Abstract
Noncoding RNA (ncRNA) constitutes a significant portion of the mammalian transcriptome. Emerging evidence suggests that it regulates gene expression in cis or trans by modulating the chromatin structure. To uncover the functional role of ncRNA in chromatin organization, we deep sequenced chromatin-associated RNAs (CARs) from human fibroblast (HF) cells. This resulted in the identification of 141 intronic regions and 74 intergenic regions harboring CARs. The intronic and intergenic CARs show significant conservation across 44 species of placental mammals. Functional characterization of one of the intergenic CARs, Intergenic10, revealed that it regulates gene expression of neighboring genes through modulating the chromatin structure in cis. Our data suggest that ncRNA is an integral component of chromatin and that it may regulate various biological functions through fine-tuning of the chromatin architecture.
Recent high-throughput transcriptomic analyses have revealed widespread transcription of the human genome (Cheng et al. 2005; The ENCODE Project Consortium 2007). A small portion of these transcripts code for proteins, while the rest are noncoding RNAs (ncRNAs). Only a limited number of ncRNAs have been assigned biological and molecular functions. However, tissue and development stage-specific expression of many of these ncRNAs indicates that they may participate in highly important biological functions linked to organismal development (Bernstein and Allis 2005; Amaral and Mattick 2008; Faghihi and Wahlestedt 2009; Ponting et al. 2009).
Recent evidence suggests a functional role for RNA-directed processes in the regulation of chromatin structure and gene expression in diverse biological phenomena (Mercer et al. 2009; Whitehead et al. 2009). For example, ncRNAs have been shown to play an important role in pluripotency and differentiation by regulating the chromatin structure through interaction with chromatin-modifying complexes (Dinger et al. 2008). Similarly, HOTAIR long ncRNA, transcribed from the human homeobox gene locus HOXC, regulates gene expression in trans of the HOXD locus by modulating the chromatin structure through targeting of the Polycomb complexes (Rinn et al. 2007). Likewise, other long ncRNAs with similar functions have also been shown to regulate classical biological pathways such as genomic imprinting and X chromosome inactivation through regulation of the chromatin structure. Kcnq1ot1 and Airn long ncRNAs regulate the genomic imprinting of multiple genes by interacting with chromatin and chromatin remodeling machinery, specifically, the G9a and PRC2 complexes, in a lineage-specific manner (Nagano et al. 2008; Pandey et al. 2008). Similarly, RepA long ncRNA also regulates X chromosome inactivation by targeting the PRC2 complexes to activate the Xist promoter on the future inactive X chromosome. This specific activation of the Xist ncRNA during initiation of X chromosome inactivation coincides with coating along the X chromosome and hierarchical recruitment of chromatin-modifying complexes (Zhao et al. 2008; Chow and Heard 2009), indicating that ncRNAs form part of complex regulatory networks regulating gene activity. Functional roles of ncRNA-directed processes are also implicated in Drosophila dosage compensation, where localization of histone acetyltransferase MOF to the male X chromosome is dependent on the roX ncRNAs (Akhtar et al. 2000). Moreover, recent findings suggest that RNA is an integral component of the chromatin (Rodriguez-Campos and Azorin 2007) and that it plays an important role in the higher-ordered chromatin structure of pericentric heterochromatin by organizing the heterochromatic components, such as HP1 and H3K9me3 marks (Maison et al. 2002). These observations collectively underline the importance of RNA as a cofactor in modifying the chromatin through recruitment of chromatin-remodeling complexes (Zhao et al. 2008).
However, barring a few exceptions in which the functional role of the RNA component in determining the chromatin structure is evident, there has been no thorough investigation addressing the identity of the chromatin-associated RNAs (CARs) on a global scale. This prompted us to identify such CARs in a genome-wide approach using high-throughput genomic platforms. In this study, CARs were purified from normal human skin fibroblast cells (HF) by isolating soluble chromatin by micrococcal nuclease (MNase) treatment followed by separation of different length chromatin fragments on a sucrose gradient. RNA was isolated from chromatin-containing fractions and was subjected to high-throughput sequencing on the Illumina platform. The sequencing of the CARs revealed association of many intronic and intergenic transcripts with chromatin, indicating that they may have structural and functional roles in chromatin organization through direct or indirect interactions with the chromatin.
Results
Isolation of RNA from the fractionated chromatin
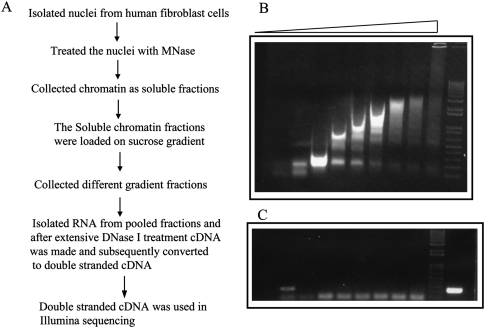
To address the functional role of ncRNA in the organization of chromatin structure and gene regulation, we purified CARs from normal HFs by isolating soluble chromatin after MNase treatment, followed by separation of chromatin fragments of different lengths on a sucrose gradient. Figure 1A schematically outlines the method we used for CAR extraction. Except for the top and bottom fractions, which contain free RNA and debris, respectively, both RNA and DNA were extracted from all gradient fractions. The extracted DNA was electrophoresed on an agarose gel to check the quality of the gradient (Fig. 1B). To rule out the possibility of contamination with free RNAs in the chromatin-bound RNA fractions, we measured beta-actin mRNA levels by semi-quantitative reverse transcriptase (RT)-PCR and found that they were below the limit of detection (Fig. 1C), suggesting that the CAR fraction is mostly devoid of free RNAs. To investigate the level of nascent transcripts in the CAR fraction, we briefly treated the HFs with actinomycin D (ActD), which inhibits transcription by blocking transcriptional elongation. The efficacy of the ActD treatment was assessed by measuring levels of MYC RNA, which has a half-life of <30 min (Supplemental Fig. S1A). The amount of the CARs purified from an equal number of the ActD-treated or untreated control cells was similar, indicating that nascent transcripts may constitute only a minor proportion of CARs (Supplemental Fig. S1B). Two small nuclear RNAs (snRNA), 7SK and U1, were enriched several thousand-fold in the chromatin fraction. Both these RNAs have been shown to regulate transcription by interacting with p-TEFb and TFIIH (Barrandon et al. 2008), respectively, indicating that CARs purified using the current method represent a true chromatin entity (Supplemental Fig. S2). Moreover, U1 snRNA has been shown to interact with one of the TATA-box-binding protein (TBP)–associated factors TAF15, indicating chromatin-dependent function for U1 snRNA (Jobert et al. 2009).
Figure 1.
Isolation of chromatin-associated RNA by sucrose gradient. (A) Flowchart showing a brief summary of the method used for isolation of chromatin RNA. (B) Chromatin isolated from MNase-treated human fibroblast nuclei was run on a 1% agarose gel after sucrose gradient fractionation. The lanes (from left to right) in the agarose gel represent DNA extracted from the chromatin fragments collected from the top to bottom fractions of the sucrose gradient. (C) Chromatin-bound RNA, isolated from the different gradient fractions, was used for semiquantitative RT-PCR with beta-actin primers, and the RT product was loaded according to the gradient fractionation in B; the extreme right lane is a positive control for RT-PCR.
High-throughput sequencing of CARs
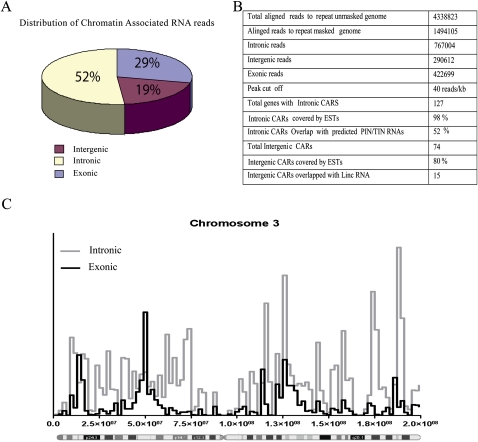
To obtain a global view of CARs, we performed deep sequencing of the purified CARs. CARs were converted into double-stranded cDNAs and sequenced using the Illumina high-throughput sequencing platform. Approximately 1.5 million reads were obtained that could be aligned to the repeat masked human genome, and further analysis of the aligned reads revealed that ∼71% of the reads originate from intergenic regions and intronic portions of protein-coding genes (19% intergenic and 52% intronic), while only 29% of the reads map to exons (Fig. 2A). This indicates that CARs are more often transcribed from the intergenic or intronic portions of the genome. An example of distribution of the sequencing reads along human chromosome 3 is depicted in Figure 2C; reads mapped to chromosome 3 are enriched mainly in the intronic regions rather than the exonic portions of the protein-coding genes.
Figure 2.
Basic characterization of chromatin-associated RNA. (A) Pie chart showing distribution of the sequencing reads over intronic, intergenic, and exonic portions of the human genome. (B) The table includes information about the aligned chromatin RNA sequencing reads and the detailed analysis of the CARs. (C) Distribution of sequencing reads in the exonic and intronic regions along chromosome 3.
A significant proportion of the CAR reads map to ribosomal RNA (rRNA) pseudo-gene repeats present on different chromosomes (Supplemental Table S1). This could be in part due to their high transcription rates. To address this issue, we analyzed expression of some of these repeat RNAs in nuclear and chromatin-bound RNA fractions and found that although these RNAs are abundantly expressed, as is beta-actin, their enrichment in the chromatin fraction was several thousand-fold higher than that of beta-actin mRNA (Supplemental Fig. S3). This indicates that they may be involved in as-yet-unknown biological functions via chromatin association. This notion is consistent with the recent observation that ribosomal DNA arrays contribute to global heterochromatin levels (Paredes and Maggert 2009).
Identification of chromatin-associated intronic and intergenic transcripts
In the next step we identified intronic and intergenic transcripts enriched in the chromatin fraction. Information from the published microarray expression data on HFs (GEO accession no. GSE1191) was used, and from this data set 10 highly expressed genes were selected for which there was no prior evidence of association with chromatin. The average number of reads per kilobase of mRNA for these genes was 20. For the identification of intergenic and intronic regions containing CARs, a cut-off of 40 reads/kb was chosen, which is twice the average read density of the abundant transcripts.
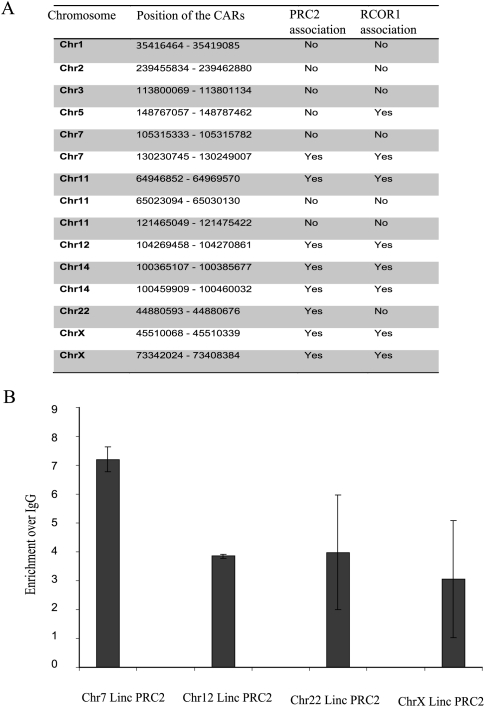
We found 141 CARs that mapped to the intronic regions of 127 protein-coding genes (Fig. 2B; Supplemental Table S2). An intronic RNA data set was used, wherein 55,000 long intronic RNAs have been mapped to introns of protein-coding genes using transcribed sequences from GenBank and termed “totally intronic” (TIN) or “partially intronic” (PIN) RNAs (Nakaya et al. 2007). Comparison of this intronic RNA data set with our intronic CARs showed that nearly 52% of the intronic CARs are part of the intronic TIN/PIN RNAs (Fig. 2B; Supplemental Table S2), which further suggests that the intronic CARs represent independent transcripts. We also found 74 CARs that map to intergenic regions of the genome. The presence of expressed sequence tags (ESTs) was investigated in all the CAR regions, and it was found that 98% of the intronic regions and 80% of the intergenic regions are covered by ESTs (Fig. 2B; Supplemental Table S3). Rather than being uniformly distributed, the sequence reads over the intronic and intergenic CARs were present in seemingly random clusters (Supplemental Tables S2, S3). This could be due to protection of the functional chromatin-interacting RNA sequences of CARs from MNase treatment, which not only cleaves linker DNA regions but also RNA. By using chromatin marks that define transcriptional activity, a recent study identified about 3300 large intervening noncoding RNAs (lincRNAs) in the human genome. Nearly 25% of these lincRNAs have been shown to interact with repressive chromatin-modifying enzymes like EZH2/SUZ12 and RCOR1 (Guttman et al. 2009; Khalil et al. 2009). We compared our intergenic CARs with the lincRNAs and found that 15 CAR regions are part of the lincRNAs, and interestingly, nine out of the 15 CARs have been shown to interact with either PRC2 or RCOR1 complexes (Fig. 3A). We verified the PRC2 interaction of the CARs that show overlap with lincRNAs by performing RNA immunoprecipitation (RIP) using an anti-EZH2 antibody. All four tested CARs showed significant enrichment in the EZH2 immunopurified RNA fraction (Fig. 3B), indicating that the CARs regulate gene expression through targeting of chromatin-remodeling complexes.
Figure 3.
Intergenic CARs overlap with lincRNAs and associate with PRC2 and RCOR1 complexes. (A) The table shows the position of the CARs, overlapping the lincRNAs present on different chromosomes as indicated, and their association with PRC2 or RCOR1 complexes. (B) The bar graph shows the enrichment of intergenic CARs in the EZH2 pull-down fraction over IgG. The analysis was performed using quantitative RT-PCR with primers corresponding to four intergenic CARs. Details of the primers are given in Supplemental Table S5.
A subset of the intronic and intergenic CARs belongs to the two major groups of small RNAs: snRNAs and small nucleolar RNAs (snoRNAs) (Supplemental Table S4). Enrichment of the small RNAs in the CAR sequences indicates that they may have additional functions other than their canonical role in RNA metabolism. Both snRNA and snoRNA protein complexes move through the different compartments in the nucleus during biogenesis so it is possible that this intranuclear transport of the RNP complexes could be guided by their chromatin association (Collins and Penny 2009).
CARs show evolutionary conservation
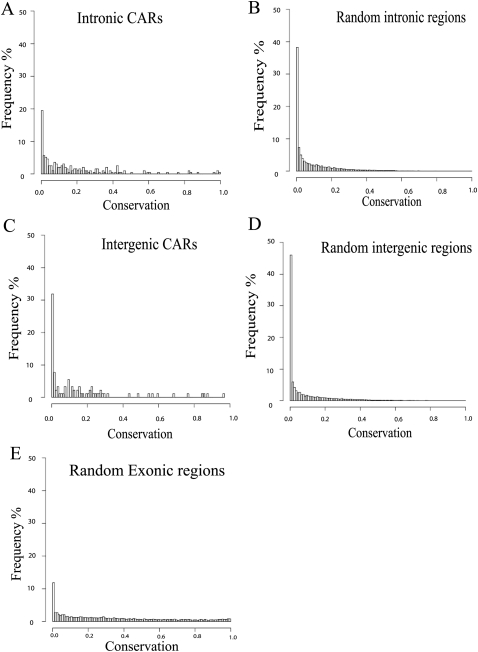
We next investigated the sequence conservation of intronic and intergenic CARs using phastCons analysis of an alignment of 44 placental mammalian genomes obtained from the UCSC Genome Browser (http://genome.ucsc.edu/). Distributions of the proportion of overlap between 1-kb CAR regions and the conserved elements are shown for intronic and intergenic CARs (Fig. 4A,C). To evaluate the significance of this conservation, we wanted to compare to the conservation of general intergenic and intronic sequences. One complication is that CARs may show conservation, not only due to conservation of their chromatin-interacting sequences, but also if they are located in regions, such as the vicinity of protein-coding genes, where regulatory elements are conserved. In order to take the genomic location of CARs into account we selected a large set of random 1-kb regions from the repeat masked genome. From these intergenic and intronic control sets were created by extracting 100,000 1-kb regions with distributions of distances to the nearest transcription start site (TSS) resembling those of intergenic and intronic CARs (Supplemental Fig. S4A,B). Many intronic and intergenic CARs appear to show a larger overlap with conserved elements compared to intronic and intergenic control sequences (Fig. 4B,D). A Mann-Whitney U test shows that sequence conservation for intronic and intergenic CARs is significantly different from the corresponding control sequences (P = 2.2 × 10−12, P = 7.1 × 10−4). It is also interesting to note that the extent of sequence conservation seen in intergenic and intronic CARs is similar to that seen in randomly selected exonic regions (Fig. 4E).
Figure 4.
CARs are conserved across species. Distribution of the proportion of CAR regions that overlap with conserved phastCons elements (44wayPlacental, UCSC Genome Browser) across intronic (A) and intergenic (C) CARs. Distributions of overlap with phastCons elements for the randomized intronic (B) and intergenic (D) control sets from matching genomic locations. (E) Distribution of overlap to randomly selected 1-kb exonic regions. Both intronic and intergenic CAR regions are significantly more conserved than control regions (P = 2.1 × 10−12, P = 7.1 × 10−4, respectively).
Intronic and intergenic CARs are enriched in the chromatin fraction
To validate the high-throughput sequencing data, we designed 15 quantitative PCR (qPCR) primers covering a random selection of 11 intronic and four intergenic CARs. First, we investigated expression from these regions using the RNA extracted from the nuclear compartment of the HFs, and all of these regions showed evidence of expression at the RNA level. Their expression levels were several-fold lower than those of the abundant beta-actin mRNA (Supplemental Fig. S5B,D). Next, we performed qPCR using the same set of primers with the CAR fraction and found that all of these RNAs were several-fold more enriched in chromatin, in comparison to beta-actin mRNA (Supplemental Fig. S5A,C). This observation is notable given that CARs are present at several-fold lower level than beta-actin mRNA in the nuclear RNA. To rule out the possibility that enrichment of the CARs in the gradient purified fraction is not due to nascent transcription, we purified the CAR fraction from an equal number of ActD treated and untreated cells and analyzed the levels of some of the validated CARs, relative to that of beta-actin. It was found that chromatin enrichment of all of the tested CARs was maintained in the sucrose gradient purification even after ActD treatment (Supplemental Fig. S6). We also investigated the subcellular localization of the validated CARs and found that all transcripts except one were clearly localized to the nuclear compartment, which is in agreement with the chromatin-associated functions of these transcripts (Supplemental Fig. S7).
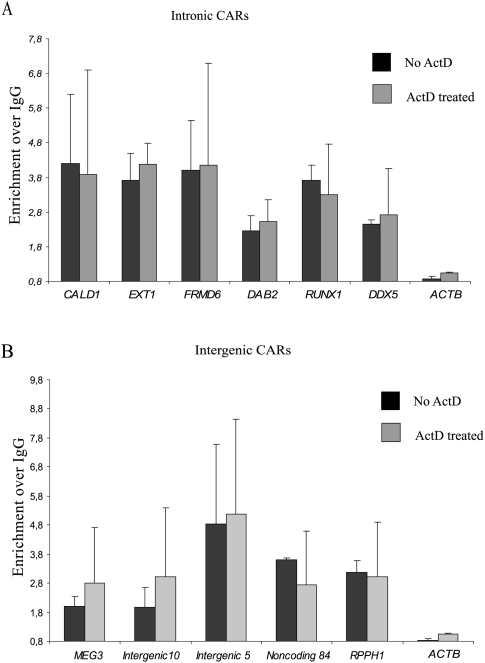
Previously, using the chromatin RNA immunoprecipitation (ChRIP) technique (Pandey et al. 2008), we demonstrated that the Kcnq1ot1 long antisense ncRNA mediates its effects through chromatin interaction. In this study, the ChRIP technique was used to further confirm the chromatin-association property of some of the intergenic and intronic CARs. Chromatin was immunopurified from HFs using an anti-histone H3 antibody, and RNA was isolated from the pull-down chromatin fraction and converted into cDNA. We then looked for enrichment of CARs identified using the sucrose gradient method. Considerable enrichment was observed for all the intergenic and intronic CARs in the immunopurified chromatin when compared to beta-actin, which did not show any enrichment over IgG (Fig. 5A,B). To reconfirm that no CARs originate from nascent transcripts, we treated the cells with the transcriptional inhibitor ActD and performed ChRIP on chromatin from the ActD-treated cells. Chromatin enrichment of all the ChRIP validated CARs was maintained even after ActD treatment (Fig. 5A,B), thus supporting the idea that CARs do not originate from nascent transcripts. By using ChRIP on the ActD-treated and untreated chromatin, we also tested the chromatin association of a few small RNAs, that were enriched in the gradient-purified chromatin fraction, and found that they were enriched in the H3 antibody purified fraction relative to IgG, whereas beta-actin and snoRNA U62A, used as negative controls, were not enriched (Supplemental Fig. S8).
Figure 5.
Validation of the intronic and intergenic CARs by ChRIP. Bar graphs show fold enrichment of the intronic (A) and intergenic (B) CARs over IgG in the histone H3 immunopurified chromatin. (Black bars) control cells; (gray bars) actinomycin D–treated cells. Beta-actin (ACTB) was used as a control, which did not show any significant chromatin enrichment in ChRIP assay.
An intergenic CAR regulates transcription in cis
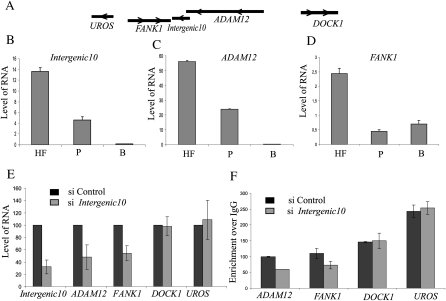
Since CARs are defined based on their chromosomal association property, they may act as transcriptional regulators. To explore this hypothesis in more detail, we chose an intergenic CAR (Intergenic10) that overlaps with an mRNA (sequences provided in Supplemental material) with no protein-coding potential. Intergenic10 is positioned between two protein-coding RNAs: fibronectin (FANK1) and ADAM metallopeptidase domain 12 (ADAM12). FANK1 is located on the 3′-side of Intergenic10, while ADAM12 is located on the 5′ side with an overlap with Intergenic10 ncRNA of a few hundred base pairs (Fig. 6A). All three transcripts were coexpressed in different tissues according to the available expression data in the UCSC Genome Browser (http://genome.ucsc.edu/). We measured expression of the Intergenic10 ncRNA and the protein-coding RNAs (FANK1 and ADAM12) by qPCR in two different tissues (placenta and brain) and in HFs; the results further supported the concept that the expression of the neighboring genes and the Intergenic10 ncRNA is coregulated (Fig. 6B–D). To address the functional role of the Intergenic10 ncRNA on the transcriptional activity of the flanking protein-coding RNAs, we first fine-mapped the 5′ and 3′ ends of the Intergenic10 ncRNA in addition to the 3′ end of the ADAM12, as it overlaps by a few hundred base pairs with the 5′ end of Intergenic10, and designed small interfering RNA (siRNA) that specifically targets the Intergenic10 ncRNA. Interestingly, specific down-regulation of the Intergenic10 ncRNA resulted in reduced expression of both FANK1 and ADAM12 mRNAs (Fig. 6E) but had no effect on the expression of two distal genes, UROS and DOCK1 (Fig. 6E). To rule out the possibility that the decrease in expression of FANK1 following Intergenic10 siRNA treatment is not because of the down-regulation of ADAM12, we used a siRNA specifically targeting the upstream exonic sequences of the ADAM12 mRNA. This siRNA treatment affected the ADAM12 expression but not the FANK1 expression (Supplemental Fig. S9). These results indicate that Intergenic10 ncRNA acts in a cis-limited manner. To check if the down-regulation of FANK1 and ADAM12 was associated with changes at the chromatin level, we performed chromatin immunoprecipitation with an anti-H3K4me2 antibody, using the chromatin obtained from the Integenic10 siRNA-treated and control cells. A significant decrease in the level of active chromatin mark H3K4me2 was observed on both FANK1 and ADAM12 promoters (Fig. 6F). Thus, these data collectively indicate that the Intergenic10 RNA positively regulates transcription of its neighboring genes by establishing active chromatin structures through interaction with the chromatin.
Figure 6.
Functional validation of an intergenic CAR. (A) The chromosomal location of the Intergenic10 transcript relative to two closely located neighboring genes FANK1 and ADAM12 and distally located UROS and DOCK1 genes. The arrows indicate the direction of the transcription. The relative levels of Intergenic10 (B), ADAM12 (C), and FANK1 (D) in human fibroblast (HF), placenta (P), and brain (B), as measured by quantitative RT-PCR. (E) siRNA knockdown of the Intergenic10 RNA and its effect on the expression of the FANK1 and ADAM12 genes. Percent expression of FANK1 and ADAM12 genes in Integenic10 (gray bars) and control (black bars) siRNA-treated cells. The percentage expression in intergenic siRNA-treated cells is presented relative to control siRNA-treated cells. The distal genes UROS and DOCK1 did not show any significant down-regulation in siRNA-treated cells. RNA amounts were measured by quantitative RT-PCR from three independent experiments, and beta-actin was used for normalization. (F) The siRNA knockdown of Intergenic10 RNA in HF cells affects the active chromatin marks H3K4me2 over ADAM12 and FANK1 promoter. Levels of H3K4me2 were measured by chromatin immunoprecipitation (ChIP) in two independent experiments using antibody against HeK4me2. The enrichment of H3K4me2 was measured by qPCR using the promoter-specific primers. (Black bars) The H3K4me3 enrichment over IgG at the indicated promoters in control siRNA-treated cells; (gray bars) enrichment over IgG in Intergenic10 siRNA-treated cells. DOCK1 and UROS promoters were used as controls, showing no change in H3K4me2 marks.
Discussion
In this study, we identified several highly conserved intergenic and intronic CARs by high-throughput sequencing of RNA purified from sucrose-gradient fractionated chromatin and verified the chromatin-association property of some of these CARs using the complementary ChRIP method. Regarding the remaining CARs, we expect them to have similar properties as the random subset that we verified with ChRIP, even though their interaction with chromatin could be direct or indirect. We also cannot determine from the sucrose-gradient purified CAR data set whether the chromatin association of these RNAs is stable or transient in nature. In addition, we observed significant enrichment of small repeat RNAs in our CAR fraction and ChRIP experiment, indicating a possible role for small RNAs in chromatin organization. However, we cannot rule out the possibility that some of the small RNP complexes could in part have been identified by copurification in the sucrose gradient.
At this point, the biological relevance of chromatin interaction of all different CARs is not clear. Since the intergenic and intronic ncRNAs identified in this study are based on their chromatin-association property, it is more likely that many CARs may mediate their actions through shaping the chromatin structure. Consistent with this notion is the observation that the Intergenic10 ncRNA positively regulates the flanking genes FANK1 and ADAM12 by establishing active chromatin structures. Given the poor homology between the ncRNA and the target genes in most cases, it is unlikely that sequence homology between the ncRNA and the target genes plays any crucial role. It is possible that some ncRNAs simply act as a scaffold for protein factors, which, in turn, could guide them to the target genes in cis or trans through protein–protein interactions, thereby modifying the associated chromatin to regulate transcription either positively or negatively. Transcriptional activation of the neighboring genes by ncRNAs could be achieved through directing transcriptional activators to the gene promoters, through their chromatin interaction.
The mechanisms by which ncRNAs target multiple genes in cis or trans are largely unknown. In the case of intronic CARs, their actions may not be restricted to the host gene but may affect nonoverlapping genes located on the same chromosome (cis) or on a different chromosome (trans). For example, Kcnq1ot1 and Airn ncRNAs, which are encoded from the intronically located differentially methylated promoters in the antisense direction to the host gene, have been shown to regulate multiple nonoverlapping genes in cis by interacting with chromatin and chromatin-modifying enzymes in a lineage-specific manner, suggesting that the intronic CARs identified in this study may use similar mechanisms for gene regulation (Nagano et al. 2008; Pandey et al. 2008). Interestingly, we have identified two intergenic CARs in the DLK1-DIO3-imprinted domain; one of them overlaps with the maternally expressed ncRNA MEG3, indicating that, like Kcnq1ot1 and Airn ncRNAs, MEG3 may play a role in regulation of imprinting of flanking genes in the DLK1–DIO3 domain.
Previously, a handful of intergenenic transcripts such as Xist, HOTAIR, and H19 have been extensively investigated for their role in gene regulation. Although H19 RNA has been shown to act as a tumor suppressor, the mechanisms underlying the RNA-mediated tumor suppressor function are unknown (Hao et al. 1993). On the other hand, the Xist and HOTAIR genes have been shown to mediate transcriptional gene silencing of an entire chromosome or a subchromosomal domain, respectively, through the recruitment of chromatin-modifying complexes (Whitehead et al. 2009). Identification of several intergenic CARs in this study provides the possibility of studying their actions in diverse biological processes. Importantly, a significant proportion of intergenic CARs that show homology with the linc ncRNAs interact with EZH2 or RCOR1, suggesting that the intergenic CARs regulate transcription at a distance through targeting of chromatin-modifying enzymes. The most challenging task ahead is to identify the target regions for the intronic and intergenic CARs, and the modes of action by which these CARs influence their target genes. In this study, we have identified the potential targets for one of the intergenic ncRNAs, Intergenic10, and this mediates transcriptional activation of the neighboring genes FANK1 and ADAM12 in cis via chromatin-level regulation. Since chromatin is a complex epigenetic regulatory framework and intergenic CARs were identified based on their chromatin interaction property, they should serve as a suitable resource for further studies on long intergenic transcripts in organismal development and disease. Characterization of the functions of these intergenic and intronic CARs would contribute to a greater understanding of the largely unexplored world of RNA-mediated functions.
Methods
Isolation of chromatin RNA by sucrose gradient
Approximately 10 million cells were resuspended in nuclei isolation buffer (0.5 M sucrose, 0.5% Triton X-100 in TES [15 mM Tris-HCl at pH 7.4, 60 mM KCl, 2 mM EDTA, 1 mM DTT]), kept on ice for 10 min, and then homogenized by 15 strokes with a dounce homogenizer. The volume was made up to 3 mL with nuclei isolation buffer and centrifuged at 4000 rpm for 15 min. After washing the nuclei once with TES, the nuclei were resuspended in 300 μL of TES containing 3 mM CaCl2, and treated with 1 unit of MNase in a 500-μL reaction volume for 1 min at 37°C. The reaction was stopped by adding EDTA to a final concentration 50 mM. The nuclei were centrifuged at 8000 rpm for 2 min at 4°C to collect the supernatant (supernatant 1) and were further resuspended in 0.2 mM EDTA to disrupt the nuclei and obtain the soluble chromatin (supernatant 2). Supernatants 1 and 2 were mixed and loaded on a 5%–30% sucrose gradient, prepared in TES, and centrifuged overnight at 28,000 rpm at 4°C. One-milliliter fractions from the sucrose gradient were collected. A sample of each fraction (200 μL) was taken for isolation of DNA, and the remainder was used for preparation of RNA, using TRIzol (Invitrogen). DNA was loaded on a 1% agarose gel to check for the separation of chromatin fragments according to length. The RNA isolated from the gradient fractions was treated extensively with DNase I to remove the contaminating DNA, and a part of the DNase-treated chromatin-bound RNA was used for reverse transcription reaction to make cDNA using Super Script II reverse transcriptase (Invitrogen). We performed a PCR reaction on the cDNA with beta-actin primers to check whether the chromatin-bound RNA was devoid of free RNAs. Based on this initial quality-control step, we have pooled the cDNA prepared from the different chromatin-bound RNA fractions. We have checked U1 SnRNA and 7SK RNA levels for their enrichment in the chromatin fractions; both showed several thousand-fold enrichment compared to beta-actin. In the next step, the pooled cDNA was converted into double-stranded cDNA using Universal Riboclone cDNA Synthesis System (Promega) following the manufacturer's protocol. Double-stranded cDNA was used for the Illumina high-throughput sequencing. A summary is presented in Figure 1A.
Preparation of the library and sequencing using the Illumina Genome Analyzer I
A sequencing library was prepared from cDNA according to the manufacturer's manual, except that the sample was not fragmented, as most of the dsDNA fragments prepared from chromatin-purified RNA were within the range of 100 to 150 bp. Briefly, the DNA was end-repaired using T4 DNA polymerase, Klenow DNA polymerase, and T4 polynucleotide kinase (PNK), followed by purification on a QIAquick PCR purification column (QIAGEN). An A-base was ligated to the blunt ends of the DNA fragments using the Klenow DNA polymerase, and the sample was purified using a MinElute PCR purification column (QIAGEN). Adapters for sequencing were ligated to the DNA fragments, and the library was size-selected on an agarose gel. A 150–200-bp fragment was excised from the gel, purified using a QIAGEN gel extraction column, and amplified for 14 or 18 cycles of PCR, followed by purification using a QIAquick or MinElute PCR Purification column. The quality and quantity of the library was evaluated using the Agilent Technologies 2100 Bioanalyzer, and a DNA 1000-kitA 3-pM solution of the purified library was used in the cluster generation on the Cluster Station (Illumina Inc.). Sequencing was performed for 36 cycles using the Genome Analyzer I, according to the manufacturer's protocols. Image analysis and base calling were performed using the analysis pipe-line supplied with the Genome Analyzer instrument.
Analysis of the Illumina reads
The Illumina sequencing reads were mapped to a repeat masked human genome (ftp://ftp.ensembl.org/pub/release-36/homo_sapiens_36_35i/data/fasta/dna), using ELAND. The alignment was performed using standard parameters, where a 32-bp seed was used for alignment, allowing a maximum of two mismatches in the seed. ELAND reports the reads that map to unique positions in the genome. A set of R scripts, available from the authors, was used to assign all reads to exonic and intronic parts of the genes and to intergenic regions. A 1-kb sliding window was used to scan the intronic and intergenic regions with the Illumina sequencing reads, and the regions where the read density exceeded a threshold of 40 reads/kb were considered as enriched regions with chromatin-associated transcripts (CARs). The threshold was based on the average number of reads per kilobase of 10 genes (∼20 reads/kb), which were selected based on their expression profiles in HFs from publicly available microarray data (GEO accession no. GSE1191). Detected regions were merged if they were within 10 kb of one another and within the same gene (if any), and checked for overlap with a list of ESTs from the UCSC Genome Browser (version 36, 2006; http://genome.ucsc.edu/). Enriched regions in the intronic portions of the genes were similarly compared to a list of partially and totally intronic transcript data sets (Nakaya et al. 2007). In Supplemental Tables S2 and S3, the start and end positions of CARs have been estimated by looking at read clusters within the regions. We have included “coverage,” which was obtained by averaging read coverage over the read clusters. In Supplemental Table S4 we have provided the list of the snRNAs and snoRNAs that overlap with the CARs.
Measuring sequence conservation in CARs
A track of conserved genomic regions based on phastCons analysis of an alignment of 44 placental mammal species was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). We observed the proportion of each CAR region that overlapped with the conserved phastCons elements. A large set of random 1-kb regions was generated from the repeat masked genome, and the distance to the nearest transcription start site (dTSS) was noted. From them, intronic and intergenic control sets were created by extracting subsets of 100,000 regions whose dTSS distribution resembled that of intronic and intergenic CARs. The control regions were then investigated for overlap with the conserved regions. We compared the distribution of overlap for the intronic and intergenic CARs with their respective control distributions using the Mann-Whitney U test.
Northern hybridization
The chromatin-associated RNA, purified from an equal number of control and ActD-treated HFs cells, was probed with α-32P radio-labeled sonicated whole genomic DNA, and blots were exposed to a PhosphorImager. Control hybridization was also performed with chromatin RNA, after treatment with RNase A, which did not show any hybridization signal (data not shown). To measure the ActD transcriptional inhibition, highly unstable MYC mRNA (half-life: 30 min) was used as a control.
Chromatin RNA immunoprecipitation (ChRIP)
ChRIP was performed using a previously described (Kuo and Allis 1999) chromatin immunoprecipitation method with the following modifications: In brief, for isolation of the chromatin-associated RNA, we have taken equal amounts of cross-linked chromatin and immunopurified it using histone H3 antibody (Abcam) or with IgG (Upstate Biotechnology; Millipore) by including RNase inhibitors (Promega) from the ActD-treated or untreated normal HF cells. The chromatin-bound RNA was extracted from the immunopurified chromatin using TRIzol (Invitrogen) and converted into cDNA after extensive DNase I treatment. The qPCR reactions were performed with each set of the primers for different intronic and intergenic CARs using H3 and IgG pull-down chromatin fractions from ActD-treated and untreated cells. To calculate the enrichment of the CARs in the H3 pull-down fraction, qPCR values for each primer set in the H3 pull-down fraction were divided with qPCR values in the IgG fraction, and the enrichment relative to IgG was plotted.
Quantitative (real-time) PCR (qPCR)
The qPCR reactions were performed to validate the enrichment of a subset of randomly selected CARs from the intergenic and intronic region, and beta-actin mRNA was used as a negative control. The relative level of each CAR was calculated compared to the beta-actin mRNA both in the sucrose-gradient purified chromatin-bound RNA fraction and in the nuclear fraction. Similarly, to check the relative abundance of CARs in nuclear or cytoplasmic fractions, we carried out RT-qPCR with nuclear and cytoplasmic RNA fractions, prepared using the protocol previously described (Chen-Kiang and Lavery 1989). To check the quality of the nuclear and cytoplasmic fractionation, we used two controls—KCNQ1OT1, which is exclusively localized in the nuclear compartment, and RPS14, which has mostly cytoplasmic localization (data not shown). The qPCR quantification was performed using the standard curve method, and the primer sequences used in various qPCR reactions are provided in Supplemental Table S5.
Rapid amplification of cDNA ends (RACE)
Both 5′ and 3′ RACE reactions were performed using Gene Racer Core Kit (Invitrogen) following the manufacturer's protocol. 5′ and 3′ RACE cDNA were used subsequently for amplification with the gene-specific RACE primers. Gene-specific RACE primer sequences are provided in Supplemental Table S5. The amplified PCR product was cloned into the pGMT-Easy vector system (Promega). Several clones were sequenced and aligned to the UCSC Genome Browser to designate the 5′ and 3′ ends of the transcripts.
siRNA transfection
siRNA transfection was performed using the N-TER Nanoparticle siRNA transfection system (Sigma). The cells were seeded at a density of 40,000 per well, 12–16 h prior to transfection in six-well plates. siRNAs 1 (duplex of si-5′-CAGGUCAAAUCAGGAGGCU and si-5′-AGCCUCCUGAUUUGACCUG) and 2 (duplex of si-5′-GAGUGACUCAUUCUCCUGU and si-5′ACAGGAGAAUGAGUCACUC; Sigma), designed for Intergenic10 RNA, were transfected in combination at a final concentration of 40 nM. Predesigned siRNA against ADAM12 mRNA was obtained from Ambion. The control siRNA (QIAGEN) was also transfected at a similar concentration. The cells were harvested 72 h post-transfection. The RNA was extracted and treated with DNase I (Promega) to synthesize random primed cDNA using reverse transcriptase (Promega). Quantitative real-time PCR reaction was performed in duplicate using SYBR green mix (Sigma). The levels of RNA in the control and Intergenic10 siRNA-treated cells were measured using different gene-specific exonic primers, and the values obtained were normalized against beta-actin. Details of the primer sequence are given in Supplemental Table S5.
Acknowledgments
This work was supported by the grants from the Swedish Medical Research Council (VR-M), the Swedish Cancer Research Foundation (Cancerfonden), and the Swedish Childhood Cancer Society (Barncancerfonden) to C.K. C.K. is a Senior Research Fellow supported by VR-M. We also thank the Department of Genetics & Pathology, Rudbeck Laboratory, and Uppsala University Medical Faculty, Uppsala University, for financial support. The sequencing was performed by the SNP Technology Platform in Uppsala, which is supported by the Knut and Alice Wallenberg Foundation, Uppsala University, and Uppsala University Hospital. We thank Marie Lindersson for technical assistance with sequencing.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE21227.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.103473.109.
References
- Akhtar A, Zink D, Becker PB 2000. Chromodomains are protein–RNA interaction modules. Nature 407: 405–409 [DOI] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS 2008. Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Barrandon C, Spiluttini B, Bensaude O 2008. Non-coding RNAs regulating the transcriptional machinery. Biol Cell 100: 83–95 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD 2005. RNA meets chromatin. Genes Dev 19: 1635–1655 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chen-Kiang S, Lavery DJ 1989. Preparation of precursors to mRNA from mammalian cell nuclei. Methods Enzymol 180: 69–82 [DOI] [PubMed] [Google Scholar]
- Chow J, Heard E 2009. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol 21: 359–366 [DOI] [PubMed] [Google Scholar]
- Collins LJ, Penny D 2009. The RNA infrastructure: Dark matter of the eukaryotic cell? Trends Genet 25: 120–128 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B 1993. Tumour-suppressor activity of H19 RNA. Nature 365: 764–767 [DOI] [PubMed] [Google Scholar]
- Jobert L, Pinzon N, Van Herreweghe E, Jady BE, Guialis A, Kiss T, Tora L 2009. Human U1 SnRNA forms a new chromatin-associated snRNP with TAF15. EMBO Rep 10: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD 1999. In vivo cross-linking and immunoprecipitation for studying protein:DNA associations in a chromatin environment. Methods 19: 425–433 [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: Insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S 2007. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol 8: R43 doi: 10.1186/gb-2007-8-3-r43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Paredes S, Maggert KA 2009. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci 106: 17829–17834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Campos A, Azorin F 2007. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One 2: e1182 doi: 10.1371/journal.pone.0001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead J, Pandey GK, Kanduri C 2009. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta 1790: 936–947 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]