Abstract
The Drosophila Enhancer of split complex [E(spl)-C] is a remarkable complex of genes many of which are effectors or modulators of Notch signaling. The complex contains different classes of genes including four bearded genes and seven basic helix-loop-helix (bHLH) genes. We examined the evolution of this unusual complex by identifying bearded and bHLH genes in the genome sequences of Arthropods. We find that a four-gene E(spl)-C, containing three bHLH genes and one bearded gene, is an ancient component of the genomes of Crustacea and Insects. The complex is well conserved in insects but is highly modified in Drosophila, where two of the ancestral genes of the complex are missing, and the remaining two have been duplicated multiple times. Through examining the expression of E(spl)-C genes in honeybees, aphids, and Drosophila, we determined that the complex ancestrally had a role in Notch signaling. The expression patterns of genes found inserted into the complex in some insects, or that of ancestral E(spl)-C genes that have moved out of the complex, imply that the E(spl)-C is a genomic domain regulated as a whole by Notch signaling. We hypothesize that the E(spl)-C is a Notch-regulated genomic domain conserved in Arthropod genomes for around 420 million years. We discuss the consequence of this conserved domain for the recruitment of novel genes into the Notch signaling cascade.
The Enhancer of split complex [E(spl)-C] of Drosophila melanogaster is a well characterized genetic locus containing 12 genes on chromosome 3R, most of which are to be effectors or modulators of Notch signaling. The E(spl)-C contains a number of different Notch responsive genes, some of which are related in sequence (Wurmbach et al. 1999). The largest class of genes in the region encodes basic helix-loop-helix (bHLH) transcription factors. There are seven of these bHLH transcription factors in the D. melanogaster E(spl)-C [HLHmβ, HLHmγ, HLHmδ, HLHm3, HLHm5, HLHm7, and E(spl)] (Delidakis and Artavanis-Tsakonas 1992; Knust et al. 1992). These bHLH proteins have a distinctive ”orange” domain and a C-terminal WRPW motif. This motif is a protein interaction domain (Fisher et al. 1996; Alifragis et al. 1997), allowing these proteins to interact with Groucho, a transcriptional corepressor that interacts with histone deacetylases to repress gene expression (Chen et al. 1999). E(spl)-C bHLH proteins act as hetero- or homodimeric transcription factors by binding either to specific enhancer sequences, or to other DNA bound transcription factors (Alifragis et al. 1997), recruiting groucho to those regions (Giagtzoglou et al. 2003). The recruitment of groucho leads to changes in chromatin conformation and transcriptional repression (Palaparti et al. 1997). E(spl)-C bHLH proteins act as transcriptional repressors and do not activate gene expression in response to Notch signaling (de Celis et al. 1996). In D. melanogaster the groucho gene lies at the telomeric end of the E(spl)-C (Hartley et al. 1988). In D. melanogaster four bearded class genes also lie in the E(spl)-C (mα, m4, m2, and m6) (Lai et al. 2000b). Bearded proteins have an N-terminal amphipathic α-helix, but overall sequence similarity is low (Lai et al. 2000b) implying rapid evolution of these genes. Three additional bearded class genes are present in the D. melanogaster genome (Tom, Brd, and Ocho) and are present in a separate complex on chromosome 3L (Lai et al. 2000b). The D. melanogaster E(spl)-C contains one other gene, m1. The function of this gene is unknown although it is similar in sequence to Kazal class protease inhibitors (Wurmbach et al. 1999).
All the genes of the E(spl)-C, except m1, are Notch responsive (Wurmbach et al. 1999). During embryonic neurogenesis the E(spl)-C bHLH genes are expressed in the neurectoderm in response to activated Notch signaling (Jennings et al. 1994) and repress key regulators of neural cell fate including proneural genes and the Notch ligand Delta (Heitzler et al. 1996). Cells expressing the E(spl)-C bHLH proteins suppress neural cell fate, allowing cells to take up a secondary epidermal fate, a process known as lateral inhibition (Tata and Hartley 1995; Nakao and Campos-Ortega 1996).
The bearded class genes of the E(spl)-C and bearded complex (Lai et al. 2000b) act in adult sensory precursor formation as antagonists of Notch signaling (Apidianakis et al. 1999; Lai et al. 2000a). Bearded proteins interact with the E3 ubiquitin ligase neuralized to promote degradation of Delta (Lai et al. 2000a; Deblandre et al. 2001; Pavlopoulos et al. 2001). Little is known about bearded class proteins from other species. There are low levels of sequence conservation between family members (Lai et al. 2000b); thus the evolution of this gene family is unclear, and no bearded class genes have been identified in vertebrates. Recently a Daphnia pulex bearded protein has been shown to interact with neuralized implying conservation of function in the absence of sequence similarity (Fontana and Posakony 2009).
In Drosophila the expression of both the E(spl)-C bHLH proteins and bearded class genes are regulated by Suppressor of Hairless [SU(H)] (Eastman et al. 1997; Nellesen et al. 1999; Lai et al. 2000b; Maeder et al. 2007), and by miRNA binding to conserved sites (GY-box, Brd-Box, and K-box) located in the 3′ untranslated region (UTR) of transcripts (Lai and Posakony 1997; Lai et al. 1998; Lai et al. 2005).
The E(spl)-C bHLH genes are related in sequence to other bHLH genes in the D. melanogaster genome. One gene, Her [hairy-E(spl)-related], is closely related to the E(spl)-C bHLH genes (Moore et al. 2000) but not linked. Other, also unlinked, more distantly related bHLH genes are hairy (h), deadpan (dpn), similar to deadpan (Side), hairy/E(spl)-related with YRPW motif (Hey), and clockwork orange (cwo). Related bHLH genes in vertebrate genomes, named the HES (hairy-enhancer of split) genes, have functions in somitogenesis, neurogenesis, and stem cell maintenance (for review, see Kageyama et al. 2007). HES genes are regulated by Notch signaling and require groucho for their activities.
Examination of the genomes of the mosquito Anopheles gambiae and the honeybee Apis mellifera has led to the suggestion that E(spl)-C in Drosophila evolved from an ancestral “Urcomplex” consisting of a single E(spl)-C bHLH gene and a single bearded class gene, as seen in the Anopheles gambiae genome (Schlatter and Maier 2005). The authors point out, however, that the Apis mellifera genome contains three E(spl)-C bHLH genes closely linked to a bearded gene and ascribe these to Apis-specific duplications (Schlatter and Maier 2005).
The E(spl)-C is unlike most other eukaryote gene complexes as it contains a number of unrelated genes involved in a single developmental process, regulating Notch signaling. How did such a complex evolve? Here we utilize the sequenced genomes of arthropods to discover the organization and origins of the E(spl)-C genes. We use the expression patterns of E(spl)-C genes in honeybee and aphid to determine the ancestral functions of these genes. We argue that the E(spl)-C is an ancient complex of genes present in the genomes of the common ancestors of insects and crustaceans, and that it is a remarkable evolutionarily conserved genomic domain that has been regulated by Notch signaling for at least 420 million yr (Myr).
Results
Phylogenetics of E(spl)-C–related genes
Phylogenetics of arthropod bHLH-orange genes
In most Drosophilid species there are 13 bHLH genes encoded in the genome. Bayseian phylogenetic analysis groups these proteins into robust phylogenetic groups, all of which are represented in D. melanogaster. This implies that this gene family has been stable since the divergence of the Drosophila lineage 60 million yr ago (Mya) (Supplemental Fig. 1). To examine more distant phylogenetic relationships, the bHLH proteins related to E(spl)-C bHLHs, h, dpn, Side, and cwo were extracted from sequenced arthropod genomes (Ixodes scapularis [chelicerate]; Daphnia pulex [crustacean]; Acyrthosiphon pisum, Pediculus humanus, Apis mellifera, Tribolium casteneum, Bombyx mori, Culex pipens, Anopheles gambiae, and D. melanogaster [insects]) and from the genome of the cephalochordate Brachiostoma floridanum (Holland et al. 2008) (as an example of a non-arthropod set of HES genes). The full-length predicted proteins were aligned and analyzed by Bayesian phylogenetic techniques (Fig. 1; Ronquist and Huelsenbeck 2003). This tree is rooted with the cephalochordate bHLH proteins, which cluster together to the exclusion of the arthropod bHLHs, indicating that these genes have arisen by duplication of an ancestral hairy/E(spl)-C protein and are independent from the duplications in arthropods; thus, chordates do not have direct orthologs of the E(spl)-C bHLH genes.
Figure 1.
Phylogram of arthropod and Branchiostoma E(spl)-C-related bHLH proteins. Bayesian phylogeny of bHLH proteins from sequenced Arthropod genomes rooted with Brachiostoma HES genes. Posterior probabilities are shown at nodes. Names of proteins and their respective species are shown in Figure 4 and in Supplemental Table 1. The phylogenetic analysis recovers six well-supported clades, a clade of Brachiostoma proteins, a clade of proteins that cluster with Drosophila CWO, a clade that contains both Hairy and Deadpan proteins, a clade clustering with Drosophila SIDE, and a final, poorly resolved clade containing proteins clustering with Drosophila E(spl)-C bHLH and HER. Drosophila HER is circled in red. Ixodes ISCW016537 and ISCW016540, apparent tandem duplications of a Deadpan-like protein, are circled in blue.
The arthropod sequences are grouped into five clades; cwo is the most deeply branching and is found in all arthropod genomes including the chelicerate Ixodes, thought to be the most distant group of arthropods to the insects (Cook et al. 2001; Hwang et al. 2001). dpn and Side are also found in all arthropod genomes examined. However, h, while present in all other genomes, appears to be missing from Ixodes. This loss is accompanied by a duplication of dpn producing two genes (ISCW016537 and ISCW016540; circled in blue in Fig. 1), that lie in a head-to-head arrangement (scaffold DS645479, ∼100 kb apart). The absence of h from the Ixodes genome is either a linage-specific loss, or is due to gaps in the genome assembly as h genes have been identified in other chelicerates including the spider Cupiennius (Damen et al. 2000, 2005). The final clade includes the E(spl)-C bHLH and HER proteins, which group together. This clade has poorly supported internal structure and a number of long branches making it difficult to assign relationships. It is clear that HER (circled in red in Fig. 1) and E(spl)-C proteins are closely related.
To better resolve relationships between the E(spl)-C bHLH proteins and HER, we extracted the bHLH and orange domains of these proteins and examined their relationships using Bayesian phylogenetics (Fig. 2; Ronquist and Huelsenbeck 2003). E(spl)-C-type bHLHs fall into three major clades with high posterior probabilities. The first of these clades we name E(spl)-C bHLH2. This clade contains E(spl)-C-related genes from honeybees, Bombyx, aphids, and Daphnia. A second, much larger clade we name E(spl)-C bHLH1; this clade contains all the Drosophila E(spl)-C bHLHs and a single bHLH protein from each of Bombyx, mosquitoes, Tribolium, honeybees, Pediculus, Acyrthosiphon, and Daphnia. The final clade contains proteins from honeybees, Tribolium, Bombyx, Pediculus, and Acyrthosiphon. This clade also contains HER, a non-E(spl)-C bHLH from Drosophila. The only protein that does not clearly fall into these clades is NCBI_GNO_130833, encoded by a Daphnia E(spl)-C gene that appears on the branch leading to E(spl)-C bHLH2. The placement of this gene close to Her, and its position in the Daphnia genome (see below) makes it most likely an ortholog of Her. While the three major clades of bHLH proteins group together robustly, the relationships between genes within the clades are either unresolved, or have low posterior probability, reflecting a lack of sequence divergence. The exception to this is the genes of the Drosophila E(spl)-C and HER, which have long branch lengths and are unusually derived members of the bHLH family.
Figure 2.
Phylogram of E(spl)-C- and HES-related proteins. Unrooted Bayesian phylogram of the bHLH and orange domains of E(spl)-C bHLHs and HER-like bHLH proteins from sequenced arthropod genomes. Names of proteins and their respective species are shown in Figure 4 and in Supplemental Table 1. Phylogenetic analysis resolves three clades: (1) a large clade with representatives from all insect and crustacean genomes, including all Drosophila E(spl)-C genes, that we designate E(spl)-C bHLH-1 (green). (2) A clade with a smaller number of members but including representatives from hemimetabolous and holometabolous insects and Crustacea; we designate this clade E(spl)-C bHLH2 (light blue). (3) The final clade contains representatives from insects and includes Drosophila HER; we designate this clade Her (blue). The Daphnia protein NCBI130833 is circled in red, as it does not cluster robustly with any of these clades; however, the position of the gene that encodes it in the Daphnia E(spl)-C complex indicates that it is most likely to be a Her gene.
Phylogenetics of non-bHLH proteins of the Drosophila E(spl)-C
The Drosophila m1 gene encodes a protein with similarity to Kazal protease inhibitors; phylogenetic analysis of all insect Kazal protease inhibitors revealed that m1 is found only in Drosophilid species (Supplemental Fig. 2).
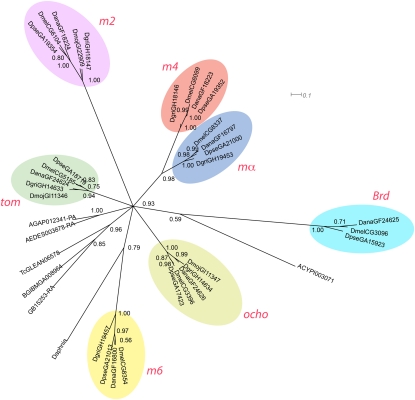
The relationships between bearded class genes are more difficult to assess due to sequence divergence. Members of this class are defined only by a basic amphipathic α-helix at the N terminus (Bailey and Posakony 1995) and a bearded motif (N-motif [NXANE(K/R)(L/M)]) (Lai et al. 2000b) and cannot be identified by sequence similarity. We identified potential bearded class genes due to their proximity to E(spl)-C bHLH1 and bHLH2 genes. ORFs and annotated genes surrounding bHLH genes were examined for bearded class characteristics (Supplemental Fig. 3). We identified bearded class genes linked with the E(spl)-C in Daphnia, Acyrthosiphon, Apis, Tribolium, Bombyx, Aedes, and Anopheles. We aligned predicted proteins from these genes, and the known bearded class proteins from Drosophila and subjected them to Bayesian phylogenetic analysis. Trees derived from this analysis separate the Drosophila bearded class proteins into seven clades, (mα, m4, m2, m6, Tom, Brd, and Ocho) (Fig. 3). This analysis produces a star-shaped phylogeny with most of the nondrosophilid sequences forming a clade, implying that the Drosophila bearded class genes all derive from duplications of the single bearded gene seen in nondrosophilid insects. Two exceptions to this, the bearded class genes from aphid and Daphnia, branch with m6 and Brd, respectively; but it is likely that these associations are spurious due to long branch attraction.
Figure 3.
Phylogram of bearded proteins in arthropod genomes. Unrooted Bayesian phylogram of bearded proteins from arthopod genomes. Names of proteins and their respective species are shown in Figure 4 and in Supplemental Table 1. Phylogenetic analysis indicates that while the individual bearded class gene from Drosophilid genomes cluster robustly together; those from other species cluster loosely together with long branches. It is likely that the relationships in this tree are distorted by long branch attraction.
Identification of Arthropod E(spl) complexes
Drosophilid E(spl) complexes
Comparisons of the E(spl)-C across Drosophila species indicates that it is very stable (data not shown). In D. melanogaster the complex consists of 12 genes. Her—the most closely related gene to the E(spl)-C bHLHs (Fig. 1)—lies on chromosome X, and the bearded cluster, Tom, Brd, and Ocho, is on chromosome 3L (Fig. 4). In other Drosophila species, the E(spl)-C is identical to that of D. melanogaster (data not shown). This conservation does not extend to Her, which is missing from the genomes of D. grimshawi, mojavensis, and yakuba.
Figure 4.
Arthropod E(spl) complexes. Genomic architecture of the arthropod E(spl)-C. E(spl) complexes from Arthropod genome sequences. Names of gene and their respective species are shown in Supplemental Table 1. To the left is a phylogenetic tree of the species represented. (Squares) bHLH genes; (hexagons) bearded class genes; (light gray heptagon) m1; (purple octagon) groucho; (ovals) intervening genes in arthropod E(spl) complexes with no similarity to Drosophila E(spl)-C genes. Genes are color coded with reference to their protein phylogeny: (light blue) E(spl)-C bHLH2-derived sequences; (dark blue) E(spl)-C bHLH1 sequences; (green) Her-derived sequences; (red) Tom/Ocho/bearded-like sequences; (dark gray) m6 sequences. Contigs are shown and labeled to the right. Breaks in the complex are denoted by breaks in the lines representing contigs. For clarity, identifiers for Daphnia genes are missing NCBI_GNO_ prefix.
Dipteran E(spl) complexes
We re-examined the E(spl)-C in the Anopheles gambiae genome. In this species, as reported previously (Schlatter and Maier 2005) only a single E(spl)-C-like bHLH gene can be identified [AGAP0012342, an E(spl)-C bHLH-1 gene]; this gene lies next to a single bearded class gene AGAP012341. This single bHLH class 1 gene next to a bearded class gene is repeated in the related Culex pipens genome and that of Aedes aegypti, although the direction of transcription of the bearded gene relative to the bHLH is reversed in Culex and Aedes (Fig. 4).
Holometabolous E(spl) clusters
The simple E(spl)-C seen in mosquito genomes is not observed in other holometabolous insects. We examined the genomes of A. mellifera, T. casteneum, Nasonia vitripennis, and Bombyx mori for E(spl)-C genes (Fig. 4). In B. mori, a cluster of four E(spl)-C-related genes lies on linkage group 3 within 240 kb. Two bHLH genes (BGIBMGA8916 [E(spl)-C bHLH2] and BGIMGA8915 [Her]) lie in a head to tail on arrangement separated by three genes [unrelated to Drosophila E(spl)-C genes] from a bearded class gene (BGIBMGA8964); next to this gene is a third bHLH gene (BGIBMGA8915 [E(spl)-C bHLH1]).
An identical arrangement of genes is present in the honeybee on linkage group 14 contained within 202 kb: two upstream bHLH genes (GB17028 [E(spl)-C bHLH2] and GB10585 [Her]; in this case separated by a gene encoding a tubulin tyrosine ligase-like protein with no similarity to any Drosophila E(spl)-C gene), a bearded class gene (GB15253), and a final bHLH gene (GB19475 [E(spl)-C bHLH1]) (Fig. 4).
The current assembly of the Nasonia genome encodes no E(spl)-C bHLH genes nor can they be found in unassembled reads. bHLHs with orange domains are present in the genome, but these are orthologs of dpn, h, and cwo. It is not clear if the E(spl)-C has been lost in the lineage leading to Nasonia, or if the absence is due to gaps in the genome sequence.
In Tribolium, the complex is 70 kb and encodes only two bHLH genes (XP_972493 [Her] and XP_972685 [E(spl)-C bHLH1]) flanking a bearded class gene (glean6579). The bHLH genes are both separated from the bearded class gene by two intervening genes with no homology with Drosophila E(spl)-C genes.
E(spl)-C in hemimetabolous insects
The conclusion that E(spl) complexes are larger and more evolutionarily stable than previously thought led us to examine the newly sequenced genomes of the pea aphid (Acyrthosiphon pisum) and louse (Pediculus humanus) (Fig. 4). In Pediculus, only two linked bHLH genes (PHUM000172 [E(spl)-C bHLH1] and PHUM9020 [Her]) could be found (on contig 484) with no evidence for a linked bearded class gene (Fig. 4).
In the aphid genome, a larger cluster of E(spl)-C genes can be found but these are distributed over two contigs. The first contig, EQ122792, contains two E(spl)-C bHLH genes (ACYPI005974 [E(spl)-C bHLH2] and ACYPI009737 [Her]); the second EQ124787 contains a bearded class gene (ACYPI003071) (Supplemental Fig. 3) and a final bHLH gene (ACYPI003697 [E(spl)-C bHLH1]) (Fig. 4).
The Daphnia cluster
As hemimetabolous insects have large E(spl)-C we examined the genome of D. pulex, a crustacean, for any evidence of an E(spl)-C. In Daphnia a four-gene E(spl)-C exists on scaffold 170. Two bHLH genes lie upstream (NCBI_GNO_234834 [E(spl)-C bHLH2] and NCBI_GNO_299514 [Her]) of a bearded class gene (NCBI_GNO_230834) and a final bHLH gene (NCBI_GNO_228834 [E(spl)-C bHLH1]) (Fig. 4).
The Ioxedes E(spl)-C genes
Phylogenetic evidence implies that chelicerates are the most distant group of arthropods from the crustacean/insect clade (Cook et al. 2001; Hwang et al. 2001). Examination of bHLH genes in the Ixodes scapularis genome did not uncover any sign of a complex of E(spl)-C genes. Two bHLH genes lie next to each other in the genome, but our phylogeny (Fig. 1) indicates that these are dpn related rather than E(spl)-C bHLHs.
This analysis reveals that the E(spl)-C is an ancient feature of insect and crustacean genomes, deriving from the common ancestor of these subphyla. The analysis also indicates that Her is ancestrally contained within the complex and has been lost from the E(spl)-C in the lineage leading to Diptera.
Noncoding sequences of the E(spl) complex
Our studies have identified a four-gene E(spl)-C as a conserved feature of both crustacean and insect genomes. In Drosophila the expression of the E(spl)-C genes is regulated by Notch signaling via SU(H) (Bailey and Posakony 1995), by proneural genes (Heitzler et al. 1996), and by miRNA binding to conserved sites (known as Brd-box, GY-box and K-box) in the 3′ UTR of these genes (Lai et al. 2005). To determine whether these regulatory mechanisms are evolutionarily conserved we identified putative regulatory motifs to the 5′ and 3′ of the Drosophila, honeybee, aphid, and Daphnia E(spl)-C genes (Supplemental Fig. 4). SU(H) sites are found upstream of the start codon in all Arthropod E(spl)-C genes with the exception of the honeybee GB17028. These SU(H) sites are usually coupled with a proneural A site, raising the possibility that these genes may be activated by Notch signaling similar to Drosophila E(spl)-C genes. In both aphid and honeybee the ortholog of Her has a paired SU(H) site and an A-box proneural site in the 5′ region of the gene, and a GY-box and K-box in the sequence to the 3′ of the stop codon. However, in the lineage leading to Drosophila these sites have been lost with only a conserved GY box. Motifs corresponding to the GY- and K-boxes were identified in the genome sequence downstream from the coding regions of a number of E(spl)-C genes in honeybee, aphid, and Daphnia, indicating that regulation of these transcripts by miRNA may be conserved. No Brd-box sequences were found associated with Daphnia genes, indicating that regulation by this miRNA may have evolved in the lineage leading to insects or that both the seed sequence and miRNA have diverged such that it cannot be detected.
Expression of E(spl)-C-related genes in honeybees
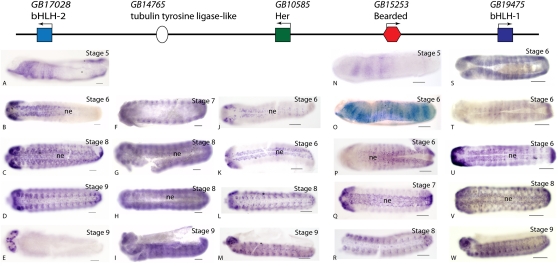
To determine whether genes of the E(spl)-C have a conserved role in Notch signaling and lateral inhibition we cloned partial coding sequences of the genes of the honeybee complex, (including GB14765, a bee-specific tubulin tyrosine ligase-like protein) and examined their expression via in situ hybridization. The four classical E(spl)-C genes of the honeybee E(spl)-C (Fig. 4) have similar expression patterns (Fig. 5). All four genes, including Her, are expressed initially between stages 5 and 6 of development and are expressed in the neuroectoderm as neurogenesis begins. Expression of these genes is limited to those neuroectodermal cells that do not take up neural cell fate, consistent with the role of E(spl)-C genes in Drosophila. All of the honeybee E(spl)-C genes are also expressed in a complex series of stripes across the ectoderm, in tracheal pits, and all except Her are expressed in the gnathal limb buds.
Figure 5.
Expression of honeybee E(spl)-C genes. Honeybee embryos stained for E(spl)-C RNA (blue) using in situ hybridization. All embryos are oriented with anterior to the left, and, unless otherwise stated, viewed ventrally. Scale bars, 100 μm. Staging as per DuPraw (1967). (ne) Neurectoderm. (A–E) Expression of GB17028, an E(spl)-C bHLH-2 ortholog. (A) Stage 6 embryo (lateral view). GB17028 RNA is expressed in ectoderm on either side of the gastrulation furrow, in the anterior and in a posterior domain. By late stage 6, after the gastrulation furrow has closed, RNA is present in a complex pattern of cells in the anterior neuroectoderm. This expression spreads from anterior to posterior by stage 7 (B), and by stage 8 (C) RNA is present in a segmentally patterned array of cells throughout the lateral neuroectoderm, and strongly in the developing brain. (D) A stage 9 embryo showing expression in paired domains on either side of the ventral midline, including the gnathal limb buds. (E) A dorsal view of the embryo shown in D. GB17028 RNA is present in the developing brain. (F–I) Expression of GB14765 RNA, a gene that encodes a tubulin tyrosine ligase-like protein. (F) Expression in a stage 7 embryo. GB14765 RNA is present in segmental paired domains at the lateral edge of the germband, probably the tracheal primordia. In stage 8 embryos (G,H [G damaged at the anterior lefthand side]) RNA is present at low levels generally but also in segmental paired domains on either side of the ventral midline. This expression fades in late stage 8 embryos (H). (I) Stage 9 embryo (lateral view) showing expression in segmental stripes, with highest expression at the dorsal edge of the germband in each segment. (J–M) Expression of GB10585 RNA, a honeybee ortholog of Her. In stage 6 embryos (J,K) GB10585 RNA is expressed in the neuroectoderm in a complex pattern of cells with the expression spreading from anterior to posterior, with the entire the pattern extending all the way to the posterior by late stage 6 (K). By stage 8 (L) this expression is lost, and GB10585 RNA is expressed in segmental paired domains on either side of the ventral midline and in the developing brain. (M) Stage 9 embryo, lateral view, showing GB10585 RNA in paired domains on either side of the midline and in patches of cells in each segment at the dorsal edge of the germband, the tracheal primordia. (N–R) Expression of GB15253 RNA, a bearded ortholog. (N) Expression in a stage 5 embryo (lateral view), just before the onset of gastrulation. GB15253 RNA is present in a series of broad stripes in the anterior lateral ectoderm and in a single narrow stripe in the posterior. (O) Stage 6 embryo (lateral view) as gastrulation begins, GB15253 RNA is present broadly in the lateral ectoderm, but expression is modulated in a series of narrow stripes. (P) Late stage 6 embryo (ventral view; embryo damaged in the posterior right side). As the gastrulation furrow closes in the posterior, GB15253 RNA expression is present in broad domains at the edge of the gastrulation furrow, the neuroectoderm; as gastrulation proceeds this broad domain fades, and in anterior to posterior sequence, leaving a complex pattern of cells expressing GB15253 RNA in the neuroectoderm at stage 7 (Q). (R) Stage 8 embryo (anterior regions damaged) showing GB15253 RNA in paired domains on either side of the midline, including in the gnathal limb buds; expression can also be seen in a small number of cells in the ventral midline. (S–W) Expression of GB19475 RNA, an E(spl)-C bHLH-1 orthologous gene. (S) Early stage 6 embryo. GB19475 RNA is present in the lateral ectoderm in broad domains modulated by possible segmental stripes. As the gastrulation furrow closes, in the anterior in this specimen, this domain becomes narrower and focuses down to just the edges of the gastrulation furrow. (T) In mid stage 6 embryos, broad lateral expression has faded, but RNA is becoming expressed in a complex set of cells on either side of the gastrulation furrow, in the neuroectoderm, and in a posterior cap. (U) In late stage 6 embryos this complex neuroectodermal staining stretches the length of the embryo to the posterior cap. (V) Stage 8 embryo. GB19475 RNA is present in a complex pattern of cells in the neuroectoderm and ventral midline. Segmental domains of expression can also be seen in cells at the dorsal edge of the germband. (W) Stage 9 embryo (lateral view) GB19475 RNA is expressed in paired groups of cells on either side of the ventral midline, including the gnathal limb buds, and faintly in the tracheal primordia at the dorsal edge of the germband.
GB14765, the intervening gene in the honeybee complex, is first expressed later in development than the other genes in the E(spl)-C, but it is expressed in a complex pattern that includes regions of the embryo that express other members of the honeybee E(spl)-C, including the neuroectoderm, and tracheal pits (Fig. 5F–I). Expression of the Drosophila ortholog of this gene, CG16833, was also determined with in situ hybridization. CG16833 RNA is expressed ubiquitously in Drosophila with a clear maternal contribution but is not specifically expressed in the neuroectoderm (Fig. 6J–L).
Figure 6.
Expression of Drosophila Her, CG16833, and aphid Her. Drosophila embryos stained for Her or CG16833 RNA (blue) using in situ hybridization. Scale bars, 50 μm for panels A–E and J–N. Embryos are oriented anterior to the left, dorsal top. (A–C) Expression of Her RNA in Drosophila embryos detected using in situ hybridization. Her RNA in a stage 5 (A), stage 11 (B), and stage 12 (C) embryo. At all stages Her RNA is ubiquitously distributed. (D,E) Sense controls for Her in situ hybridization stained under the same conditions as A–C. Weak staining is seen in early embryos, up to stage 4 (D); later embryos (E, stage 11) show no significant expression. (F–H) Viviparous aphid embryos stained for Her RNA (ACYPI009737) (blue) using in situ hybridization. All embryos are oriented with anterior to the left. Scale bars, 200 μm for panels F–I. Embryos are staged according to the scheme of Miura et al. (2003) and Chang et al. (2007). (F) At stages 11 and 12 (lateral view) Ap-Her is expressed diffusely throughout the embryonic germband during germband elongation including the presumptive head region. (G) By stage 13 and 14 (ventral view) limb buds are formed and diffuse Ap-Her staining is observed in the head region, with stronger staining observed in cells of the ventral midline as well as some surrounding cells. Expression of Ap-Her is not detected in the fully differentiated CNS. (H) At stage 18 (ventral view) expression of Ap-Her is restricted to tissues associated with the differentiating compound eyes. (I) Ap-otd (also known as oc) expression is shown for comparison. At stage 14 (ventral view) Ap-otd is strongly expressed in the presumptive cephalic regions of the head and in cells of the ventral midline in the developing CNS (Huang et al. 2010). (J–L) Expression of Drosophila CG16833 RNA detected by in situ hybridization. Expression is ubiquitous in all embryos, shown are stage 4 (J), stage 5 (K), and stage 12 (L). (M,N) Sense negative control in situ hybridization for CG16833. No staining is seen at any stage; shown are stage 6 (M, dorsal view) and stage 12 (N).
This analysis shows that genes within the honeybee E(spl)-C (including Her and the tubinyl tyrosine ligase) are expressed in a pattern consistent with a role in lateral inhibition in the neuroectoderm.
Expression of Aphid and Drosophila Her
Honeybee Her, unlike Drosophila Her, lies within the E(spl)-C (Fig. 4) and is regulated like the other bHLH genes in the complex (Fig. 5). To determine if this expression pattern is unusual in insects or a honeybee-specific pattern we re-examined the expression of Drosophila Her, previously reported as ubiquitously expressed (Moore et al. 2000) and compared this with the expression of Her in the aphid. Drosophila Her RNA is expressed in all cells of the embryo (Fig. 6A–C), from very early stages where RNA appears to be maternally provided, though weak staining can be seen in negative controls at this stage (Fig. 6 D,E). This expression pattern is markedly different from that seen with honeybee Her (Fig. 5J–M).
In contrast, aphid Her (ACYPI009737) RNA is not ubiquitously distributed (Fig. 6F–G). Coincident with the specification of the CNS aphid Her RNA is detected in cells of the ventral midline and in cells surrounding the midline, with diffuse staining throughout the head of the developing aphid (Fig. 6G). This expression pattern is similar to that seen for aphid orthodenticle (otd), which we have used to help describe the embryo (Fig. 6I; Huang et al. 2010), although there are differences; expression of Ap-Her is more diffuse and there is clear evidence for staining in cells either side of the midline. No expression of Her RNA can be detected in the mature CNS.
Together the expression of Her in aphids and honeybees implies that the ancestral expression of Her is neuroectodermal, with a probable role in lateral inhibition. The ubiquitous expression seen in Drosophila is clearly unusual and may be related to its location outside of the E(spl)-C.
Discussion
Evolution of the arthropod E(spl) complex
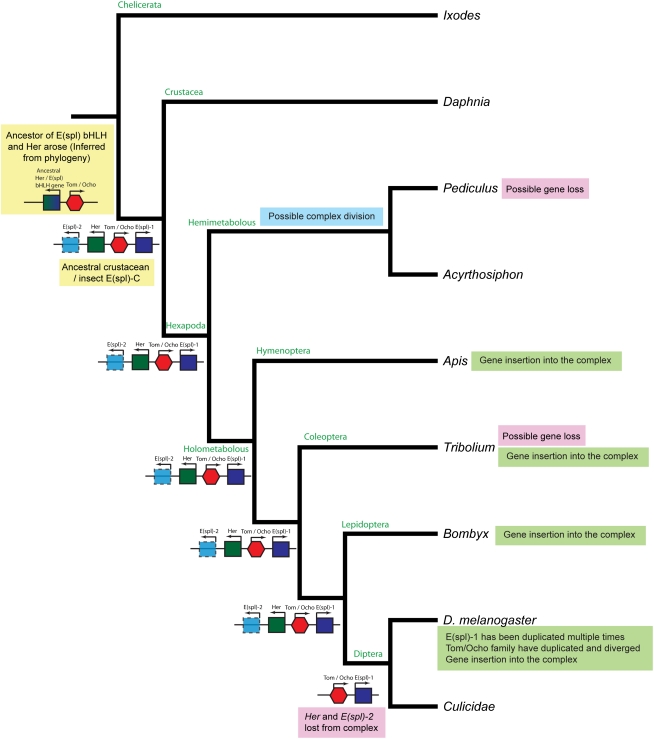
Our analysis of the architecture of the E(spl)-C across insect and non-insect arthropods clearly contradicts the previous assertion that the E(spl)-C of Drosophila has evolved from a simple two-gene Urcomplex (Schlatter and Maier 2005). Indeed this four-gene complex has a long evolutionary history predating the divergence of insects from crustaceans ∼420 Mya (Gaunt and Miles 2002). This “ancestral” complex is not present in the chelicerate Ixodes, and so likely arose between 550 and 420 Mya (Gaunt and Miles 2002) after the split of myriapod and chelicerate lineages. This makes the E(spl)-C both more ancient, and more complex, than previously described. Parsimony ancestral state reconstruction was used to visualize the key steps in the evolution of the E(spl)-C (Fig. 7). Ancestrally the complex was likely to consist of two E(spl)-C–type bHLH genes [E(spl) bHLH1 and E(spl) bHLH2], a single Her-type bHLH gene, and a bearded class gene (Fig. 7). Although, as bHLH2 has been lost in the Diptera, Tribolium, and Pediculus the parsimony reconstruction is ambivalent as to whether the ancestral complex included bHLH2. The phylogenetic evidence (Fig. 2), however, precludes the reevolution of bHLH2 from a duplication of bHLH1; thus, repeated losses of this gene have occurred in these lineages. The conservation of gene organization and direction of transcription of genes in the E(spl)-C in Crustacea, hemimetabolous insects (with the caveat that the Acyrthosiphon contigs are not currently joined), and holometabolous insects is strong evidence that this four-gene structure is the ancestral organization of this complex and was present in the last common ancestor of insects and crustaceans.
Figure 7.
Parsimony ancestral state reconstruction for the evolution of arthropod E(spl) complexes. Ancestral character states were inferred using parsimony analysis under an Mk1 model in Mesquite. Character states were mapped onto a cladogram representing the currently accepted phylogenetic relationships between insect species (Krauss et al. 2005; Robertson 2005; Savard et al. 2006). Reconstructions of the ancestral E(spl)-C are shown at the nodes and genes are color coded as for Figure 4. bHLH2 is absent from the Diptera, Tribolium, and Pediculus and the parsimony reconstruction is ambivalent as to whether the ancestral complex included bHLH2. However, we argue that the phylogenetic evidence (Fig. 2) precludes the re-evolution of bHLH2 from a duplication of bHLH1, and it is likely that bHLH2 has been lost independently. However, this uncertainty is represented by the dotted line surrounding bHLH2 in the diagrams.
Drosophila E(spl) complexes
The E(spl) complex of Drosophilids is unusual. Most surprisingly it is missing two of the ancestral genes: Her, which has moved out of the complex and is now expressed in a ubiquitous manner in D. melanogaster but is missing from the genomes of D. grimshawi, D. mojavensis, and D. yakuba; and E(spl)-C bHLH2, which is not present in any Drosophilid genome. Based on our analyses these genes have remained in the E(spl)-C throughout insect evolution before being lost in Diptera. This loss of the Her and E(spl)-C bHLH2 genes is associated with the expansion of the E(spl)-C bHLH1 class from one to eight genes in Drosophila. Bearded genes have also duplicated with four partially redundant copies in the complex, and three others in another genomic location (the bearded complex) (Lai et al. 2000b). These duplication events appear to be associated with significant sequence divergence with both bHLH and bearded class genes having long branch lengths in the phylogenetic analysis. It is unclear why the E(spl)-C has undergone such duplication and divergence in Drosophila. This expansion has not been driven solely by the loss of Her and E(spl)-C bHLH2 in Drosophila as both genes are also missing from mosquito genomes, where there no evidence for complex expansion. None of the Drosophila E(spl)-C genes (except m1) contain introns, while those in other species do; perhaps the loss of introns has facilitated repeated gene duplication. It is possible that the increase in E(spl)-C genes in Drosophila, mirrored as it is by an increase in the number of proneural genes (Schlatter and Maier 2005), may provide more specific control over neurogenesis in Drosophila and may reflect a need to place neural elements such as bristles in more stereotyped locations in the Drosophilidae than in mosquitoes and other holometabolous insects (Simpson et al. 1999). Consistent with this idea, polymorphisms in the Drosophila E(spl)-C are known to be associated with bristle number variation (Macdonald et al. 2005).
The Drosophila E(spl)-C complex is tightly linked to two other genes, m1 and groucho. m1 is not Notch responsive (Wurmbach et al. 1999), while Groucho is a key component of transcriptional repression mediated by Notch (Paroush et al. 1994). These genes do not appear in the complex in any other species and are likely to be serendipitous insertions into, or near, the complex. It is interesting to speculate, however, that given the key role of groucho in Notch signaling and the function of the E(spl) bHLH genes, it is extraordinary that this gene now sits next to the E(spl) complex. Perhaps this close linkage to the E(spl)-C represents the incipient capture of another Notch signaling component by the E(spl)-C.
GB14765 is a honeybee-specific insertion into the E(spl)-C and is expressed in a pattern consistent with it being regulated, in part, by Notch signaling, while its ortholog in D. melanogaster is not. Conversely bHLH genes, like Her in Drosophila, that move out of the complex, lose their “E(spl)”-type expression pattern and presumably their ability to be Notch responsive. These data lead us to hypothesize that the E(spl)-C, at least in early embryos, may act as a genomic regulatory domain that can be regulated as a whole by Notch signaling. Support for this hypothesis also comes from the recent finding that the E(spl)-C may be regulated in an unusual way (Schaaf et al. 2009). Chromatin immunoprecipitation experiments demonstrate that the E(spl)-C is bound by both cohesin and polycomb group proteins, and knock-down of the VTD (also known as RAD21) subunit of cohesin in these cells causes many of the E(spl)-C genes' expression to be up-regulated (Schaaf et al. 2009). This is consistent with the idea that the complex is a genomic domain, regulated in concert by cohesin and polycomb in some developmental contexts. Consistent with this Her, the E(spl)-C like bHLH that has moved out of the complex in Drosophila is not sensitive to cohesin knockdown (Schaaf et al. 2009).
Function of the E(spl) complex
The function of the E(spl)-C genes in the honeybee embryo, based on their expression pattern, is similar to their role in Drosophila. Expression during neurogenesis, in cells of the neuroectoderm that do not delaminate, implies that honeybee E(spl)-C genes are acting to repress neural cell fate. This expression pattern is the same for bHLH genes and bearded genes.
Earlier, during gastrulation (stage 6), expression of RNA from these genes is in a complex series of stripes across the ectoderm, which are not of a periodicity, or at a stage, that suggests a role in segmentation. Yet Notch signaling triggered by Delta does not regulate segmentation, despite both Delta and fringe being expressed in clear segmental stripes (MJ Wilson, BH McKelvey, S van der Heide, and PK Dearden, unpubl.). The expression patterns of the E(spl)-C genes may reflect a general tendency for genes to be expressed in stripes during segmentation with no functional consequence, as seen for many Drosophila genes (Liang and Biggin 1998). Later expression of honeybee E(spl)-C genes indicates a role in brain, mouthpart, and tracheal development.
The conservation of expression patterns of these genes across the 300 Myr divergence between honeybees and Drosophila implies that the function of the E(spl)-C is stable over evolutionary time. Phylogenetic and phylogenomic evidence supports the idea that the Hymenoptera are the most basally branching group of the holometabolous insects (Krauss et al. 2005; Savard et al. 2006). This phylogenetic placement implies that the function of the E(spl)-C is conserved in holometabolous insects. Aphid Her is also expressed in a pattern implying a role in lateral inhibition, suggesting that the E(spl)-C in aphids, and possibly all holometabolous insects, also acts in neurogenesis.
Gene complexes and gene co-option
The recent sequencing of a number of arthropod genomes has revealed a significant difference between arthropod and vertebrate genome evolution. In vertebrates, gene organization and synteny is often conserved over long evolutionary distances (Barbazuk et al. 2000; Kohn et al. 2006; Kikuta et al. 2007). In arthropods this is not the case. The relationships between genes changes more rapidly in arthropod lineages, perhaps reflecting shorter average generation times. Gene complexes in insects are thus less likely to remain together by serendipity than in vertebrates. Therefore, it is likely that any highly conserved complex in arthropods must be functionally constrained. In arthropods few such complexes exist, the best described being the Hox complex, in which gene organization and function are linked (Hughes and Kaufman 2002), the Runt complex, where it is not clear why the genes remain together over long evolutionary periods (Duncan et al. 2008), and now the E(spl)-C.
The E(spl)-C is a remarkable example of a conserved gene complex as it contains, at its most simple, two functionally different classes of genes. This structure suggests that the complex is maintained for functional reasons.
Data presented here show that genes that are inserted in the complex appear to become controlled by Notch signaling: GB14765 has moved into the honeybee E(spl)-C and is now expressed in an E(spl)-C-like way, unlike its ubiquitous expression in Drosophila. Those that move out seem to lose that Notch responsiveness: Her has moved out of the E(spl)-C in Drosophila and is now expressed ubiquitously, a pattern of expression dissimilar to either GB10585 or ACYPI009737, its honeybee and aphid orthologs, and not consistent with a role in neurogenesis. This, and the coordinated cohesin regulation of the complex (Schaaf et al. 2009), implies that the E(spl)-C is an evolutionarily conserved functional genomic domain, a region of the genome that is coordinately regulated by Notch signaling, a cell signaling system used pleiotropically in arthropods, and that this domain explains the maintenance of the E(spl)-C for at least 420 Myr of arthropod evolution.
More importantly, however, is the implication of this hypothesis for co-option of genes into the Notch signaling pathway. It seems that any gene that is inserted into the E(spl)-C becomes regulated by Notch signaling because the complex acts as a coordinated regulatory domain. Such inserted genes have the opportunity to become part of the Notch signaling pathway. We suggest that this may be how the complex was built, that an ancestral E(spl)-C–like bHLH gene regulated by Notch became close to a bearded sequence and captured it, causing it to become regulated by Notch signaling, and eventually to act in it. Such capture of a sequence into a signaling pathway due to a local chromatin domain may be a general mechanism whereby novel genes are recruited into ancient cell signaling pathways.
Methods
Gene identification
E(spl)-C genes were identified using BLASTP or TBLASTN searches (Altschul et al. 1990) on whole insect genome sequence databases (BeetleBase, http://www.bioinformatics.ksu.edu/BeetleBase; I. scapularis VectorBase, http://www.vectorbase.org; Nasonia Genome Project, http://www.hgsc.bcm.tmc.edu/projects/nasonia/; P. humanus VectorBase, http://www.vectorbase.org; wFleaBase: Daphnia Genome project, http://wfleabase.org/; Colbourne et al. 2005; Wang et al. 2005, 2007; Nene et al. 2007; Drysdale 2008; Tribolium Genome Sequencing Consortium 2008; Lawson et al. 2009; The International Aphid Genomics Consortium 2010; Legeai et al. 2010; The Nasonia Genome Working Group 2010). When E(spl)-C bHLH genes were identified we searched the regions around those genes using BLAST searches against the Drosophila genome to identify other possible components of a complex. For identification of bearded class genes, the secondary structure of the predicted protein sequence was assessed using the multivariate linear regression combination (MLRC) algorithm (Guermeur et al. 1999) at the Network Protein Sequence Analysis website (NPS) (Combet et al. 2000). The nature of predicted helical regions was determined using Heliquest (Gautier et al. 2008).
Phylogenetics
Species names, sequences, and identifiers for genes and proteins used in this study are provided in Supplemental Table 1. Protein sequences were aligned using ClustalX (Thompson et al. 1994) and subjected to Bayesian phylogenetic analysis using MrBayes (Ronquist and Huelsenbeck 2003). Phylogenetic relationships were reconstructed using the Jones (bHLH proteins; Jones et al. 1992) or WAG model (Whelan and Goldman 2001), which were found to be the most appropriate after preliminary investigations using mixed models. The first 25% of trees were discarded as burn-in and the remaining trees summarized and visualized using Dendroscope (Huson et al. 2007).
Ancestral state reconstruction of the E(spl)-C was performed with Mesquite (v2.72) (http://mesquiteproject.org). Reconstruction was performed using parsimony methods with the Mk1 model (Markov K-state 1 parameter model), which assumes equal probability for changes between states. Character states were mapped onto the currently accepted phylogeny of arthropods (Krauss et al. 2005; Robertson 2005; Savard et al. 2006) assuming equal branch lengths.
In situ hybridization
Fragments of E(spl)-C genes were amplified using PCR (see Supplemental Table 2 for primer sequences) and cloned into PCRII Topo (Invitrogen) following the manufacturer's instructions.
Honeybee embryo in situ hybridization was performed as described previously (Osborne and Dearden 2005; Dearden et al. 2009).
Aphid ovaries and nymphs were dissected into cold PBS (phosphate-buffered saline) and fixed for 1 h in a 1:1 mix of 4% formaldehyde: heptane in PBS. Samples were stored in methanol at −20°C until required. Aphids were rehydrated through a methanol/0.3% PTw series (PBS with 0.3% Tween-20), fixed for 20 min in 4% formaldehyde, washed three times in 0.3% PTw, and then incubated for 45 min in detergent solution (1% SDS, 0.5% Tween-20, 50 mM Tris-HCl at pH 7.5, 1 mM EDTA at pH 8.0, 150 mM NaCl) (Shigenobu et al. 2010). Embryos/ovaries were washed seven times in 0.3% PTw, and hybridization was performed as described for honeybees (Osborne and Dearden 2005; Dearden et al. 2009). Anti-DIG antibody (Roche) was used at a 1:2000 dilution, samples were incubated overnight at 4°C, and color development was performed using standard protocols.
Acknowledgments
We thank Megan Wilson and Nathan Kenny for critical comments on this manuscript. P.K.D. thanks David Hartley for first introducing him to the E(spl)-C and for patiently waiting for a paper. We also thank Gabrielle Drayton and David Teulon from Plant and Food Research for providing us with the pea aphid cultures. This work was supported by a grant to P.K.D. from the National Research Centre for Growth and Development (MP04) and by a UORG grant from the University of Otago.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.104794.109.
References
- Alifragis P, Poortinga G, Parkhurst SM, Delidakis C 1997. A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc Natl Acad Sci 94: 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Apidianakis Y, Nagel AC, Chalkiadaki A, Preiss A, Delidakis C 1999. Overexpression of the m4 and mα genes of the E(spl)-Complex antagonizes Notch mediated lateral inhibition. Mech Dev 86: 39–50 [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW 1995. Suppressor of hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9: 2609–2622 [DOI] [PubMed] [Google Scholar]
- Barbazuk WB, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell JA, McPherson JD, Johnson SL 2000. The syntenic relationship of the zebrafish and human genomes. Genome Res 10: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lin GW, Cook CE, Horng SB, Lee HJ, Huang TY 2007. Apvasa marks germ-cell migration in the parthenogenetic pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Dev Genes Evol 217: 275–287 [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ 1999. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev 13: 2218–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Singan VR, Gilbert DG 2005. wFleaBase: The Daphnia genome database. BMC Bioinformatics 6: 45 doi: 10.1186/1471-2105-6-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G 2000. NPS@: Network protein sequence analysis. Trends Biochem Sci 25: 147–150 [DOI] [PubMed] [Google Scholar]
- Cook CE, Smith LM, Telford MJ, Bastianello A, Akam ME 2001. Hox genes and the phylogeny of the arthropods. Curr Biol 11: 759–763 [DOI] [PubMed] [Google Scholar]
- Damen WG, Weller M, Tautz D 2000. Expression patterns of hairy, even-skipped, and runt in the spider Cupiennius salei imply that these genes were segmentation genes in a basal arthropod. Proc Natl Acad Sci 97: 4515–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen WG, Janssen R, Prpic NM 2005. Pair rule gene orthologs in spider segmentation. Evol Dev 7: 618–628 [DOI] [PubMed] [Google Scholar]
- Dearden PK, Duncan EJ, Wilson MJ 2009. The Honeybee Apis mellifera. Cold Spring Harb Protoc 2009: doi: 10.1101/pdb.emo123 [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C 2001. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell 1: 795–806 [DOI] [PubMed] [Google Scholar]
- de Celis JF, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: The E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122: 2719–2728 [DOI] [PubMed] [Google Scholar]
- Delidakis C, Artavanis-Tsakonas S 1992. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci 89: 8731–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R 2008. FlyBase: A database for the Drosophila research community. Methods Mol Biol 420: 45–59 [DOI] [PubMed] [Google Scholar]
- Duncan EJ, Wilson MJ, Smith JM, Dearden PK 2008. Evolutionary origin and genomic organisation of runt-domain containing genes in arthropods. BMC Genomics 9: 558 doi: 10.1186/1471-2164-9-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPraw EJ 1967. The honeybee embryo. In Methods in developmental biology (ed. FH Wilt and NK Wessells), pp. 183–217 Thomas Y. Crowell Company, New York [Google Scholar]
- Eastman DS, Slee R, Skoufos E, Bangalore L, Bray S, Delidakis C 1997. Synergy between Suppressor of Hairless and Notch in regulation of Enhancer of split mγ and mδ expression. Mol Cell Biol 17: 5620–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M 1996. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol 16: 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana JR, Posakony JW 2009. Both inhibition and activation of Notch signaling rely on a conserved Neuralized-binding motif in Bearded proteins and the Notch ligand Delta. Dev Biol 333: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA 2002. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol 19: 748–761 [DOI] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, Drin G 2008. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 24: 2101–2102 [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Alifragis P, Koumbanakis KA, Delidakis C 2003. Two modes of recruitment of E(spl) repressors onto target genes. Development 130: 259–270 [DOI] [PubMed] [Google Scholar]
- Guermeur Y, Geourjon C, Gallinari P, Deleage G 1999. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15: 413–421 [DOI] [PubMed] [Google Scholar]
- Hartley DA, Preiss A, Artavanis-Tsakonas S 1988. A deduced gene-product from the Drosophila neurogenic locus, Enhancer of split, shows homology to mammalian G-protein β subunit. Cell 55: 785–795 [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122: 161–171 [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutierrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. 2008. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res 18: 1100–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-Y, Cook CE, Davis GK, Shigenobu S, Chen RP-Y, and Chang C-C 2010. Anterior development in the parthenogenetic and viviparous form of the pea aphid, Acyrthosiphon pisum: hunchback and orthodenticle expression. Insect Mol Biol 19: 75–85 [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC 2002. Hox genes and the evolution of the arthropod body plan. Evol Dev 4: 459–499 [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R 2007. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460 doi: 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang U, Friedrich M, Tautz D, Park C, Kim W 2001. Mitochondrial protein phylogeny joins myriapods with chelicerates. Nature 413: 154–157 [DOI] [PubMed] [Google Scholar]
- The International Aphid Genomics Consortium 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8 doi: 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S 1994. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120: 3537–3548 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Kobayashi T 2007. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development 134: 1243–1251 [DOI] [PubMed] [Google Scholar]
- Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, Akalin A, Caccamo M, Sealy I, Howe K, et al. 2007. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E, Schrons H, Grawe F, Campos-Ortega JA 1992. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132: 505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn M, Hogel J, Vogel W, Minich P, Kehrer-Sawatzki H, Graves JA, Hameister H 2006. Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet 22: 203–210 [DOI] [PubMed] [Google Scholar]
- Krauss V, Pecyna M, Kurz K, Sass H 2005. Phylogenetic mapping of intron positions: A case study of translation initiation factor eIF2γ. Mol Biol Evol 22: 74–84 [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW 1997. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 124: 4847–4856 [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW 1998. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development 125: 4077–4088 [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Kavaler J, Freschi G, Posakony JW 2000a. Antagonism of Notch signaling activity by members of a novel protein family encoded by the Bearded and Enhancer of split gene complexes. Development 127: 291–306 [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Posakony JW 2000b. The Enhancer of split Complex of Drosophila includes four Notch-regulated members of the Bearded gene family. Development 127: 3441–3455 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 19: 1067–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. 2009. VectorBase: A data resource for invertebrate vector genomics. Nucleic Acids Res 37: D583–D587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legeai F, Shigenobu S, Gauthier JP, Colbourne J, Rispe C, Collin O, Richards S, Wilson ACC, Murphy T, Tagu D 2010. AphidBase: A centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol Biol 19: 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Biggin MD 1998. Eve and ftz regulate a wide array of genes in blastoderm embryos: The selector homeoproteins directly or indirectly regulate most genes in Drosophila. Development 125: 4471–4482 [DOI] [PubMed] [Google Scholar]
- Macdonald SJ, Pastinen T, Long AD 2005. The effect of polymorphisms in the Enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics 171: 1741–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Polansky BJ, Robson BE, Eastman DA 2007. Phylogenetic footprinting analysis in the upstream regulatory regions of the Drosophila Enhancer of split genes. Genetics 177: 1377–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL 2003. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J Exp Zoolog B Mol Dev Evol 295: 59–81 [DOI] [PubMed] [Google Scholar]
- Moore AW, Barbel S, Jan LY, Jan YN 2000. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc Natl Acad Sci 97: 10436–10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K, Campos-Ortega JA 1996. Persistent expression of genes of the Enhancer of split complex suppresses neural development in Drosophila. Neuron 16: 275–286 [DOI] [PubMed] [Google Scholar]
- The Nasonia Genome Working Group 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW 1999. Discrete enhancer elements mediate selective responsiveness of Enhancer of split complex genes to common transcriptional activators. Dev Biol 213: 33–53 [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne P, Dearden PK 2005. Non-radioactive in situ hybridisation to honeybee embryos and ovaries. Apidologie 36: 113–118 [Google Scholar]
- Palaparti A, Baratz A, Stifani S 1997. The groucho/transducin-like Enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem 272: 26604–26610 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ishhorowicz D 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C 2001. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell 1: 807–816 [DOI] [PubMed] [Google Scholar]
- Robertson HM 2005. Insect genomes. Am Entomol 51: 166–171 [Google Scholar]
- Ronquist F, Huelsenbeck JP 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, Tettelin H, Lercher MJ 2006. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res 16: 1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D 2009. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One 4 doi: 10.1371/journal.pone.0006202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatter R, Maier D 2005. The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol Biol 5: 67 doi: 10.1186/1471-2148-5-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Bickel RD, Brisson J, Butts T, Chang C-c, Davis GK, Duncan EJ, Janssen R, Ferrier D, Lu H-L, et al. 2010. Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: Frequent lineage-specific duplications and losses of developmental genes. Insect Mol Biol 19: 47–62 [DOI] [PubMed] [Google Scholar]
- Simpson P, Woehl R, Usui K 1999. The development and evolution of bristle patterns in Diptera. Development 126: 1349–1364 [DOI] [PubMed] [Google Scholar]
- Tata F, Hartley DA 1995. Inhibition of cell fate in Drosophila by Enhancer of split genes. Mech Dev 51: 305–315 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955 [DOI] [PubMed] [Google Scholar]
- Wang J, Xia Q, He X, Dai M, Ruan J, Chen J, Yu G, Yuan H, Hu Y, Li R, et al. 2005. SilkDB: A knowledgebase for silkworm biology and genomics. Nucleic Acids Res 33: D399–D402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LJ, Wang S, Li Y, Paradesi MS, Brown SJ 2007. Beetlebase: The model organism database for Tribolium castaneum. Nucleic Acid Res 35: D476–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18: 691–699 [DOI] [PubMed] [Google Scholar]
- Wurmbach E, Wech I, Preiss A 1999. The Enhancer of split complex of Drosophila melanogaster harbors three classes of Notch responsive genes. Mech Dev 80: 171–180 [DOI] [PubMed] [Google Scholar]