SUMMARY
PRDM16 is a member of the PR domain-containing protein family and is associated with various disease states including myelodysplastic syndrome and adult T cell leukemia, as well as developmental abnormalities such as cleft palate. It is also known to act as a regulator of cell differentiation. Expression analysis of PRDM16 is limited, especially within the developing embryo. The current study evaluated the temporal and spatial localization of PRDM16 during early mouse development (embryonic days 8.5–14.5). PRDM16 was first detected on E9.5 in a limited number of tissues and by E14.5, was expressed in a broad range of developing tissues including those of the brain, lung, kidney, and gastrointestinal tract. The expression pattern is consistent with a role for PRDM16 in the development of multiple tissues. Collectively, these studies are the first to characterize the expression of the PRDM16 gene during early murine development.
Keywords: embryo, development, MEL1, PRDM16, transcription factor
INTRODUCTION
PRDM16 (also known as MDS1/EVI1-like gene 1 [MEL1] or PFM13) was discovered from studies of (1;3)(p36;q21)-positive myeloid leukemias where it was found to be inappropriately expressed and was hypothesized to promote hyperproliferation (Mochizuki et al. 2000; Nishikata et al. 2003a). Most of the published studies to date on PRDM16 have focused on its role in these specific leukemias and have resulted in the emergence of the notion that PRDM16 is a tumor suppressor (Yoshida et al. 2004; Shing et al. 2007). PRDM16 belongs to a 17- (human) or 16- (mouse) member family of PR (PRDI-BF1 and RIZ1 homologous) domain containing proteins (PRDMs). It is a 140 kDa protein that, in addition to the PR domain, includes two DNA binding domains of either 7- or 3-C2H2 type zinc fingers. The PRDM16 gene is located on chromosome 1p36.3 and shares 56% sequence homology and a similar domain structure to MDS1/EVI1 (Mochizuki et al. 2000). MDS1/EVI1, in turn, is similar in structure to the known oncogene, EVI1. The PR domain was first identified based on its homology to a 100 amino acid sequence shared between positive regulatory domain I binding factor 1 (PRDI-BF1; PRDM1) and retinoblastoma-interacting zinc finger protein (RIZ; PRDM2) (Keller and Maniatis 1991; Buyse et al. 1995). It shares 20–30% identity with the SET domains (Suvar3–9, Enhancer-of-zeste, Trithorax) found on a class of histone methyltransferases. PRDM16 has two known splice variants: a 170 kDa isoform and a short form (sPRDM16) truncated at the amino terminus resulting in the absence of most of the PR domain (Nishikata et al. 2003b).
Recent studies have demonstrated that PRDM16 is highly expressed in brown adipose tissue (BAT) and that forced expression in preadipocytes leads to induction of the BAT cell program (Seale et al. 2007). This is accomplished, in part, by formation of a transcriptional complex that includes CtBP-1 and CtBP-2 that represses the transcription of genes associated with formation of white adipose tissue (Kajimura et al. 2008). Thus, in addition to its proposed role in cell proliferation, PRDM16 can drive certain tissue-specific differentiation. Given its size and complex domain structure, it is likely that PRDM16 forms complexes with numerous proteins, which may, in part, explain its diverse functions. We have previously demonstrated that PRDM16 forms a complex with various Smad proteins, important mediators of TGFß and BMP signaling (Warner et al. 2007). While the functional role of this interaction is unclear, regulation of TGFß-induced gene expression through modulation of the strength and/or extent of the TGFß response may be mediated by PRDM16/Smad interactions. Because Smad 3 was identified as a PRDM16 binding protein in orofacial tissue, we have proposed a role for this protein in development of this tissue, and specifically, the secondary palate (Warner et al. 2007). In support of this hypothesis is the observation that a missense mutation of PRDM16 results in an isolated cleft palate in mice (Bjork et al. 2006).
To date, studies investigating the expression of PRDM16 have been limited. Mochizuki et al. (2000) reported that PRDM16 is expressed in the uterus, fetal kidney, and leukemia cells with the t(1;3) translocation. Moreover, PRDM16 expression has also been described in adult human heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, peripheral blood, and bone marrow (Lahortiga et al. 2004). Van Campenhout et al. (2006) described PRDM16 expression in neural crest cells migrating into the second branchial arch, the otic vesicle, the brain, the retinal pigment epithelium of eye, the heart, and the kidney at various stages of Xenopus development. More recently, we observed PRDM16 expression in the heart, brain, liver, limb bud, palate, nasal septum, and upper lip within mid- to late- staged (E13.5–E14.5) mouse embryos (Warner et al. 2007). Delineation of the role of PRDM16 during embryonic development necessitates knowledge of its spatial and temporal patterns of expression. The purpose of the current study was to assess the spatial and temporal localization of PRDM16 during early and midgestational development of the mouse.
MATERIALS AND METHODS
Animals
ICR mice (Harlan Laboratories; Indianapolis, IN) were utilized for these studies. The use of animals for these experiments was approved by the University of Louisville Institutional Animal Care and Use Committee. Mice were maintained on a 12-hour light/dark cycle in an American Association for Accreditation of Laboratory Animal Care (AAALAC) approved facility at the University of Louisville. For timed matings, two females were housed overnight with a single, mature male and the presence of a vaginal plug the following morning was taken as evidence of copulation. This time point was designated as embryonic day 0.5 (E0.5). Embryos were collected on each embryonic day between E8.5 and E12.5, and on E14.5 and processed for in situ hybridization analysis as described below.
Riboprobe Synthesis
The generation of sense and antisense riboprobes to PRDM16 have been described previously (Warner, et al., 2007). Briefly, a 508 bp fragment of murine PRDM16 was cloned into plasmid pSPT-18 (Roche Diagnostics, Indianapolis, IN). The resulting plasmid was linearized with either EcoRI (for antisense probe) or HindIII (sense probe) and riboprobes synthesized with the DIG RNA labeling kit (Roche Diagnostics) followed by a series of lithium chloride precipitations to remove free nucleotides. Probe yields were determined via a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and probe quality was assessed by agarose gel electrophoresis. Only transcripts demonstrating a single, compact band were utilized for these experiments.
Sectional In situ Hybridization
Mouse embryos of the appropriate gestational age were collected into cold calcium/magnesium-free phosphate buffered saline (PBS) (Gibco Invitrogen Corp., Gaithersburg, MD) and crosslinked overnight in fresh 4% (w/v) paraformaldehyde (Polysciences, Warrington, PA) / PBS. Following crosslinking, embryos were processed through a series of overnight sucrose washes (30% [w/v] sucrose (Fisher Scientific, Pittsburgh, PA) / PBS then 1:1 30% [w/v] sucrose/PBS, and finally Optimum Cutting Temperature compound [OCT, Sakura Finetek, Torrence, CA]), frozen in OCT, and stored at −80 °C until sectioned. Frozen serial sections, eight to twelve microns (8–12 um) thick, were prepared using a cryostat (Leica CM1900; Leica Microsystems Nussloch GmbH, Nussloch, Germany), collected onto RNase-free glass slides (VWR, Media, PA), and dried overnight at room temperature to ensure adherence of the sections. Slide-mounted sections were then crosslinked with 4% (w/v) paraformaldehyde/PBS and endogenous peroxidases were quenched with 6% (v/v) hydrogen peroxide (Fisher Scientific). Tissue sections were permeabilized with 1 μg/ml proteinase K (Roche Diagnostics) for 15 minutes. Digestion was stopped by incubation for 10 minutes in 2 mg/ml glycine (Fisher Scientific). Sections were then washed with PBT (PBS + 0.1% (v/v) Tween-20 [Fisher Scientific]) and again crosslinked in 4% (w/v) paraformaldehyde/0.2% (v/v) glutaraldehyde (Fisher Scientific) / PBT for 15 minutes at room temperature. Sections were then prehybridized at 65°C for 1 hour (50% formamide; 5X SSC, pH 4.5; 50 μg/ml yeast tRNA, 1% SDS; 50ug/ml heparin, all from Sigma Chemical Co., St. Louis, MO) and then incubated overnight at 65°C in the same solution containing 500–1000 ng/ml DIG-labeled riboprobe. Sections were then washed, blocked for one hour in 5% sheep serum (Sigma Chemical Co.), and again incubated overnight at 4°C with alkaline phosphatase-labeled anti-digoxigenin antibody (0.375 units/ml, Roche Diagnostics). Riboprobe-specific hybridization was detected by exposing sections to (188 μg/ml NBT / 94 μg/ml BCIP (nitro blue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl phosphate) (Roche Diagnostics) and 2 mM tetramisole (Sigma Chemical Co.) for three to four days at in the dark at room temperature. Color development was quenched by washing with PBS (pH 5.5), and sections mounted in Biomedia™Crystal/Mount (Electron Microscopy Sciences, Hatfield, PA). For each embryonic day analyzed, at least two embryos were fully sectioned and 5–6 slides from each embryo were analyzed with either the sense- or antisense-PRDM16 riboprobe, with similar patterns of expression observed.
Image acquisition
Images were taken with a Nikon DXM1200F digital camera controlled by a computer running ACT-1 acquisition software and mounted on a Nikon Eclipse E600 microscope (Nikon, Inc., Melville, NY). All images were collected under similar lighting and are presented without digital alteration.
RESULTS
To gain insight into the role that PRDM16 may play in embryonic development, we evaluated the temporal and spatial localization of PRDM16 mRNA during early and midgestational development of the mouse (E8.5–E14.5).
Embryonic day 9.5
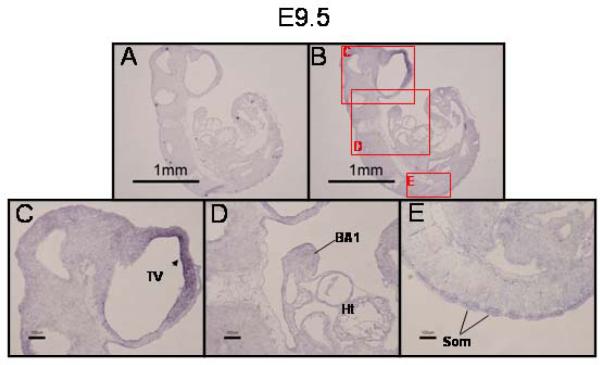
While expression of PRDM16 mRNA was not observed by in situ hybridization until E9.5, we detected PRDM16 by RT-PCR in earlier embryos (E8.5, data not shown). This is likely because the level of expression on E8.5 may be below the limit of detection by in situ hybridization. On E9.5, however, expression of PRDM16 was observed predominately in the neuroepithelium lining the telencephalic vesicle (TV), within the mesenchyme of the first branchial arch (BA1), and in the dorsal aspect of the somites (Som) (Fig. 1, panels B–E). No signal was detected when a PRDM16 sense riboprobe was used (Fig. 1, panel A).
Figure 1. Expression of PRDM16 in the E9.5 mouse embryo.
Fixed, frozen E9.5 embryos were sectioned and subjected to in situ hybridization with a PRDM16-specific riboprobe. A) PRDM16 sense riboprobe negative control. B) PRDM16 antisense riboprobe. Lettered red boxes in panel B indicate areas shown in higher magnification in panels C–E. C) PRDM16 was predominately expressed in the neuroepithelial lining of the telencephalic vesicle (TV, arrowhead). D) Expression was also detected in the first branchial arch (BA1) and in the dorsal aspect of the somites (Som) (panel E). Ht, heart. Scale bar = 1 mm in panels A and B and 100 μm in panels C–E. Panel E was taken from a section adjacent to the one presented in panel B.
Embryonic day 10.5
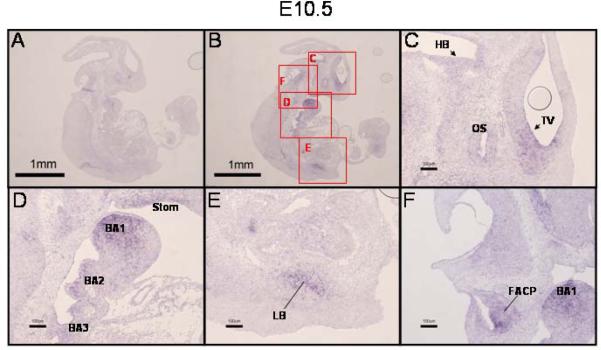
As development proceeds, expression of PRDM16 is found in an increasing number of tissues. On E10.5 expression remained in the telencephalic vesicle (TV) and was also found in the neuroepithelium of the hindbrain (HB) and in the optic stalk (OS) (Fig. 2C). Expression continued in the first branchial arch (BA1) and was seen to have expanded into the mesenchyme of the second and third branchial arches (BA2 and BA3, respectively, Fig. 2D) and into the epithelium of the stomadeal roof (Stom) (Fig. 2D). The limb bud (LB) (Fig. 2E) and the facio-acoustic preganglion complex (FACP) (Fig. 2F) were also PRDM16-positive.
Figure 2. Expression of PRDM16 in the E10.5 mouse embryo.
A,B) In situ hybridization with PRDM16 sense or antisense riboprobe, respectively (low magnification, 20X). Lettered red boxes in panel B indicate areas shown in higher magnification in panels C–F (100X). C) Similar to E9.5, expression of PRDM16 was detected in the neuroepithelium lining telencephalic vesicle (TV, arrowhead). (Note: the circle within the TV in panel C is an artifact (air bubble)). Expression of PRDM16 was also detected in the neuroepithelium of the hindbrain (HB, arrowhead) and in the optic stalk (OS). D) Notable expression of PRDM16 was seen in the first, second and third branchial arches (BA1, BA2, BA3) and in the epithelium of the stomadeal roof (Stom). PRDM16 was expressed in the developing lung bud (LB, panel E) and in the facio-acoustic preganglion complex (FAPC, panel F). Scale bar = 1 mm in panel A and B and 100 μm in panels C–F.
Embryonic day 11.5
By E11.5, expression of PRDM16 was found in the medial and lateral nasal processes (MNP and LNP, respectively) (Fig. 3C) and continued to be present in the first branchial arch (BA1) (Fig. 3C and 3D). PRDM16 expression was seen in the developing liver (L) and heart (Ht) (Fig. 3E), as well as in the trigeminal ganglion (TNG) (Fig. 3F) and the vertebral region of the sclerotome (VR) (Fig. 3G).
Figure 3. PRDM16 expression in the E11.5 mouse embryo.
A) In situ hybridization of sagittal sections of an E11.5 mouse embryo with a PRDM16 sense riboprobe (negative control) demonstrating the extent of background staining. B) A section similar to that shown in panel A probed with a PRDM16 antisense probe revealing staining in multiple tissues. Lettered boxes in panel B indicate areas shown in higher magnification in panels C–G. PRDM16 expression was detected in the medial nasal process (MNP) and the first branchial arch (BA1) (panels C and D). Expression was also detected in the liver (L) and heart (Ht) (panel E), as well as in the ganglion of the trigeminal nerve (TNG, panel F), and vertebral region (VR, panel G). Scale bar = 1 mm in panels A and B and 100 μm in panels C–G.
Embryonic day 12.5
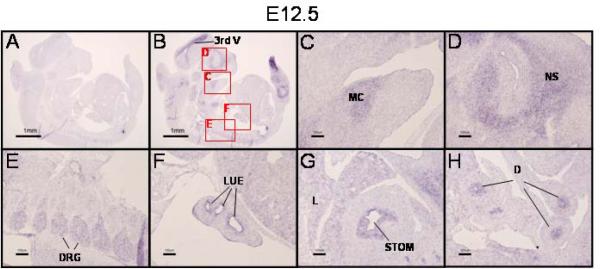
On E12.5, PRDM16 expression continued in the brain, specifically in the neuroepithelium lining the third ventricle (3rd V, Fig. 4B). In the orofacial region, PRDM16 was found in Meckel's cartilage (MC) (Fig. 4C) and the future nasal septum (NS) (Fig. 4D). In addition, PRDM16 was also expressed in the dorsal root ganglia (DRG) (Fig. 4E), lung epithelium (LUE) (Fig. 4F), liver (L), stomach (STOM) (Fig. 4G) and duodenum (D)(Fig. 4H).
Figure 4. PRDM16 is expressed in multiple tissues on E12.5.
A,B) In situ hybridization with PRDM16 sense or antisense riboprobe, respectively. Lettered boxes in panel B indicate areas shown in higher magnification in panels C–H. On E12.5 PRDM16 was still expressed in the neuroepithelium lining the third ventricle (3rd V, panel B). In addition, PRDM16 is expressed in Meckel's cartilage (MC, panel C), the nasal septum (NS, panel D), dorsal root ganglia (DRG, panel E) and lung epithelium (LUE, panel F). Expression is now also seen in the developing gastrointestinal system in the stomach (STOM, panel G) and duodenum (D, panel H). Panels G and H were photographed from sections adjacent to that presented in panel B in order to demonstrate expression of PRDM16 in the developing gut. Scale bar = 1 mm in panels A and B and 100 μm in panels C–H.
Embryonic day E14.5
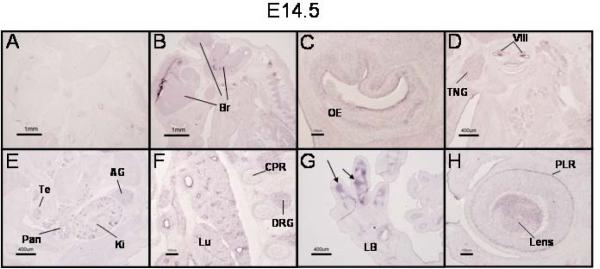
On E14.5, there was widespread expression of PRDM16. The brain (Br) maintained PRDM16 expression (Fig. 5B). In the orofacial region, PRDM16 was expressed in the oral epithelium (OE), vestibulocochlear ganglion (VIII), trigeminal ganglion (TNG), and submandibular glands (SMG) (Figs. 5C and 5D). Additional tissues in which PRDM16 expression was detected included the pancreatic primordium (Pan), testes (Te), kidney (Ki), and adrenal gland (AG) (Fig. 5E). Expression continued to be seen in the developing lung (Lu) and dorsal root ganglia (DRG) (Fig. 5F). Expression was also detected in the cartilage primordia of the developing rib (CPR, Fig. 5F). In the developing limbs (limb bud, LB), PRDM16 expression was restricted to the perichondrial region (Fig. 5G, arrows). Finally, expression in the eye was found in the pigment layer of the retina and in the developing lens (PLR and Lens, respectively, Fig. 5H).
Figure 5. Broad expression of PRDM16 in E14.5 mouse embryos.
In situ hybridization of sagittal sections of an E14.5 mouse embryo with a PRDM16 sense (A) and antisense (B–H) riboprobes. Expression of PRDM16 during this stage of development encompasses multiple tissues including widespread signals in the developing brain (Br, panel B) and olfactory epithelium (OE, panel C). Panel D demonstrates expression of PRDM16 in the trigeminal ganglion (TNG), the vestibulocochlear ganglion (VIII), and submandibular glands (SMG). Expression was also observed in the developing testis (Te), pancreas (Pan), kidney (Ki) and adrenal gland (AG) (panel E). The developing lung (Lu) also expressed PRDM16 as well as the dorsal root ganglia (DRG) and cartilage primordia of the ribs (CPR) (panel F). In the hindlimb, PRDM16 expression was restricted to the perichondrium surrounding the developing tarsal bones (panel G, arrows). Expression was detected in the eye in the pigment layer of the retina (PLR) and in the lens (Lens, panel H). Scale bars: panels A and B = 1 mm; C, F, and H = 100 μm; and D, E, and G = 400 μm.
DISCUSSION
The expression of PRDM16 was first detected by in situ hybridization on E9.5. Between E9.5 and E14.5, the expression pattern of PRDM16 quickly expanded to include many types of tissue. There was consistent expression in the developing brain and other neural tissue, branchial arches, and nascent lung throughout the gestational period examined. In addition, PRDM16 was expressed in the developing liver, gut, and eye. Based upon previous analyses (Lahortiga et al. 2004), much of this pattern of expression persists into adulthood. Because PRDM16 is a putative transcription factor capable of interacting with many different proteins, it may have unique functions, depending upon the cell type in which it is expressed and thus may be critical for the modulation of multiple signaling pathways. We have recently demonstrated that PRDM16 interacts with multiple members of the Smad family of nucleocytoplasmic proteins that transmit extracellular signals from TGFßs and BMPs to the nucleus (Warner et al. 2007). Therefore it is possible that PRDM16 modulates these two pathways in embryonic development. Recently, it was shown that PRDM16 is necessary for differentiation of muscle precursor cells into a brown fat cell lineage (Seale et al. 2008), in part through interaction with CtBP (Seale et al. 2007). The fact that BMP-7 regulates brown fat cell differentiation through up-regulation of PRDM16 (Tseng et al. 2008) further supports the association between PRDM16 and BMP signaling. In a recent paper, Kinameri, et al. examined the expression of several members of the PRDM gene family, including PRDM16, during mouse neurogenesis (Kinameri et al. 2008). They describe a brain expression pattern for PRDM16 very similar to what we have observed and reported here.
One of the hallmarks of early embryonic development is rapid growth due, in part, to robust cell proliferation. The broad expression of PRDM16 in multiple cell types, in addition to its demonstrated role in cell differentiation, as described above, supports the notion of a role for the PRDM protein in cell proliferation. Indeed, because of the association of PRDM16 with specific cancers and similarities to MDS1/EVI1, early work focused on its role in regulating cell proliferation. Like PRDM16, MDS1/EVI is a PR-domain containing protein that results from fusion of the MDS1 gene (containing the PR-domain) to the oncogene, EVI1 (Fears et al. 1996). In addition to MDS1/EVI1 (Sood et al. 1999), EVI1 itself has been linked to myeloid malignancies (Trubia et al. 2006). It is likely that a balance between the PR-domain-positive isoforms vs. PR-domain-minus isoforms is important for the control of cell proliferation, as has been previously suggested for other PR-domain containing proteins (Nucifora et al. 2006). Two isoforms of PRDM16 have been reported, with a shorter variant missing most of the PR domain. Aberrant expression of this variant may lead to myelodysplastic syndrome, suggesting that the PR-domain confers tumor suppressive activity (Xiao et al. 2006). We were unable to distinguish between the two isoforms of PRDM16 in our in situ hybridization analyses. Evi1 has been shown to interact with several proteins such as Smad3, GATA1, HDAC, P/CAF, JNK, CBP, and CtBP that are involved in transcriptional regulation (Wieser 2007). Based on sequence conservation, it is possible that PRDM16 binds a similar repertoire of proteins.
Given the similarity of the PR domain to the SET domain found in histone methyltransferases, it is also possible that PRDM16 may modulate epigenetic control of gene transcription. This adds another layer of complexity to the possible roles for this protein. The identification of both upstream effectors and downstream targets are necessary to provide clarification of the specific role of PRDM16 in modulation of developmental signaling pathways, and tissue homeostasis.
In summary, the results reported in this study represent the first to describe the temporal and spatial expression of PRDM16 in the developing murine embryo and are an important first step towards unraveling the functional roles for this protein.
ACKNOWLEDGEMENTS
This work was supported in part by PHS grants HD53509 and DE018215 to RMG, AA13205 to MMP, and a COBRE grant (P20RR017702) from the National Center for Research Resources. Additional support was provided by the Commonwealth of Kentucky Research Challenge Trust Fund and grant to DRW from the Kentucky Science and Engineering Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bjork BC, Vieira AR, Faust S, Camper SA, Murray JC, Beier DR. Phenotypic, Genetic, and Developmental Characterization of CPO1, a Recessive ENU-induced Mouse Model of Cleft Palate. Mouse Molecular Genetics Cold Spring Harbor Press; Woodbury, NY: 2006. p. 27. [Google Scholar]
- Buyse I, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci U S A. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley JD, Nucifora G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc Natl Acad Sci U S A. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes and Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Kinameri E, Inoue T, Aruga J, Imayoshi I, Kageyama R, Shimogori T, Moore AW. Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE. 2008;3:e3859. doi: 10.1371/journal.pone.0003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahortiga I, Agirre X, Belloni E, Vazquez I, Larrayoz MJ, Gasparini P, Lo Coco F, Pelicci PG, Calasanz MJ, Odero MD. Molecular characterization of a t(1;3)(p36;q21) in a patient with MDS. MEL1 is widely expressed in normal tissues, including bone marrow, and it is not overexpressed in the t(1;3) cells. Oncogene. 2004;23:311–316. doi: 10.1038/sj.onc.1206923. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003a;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, Nakamura T, Morishita K. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. 2003b;102:3323–3332. doi: 10.1182/blood-2002-12-3944. [DOI] [PubMed] [Google Scholar]
- Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: history and perspectives. Gene. 2006;368:1–11. doi: 10.1016/j.gene.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork BC, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing DC, Trubia M, Marchesi F, Radaelli E, Belloni E, Tapinassi C, Scanziani E, Mecucci C, Crescenzi B, Lahortiga I, Odero MD, Zardo G, Gruszka A, Minucci S, DiFiore PP, Pelicci PG. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Invest. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Talwar-Trikha A, Chakrabarti SR, Nucifora G. MDS1/EVI1 enhances TGF-beta1 signaling and strengthens its growth-inhibitory effect but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-beta1. Leukemia. 1999;13:348–357. doi: 10.1038/sj.leu.2401360. [DOI] [PubMed] [Google Scholar]
- Trubia M, Albano F, Cavazzini F, Cambrin GR, Quarta G, Fabbiano F, Ciambelli F, Magro D, Hernandezo JM, Mancini M, Diverio D, Pelicci PG, Coco FL, Mecucci C, Specchia G, Rocchi M, Liso V, Castoldi G, Cuneo A. Characterization of a recurrent translocation t(2;3)(p15–22;q26) occurring in acute myeloid leukaemia. Leukemia. 2006;20:48–54. doi: 10.1038/sj.leu.2404020. [DOI] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Campenhout C, Nichane M, Antoniou A, Pendeville H, Bronchain OJ, Marine JC, Mazabraud A, Voz ML, Bellefroid EJ. Evi1 is specifically expressed in the distal tubule and duct of the Xenopus pronephros and plays a role in its formation. Dev. Biol. 2006;294:203–219. doi: 10.1016/j.ydbio.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Warner DR, Horn KH, Mudd L, Webb CL, Greene RM, Pisano MM. PRDM16/MEL1: A novel Smad binding protein expressed in murine embryonic orofacial tissue. Biochim Biophys Acta. 2007;1773:814–820. doi: 10.1016/j.bbamcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Wieser R. The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene. 2007;396:346–357. doi: 10.1016/j.gene.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Zhang M, Liu X, Zhang Y, Yang L, Hao Y. MEL1S, not MEL1, is overexpressed in myelodysplastic syndromes patients with t(1;3)(p36;q21) Leuk. Res. 2006;30:332–334. doi: 10.1016/j.leukres.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Nosaka K, Yasunaga J, Nishikata I, Morishita K, Matsuoka M. Aberrant expression of the MEL1S gene identified in association with hypomethylation in adult T-cell leukemia cells. Blood. 2004;103:2753–2760. doi: 10.1182/blood-2003-07-2482. [DOI] [PubMed] [Google Scholar]