Abstract
The accumulation of base substitutions (mutations) not subject to natural selection is the neutral mutation rate. Because this rate reflects the in vivo processes involved in maintaining the integrity of genetic information, the factors that affect the neutral mutation rate are of considerable interest. Mammals exhibit two dramatically different neutral mutation rates: the CpG mutation rate, wherein the C of most CpGs (i.e., methyl-CpG) mutate at 10–50 times that of C in any other context or of any other base. The latter mutations constitute the non-CpG rate. The high CpG rate results from the spontaneous deamination of methyl-C to T and incomplete restoration of the ensuing T:G mismatches to C:Gs. Here, we determined the neutral non-CpG mutation rate as a function of CpG content by comparing sequence divergence of thousands of pairs of neutrally evolving chimpanzee and human orthologs that differ primarily in CpG content. Both the mutation rate and the mutational spectrum (transition/transversion ratio) of non-CpG residues change in parallel as sigmoidal (logistic) functions of CpG content. As different mechanisms generate transitions and transversions, these results indicate that both mutation rate and mutational processes are contingent on the local CpG content. We consider several possible mechanisms that might explain how CpG exerts these effects.
DNA base substitutions (mutations) are the most frequent class of genetic variants. Thus, determining the factors that affect the base mutation rate (i.e., the number of base substitutions over time) remains a major concern of geneticists and molecular evolutionists (e.g., Nachman and Crowell 2000; Hwang and Green 2004; Duret 2009). Mutations not subject to natural selection are considered neutral and the neutral mutation rate is considered to closely reflect or equal the actual mutation rate (Ochman 2003). Thus, the neutral mutation rate is a basic biological parameter, which can be estimated from the number of interspecies base differences (sequence divergence) between neutrally evolving orthologous sequences (i.e., those sharing a common ancestral sequence, e.g., Nachman and Crowell 2000).
The neutral mutation rate varies considerably between and within chromosomes. Although numerous factors have been correlated with the neutral mutation rate (e.g., Krawczak et al. 1998; Hardison et al. 2003; Hwang and Green 2004; Chimpanzee Sequencing Analysis Consortium 2005; Gaffney and Keightley 2005; Hellmann et al. 2005; Taylor et al. 2006), the mechanism(s) accounting for these correlations remain elusive (e.g., Hodgkinson et al. 2009; for review, see Duret 2009).
One of the more intriguing covariates of the neutral mutation rate is its positive correlation with CpG content. In part, this correlation is not surprising because most CpGs in mammals are uniquely hypermutable (e.g., Hwang and Green 2004). The Cs of most CpGs are methylated (Ehrlich and Wang 1981), which enhances the deamination of C, in this case producing a T:G mismatch. The net result is that methyl-CpGs mutate at 10–50 times the rate of C in any other context (Coulondre et al. 1978; Duncan and Miller 1980; Bulmer 1986; Sved and Bird 1990), or of any other base (Hwang and Green 2004). Consequently, CpGs not under selection are replaced over time by TpG/CpAs.
Inexplicably, however, the positive correlation between CpG content and the neutral mutation rate persists even if mutations at CpG sites are not counted, i.e., if only non-CpG mutations are measured (Chimpanzee Sequencing Analysis Consortium 2005; Gaffney and Keightley 2005; Hellmann et al. 2005; Tyekucheva et al. 2008). A prevailing reasonable explanation for this odd result was that the non-CpG mutation rate and CpG content were joint manifestations of the chromosomal environment (Hellmann et al. 2005; Tyekucheva et al. 2008).
We recently considered an alternative, that CpGs (i.e., methyl-CpGs), or mutations thereof, somehow directly affect the mutation of flanking non-CpG DNA. This explanation would have far-reaching implications given the epigenetic role of CpG methylation in gene regulation, chromatin structure, imprinting, and the silencing of transposable elements and other genomic insertions (Lees-Murdock and Walsh 2008; Cedar and Bergman 2009). We addressed this issue by examining the sequence divergence of thousands of neutrally evolving orthologous sequences in the chimpanzee and human genomes that differed primarily in CpG content (Walser et al. 2008).
These orthologs were the repeated DNA fossils that had been interspersed throughout the genome at different times in the primate lineage of humans and chimpanzees by six now extinct families of L1 non-LTR retrotransposons. As L1 fossils are not under selection, the CpG content of these otherwise very similar sequences should differ. Thus, the CpG content of the younger L1 fossils should be higher than that of the older ones and, if our supposition was correct, so should their mutation rate, regardless of chromosomal location. And this is the result we obtained (Walser et al. 2008).
Our current examination of non-CpG mutations over a considerably wider range of CpG content than previously (Walser et al. 2008) revealed two unexpected findings: First, the correlation between the two is best fit by a sigmoid (logistic) function. Thus, both a certain threshold CpG content is required to have a substantial effect on the overall non-CpG mutation rate, and “saturation” is reached—increases of CpG content above a certain level are not accompanied by marked increases in non-CpG mutations.
Second, and most provocatively, the mutational spectrum closely parallels the changes in non-CpG mutation rate. In particular, changes in the ratio of transitions (purine or pyrimidine interchanges) to transversions (purine/pyrimidine interchanges) parallel the change in overall non-CpG mutation rate.
These results substantiate the idea that CpGs, or mutations thereof, directly affect the mutational environment of neighboring non-CpG DNA (Walser et al. 2008). As importantly, this more precise delineation of the “CpG effect” on mutation rates permits the framing of experimental approaches that could reveal its biochemical basis.
Results
Using L1 orthologs to determine the neutral mutation rate as a function of CpG content
We, and others, have previously described the benefits of using the interspecies sequence divergence of the assuredly neutrally evolving, interspersed repeated DNA fossils generated by transposable elements for estimating neutral mutation rates (Hardison et al. 2003; Chimpanzee Sequencing Analysis Consortium 2005; Gaffney and Keightley 2005; Hellmann et al. 2005; Tyekucheva et al. 2008; Walser et al. 2008; Mugal et al. 2009). The well-defined lineage of L1 non-LTR retrotransposons in primates (International Human Genome Sequencing Consortium 2001; Khan et al. 2006; Furano and Boissinot 2008) comprises a set of closely related but nonetheless distinct L1 families that amplified and then went extinct at different times during the last ∼80 million years (Myr) of primate evolution. Thus, L1 DNA is particularly suitable for examining the correlation between CpG content and non-CpG mutation rate.
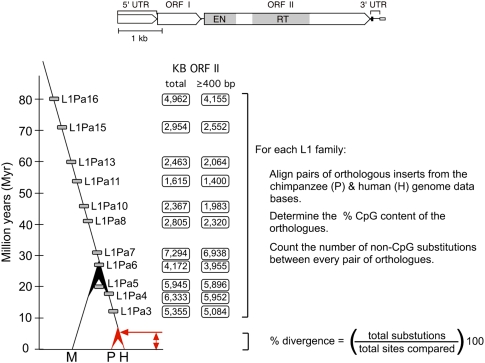
Figure 1 shows the 11 L1 families that we examined placed on a simplified primate phylogenetic tree according to their average age (see Methods). None of these long-extinct families contain active members. Despite their age differences these distinct families are highly similar as exemplified by both their dinucleotide composition and primary sequence (Supplemental Table S1, Supplemental Fig. S8; Walser et al. 2008; Furano and Walser 2009). Thus, the widely dispersed members of these families provide a common substrate for mutation despite their chromosomal location. However, because of their different times in the genome, the CpG content of the different families differ due to the clock-like conversion of CpG to TpG/CpA (Hwang and Green 2004; Walser et al. 2008; Furano and Walser 2009).
Figure 1.
Primate L1 families. A full-length generic primate L1 element is shown. The primate-specific L1Pa families examined here are placed on a simplified primate tree according to their average ages (see Methods). The columns give the KB of ORFII orthologs of the various families used for the various analyses in this paper (see Methods). M, P, and H indicate Macaca mulatta (macaque, Old World monkey), Pan troglodytes (chimpanzee), and Homo sapiens (human), respectively. The ages for the divergences of theses species were derived as described earlier (Walser et al. 2008). See Methods and Results, for details on the various steps outlined on the right side of the figure.
Figure 1 shows our procedure for determining the non-CpG mutation rate as a function of CpG content (explained in detail in Methods and Furano and Walser 2009; Walser et al. 2008). In summary, we collected and aligned pairs of orthologous inserts of each L1 family from the chimpanzee (P) and human (H) genome databases. We then determined both their % CpG content and % non-CpG nucleotide differences. As orthologous sequences are identical by descent, the nucleotide differences represent only the substitutions that occurred since chimpanzees and human diverged from their common ancestor (red double arrow, Fig. 1).
In contrast to earlier work (Walser et al. 2008), here we limited our divergence measurements to the ORF2 sequence: We thereby could use the highly conserved ORF2 protein sequence as a scaffold for aligning the DNA sequences of the older L1 families. Additionally, 5′-truncated members dominate the older families. To provide sufficient DNA sequence for robust statistical analysis we limited ourselves to families from which we could obtain at least 1.5 megabases of ORF2 sequence (Fig. 1).
CpG content and the age and divergence of L1 orthologs
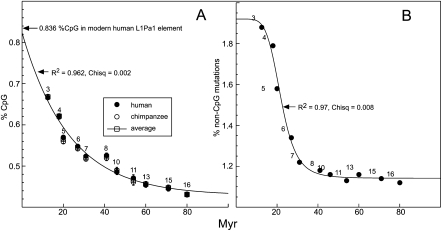
Figure 2A shows that the % CpG content of the orthologs decays exponentially with time (i.e., with the ages of the L1 families) and extrapolates at zero time to about that of the 0.836% CpG content of the ORF2 of the modern active L1Pa1 family (i.e., Ta1, Boissinot et al. 2000). However, Figure 2B shows the relationship between non-CpG divergence and family age is dramatically different. In this case the change in divergence fits a sigmoid (dose response) curve.
Figure 2.
Percent CpG content and non-CpG mutations as a function of L1 family age. The percent CpG content and non-CpG mutations were determined as described in the Methods. The relationship of CpG content to age is best fit by an exponential decay function (A), but that of the non-CpG mutations is best fit by a sigmoid (logistic) function (B; see text). Numbers indicate L1Pa families.
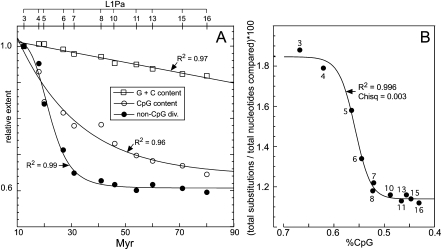
Figure 3 shows the CpG content, non-CpG divergence, and % (G + C) of each L1 family normalized to that of L1Pa3 and illustrates three important findings: First, panel A highlights the discordance between the decreases in non-CpG divergence and CpG content. For example, relative to the L1Pa3 orthologs the non-CpG mutation rate of the L1Pa7 orthologs has decreased by ∼38%, but their CpG content by only ∼18%.
Figure 3.
CpG, G + C, and non-CpG mutations. (A) The % (G + C) and CpG contents of the various L1 orthologs and their % non-CpG mutations are normalized to those of L1Pa3 (set as 1) and plotted as a function of L1 family age. The axis at the top of the panel indicates which L1Pa families correspond to the data points in the body of the panel. (B) The % non-CpG mutations are plotted as a function of % CpG content. The non-CpG divergences fit a sigmoidal function of CpG content better than a linear function using either all or subsets of the data (see text and Supplemental Fig. S7). Numbers indicate L1Pa families.
Second, following from the first, the progressive decrease in CpG content of L1 families older than L1Pa7 was accompanied by only modest changes in their overall non-CpG divergence. These results are exemplified in Figure 3B where the change in ortholog divergence is plotted as a function of their CpG content. As L1 age is a proxy for CpG content (Fig. 2), the correlation between overall divergence and CpG content also best fits a sigmoid (“dose response”) function. Thus, the correlation between overall ortholog divergence and CpG content above ∼0.63% or below ∼0.53% CpG content is greatly attenuated compared to the divergence changes between these values. Various linear fits between non-CpG divergence and CpG content (Supplemental Fig. 7S) were not as robust as the sigmoid fit. However, a bilinear fit substantiated the dramatic difference in the relationship between divergence and CpG content above and below 0.53% CpG (Supplemental Fig. 7S).
Third, Figure 3A shows that the pronounced decrease in non-CpG divergence between the L1Pa3 and L1Pa7 orthologs occurred in the face of minimal change in the % (G + C) content (see also Supplemental Table S1). This latter finding recapitulates our previous ones (Walser et al. 2008), which showed that CpG content was the only substantive covariate of non-CpG divergence that we identified among other previously noted ones; e.g., recombination rate, transcription, G + C content (Krawczak et al. 1998; Green et al. 2003; Hardison et al. 2003; Hwang and Green 2004; Chimpanzee Sequencing Analysis Consortium 2005; Gaffney and Keightley 2005; Hellmann et al. 2005; Taylor et al. 2006; Mugal et al. 2009).
Also, here we found no correlation between non-CpG divergence of ortholog pairs with recombination rate—as assessed by either linkage analysis or SNP content of their flanking sequences (0.5 Mb), their transcriptional orientation, or their G + C content or that of their flanking sequences (0.5 Mb) (Fig. 3A; Supplemental Data; see also Walser et al. 2008).
The mutational spectrum is correlated with CpG content
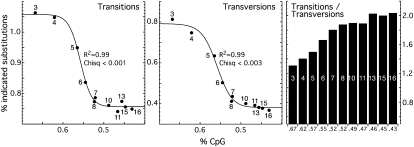
We determined whether the mutational spectrum varies with CpG content by identifying the base changes undergone by each mutated base, using the consensus sequence as a proxy for the ancestral sequence (see Methods). Figure 4 shows the total transitions and transversions that occurred between the ortholog pairs of the L1 families as a function of their CpG content. The changes in transition and transversion mutations with CpG content closely parallel that of the total non-CpG mutations (Fig. 2B). But the transversion rate falls about twice as much as the transition rate. This difference is reflected in an increase in the transition/transversion rate from ∼1.3 for L1Pa3 to ∼2 for the oldest L1 families. These changes are sufficient to produce statistically significant differences between the proportions of these transition and transversion mutations for the various L1 orthologs.
Figure 4.
Percent total transitions, transversions, and transition/transversion ratio as a function of CpG content. Mutations undergone by the chimpanzee and human members of the ortholog pairs from each L1Pa family were classified as transitions or transversions as described in the Methods (Determination of the Mutational Spectrum) and plotted as a function of the % CpG content. The transition/transversion ratio for each L1Pa family and its corresponding CpG content are plotted in the right most panel.
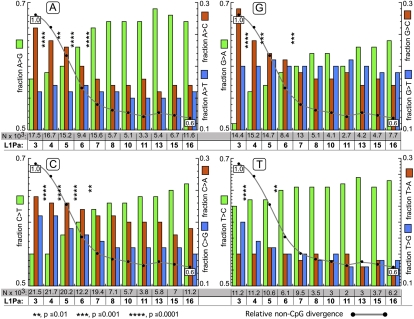
Figure 5 shows the fraction of transition and transversion mutations for each base for the L1Pa3–L1Pa16 orthologs. Although the Figure seems complex it is relatively easy to follow: The mutations undergone by each base are presented in separate panels (A, G, C, T). Each panel contains two x-axes: The one outlined in the gray box gives the total number of mutations (N × 103) and the one below it gives the name of the L1 family. For example panel A shows that L1Pa3 underwent ∼17,500 A mutations. The fate of these mutations is given on the two y-axes: On the left, the green bar gives the fraction of transitions to G, and on the right the fractions of transversions to C (red bars) or T (blue bars). The same pattern is shown for each of the other panels (transitions on the left y-axis, transversions on the right). In all panels the gray line shows the change in overall mutation rate for each family relative to L1Pa3 set to 1.
Figure 5.
Mutational fate of each of the four bases for different L1 families. Panel A shows the distribution of A mutations to G, C, and T that occurred between the human and chimpanzee lineages for each L1Pa family. There are two x-axes on the bottom of panel A: the top one (boxed in gray) gives the total mutations (N × 103) that occurred and the bottom one (in bold) indicates the L1Pa family. For example, we found (Methods, Determination of the Mutational Spectrum) that 17,458 substitutions of A occurred between the chimpanzee and human L1Pa3 orthologs (rounded to 17.5 × 103 in Fig. 5): 8387 and 9071, respectively, for the human and chimpanzee orthologs. Of the total, 10,095 (0.58) were G transitions (green bar, left y-axis), 4595 (0.26) were C transversions (red bar, right y-axis), and 2768 (0.16) were T transversions (blue bar, right y-axis). Note that the left (transitions) and right (transversions) axes cover different ranges. The numbers of A mutations to G, and A transversions to C or T were about the same for chimpanzee and human (results not shown). For L1Pa4, 16,678 (16.7 × 103) A mutations occurred, again with about one-half occurring in chimpanzee and human, and again the numbers of G transitions and C or T transversions were about the same in chimpanzees and human. And so on for the rest of the families in panel A and for the mutations of G, C, and T presented in panels G, C, and T respectively. In each case the green bar (left axis) shows transitions and the red and blue bars (right axis) show transversions. The gray line is the total non-CpG divergence for each L1Pa family normalized to that of L1Pa3, set to 1.0. Families that differ in total non-CpG divergence generally differ in their proportions of transitions and transversions, especially in regard to mutations of A, G, and C (much less so for T). For example, chi-square comparisons in panel A showed that the proportion of transitions and transversions in L1Pa3 are significantly different from that of L1Pa4 (indicated by the asterisks between these families). Likewise the distribution of transitions and transversions in L1Pa4 is significantly different from that of L1Pa5, but this is not the case for proportions of transitions and transversion between L1Pa7 and L1Pa8.
Statistically significant changes in the mutational spectrum parallel the decrease in total mutations that accompany the decrease of CpG content exhibited by each family up to the L1Pa7 family. Neither the mutational spectrum nor the total mutation rate for L1 orthologs changes substantially at lower (≲0.53%) contents of CpG (see also Figs. 3, 4). As the mutational mechanisms involved in transitions and transversions differ, these results further support the idea that CpGs, or mutations thereof, directly influence basic mutational processes.
Discussion
Extending our analysis of non-CpG divergence to L1 orthologs pairs in chimpanzee and humans over a wide range of CpG content produced two unexpected and provocative findings.
First, the correlation between CpG content and non-CpG mutation rate is best fit by a sigmoid (logistic) function. Interpreting this result in terms of a dose response curve implies that a “threshold” CpG content (∼0.53%) must be attained before the non-CpG mutation rate is markedly affected, and that the CpG “effect” reaches “saturation” at levels above ∼0.63% CpG. However, the latter statement is only supported by two sets of orthologs (L1Pa3 and L1Pa2; Walser et al. 2008) that contain the putative “saturating” levels of CpG. Linear fits of non-CpG divergence to CpG content, though not as robust as the sigmoid fit, nonetheless reveal the dramatic decrease of non-CpG divergence as a function of CpG contents below ∼0.53% (Supplemental Fig. 7S).
Second, the ratio of transition to transversion mutations changes with CpG content and closely parallels that of the total non-CpG mutation rate (Figs. 4, 5). As different mutational mechanisms produce transitions and transversions, this correlation corroborates our earlier contention (Walser et al. 2008) that the covariation of CpG content and non-CpG mutation is an intrinsic property of the DNA sequence and not a joint manifestation of the chromosomal location or environment. The following observations support this conclusion.
Except for CpG content, both the DNA sequence and chromosomal distribution of the L1 orthologs are highly similar (Supplemental Data; Chimpanzee Sequencing Analysis Consortium 2005; Walser et al. 2008). In addition, the non-CpG mutation rates were not correlated with either the G + C content or the recombination rates of the genomic environment of the orthologs (Fig. 3; Supplemental Data; Walser et al. 2008). And finally, the orientation of L1 orthologs located in transcriptional units was unbiased (Supplemental Table S1). As the nontranscribed strand is more prone to mutation than the transcribed one (Green et al. 2003; Mugal et al. 2009), differences in non-CpG mutation rates cannot be ascribed to an effect of transcription.
Most CpG sites, particularly those in transposable elements, are preferred sites of C methylation (Ehrlich et al. 1982; Nur et al. 1988; Yoder et al. 1997; Branscombe Miranda and Jones 2007). Therefore, the correlation between CpG content and non-CpG mutations could be due to an effect of methyl-CpG per se, to its spontaneous deamination to produce a T:G mismatch and subsequent recruitment of error-prone DNA repair mechanisms, or both (Walser et al. 2008).
Methyl-CpG can either mark DNA sequences for subsequent chromatin modification (i.e., closed or heterochromatin formation) (Jaenisch and Bird 2003; Pennings et al. 2005) or occur subsequent to such modifications (Bird 2002; Cedar and Bergman 2009). In either case, methyl-CpG may mediate the recruitment of various DNA- or histone-binding proteins and other factors (Martens et al. 2005; Cedar and Bergman 2009), which could conceivably affect the susceptibility of the DNA to mutation. In this case the correlation between non-CpG mutations and CpG content would mean that chromatin states promoted by CpG methylation, or that result in it, render DNA more susceptible to mutation than DNA not in such states. There is some evidence that the mutation rate of compact heterochromatin is higher than open, i.e., euchromatin (Prendergast et al. 2007).
However, it seems unlikely that the heterochromatin state (as defined in Prendergast et al. 2007) of L1 orthologs accounts for the correlation between their CpG content and non-CpG mutation rate. For this would mean that the chromatin state of the L1 orthologs is solely a function of their age in the genome. But the members of the different L1 families are intermingled throughout the genome (International Human Genome Sequencing Consortium 2001; Chimpanzee Sequencing Analysis Consortium 2005), sometimes to the point where younger copies are inserted into older ones (Giordano et al. 2007). We know of no evidence for heterochromatin states varying at such a fine granularity. Additionally, compact heterochromatin formation is not an inevitable consequence of CpG methylation (e.g., Bird 2002; Martens et al. 2005; Branscombe Miranda and Jones 2007) and, at least in mouse, little if any of the L1-containing chromatin bears the histone marks of compact heterochromatin (Martens et al. 2005).
Compact heterochromatin aside, methyl-CpG-containing DNA does recruit proteins that could conceivably affect its mutagenic environment. Thus, DNA methyl transferases, which are bound to nucleosomes of methylated DNA (Jeong et al. 2009), can also bind PCNA (proliferating cell nuclear antigen, the eukaryotic DNA replication beta clamp; Chuang et al. 1997; Moldovan et al. 2007). PCNA, in a process that involves its ubiquitination (Stelter and Ulrich 2003; Pfander et al. 2005; Andersen et al. 2008; Lee and Myung 2008), can recruit various error-prone DNA polymerases (for recent reviews, see Loeb and Monnat 2008; McCulloch and Kunkel 2008) at the expense of high fidelity replicative DNA polymerases. However, a more likely scenario has PCNA recruiting error-prone polymerases in response to the repair of the T/G mismatches that result from the spontaneous deamination of methyl-CpG.
Eukaryotes contain T/G-specific mismatch repair systems that restore this mismatch to C/G at ∼90% efficiency (Walsh and Xu 2006). One involves removal of the mismatched thymine by a glycosylase to produce an abasic site; a reaction that could also conceivably excise thymines from normally paired A/T sites (Li et al. 2007). The abasic site could then be repaired by excision repair. However, pausing of a replication fork at such sites would induce the above-mentioned modification of PCNA and its subsequent recruitment of error-prone Y family DNA polymerases.
PCNA has the capacity to simultaneously bind several DNA polymerases. This property, which would facilitate switching between high and low fidelity polymerases during DNA synthesis (for reviews, see Lehmann et al. 2007; Andersen et al. 2008; Lee and Myung 2008), has been demonstrated in vitro for prokaryotic DNA replication (Indiani et al. 2005). However, if the PCNA modification persists after the replication fork has passed the abasic site, then replication by the error-prone polymerase could persist beyond the lesion, thereby introducing mutations into normal flanking DNA (Andersen et al. 2008).
A role for error-prone DNA replication or repair has long been suggested to explain the occurrence of simultaneous multiple base mutations in certain mammalian cell lines (Seidman et al. 1987; Harwood et al. 1991) and almost certainly participates in generating the somatic hypermutation (SHM) that underlies immunoglobulin diversity (for review, see Teng and Papavasiliou 2007). These mutations occur in somatic cells and SHM depends on additional factors unique to immunoglobulin gene transcription (Teng and Papavasiliou 2007). However, these studies establish the precedent for the occurrence of multiple simultaneous mutations in vivo, a prediction of our proposal that mutations at non-CpG sites could accompany the repair of T/G mismatches.
Although non-CpG mutations produced by error-prone DNA repair processes recruited to repair T/G mismatches could explain the correlation between CpG content and non-CpG mutations, several issues remain. First, the mutational spectrum exhibited by L1 orthologs, including the “excess” of transversions (Fig. 4), is not congruent with the in vitro base substitutions exhibited by the known error-prone DNA polymerases (e.g., McCulloch and Kunkel 2008). However, and without implying any connection, we do note that an increase in transversions has been associated with the so-called mutator phenotype thought to be prerequisite for carcinogenesis (Liu et al. 2002; Bielas et al. 2006; Venkatesan et al. 2006).
Second, the nonlinear (sigmoid) correlation between CpG content and non-CpG mutation rate, which at lower CpG contents is accompanied by a stabilization of the transversion/transition ratio, suggests that the density of T/G mismatches may affect the mutational environment. One might expect a “saturation” effect on the non-CpG mutation rate at the higher CpG content. Presumably at some given density of CpGs, a T/G mismatch generated at any one, or just a subset, of CpGs could be sufficient to affect the non-CpG mutation rate of an entire region. On the other hand, several explanations could possibly produce the marked decrease in non-CpG as a function of CpG levels below the “threshold” 0.53% value (Figs. 3–5).
One possibility is that the lower non-CpG mutation rate of the CpG-poor older L1 orthologs reflects the time-dependent depletion of a class of particularly “mutable” non-CpG sites. However, we know of no evidence for such sites: Only methyl-C of CpG mutates with a clock-like rate while the mutation rate of C in other contexts and all other nucleotides are contingent on factors other than time (Hwang and Green 2004). Furthermore, no dinucleotide other than CpG is underrepresented in mammalian DNA (Duret and Galtier 2000), which would not be the case if given classes of mutable sites are irreversibly lost over time. Finally, there is no a priori reason why any putative class of “mutable” sites would not be regenerated by random mutations.
Another possibility is that the extent (efficiency) of CpG methylation is not linearly related to CpG content; e.g., below some threshold level, some CpGs may escape methylation. This would occur if the tuning of methylation efficiency varies over a fairly short range. However, as discussed above, L1 orthologs of different ages (and CpG content) are intermingled in the genome.
On the other hand, differences between our findings and those expected from the known biochemistry of DNA replication and repair may not be so surprising. The parameters affecting the accumulation of mutations in germline cells (only these can accumulate in the population and contribute to the neutral mutation rate) are likely to differ from those that affect mutation rates or patterns in somatic cells or in vitro (or in cell culture experiments).
In particular, our knowledge of the biochemical and cell biological determinants of mutation rate is undoubtedly incomplete (e.g., see Loeb and Monnat 2008). In fact, the novel and unexpected features of the CpG “effect” that we report here support this contention. As importantly, our findings, unlikely to have been revealed without analyzing L1 DNA fossils as described here, provide a rationale and context for experimental analysis of the biochemical basis of the CpG effect on the neutral mutation rate.
Methods
Isolation of ortholog L1 sequence pairs
Sequence and annotation data were retrieved from the UCSC Genome Browser download site (http://genome.ucsc.edu/). The following assemblies were used here: human genome-freeze March 2006 (UCSC hg18, NCBI Build 36.1) and chimpanzee-freeze March 2006 (UCSC panTro2). RepeatMasker track files based on the RepeatMasker program and RepBase library were used to obtain L1 family information and genome coordinates (ftp://hgdownload.cse.ucsc.edu/goldenPath/). Based on the RepeatMasker track file information sequences of L1 elements in the human genome were retrieved. L1 sequences <100 bp and records with ambiguous information (e.g., insertions that could not be precisely located on a chromosome) were removed from the data set. Orthologous insertions (i.e., those identical-by-descent) between the human and chimpanzee genome were obtained by converting the human genome coordinates for L1 insertions between assemblies using the command line tool liftOver (Version 134 for Mac OSX, http://genome.ucsc.edu/). The following parameters were used: The minimum ratio of bases that must remap was set to 0.85 (–minMatch) and multiple output regions were not allowed. The program and the appropriate chain files can be downloaded from the UCSC Genome Browser website (http://genome.ucsc.edu/). The converted L1 element insertions were compared with the RepeatMasker track files for the target species' L1 families (i.e., chimpanzee). Records that did not correspond to the query L1 family, L1 sequences >10 kb, and multiple hits with overlapping regions or ambiguous coordinate information in the target genome were removed from the data set.
Consensus sequences of L1 families
These were derived as described earlier (Walser et al. 2008) except we limited ourselves to the ∼3800 bp (depending on the family) ORF2 sequence because we could use the highly conserved amino acid sequence of the ORF2 protein as a guide for aligning the base sequences. In addition, as most L1 sequences are 5′ truncated, maximal statistical support is obtained from the more 3′ region of the L1 sequences. Alignments of ORF2 sequences corresponding to the L1 families indicated in Figure 1 isolated from the human and chimpanzee databases were manually adjusted using the Seaview multiple sequence alignment editor (Galtier et al. 1996). As the sequences of the youngest and oldest elements are highly similar (Boissinot et al. 2000; Khan et al. 2006), we could put all of the consensus sequences in the same register by alignment to the modern active human L1.3 element (Sassaman et al. 1997). The consensus sequences can be considered “current” because they are built from the members of each family present in the modern human and chimpanzee genomes. We used a 60% plurality to assign the consensus base, and the separately derived chimpanzee and human consensus ORF2 sequences for any given family were identical. Thus, the current consensus for a given L1 family can serve as a reasonable facsimile of the common ancestor of its ortholog pairs (see below, Determination of the Mutational Spectrum). An alignment of these sequences is presented in Supplemental Figure S8.
Ortholog alignments, determination of % CpG content, and other sequence manipulations
We used the multiple sequence alignment application MUSCLE (Edgar 2004) to align each human and chimpanzee ortholog pair, using the relevant ORF2 consensus sequence as a guide. A site was considered if it contained a nonambiguous nucleotide (i.e., A, T, C, or G) in the human and chimpanzee ortholog and any nucleotide (A, T, C, G, or N) in the consensus sequence. Gaps and non-L1 insertions were ignored. The combined lengths of the ORF2 ortholog pairs for each family that fulfilled these criteria are shown in Figure 1 (“total” column). For calculating % CpG content we used the CpG content of ortholog pairs that were ≥400 bp, which eliminated an expected bias toward high % CpG contents of the shorter fragments. Because the maximal % CpG content of even a modern element is ≤0.9%, the biases toward higher and lower than expected values of % CpG due to the nonrandom distribution of CpGs in ORF2 cannot offset each other in the shorter ortholog pairs; 0 bounds the lowest values but the upper ones are unbounded. The combined length of the ≥400-bp orthologs is also given in Figure 1. We used EMBOSS (European Molecular Biology Open Software Suite) (Rice et al. 2000) for general sequence handling and sequence comparisons and generated custom UNIX, Perl, and Python scripts as needed.
Determination of non-CpG mutations
We defined non-CpG sites using the stringent criteria defined by others (Keightley and Gaffney 2003; Meunier and Duret 2004; Kondrashov et al. 2006). Thus, non-CpG mutations were counted only at [A,G,T]N[A,C,T] sites. Or put another way, sites such as CAG, CTG (in addition to CCG and CGG) are excluded, as the CA and the TG of the first two trinucleotides could have been derived from ancestral CGs. Simulation studies (Meunier and Duret 2004; Gaffney and Keightley 2008) showed that these criteria remove biases that can be introduced into non-CpG divergence calculations by ancestral CpG sites (not recognized in the current sequence). We only considered sites where at least the human or chimpanzee ortholog is identical to its consensus sequence (see Determination of the Mutational Spectrum for our handling of sites where both orthologs differed from the consensus). We also eliminated sites directly flanked by an insertion or a deletion in either member of the ortholog pair or the consensus sequence. Thus, divergence measurements are not skewed by arbitrary placement of nucleotides on either side of a gap (Khelifi et al. 2006). We emphasize that we only used the consensus sequence to help align the ortholog pairs and not to calculate their divergence. Thus, the divergence of each L1 family is simply the number of non-CpG nucleotide differences (substitutions) between each ortholog pair divided by the total number of nucleotides compared summed over all the ortholog pairs (see Fig. 1). Divergence values were not corrected for superimposed or back mutations as the number of substitutions was small.
Estimation of L1 family ages
We used ages of the L1Pa3–L1Pa16 families that were based on their sequence divergence and estimates of the primate molecular clock (i.e., the % neutral substitutions/Myr; Boissinot et al. 2000; Khan et al. 2006; Furano and Boissinot 2008). The ordering of the families by this method agreed completely with an analysis based on an entirely different method for determining the relative age of L1 families in primates (Giordano et al. 2007). The ages of some of the families given here differ from those used earlier (Walser et al. 2008) but the ordering was the same. The relative ordering of the families and their CpG content is far more important for the analysis here than their precise age which will always be beset by uncertainties in the estimates of the molecular clock over the ∼80 Myr of primate evolution covered by the L1 families used here.
Determination of the mutational spectrum
We determined the fate of each mutated base by using the relevant current consensus sequence as a proxy for the ancestor of the chimpanzee and human orthologs. Consensus sequences have long been used as a reasonable approximation of ancestral sequences and are the only choice for some of the L1 families (L1Pa3–L1Pa5) for which orthologous outgroup sequences are not available (or present in insufficient quantity) because these families are confined only, or largely, to chimpanzee and humans (Walser et al. 2008). By comparing ortholog pairs we counted only the base substitutions that occurred during the ∼6 Myr since chimpanzees and humans diverged (see double-headed arrow, Fig. 1). Thus, in terms of using the consensus as the ancestral sequence, the prior history of the members of the various families matters only in as much as the base sequence at some positions in the current consensus sequence may not be relevant to the corresponding sites in a given ortholog pair. This could be the case for two reasons: First, the divergence of the family could produce inaccuracies in the consensus. As mutations accumulate randomly at non-CpG sites, one measure of imprecision of the consensus sequence would be the number of positions in an alignment where both orthologs differ from each other and the consensus sequence. Mutations to different bases at a given site in each ortholog should be very rare given the short evolutionary time between chimpanzees and humans. Therefore, the consensus base could be incorrect at such sites. (In fact, if the consensus base was not incorrect it might actually match the base of one of the orthologs.) In any event, one would expect the percent of positions where both orthologs and the sequence contain a different base to increase with family age, which is what we found. The family and percent of such sites are: L1Pa3, 0.9; L1Pa4, 1.2; L1Pa5, 1.6; L1Pa6, 2.3; L1Pa7, 2.8; L1Pa8, 3.6; L1Pa10, 4.5; L1Pa11, 4.8; L1Pa13, 5.4; L1Pa15, 6.7; L1Pa16, 7.3. The second reason is that the actual ancestor of a given ortholog pair contains a variant not represented by the consensus. But such positions would not be counted as substitutions if the base in both orthologs is the same and, if they are not, they would be indistinguishable from the first category (i.e., sites where both orthologs and the consensus differ), which we excluded from our determinations.
Statistical analysis
Statistical analyses were carried out using R (R Development Core Team 2007; http://www.R-project.org) and an online statistic site, http://department.obg.cuhk.edu.hk/ResearchSupport/statstesthome.asp. χ2 tests were used to determine if differences between the distributions of transitions and transversions between successive L1Pa families were statistically significant.
Acknowledgments
We thank Michael Seidman (National Institutes of Health) for his comments. This research was supported by the Intramural Research Program of the NIH, NIDDK.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103283.109.
References
- Andersen PL, Xu F, Xiao W 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 18: 162–173 [DOI] [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA 2006. Human cancers express a mutator phenotype. Proc Natl Acad Sci 103: 18238–18242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17: 915–928 [DOI] [PubMed] [Google Scholar]
- Branscombe Miranda T, Jones PA 2007. DNA methylation: The nuts and bolts of repression. J Cell Physiol 213: 384–390 [DOI] [PubMed] [Google Scholar]
- Bulmer M 1986. Neighboring base effects on substitution rates in pseudogenes. Mol Biol Evol 3: 322–329 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y 2009. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Chuang LSH, Ian H-I, Koh T-W, Ng H-H, Xu G, Li BFL 1997. Human DNA-(cytosine-5) methyltransferase–PCNA complex as a target for p21WAF1. Science 277: 1996–2000 [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780 [DOI] [PubMed] [Google Scholar]
- Duncan BK, Miller JH 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561 [DOI] [PubMed] [Google Scholar]
- Duret L 2009. Mutation patterns in the human genome: More variable than expected. PLoS Biol 7: e1000028 doi: 10.1371/journal.pbio.1000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N 2000. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol Biol Evol 17: 1620–1625 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Wang RY 1981. 5-Methylcytosine in eukaryotic DNA. Science 212: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV, Boissinot S 2008. Long Interspersed Nuclear Elements (LINEs): Evolution. In Encyclopedia of Life Sciences (ELS) John Wiley, Chichester, UK: doi: 10.1002/9780470015902.a0005304.pub2 [Google Scholar]
- Furano AV, Walser JC 2009. Mutation rate of non-CpG DNA. In Encyclopedia of Life Sciences (ELS) John Wiley, Chichester, UK: doi: 10.1002/9780470015902.a0021740 [Google Scholar]
- Gaffney DJ, Keightley PD 2005. The scale of mutational variation in the murid genome. Genome Res 15: 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, Keightley PD 2008. Effect of the assignment of ancestral CpG state on the estimation of nucleotide substitution rates in mammals. BMC Evol Biol 8: 265 doi: 10.1186/1471-2148-8-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C 1996. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548 [DOI] [PubMed] [Google Scholar]
- Giordano J, Ge Y, Gelfand Y, Abrusan G, Benson G, Warburton PE 2007. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Comput Biol 3: e137 doi: 10.1371/journal.pcbi.0030137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Green ED 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet 33: 514–517 [DOI] [PubMed] [Google Scholar]
- Hardison RC, Roskin KM, Yang S, Diekhans M, Kent WJ, Weber R, Elnitski L, Li J, O'Connor M, Kolbe D, et al. 2003. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res 13: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood J, Tachibana A, Meuth M 1991. Multiple dispersed spontaneous mutations: A novel pathway of mutation in a malignant human cell line. Mol Cell Biol 11: 3163–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Prufer K, Ji H, Zody MC, Paabo S, Ptak SE 2005. Why do human diversity levels vary at a megabase scale? Genome Res 15: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Ladoukakis E, Eyre-Walker A 2009. Cryptic variation in the human mutation rate. PLoS Biol 7: e1000027 doi: 10.1371/journal.pbio.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci 101: 13994–14001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M 2005. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell Biol 19: 805–815 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33(Suppl): 245–254 [DOI] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA 2009. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29: 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Gaffney DJ 2003. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc Natl Acad Sci 100: 13402–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A, Meunier J, Duret L, Mouchiroud D 2006. GC content evolution of the human and mouse genomes: Insights from the study of processed pseudogenes in regions of different recombination rates. J Mol Evol 62: 745–752 [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Ogurtsov AY, Kondrashov AS 2006. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J Theor Biol 240: 616–626 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Ball EV, Cooper DN 1998. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet 63: 474–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-y, Myung K 2008. PCNA modifications for regulation of post-replication repair pathways. Mol Cells 26: 5–11 [PMC free article] [PubMed] [Google Scholar]
- Lees-Murdock DJ, Walsh CP 2008. DNA methylation reprogramming in the germ line. Epigenetics 3: 5–13 [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6: 891–899 [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL 2007. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res 35: 390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu W, Jakubczak JL, Erexson GL, Tindall KR, Chan R, Muller WJ, Adhya S, Garges S, Merlino G 2002. Genetic instability favoring transversions associated with ErbB2-induced mammary tumorigenesis. Proc Natl Acad Sci 99: 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ 2008. DNA polymerases and human disease. Nat Rev Genet 9: 594–604 [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24: 800–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res 18: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J, Duret L 2004. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol 21: 984–990 [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Mugal CF, von Grunberg HH, Peifer M 2009. Transcription-induced mutational strand bias and its effect on substitution rates in human genes. Mol Biol Evol 26: 131–142 [DOI] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur I, Pascale E, Furano AV 1988. The left end of rat L1 (L1Rn, long interspersed repeated) DNA which is a CpG island can function as a promoter. Nucleic Acids Res 16: 9233–9251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H 2003. Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol 20: 2091–2096 [DOI] [PubMed] [Google Scholar]
- Pennings S, Allan J, Davey CS 2005. DNA methylation, nucleosome formation and positioning. Brief Funct Genomics Proteomics 3: 351–361 [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Prendergast JG, Campbell H, Gilbert N, Dunlop MG, Bickmore WA, Semple CA 2007. Chromatin structure and evolution in the human genome. BMC Evol Biol 7: 72 doi: 10.1186/1471-2148-7-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2007. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rice P, Longden I, Bleasby A 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr 1997. Many human L1 elements are capable of retrotransposition. Nat Genet 16: 37–43 [DOI] [PubMed] [Google Scholar]
- Seidman MM, Bredberg A, Seetharam S, Kraemer KH 1987. Multiple point mutations in a shuttle vector propagated in human cells: Evidence for an error-prone DNA polymerase activity. Proc Natl Acad Sci 84: 4944–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Sved J, Bird A 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci 87: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Tyekucheva S, Zody M, Chiaromonte F, Makova KD 2006. Strong and weak male mutation bias at different sites in the primate genomes: Insights from the human-chimpanzee comparison. Mol Biol Evol 23: 565–573 [DOI] [PubMed] [Google Scholar]
- Teng G, Papavasiliou FN 2007. Immunoglobulin somatic hypermutation. Annu Rev Genet 41: 107–120 [DOI] [PubMed] [Google Scholar]
- Tyekucheva S, Makova KD, Karro JE, Hardison RC, Miller W, Chiaromonte F 2008. Human-macaque comparisons illuminate variation in neutral substitution rates. Genome Biol 9: R76 doi: 10.1186/gb-2008-9-4-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan RN, Bielas JH, Loeb LA 2006. Generation of mutator mutants during carcinogenesis. DNA Repair 5: 294–302 [DOI] [PubMed] [Google Scholar]
- Walser JC, Ponger L, Furano AV 2008. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res 18: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Xu GL 2006. Cytosine methylation and DNA repair. Curr Top Microbiol Immunol 301: 283–315 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13: 335–340 [DOI] [PubMed] [Google Scholar]