidisciplinary research activities, ranging from cell biology to medicinal chemistry, would be a logical way to train future scientists that are interested in drug discovery. In such a context, a good compromise between basic and applied research could also include adopting more detailed studies in which NMR is used to decompose the overall thermodynamics of binding for a given ligand–protein interaction into the enthalpic and entropic contributions from the ligand, protein and solvent50,51.
Finally, one common pitfall of the implementation of NMR in industrial drug discovery pipelines is that it is often brought in too late. Although many examples exist of successful drug discovery projects that are entirely jump-started by NMR-based approaches, it is clear that when applied in isolation, these methodologies, much like any other technique, cannot be fully effective. The successful implementation of NMR in the drug discovery process is often based on the early and effective integration of medicinal chemistry, computational approaches and biology. Training the scientists of the future based on these observations might be the long-term solution for these problems; establishing large collaborative efforts with academia or coordinating NMR-based infrastructures in industrial settings may represent short-term solutions.
FURTHER INFORMATION.
Burnham Center for Chemical Genomics: http://www.sdccg.burnham.org/
The University of California at San Diego School of Pharmacy: http://pharmacy.ucsd.edu/
The University of California at San Francisco School of Pharmacy: http://pharmacy.ucsf.edu/
Burnham Institute for Medical Research, Graduate Programs in Molecular Medicine and Integrated and Applied Biosciences: http://www.burnham.org/
Molecular Libraries Screening Centers Network initiative: http://nihroadmap.nih.gov/molecularlibraries
NIAID’s Antimicrobial Acquisition and Coordinating Facility: http://niaid-aacf.org
NCI’s Developmental Therapeutics Program: http://dtp.nci.nih.gov
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Acknowledgements
This article is based on a document drafted by I. B., C. L. and M. P., during the workshop ‘Perspectives of NMR in Drug Discovery’ held in Florence April 2007. Financial support by the NMR-Life Coordination Action LSHG-CT-2005-018, 758, by Ente CR Firenze and by MCYT-FEDER (Bio2005-00295) is gratefully acknowledged. We thank M. Fragai and C. Lipinski for suggestions and comments. We also thank L. Slivka for careful assistance in the preparation of the manuscript.
Glossary
- Druggability
The ability of a target to be modulated by a lead candidate that has the requisite physicochemical and absorption, distribution, metabolism and excretion properties for development as a drug candidate.
- Drug-like
Sharing certain characteristics — such as size, shape and solubility in water and organic solvents — with other molecules that act as drugs.
- Hit
A compound that satisfies an initial set of criteria (for example, minimum potency and solubility), but which requires elaboration or validation through further detailed analysis of performance or additional iterations to become a lead.
- Hot spots
Compact, centralized regions of residues at a protein–protein interface that are crucial for the affinity of the interaction.
- Lipinski’s “Rule of Five”
This identifies several key properties that should be considered for small molecules that are intended to be orally administered. These properties are: molecular mass <500 Da; number of hydrogen-bond donors <5; number of hydrogen-bond acceptors <10; calculated octanol–water partition coefficient (an indication of the ability of a molecules to cross biological membranes) <5.
- Relaxation rate
The terms longitudinal and transverse relaxation rates describe the rates at which nuclear magnetization returns to the equilibrium after perturbation in a non-equilibrium state.
- NMR chemical shift
The chemical shift of a particular nucleus is a measure of the dependence of the resonance frequency of the nucleus on its chemical environment.
- Nuclear Overhauser effect (NOE)
Change in the intensity of the NMR signal, which is caused by through-space dipole–dipole coupling. Upper distance constraints obtained from 1H–1H NOEs are used to determine the structure of biological macromolecules.
- Nuclear spin-relaxation
This term describes several physical processes by which nuclear magnetization that is perturbed in a non-equilibrium state returns to equilibrium. Nuclear spin relaxation rates depend on the overall rotational correlation time of the molecule and on the number and nature of interacting spins.
- Paramagnetically labelled
Nuclear spin relaxation rates are enhanced for a given nucleus when it is in close proximity to a molecule containing an electron spin (a paramagnetic molecule). Labelling a reference ligand or a target with a paramagnetic molecule can provide spatial information on the binding of a test ligand.
- Pharmacophore
The steric and electronic features of a ligand that are necessary to ensure optimal interactions with a biological target structure and to trigger (or to block) its biological response.
- Saturated ligand
NMR relaxation phenomena on the nuclei of a bound ligand result in the attenuation of its NMR signal intensities. When the signal is nearly completely suppressed, the ligand is said to be saturated.
- Selective irradiation
Application of radio frequency energy at a particular narrow frequency. This will cause the selective saturation of the resonance lines in the spectrum of nuclei that resonate at that frequency.
- Solid-state NMR
NMR measurement of the magnetic properties of nuclei in solid samples rather than of samples in solution. They are characterized by anisotropic and directionally dependent interactions that can be useful to obtain structural information.
- Two-dimensional correlation spectra
NMR experiments that exploit nuclear coupling to correlate the chemical shifts of protons with other NMR-active nuclei, most often 13C or 15N.
Footnotes
Competing interests statement
The authors declare competing financial interests: see web version for details.
Contributor Information
Maurizio Pellecchia, Burnham Institute for Medical Research, La Jolla, 92037 California, USA..
Ivano Bertini, Magnetic Resonance Center, The University of Florence, 50019 Sesto Fiorentino, Italy..
David Cowburn, New York Structural Biology Center, New York, 10027 New York, USA..
Claudio Dalvit, Experimental Therapeutics Programme, Centro Nacional de Investigaciones Oncológicas (CNIO), 28029 Madrid, Spain..
Ernest Giralt, Institute for Research in Biomedicine, Barcelona and Department of Organic Chemistry, University of Barcelona, 08028 Barcelona, Spain..
Wolfgang Jahnke, Novartis Institute for BioMedical Research, 4002 Basel, Switzerland..
Thomas L. James, School of Pharmacy, University of California at San Francisco, 94158 California, USA.
Steve W. Homans, Institute of Molecular and Cellular Biology, University of Leeds, LS2 9JT UK.
Horst Kessler, Center of Integrated Protein Science at the Technische Universität München, Department Chemie, 85747 Munich, Germany..
Claudio Luchinat, Magnetic Resonance Center, The University of Florence, 50019 Sesto Fiorentino, Italy..
Bernd Meyer, Institute for Organic Chemistry University of Hamburg, 20146 Hamburg, Germany..
Hartmut Oschkinat, Forschungsinstitut für Molekulare Pharmakologie, Berlin 13125, Germany..
Jeff Peng, University of Notre Dame, 46556 Indiana, USA..
Harald Schwalbe, Center for Biomolecular Magnetic Resonance, 60438 Frankfurt, Germany..
Gregg Siegal, Institute of Chemistry, Leiden University, 2300RA Leiden, The Netherlands..
References
- 1.Pellecchia M, Sem DS, Wuthrich K. NMR in drug discovery. Nature Rev. Drug Discov. 2002;1:211–219. doi: 10.1038/nrd748. [DOI] [PubMed] [Google Scholar]
- 2.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angewandte Chemie International Edition. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Meyer B, et al. Saturation transfer difference NMR spectroscopy for identifying ligand epitopes and binding specificities. Ernst Schering Res. Found. Workshop. 2004:149–167. doi: 10.1007/978-3-662-05397-3_9. [DOI] [PubMed] [Google Scholar]
- 4.Hajduk PJ, Olejniczak ET, Fesik SW. One-dimensional relaxation- and diffusion-edited NMR methods for screening compounds that bind to macromolecules. J. Am. Chem. Soc. 1997;119:12257–12261. [Google Scholar]
- 5.Jahnke W, Erlanson DA, editors. Fragment-based Approaches in Drug Discovery. Wiley-VCH; 2006. [Google Scholar]
- 6.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nature Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 7.Klages J, Coles M, Kessler H. In: NMR-Based Screening in Exploiting Chemical Diversity for Drug Discovery. Bartlett PA, Etzeroth M, editors. RSC Publishing; 2006. pp. 263–290. [Google Scholar]
- 8.Hajduk PJ, Burns DJ. Integration of NMR and high-throughput screening. Comb. Chem. High Throughput Screen. 2002;5:613–621. doi: 10.2174/1386207023329996. [DOI] [PubMed] [Google Scholar]
- 9.Huth JR, et al. ALARM NMR: a rapid and robust experimental method to detect reactive false positives in biochemical screens. J. Am. Chem. Soc. 2005;127:217–224. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- 10.Dalvit C, Caronni D, Mongelli N, Veronesi M, Vulpetti A. NMR-based quality control approach for the identification of false positives and false negatives in high throughput screening. Curr. Drug Discov. Technol. 2006;3:115–124. doi: 10.2174/157016306778108875. [DOI] [PubMed] [Google Scholar]
- 11.Pellecchia M, et al. NMR-based structural characterization of large protein-ligand interactions. J. Biomol. NMR. 2002;22:165–173. doi: 10.1023/a:1014256707875. [DOI] [PubMed] [Google Scholar]
- 12.Pellecchia M, et al. NMR-based techniques in the hit identification and optimisation processes. Expert Opin. Ther. Targets. 2004;8:597–611. doi: 10.1517/14728222.8.6.597. [DOI] [PubMed] [Google Scholar]
- 13.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 14.Leone M, Freeze HH, Chan CS, Pellecchia M. The Nuclear Overhauser Effect in the lead identification process. Curr. Drug Discov. Technol. 2006;3:91–100. doi: 10.2174/157016306778108884. [DOI] [PubMed] [Google Scholar]
- 15.Luy B, Frank A, Kessler H. Conformational analysis of drugs by nuclear magnetic resonance spectroscopy. In: Mannhold R, Kubinyi H, Volkers G, editors. Methods and Principles in Medicinal Chemistry. 2008. pp. 207–254. [Google Scholar]
- 16.Dalvit C, et al. A general NMR method for rapid, efficient, and reliable biochemical screening. J. Am. Chem. Soc. 2003;125:14620–14625. doi: 10.1021/ja038128e. [DOI] [PubMed] [Google Scholar]
- 17.Dalvit C, Ardini E, Fogliatto GP, Mongelli N, Veronesi M. Reliable high-throughput functional screening with 3-FABS. Drug Disc. Today. 2004;9:595–602. doi: 10.1016/S1359-6446(04)03161-7. [DOI] [PubMed] [Google Scholar]
- 18.Manzenrieder F, Frank AO, Kessler H. Phosphorus NMR spectroscopy as a versatile tool for compound library screening. Angewandte Chemie (International ed.) 2008;47:2608–2611. doi: 10.1002/anie.200705256. [DOI] [PubMed] [Google Scholar]
- 19.Becattini B, Pellecchia M. SAR by ILOEs: an NMR-based approach to reverse chemical genetics. Chemistry (Weinheim an der Bergstrasse, Germany) 2006;12:2658–2662. doi: 10.1002/chem.200500636. [DOI] [PubMed] [Google Scholar]
- 20.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 21.Jahnke W, et al. Second-site NMR screening with a spin-labeled first ligand. J. Am. Chem. Soc. 2000;122:7394–7395. [Google Scholar]
- 22.Becattini B, et al. Structure-activity relationships by interligand NOE-based design and synthesis of antiapoptotic compounds targeting Bid. Proc. Natl Acad. Sci. USA. 2006;103:12602–12606. doi: 10.1073/pnas.0603460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajduk PJ, Bures M, Praestgaard J, Fesik SW. Privileged molecules for protein binding identified from NMR-based screening. J. Med. Chem. 2000;43:3443–3447. doi: 10.1021/jm000164q. [DOI] [PubMed] [Google Scholar]
- 24.Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc. Natl Acad. Sci. USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Disc. Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 26.Cele AZ, Metz JT. Ligand efficiency indices as guideposts for drug discovery. Drug Disc. Today. 2005;10:464–469. doi: 10.1016/S1359-6446(05)03386-6. [DOI] [PubMed] [Google Scholar]
- 27.Hajduk PJ. Fragment-based drug design: how big is too big? J. Med. Chem. 2006;49:6972–6976. doi: 10.1021/jm060511h. [DOI] [PubMed] [Google Scholar]
- 28.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 29.Szczepankiewicz BG, et al. Discovery of a potent, selective protein tyrosine phosphatase 1B inhibitor using a linked-fragment strategy. J. Am. Chem. Soc. 2003;125:4087–4096. doi: 10.1021/ja0296733. [DOI] [PubMed] [Google Scholar]
- 30.Mayer M, James TL. NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 2004;126:4453–4460. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 31.Koglin A, et al. Combination of cell-free expression and NMR spectroscopy as a new approach for structural investigation of membrane proteins. Magn. Reson. Chem. 2006;44:S17–S23. doi: 10.1002/mrc.1833. [DOI] [PubMed] [Google Scholar]
- 32.Selenko P, Serber Z, Gadea B, Ruderman J, Wagner G. Quantitative NMR analysis of the protein G. B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl Acad. Sci. USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reckel S, Lohr F, Dotsch V. In-cell NMR spectroscopy. Chembiochem. 2005;6:1601–1606. doi: 10.1002/cbic.200500076. [DOI] [PubMed] [Google Scholar]
- 34.Bertini I, et al. Combining in silico tools and NMR data to validate protein-ligand structural models: application to matrix metalloproteinases. J. Med. Chem. 2005;48:7544–7559. doi: 10.1021/jm050574k. [DOI] [PubMed] [Google Scholar]
- 35.Bertini I, et al. Exploring the subtleties of drug-receptor interactions: the case of matrix metalloproteinases. J. Am. Chem. Soc. 2007;129:2466–2475. doi: 10.1021/ja065156z. [DOI] [PubMed] [Google Scholar]
- 36.Hajduk PJ, et al. SOS-NMR: a saturation transfer NMR-based method for determining the structures of protein-ligand complexes. J. Am. Chem. Soc. 2004;126:2390–2398. doi: 10.1021/ja039480v. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Westerhoff LM, Merz KM., Jr A critical assessment of the performance of protein-ligand scoring functions based on NMR chemical shift perturbations. J. Med. Chem. 2007;50:5128–5134. doi: 10.1021/jm070484a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy MA, Wyss DF. Spatial localization of ligand binding sites from electron current density surfaces calculated from NMR chemical shift perturbations. J. Am. Chem. Soc. 2002;124:11758–11763. doi: 10.1021/ja026166c. [DOI] [PubMed] [Google Scholar]
- 39.Zech SG, Olejniczak E, Hajduk P, Mack J, McDermott AE. Characterization of protein-ligand interactions by high-resolution solid-state NMR spectroscopy. J. Am. Chem. Soc. 2004;126:13948–13953. doi: 10.1021/ja040086m. [DOI] [PubMed] [Google Scholar]
- 40.Bertini I, et al. Conformational variability of matrix metalloproteinases: beyond a single 3D structure. Proc. Natl Acad. Sci. USA. 2005;102:5334–5339. doi: 10.1073/pnas.0407106102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogtherr M, et al. NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angewandte Chemie (International ed. 2006;45:993–997. doi: 10.1002/anie.200502770. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez C, Wuthrich K. NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 2003;555:144–150. doi: 10.1016/s0014-5793(03)01155-4. [DOI] [PubMed] [Google Scholar]
- 43.Hong M. Oligomeric structure, dynamics, and orientation of membrane proteins from solid-state NMR. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Luca S, Heise H, Baldus M. High-resolution solid-state NMR applied to polypeptides and membrane proteins. Acc. Chem. Res. 2003;36:858–865. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- 45.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn. Reson. Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 46.Luca S, Heise H, Lange A, Baldus M. Investigation of ligand-receptor systems by high-resolution solid-state NMR: recent progress and perspectives. Arch. Pharm. (Weinheim) 2005;338:217–228. doi: 10.1002/ardp.200400991. [DOI] [PubMed] [Google Scholar]
- 47.Castellani F, et al. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 48.Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H. Determination of solid-state NMR structures of proteins by means of three-dimensional 15N-13C-13C dipolar correlation spectroscopy and chemical shift analysis. Biochemistry. 2003;42:11476–11483. doi: 10.1021/bi034903r. [DOI] [PubMed] [Google Scholar]
- 49.Werner K, et al. Combined solid state and solution NMR studies of alpha, epsilon-15N labeled bovine rhodopsin. J. Biomol. NMR. 2007;37:303–312. doi: 10.1007/s10858-007-9143-0. [DOI] [PubMed] [Google Scholar]
- 50.Shimokhina N, Bronowska A, Homans SW. Contribution of ligand desolvation to binding thermodynamics in a ligand-protein interaction. Angewandte Chemie (International ed.) 2006;45:6374–6376. doi: 10.1002/anie.200602227. [DOI] [PubMed] [Google Scholar]
- 51.Homans SW. Water, water everywhere — except where it matters? Drug Disc. Today. 2007;12:534–539. doi: 10.1016/j.drudis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Valler MJ, Green D. Diversity screening versus focussed screening in drug discovery. Drug Disc. Today. 2000;5:286–293. doi: 10.1016/s1359-6446(00)01517-8. [DOI] [PubMed] [Google Scholar]
- 53.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 54.Lipinski CA. Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Disc. Today. 2003;8:12–16. doi: 10.1016/s1359-6446(02)02556-4. [DOI] [PubMed] [Google Scholar]
- 55.Veber DF, et al. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 56.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 57.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Disc. Today. 2005;10:987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 58.Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three’ for fragment-based lead discovery? Drug Disc. Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 59.Erlanson DA, Hansen SK. Making drugs on proteins: site-directed ligand discovery for fragment-based lead assembly. Curr. Opin. Chem. Biol. 2004;8:399–406. doi: 10.1016/j.cbpa.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Erlanson DA, McDowell RS, O’Brien T. Fragment-based drug discovery. J. Med. Chem. 2004;47:3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 61.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu. Rev. Biophys. Biomol. Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 62.Gill A, Cleasby A, Jhoti H. The discovery of novel protein kinase inhibitors by using fragment-based high-throughput x-ray crystallography. Chembiochem. 2005;6:506–512. doi: 10.1002/cbic.200400188. [DOI] [PubMed] [Google Scholar]
- 63.Hartshorn MJ, et al. Fragment-based lead discovery using X-ray crystallography. J. Med. Chem. 2005;48:403–413. doi: 10.1021/jm0495778. [DOI] [PubMed] [Google Scholar]
- 64.Moore WR., Jr Maximizing discovery efficiency with a computationally driven fragment approach. Curr. Opin Drug Discov. Devel. 2005;8:355–364. [PubMed] [Google Scholar]
- 65.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nature Rev. Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 66.Schade M, Oschkinat H. NMR fragment screening: tackling protein-protein interaction targets. Curr. Opin Drug Discov. Devel. 2005;8:365–373. [PubMed] [Google Scholar]
- 67.Villar HO, Yan J, Hansen MR. Using NMR for ligand discovery and optimization. Curr. Opin Chem. Biol. 2004;8:387–391. doi: 10.1016/j.cbpa.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Zartler ER, Shapiro MJ. Fragonomics: fragment-based drug discovery. Curr. Opin Chem. Biol. 2005;9:366–370. doi: 10.1016/j.cbpa.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 69.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 70.Langer T, et al. NMR backbone assignment of a protein kinase catalytic domain by a combination of several approaches: application to the catalytic subunit of cAMP-dependent protein kinase. Chembiochem. 2004;5:1508–1516. doi: 10.1002/cbic.200400129. [DOI] [PubMed] [Google Scholar]
- 71.Allen KN, Lavie A, Petsko GA, Ringe D. Design, synthesis, and characterization of a potent xylose isomerase inhibitor, D-threonohydroxamic acid, and high-resolution X-ray crystallographic structure of the enzyme-inhibitor complex. Biochemistry. 1995;34:3742–3749. doi: 10.1021/bi00011a032. [DOI] [PubMed] [Google Scholar]
- 72.Rutenber E, et al. Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design. J. Biol. Chem. 1993;268:15343–15346. [PubMed] [Google Scholar]
- 73.Rutenber E, et al. A new class of HIV-1 protease inhibitor: the crystallographic structure, inhibition and chemical synthesis of an aminimide peptide isostere. Bioorg. Med. Chem. 1996;4:1545–1558. doi: 10.1016/0968-0896(96)00147-2. [DOI] [PubMed] [Google Scholar]
- 74.Shoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- 75.Verlinde CLMJ, et al. In: Perspectives in Medicinal Chemistry. Testa B, Kyburz E, Fuhrer W, Giger R, editors. Verlag Helvetica Chimica Acta Basel; 1993. pp. 135–148. [Google Scholar]
- 76.Bertini I, et al. Experimentally exploring the conformational space sampled by domain reorientation in calmodulin. Proc. Natl Acad. Sci. USA. 2004;101:6841–6846. doi: 10.1073/pnas.0308641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hajduk PJ, et al. High-throughput nuclear magnetic resonance-based screening. J. Med. Chem. 1999;42:2315–2317. doi: 10.1021/jm9901475. [DOI] [PubMed] [Google Scholar]
- 78.Jahnke W. Perspectives of biomolecular NMR in drug discovery: the blessing and curse of versatility. J. Biomol. NMR. 2007;39:87–90. doi: 10.1007/s10858-007-9183-5. [DOI] [PubMed] [Google Scholar]
- 79.Kessler H, Klages J. In: Comprehensive Medicinal Chemistry II. Triggle DJ, Taylor JB, editors. Vol. 3. Oxford: Elsevier; 2006. pp. 901–920. [Google Scholar]
- 80.Klages J, Coles M, Kessler H. NMR-based Screening. Vol. 12. RSC Publishing; 2006. [Google Scholar]
- 81.Klages J, Coles M, Kessler H. NMR-based screening: a powerful tool in fragment-based drug discovery. Analyst. 2007;132:693–705. doi: 10.1039/b709658p. [DOI] [PubMed] [Google Scholar]
- 82.Coles M, Heller M, Kessler H. NMR-based screening technologies. Drug Disc. Today. 2003;8:803–810. doi: 10.1016/s1359-6446(03)02796-x. [DOI] [PubMed] [Google Scholar]
- 83.Medek A, Olejniczak ET, Meadows RP, Fesik SW. An approach for high-throughput structure determination of proteins by NMR spectroscopy. J. Biomol. NMR. 2000;18:229–238. doi: 10.1023/a:1026544801001. [DOI] [PubMed] [Google Scholar]
- 84.Hajduk PJ, Meadows RP, Fesik SW. NMR-based screening in drug discovery. Q. Rev. Biophys. 1999;32:211–240. doi: 10.1017/s0033583500003528. [DOI] [PubMed] [Google Scholar]
- 85.Frutos S, et al. Disruption of the HIV-1 protease dimer with interface peptides: structural studies using NMR spectroscopy combined with [2-(13)C]-Trp selective labeling. Biopolymers. 2007;88:164–173. doi: 10.1002/bip.20685. [DOI] [PubMed] [Google Scholar]
- 86.Salvatella X, et al. A tetraguanidinium ligand binds to the surface of the tetramerization domain of protein P53. Angewandte Chemie (International ed.) 2004;43:196–198. doi: 10.1002/anie.200352115. [DOI] [PubMed] [Google Scholar]
- 87.Mayer M, et al. Synthesis and testing of a focused phenothiazine library for binding to HIV-1 TAR RNA. Chem. Biol. 2006;13:993–1000. doi: 10.1016/j.chembiol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 88.Mayer M, James TL. Detecting ligand binding to a small RNA target via saturation transfer difference NMR experiments in D(2)O and H(2)O. J. Am. Chem. Soc. 2002;124:13376–13377. doi: 10.1021/ja027526z. [DOI] [PubMed] [Google Scholar]
- 89.Yan J, Kline AD, Mo H, Shapiro MJ, Zartler ER. The effect of relaxation on the epitope mapping by saturation transfer difference NMR. J. Magn. Reson. 2003;163:270–276. doi: 10.1016/s1090-7807(03)00106-x. [DOI] [PubMed] [Google Scholar]
- 90.Dalvit C, et al. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR. 2000;18:65–68. doi: 10.1023/a:1008354229396. [DOI] [PubMed] [Google Scholar]
- 91.Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman B. WaterLOGSY as a method for primary NMR screening: practical aspects and range of applicability. J. Biomol. NMR. 2001;21:349–359. doi: 10.1023/a:1013302231549. [DOI] [PubMed] [Google Scholar]
- 92.Jahnke W, Rudisser S, Zurini M. Spin label enhanced NMR screening. J. Am. Chem. Soc. 2001;123:3149–3150. doi: 10.1021/ja005836g. [DOI] [PubMed] [Google Scholar]
- 93.Vanwetswinkel S, et al. TINS, target immobilized NMR screening: an efficient and sensitive method for ligand discovery. Chem. Biol. 2005;12:207–216. doi: 10.1016/j.chembiol.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 94.Stebbins JL, Jung D, Leone M, Zhang XK, Pellecchia M. A structure-based approach to retinoid X receptor-α inhibition. J. Biol. Chem. 2006;281:16643–16648. doi: 10.1074/jbc.M600318200. [DOI] [PubMed] [Google Scholar]
- 95.Fejzo J, et al. The SHAPES strategy: an NMR-based approach for lead generation in drug discovery. Chem. Biol. 1999;6:755–769. doi: 10.1016/s1074-5521(00)80022-8. [DOI] [PubMed] [Google Scholar]
- 96.Lepre CA, et al. Applications of SHAPES screening in drug discovery. Comb. Chem. High Throughput Screen. 2002;5:583–590. doi: 10.2174/1386207023329950. [DOI] [PubMed] [Google Scholar]
- 97.Johnson EC, Feher VA, Peng JW, Moore JM, Williamson JR. Application of NMR SHAPES screening to an RNA target. J. Am. Chem. Soc. 2003;125:15724–15725. doi: 10.1021/ja037499s. [DOI] [PubMed] [Google Scholar]
- 98.Tarrago T, Kichik N, Segui J, Giralt E. The natural product berberine is a human prolyl oligopeptidase inhibitor. ChemMedChem. 2007;2:354–359. doi: 10.1002/cmdc.200600303. [DOI] [PubMed] [Google Scholar]
- 99.Tarrago T, Frutos S, Rodriguez-Mias RA, Giralt E. Identification by 19F NMR of traditional Chinese medicinal plants possessing prolyl oligopeptidase inhibitory activity. Chembiochem. 2006;7:827–833. doi: 10.1002/cbic.200500424. [DOI] [PubMed] [Google Scholar]
- 100.Frutos S, Tarrago T, Giralt E. A fast and robust 19F NMR-based method for finding new HIV-1 protease inhibitors. Bioorg. Med. Chem. Lett. 2006;16:2677–2681. doi: 10.1016/j.bmcl.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 101.Forino M, et al. Efficient synthetic inhibitors of anthrax lethal factor. Proc. Natl Acad. Sci. USA. 2005;102:9499–9504. doi: 10.1073/pnas.0502733102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fattorusso R, Frutos S, Sun X, Sucher NJ, Pellecchia M. Traditional Chinese medicines with caspase-inhibitory activity. Phytomedicine. 2006;13:16–22. doi: 10.1016/j.phymed.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 103.Fattorusso R, Jung D, Crowell KJ, Forino M, Pellecchia M. Discovery of a novel class of reversible non-peptide caspase inhibitors via a structure-based approach. J. Med. Chem. 2005;48:1649–1656. doi: 10.1021/jm0493212. [DOI] [PubMed] [Google Scholar]
- 104.Dalvit C. Ligand- and substrate-based 19F NMR screening: Principles and applications to drug discovery. Prog. Nuclear Magn. Reson. Spectrosc. 2007;51:243–271. [Google Scholar]
- 105.Dalvit C, Fagerness PE, Hadden DT, Sarver RW, Stockman BJ. Fluorine-NMR experiments for high-throughput screening: theoretical aspects, practical considerations, and range of applicability. J. Am. Chem. Soc. 2003;125:7696–7703. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- 106.Dalvit C, Flocco M, Veronesi M, Stockman BJ. Fluorine-NMR competition binding experiments for high-throughput screening of large compound mixtures. Comb. Chem. High Throughput Screen. 2002;5:605–611. doi: 10.2174/1386207023329923. [DOI] [PubMed] [Google Scholar]
- 107.Taylor JD, Gilbert PJ, Williams MA, Pitt WR, Ladbury JE. Identification of novel fragment compounds targeted against the pY pocket of v-Src SH2 by computational and NMR screening and thermodynamic evaluation. Proteins. 2007;67:981–990. doi: 10.1002/prot.21369. [DOI] [PubMed] [Google Scholar]
- 108.Price SW. Pulsed-field gradient nuclear magnetic resonance as a tool for studying translational diffusion: Part 1. Basic theory. Concepts Magn. Reson. 1997;9:299–336. [Google Scholar]
- 109.Price SW. Pulsed-field gradient nuclear magnetic resonance as a tool for studying translational diffusion: Part II. Experimental aspects. Concepts Magn. Reson. 1998;10:197–237. [Google Scholar]
- 110.Dehner A, Kessler H. Diffusion NMR spectroscopy: folding and aggregation of domains in p53. Chembiochem. 2005;6:1550–1565. doi: 10.1002/cbic.200500093. [DOI] [PubMed] [Google Scholar]
- 111.Hajduk PJ, et al. Discovery of potent nonpeptide inhibitors of stromelysin using SAR by NMR. J. Am. Chem. Soc. 1997;119:5818–5827. [Google Scholar]
- 112.Petros AM, et al. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J. Med. Chem. 2006;49:656–663. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]
- 113.Vazquez J, et al. Development of molecular probes for second-site screening and design of protein tyrosine phosphatase inhibitors. J. Med. Chem. 2007;50:2137–2143. doi: 10.1021/jm061481l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Becattini B, et al. Targeting apoptosis via chemical design: inhibition of bid-induced cell death by small organic molecules. Chem. Biol. 2004;11:1107–1117. doi: 10.1016/j.chembiol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 115.Chen J, et al. A fragment-based approach for the discovery of isoform-specific p38alpha inhibitors. ACS Chem. Biol. 2007;2:329–336. doi: 10.1021/cb700025j. [DOI] [PubMed] [Google Scholar]
- 116.Carulla N, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 117.Sanchez-Pedregal VM, et al. The INPHARMA method: protein-mediated interligand NOEs for pharmacophore mapping. Angewandte Chemie (International ed.) 2005;44:4172–4175. doi: 10.1002/anie.200500503. [DOI] [PubMed] [Google Scholar]