Abstract
Gene expression becomes more variable with age, and it is widely assumed that this is due to a decrease in expression regulation. But currently there is no understanding how gene expression regulatory patterns progress with age. Here we explored genome-wide gene expression variation and regulatory loci (eQTL) in a population of developing and aging C. elegans recombinant inbred worms. We found almost 900 genes with an eQTL, of which almost half were found to have a genotype-by-age effect (gxaeQTL). The total number of eQTL decreased with age, whereas the variation in expression increased. In developing worms, the number of genes with increased expression variation (1282) was similar to the ones with decreased expression variation (1328). In aging worms, the number of genes with increased variation (1772) was nearly five times higher than the number of genes with a decreased expression variation (373). The number of cis-acting eQTL in juveniles decreased by almost 50% in old worms, whereas the number of trans-acting loci decreased by ∼27%, indicating that cis-regulation becomes relatively less frequent than trans-regulation in aging worms. Of the 373 genes with decreased expression level variation in aging worms, ∼39% had an eQTL compared with ∼14% in developing worms. gxaeQTL were found for ∼21% of these genes in aging worms compared with only ∼6% in developing worms. We highlight three examples of linkages: in young worms (pgp-6), in old worms (daf-16), and throughout life (lips-16). Our findings demonstrate that eQTL patterns are strongly affected by age, and suggest that gene network integrity declines with age.
The expression level of most genes changes with age in all species studied so far. Some genes are expressed during development, whereas others are expressed during or after the reproductive period (Hill et al. 2000; Lund et al. 2002; Ramoni et al. 2002; Geigl et al. 2004; Zhan et al. 2007). For example, McCarroll et al. (2004) studied the temporal dynamics of gene expression throughout the life cycle in Caenorhabditis elegans and Drosophila melanogaster. They reported that genes involved in mitochondrial metabolism, DNA repair, catabolism, peptidolysis, and cellular transport are expressed during adulthood, whereas other transcriptional signatures were observed during larval development, embryogenesis, and gametogenesis.
In addition to the expression level, the variation in gene expression also changes with age. For instance, Somel et al. (2006) reported that gene expression becomes heterogeneous with age in humans and rats. The age-correlated heterogeneity of gene expression had a minor effect on individual genes but was widespread throughout the transcriptome (Somel et al. 2006). These, and other, studies have led to the assumption that regulation of gene expression decreases with age, leading to more variation in gene expression (Lu et al. 2004; McCarroll et al. 2004). But currently there is no understanding on how genome-wide gene expression regulatory patterns progress with age.
Genetic mapping of gene expression regulatory loci (eQTL) provides a means to address this problem (Jansen and Nap 2001; Rockman and Kruglyak 2006). eQTL can be cis- or trans-acting, reflecting local and distant regulation of gene expression, respectively. They have been thoroughly studied in humans, yeast, plants, rats, mice, and worms (Brem et al. 2002; Schadt et al. 2003; Li et al. 2006; Petretto et al. 2006; Keurentjes et al. 2007; Farber et al. 2009). But, as far as we are aware of, all eQTL studies only focused on one time point or age group, whereas most complex traits and diseases such as cancer are age-related. Surprisingly, we have no understanding of the dynamics of eQTL throughout the life of an organism, although it is long known that gene expression itself is strongly affected by age.
Here we present the first genome-wide study of heritable gene expression regulation as a function of age using a set of 36 single nucleotide polymorphism (SNP)-genotyped recombinant inbred lines (RILs) (Supplemental Table 1) derived from C. elegans wild types N2 and CB4856 (Li et al. 2006). The CB4856 × N2 RIL population displays large genetic and phenotypic differences for a wide range of traits such as reproduction, growth (Gutteling et al. 2007; Kammenga et al. 2007), gene expression (Li et al. 2006), and copulatory plug formation (Palopoli et al. 2008), to name a few. We used microarrays to measure genome-wide gene expression using microarrays from the RILs reared at 24°C at three different ages: For young worms, t1, age was 40 h; for reproductive worms, t2, age was 96 h; and for old worms, t3, age was 214 h. In this way we obtained 108 observations perturbed by age and genotype per gene. First, we investigated whether genome-wide gene expression was affected by age across all recombinant genotypes; then, we asked if age-specific gene expression can be attributed to regulatory loci (eQTL) and whether eQTL patterns change during aging.
Results
Variation in gene expression level increases in aging worms
First, we assessed the influence of age on gene expression levels across all RILs. We found that more than 14,000 gene transcript levels, ∼75% of all genes, were affected by age (P ≤ 0.0001). This proportion is congruent with findings by Jiang et al. (2001), who reported ∼69% of all genes being dynamic in the larval stages of wild-type N2 worms. Yet, we demonstrate that dynamic patterns of gene expression proceed at older ages for many different genotypes. We could identify eight different dynamic expression patterns, each containing about 1200 or more genes. More information about these gene expression patterns can be found in the Supplemental Text and Supplemental Tables 2 and 3.
Then we investigated which genes had increased or decreased variation in expression levels during aging. This was done for the differences both between young and reproductive worms (called developing worms) and between the reproductive and old worms (called aging worms). In developing worms, the transcript level variation of a similar number of genes increased and decreased (1282 and 1328, respectively). However, after reproduction, in aging worms the number of genes with an increased variation was almost five times larger than the group with decreased variation (1772 and 373, respectively). Gene Ontology (GO) term analysis revealed that this group of genes is mainly related to protein kinase activity, while genes with decreased variation showed enrichment of monooxygenase activity and transferase activity (Supplemental Table 4).
Regulatory loci of gene expression depend on age
To investigate if regulatory loci affecting gene expression also depended on age, we first mapped eQTL for each of the three age classes separately. In Figure 1, the upper panels show the peak position of the eQTL (x-axis) against the genomic position of genes having an eQTL (y-axis) at each age group. Overall, the number of eQTL decreased with age; we detected 978, 864, 577 eQTL in young, reproductive, and old worms, respectively. The lower panels of Figure 1 show the frequency of eQTL per marker for each age. A distinction was made between cis- and trans-acting loci to show that both types of heritable regulation are very dynamic throughout the life of C. elegans. We detected 685, 642, and 373 cis-acting and 293, 222, and 204 trans-acting for the young, reproductive, and old worms, respectively. At all age groups, a major part of the cis-eQTLs was caused by genes deleted from the CB4856 genome (Maydan et al. 2007), ∼24%, ∼30%, and ∼31% for the young, reproductive, and old worms, respectively. To analyze the cis- and trans-eQTL frequency at the three age groups, we left out the CB4856 deleted genes, because of their high probability to generate a cis-eQTL. In this case, the number of cis-acting eQTL decreased in old worms with ∼50% (compared with juveniles), whereas the number of trans-acting loci decreased with ∼27%. This indicates that cis-regulation becomes relatively less frequent than trans-regulation in aging worms.
Figure 1.
Heritable transcript-level differences at three different time points. The time points represent juvenile, reproductive and old worms. The upper graphs show the position of the genes with an eQTL (−log10 P > 3.8 ; FDR = 0.01) on the y-axis, and on the x-axis the marker at which the peak of the eQTL was found. (From left to right) Young worms in black, late reproductive worms in green, and old worms in red. The lower graphs show the distribution of eQTL per marker. The height of the bar is the total number of eQTL at that marker; cis-acting eQTLs are indicated in the darker and trans-acting in the lighter color. The constitutive and age specific hotspots can easily be identified by comparing the three panels. Any deleted genes in CB4856 were left out of the analysis. The horizontal yellow lines are the trans-band thresholds to 0.01, as determined by permutation.
A two-time-point model identifies genotype-by-age interaction eQTL
Previously, we found that post-reproductive lifespan varied strongly among N2 × CB4856 recombinant lines, with some lines living almost twice as long than others (Doroszuk et al. 2009). We found similar differences in lifespan among the RILs used in this study (Supplemental Fig. 1). This variation in lifespan across the RILs, ranged from a minimum of ∼13 d to a maximum of ∼20 d. The average lifespan for some of the RILs transgressed beyond the mean value of either parental strain (N2, ∼16 d; CB4856, ∼13 d). This is often observed for many phenotypic traits in recombinants derived from genetically divergent parental strains (Kammenga et al. 2007). The differences among RILs means that, after reproduction, the physiological age differs between the RILs and that sampling mRNA at an older post-reproductive age, the third time point, t3, would implicitly result in different physiological stages being sampled. Therefore we analyzed gene expression in relation to relative age, which we defined for each RIL as the age at the time of mRNA extraction divided by the average lifespan of that RIL. In this way, the age-physiological differences among the RILs were taken into account when comparing the gene expression profiles.
In order to find the causal loci for heritable differences in transcript levels and possible interactions between age and genotype, we applied a two-time-point model. In this model, we used three factors—(1) relative age, (2) genotype (marker), and (3) the interaction between factors 1 and 2—to explain the differences in gene expression between RILs and age groups. With this mapping procedure, we found almost 900 genes that had an eQTL or gxaeQTL in developing and/or aging worms (P < 0.0001; Fig. 2). Almost half of these genes with heritable transcript differences were found to have a genotype-by-age effect (396 at P < 0.0001; Table 1) allocated to a specific marker, which we coined genotype-by-age expression-QTL (gxaeQTL). One specific hotspot (trans-band) for gxaeQTL was found on chromosome IV for aging worms and a trans-band for eQTL on chromosome I was detected in developing worms (Fig. 2).
Figure 2.
Gene position (y-axis) plotted against the marker (x-axis) of the peak of the eQTL (P ≤ 0.0001) of genotype-by-age eQTL (gxaeQTL) for that gene. The size of the triangles is relative to the significance of the positive effects of the allele; (red) N2 effect; (blue) CB4856 effect. Chromosomes are separated by the gray dashed lines. Chromosome I is located at the bottom and left of the panels, and chromosome X at the top and right of the panels. (A) Developing worms. (Left) Shows the significant (independent from age) marker (eQTLs) for the developing worms (the juvenile and reproductive worms in one model); (right) shows the significant genotype (marker) by age effects (gxaeQTL) for the developing worms (the juvenile and reproductive worms in one model). (B) Aging worms. (Left) Shows the significant (independent from age) marker (eQTLs) for the aging worms (the reproductive and old worms in one model); (right) shows the significant genotype (marker) by age effects (gxaeQTL) for the aging worms (the reproductive and old worms in one model). Two distinct trans-bands (hotspots) can be observed in the developing eQTLs (upper left) and in the aging gxaeQTLs (lower right), showing that hotspots can be found for genotype-independent eQTLs as well as genotype-by-age interaction gxaeQTLs.
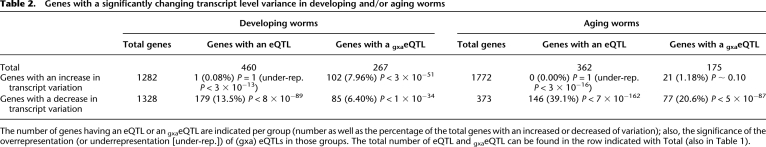
Table 1.
Type of eQTL (P < 0.0001 = −log10 P ≥ 4) at different ages calculated with the two-time-point model
Significant linkage to either marker (eQTL) or the genotype-by-age interaction (gxaeQTL) is indicated, as well as the overlap between groups. In total, we identified 893 unique genes with at least one significant feature.
Increased gene expression variation in old worms co-occurs with a lack of regulatory loci
Increased variation of gene expression during aging is often assumed to be the result of decreased gene expression regulation (Lu et al. 2004; McCarroll et al. 2004). We found that genes with decreased expression level variation had a much higher percentage of eQTL (i.e., regulatory loci) and gxaeQTL in aging worms compared with developing worms (39.1% and 13.5%, respectively, for eQTLs; 20.6% and 6.4%, respectively, for gxaeQTLs). We found hardly any eQTLs or gxaeQTL in the group of genes that had increased expression level variation in aging worms (Table 2; Supplemental Fig. 2). Although we found a small but significant group that showed less transcript variation and increased heritable regulation at old age (Supplemental Table 5; Supplemental Fig. 2), these results show that a large group of genes are more variably expressed at old-age and had a lack of eQTL, which indicates a reduction of heritable regulation.
Table 2.
Genes with a significantly changing transcript level variance in developing and/or aging worms
The number of genes having an eQTL or an gxaeQTL are indicated per group (number as well as the percentage of the total genes with an increased or decreased of variation); also, the significance of the overrepresentation (or underrepresentation [under-rep.]) of (gxa) eQTLs in those groups. The total number of eQTL and gxaeQTL can be found in the row indicated with Total (also in Table 1).
Examples of genes having a gxaeQTL
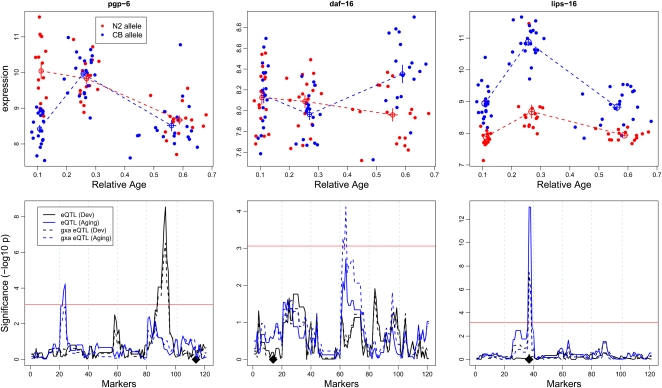
We show three examples of genes that have characteristic heritable expression patterns (Fig. 3). These examples show (1) differential gene expression in young worms, (2) differential gene expression in old worms, and (3) differential gene expression throughout the worm's life.
Figure 3.
Three gene examples in which gene expression depends on age, genotype, and their interaction. (Upper panel) The relative expression (y-axis) of the N2 allele (red) and the CB4856 allele (blue) of all the RILs at the peak of the major eQTL average and standard errors. Relative age (x-axis) is the time at which the specific RIL was sampled divided by the average age of that RIL. (Lower panels) The eQTL profiles of the different factors affecting gene expression. Gene position indicated by a black diamond. (Black) The eQTL profiles obtained with juvenile/reproductive age groups; (blue) the eQTL profiles found with the reproductive/old age groups. (Solid lines) Independent (from age) genotype effects; (dashed lines) genotype-by-age interaction effects. Chromosomes are separated by vertical dotted lines. (X-axis) Marker positions: (left) chromosome I; (right) chromosome X. (Y-axis) significance level (−log10 P). (Left) pgp-6; (middle) daf-16; (right) lips-16. Thresholds are designated by the red horizontal line.
Example 1 was found for pgp-6, for which we detected a trans-acting gxaeQTL at chromosome V in developing worms. The gxaeQTL disappeared in the aging worms. The CB4856 allele had a much lower expression in young worms. In reproductive worms, the expression of the CB4856 allele was strongly up-regulated and climbed to “N2”-levels. After reproduction, in old worms, the expression of both alleles decreased. A smaller, yet significant eQTL was also detected on chromosome II in aging worms. Example 2 was found for the forkhead family transcription factor encoding gene daf-16, which had a gxaeQTL on chromosome IV in aging worms. Example 3 was found for lips-16: This cis-acting gxaeQTL was found in all age groups, although the difference between N2 and CB4856 was largest by the end of the reproductive phase. Both alleles showed a similar pattern, but the CB4856 allele of this gene had a much higher expression throughout the worm's life. The gxaeQTL in this case is caused by a much steeper increase in developing worms and a steeper decrease in aging worms of the CB4856 allele expression.
Discussion
We showed that variation in genome-wide gene expression increases in aging worms. This agrees with findings by Bahar et al. (2006), who reported increased cell-to-cell variation in gene expression in aging mouse heart. Somel et al. (2006) used a number of microarray gene expression tissue data sets from humans and rats and found that gene expression becomes more variable with advancing age. Recently, a meta-analysis of age-related gene expression profiles with microarrays, including 27 data sets from mice, rats, and humans, revealed evidence of up-regulation of gene transcription repression and negative regulation of transcription, suggesting that transcriptional activity decreases with age (de Magalhães et al. 2009). Southworth et al. (2009) used a coexpression network approach to detect age-dependent differences in mice based on AGEMAP, a large DNA microarray study of gene expression as a function of age. They reported a decrease in gene coexpression in old mice and identified potential transcriptional mechanisms as a possible cause underlying the correlation decline. We provide for the first time insight into the regulatory loci of gene expression as a function of age and show that the number of eQTL decreases with age in C. elegans.
As an extension of the three single age-group models, we used a two-time-point model in which we tested if a time and/or marker(genotype) and/or an interaction between the two is present. If an eQTL is present at both time points, a significant marker effect will be found. If the eQTL is only present at one time point, we will find a significant marker-by-time (GxA) interaction effect. In fact, this approach increases the detection power in respect to eQTL appearing or disappearing at the different ages. Therefore we could work with a very strict significance threshold (P < 0.0001; false discovery rate [FDR] = 0.006). Furthermore, the same conclusion could be drawn when we repeated our analysis at P < 0.001. This will not guarantee that eQTLs at all age groups are detected for every individual gene, but our general conclusions will not depend on the thresholds used.
When juveniles were compared with old worms, the decline in number of cis-acting eQTL was stronger than the decline of trans-acting loci. Previous studies have shown that cis-acting linkages are less prone to variation induced by different environments (Li et al. 2006) and tissues (Petretto et al. 2006). We demonstrate that the proportion of cis-acting linkages is not stable during aging but decreases. Apparently, aging influences the efficacy of cis-regulatory sites for controlling local gene expression. If we assume that the trans-acting loci influence gene expression levels via protein intermediates, as suggested by Smith and Kruglyak (2008), than these protein regulators become relatively more influential during aging. In contrast to cis-regulation, trans-acting intermediates can affect the expression of many genes, thus allowing a coordinated gene expression response. Our results show that although variation in genes expression increases at old ages and the number of eQTL decreases, there is a substantial contribution of trans-acting loci at old ages. This suggests that studies focusing on regulatory patterns underlying complex traits at older ages, such as progressive diseases, should focus on trans-acting loci late in life rather than cis-acting.
The trans-band for eQTL in developing worms could be due to segregation distortion, which results in unequal marker distribution at this locus on chromosome I (Li et al. 2006). This distortion is caused by a partial genome incompatibility as described by Seidel et al. (2008). However, if this was a false positive caused by population structure, it would also be present in aging worms, where it was absent. Although one needs to be cautious with the conclusions about this locus, an important/influential polymorphic regulator in developing worms could be located at this genomic region on chromosome I. The trans-band for gxaeQTL in aging worms on chromosome IV shows the genomic position of a polymorphic regulator that becomes more influential as the worms age. Li et al. (2006) found a temperature dependent trans-band on chromosome V when they mapped eQTLs at two different temperatures, 16°C and 24°C. The worms grown at 24°C are physiologically older than those grown at 16°C, although their gene expression profiles were measured at the same stage. A re-examination by scoring the frequency of eQTLs per marker in this experiment revealed a second trans-band at the top of chromosome IV in 24°C grown worms, but not in the 16°C grown worms. This trans-band colocates with our gxaeQTL trans-band in aging worms. This suggests that specifically physiological age dependent gene expression is affected by a polymorphic gene on the top of chromosome IV.
Together, our results show that the increase in transcript variation in aging worms is due to a lack of large-effect regulatory loci. Alternatively, this increased expression variation could also be the result of an increase of small-effect eQTL. To get an indication for multiple small effect regulators, we investigated if the number of genes with two eQTLs would be larger in aging worms. This was not the case (Supplemental Table 6) and suggests that a lack of heritable regulation instead of more regulators is causal to the larger variation in a gene expression levels at older age.
The results allow us to infer the genetic architecture of heritable gene regulation with age. We highlighted three genes, pgp-6, daf-16, and lips-16, each displaying different differential expression patterns during development and aging. Figure 3 illustrates that pgp-6 is under the control of different factors at different ages. We detected a major QTL on chromosome V in developing worms. We also detected a small but significant eQTL at chromosome II in aging worms. This indicates that the expression of a gene can be regulated by different regulators depending on the age of a worm. In C. elegans, pgp-6, an ATP-binding cassette (ABC) transporter is expressed in the larval and adult intestine and adult amphids in N2 (Zhao et al. 2004). ABC transporters are one of the largest protein families in almost all species. They bind ATP and use its energy for transporting various molecules across membranes of organelles, such as the endoplasmic reticulum, peroxisomes, and mitochondria (Dean and Allikmets 1995). Mutation of ABC transporters can result in various diseases in humans or in hyper-sensitivity to drugs. Moreover, pgp-genes have been shown to be involved in multidrug resistance (Mahajan-Miklos et al. 1999) and resistance against Pseudomonas aeruginosa (Kurz et al. 2007). In our experiment, the expression level of pgp-6 is much higher in N2 compared with CB4856 in young worms and decreases in older worms. Our results imply that the regulatory control of pgp-6 confers differential resistance between N2 and CB4856 in young worms through its trans-eQTL on chromosome V.
In contrast to pgp-6, expression of daf-16 is differentially controlled in old worms and not in younger ones. Its gxaeQTL maps to the top of chromosome IV, where a trans-regulatory band for gxaeQTL was found in aging worms. daf-16 controls the insulin/IGF-1 signaling pathway underlying lifespan determination in C. elegans. This pathway operates during adulthood, affecting lifespan, and not during the juvenile period (Dillin et al. 2002). Nevertheless, daf-16 cannot be the causal gene for this trans-regulatory band in aging worms since it is not physically located at the genomic position of the trans-regulatory band. Still, it is intriguing to think that a polymorphic regulator operating through the daf-16 pathway must be present at this locus.
The third example was provided for lips-16, encoding a tri-acylglycerol lipase. Lipases break down lipids and possible mobilization of stored tri-acylglycerides. Our results show that in both wild types N2 and CB4856, lips-16 is expressed more in reproductive worms compared with juveniles and old worms; but CB4856 displays a higher lips-16 expression throughout its life than N2. This suggests that lipid catabolism mediated by lips-16 is more intense in CB4856 than in N2.
Although it is tempting to search the detected eQTL for candidate regulators of these three transcripts, the current study was not designed for this purpose; but this would be a likely next step. The primary goal was to explore the landscape of regulatory loci during aging rather than pinpointing potential causal genes. Future mapping studies, using N2 × CB4856 introgression lines (Doroszuk et al. 2009) and complementation studies (Kammenga et al. 2007) would be most suitable for finding the causal polymorphic genes at the regulatory loci.
In our two-time-point model, we corrected for physiological differences among the RILs during aging because we assumed different physiological states associated with post-reproductive ages. This raises the question as to whether the explanatory variable here is age in a continuous sense or physiological state in a categorical sense. As pointed out by Collins et al. (2008), it is important to make a distinction between age-related changes that represent development and growth and age-related changes that represent senescence. A criterion they put forward is the life-cycle stage, during which the change occurs. But development and senescence are arbitrary definitions of two periods within the same continuous process, namely, aging. The lack of heritable regulation later in life is an age-related change, since we can associate this process to old worms. The physiological age is related with genotype-by-environment interactions, and the absolute age (in days) is an arbitrary measurement, as it is time. We measured gene expression in juveniles, late reproductive, and post-reproductive old worms. Clearly these are defined stages, each with their own physiology. Correcting for these physiological differences allows for a more precise measurement related to age rather than stage. Yet, there is no clear moment during the life cycle that clearly separates development from aging; indeed, it seems very likely that developmental changes and aging changes occur simultaneously (Collins et al. 2008).
Our results show that the heritable regulation of genome-wide gene expression decreases with age. What could be the reason why the regulation is no longer genetically determined in old worms for most genes but is stronger for a small group of genes? One explanation might be found in our GO analysis, which revealed that monooxygenase activity was highly significant within the group of genes that displayed decreased variation in old worms. This points at increased regulation of stress pathways. Regulation of the functions of monooxygenase enzymes and their catalytic reactions has important significance for cell homeostasis. Halaschek-Wiener et al. (2005) identified longevity-associated genes in a long-lived C. elegans daf-2 (insulin/IGF receptor) mutant using serial analysis of gene expression and reported that expression of metabolism-associated genes diminished and stress response genes increased during aging.
Our work provides new insights into the regulatory control of gene expression as a function of age. So far, only a few mapping studies of regulatory loci of gene expression have focused on the dynamics of eQTL, for example, our study of the environmental influence on eQTL distribution (Li et al. 2006) and the studies of Schadt et al. (2008) and Gerrits et al. (2009), who focus on the spatial distribution of eQTL among different tissues and cell types. With this study, we have shown that age influences the distribution of eQTLs and that the temporal dynamics of regulatory loci are heritable and can become more prominent at older ages. We believe our results have important implications for the identification of loci that are linked to complex progressive and geriatric disease phenotypes.
Methods
A description of nematode culturing methods, lifespan assays, microarray experiments, and Gene Ontology analysis can be found in the Supplemental Methods.
Assigning cis- or trans-acting regulation to eQTLs
We assigned “trans-acting” regulation to any eQTL of which the peak was found at least five markers from the position of the gene the eQTL was calculated for. The rest were assigned “cis-acting.”
eQTL determination for daf-16
A closer inspection of daf-16 was needed because it is represented with seven different probes on the microarray (Washington University; array ID 3998, 5934, 7870, 8690, 9806, 17494, and 20336). To (re)determine the quality of the probes, we blasted them against the C. elegans genome. Two groups were found: Three probes (3998, 7870, and 17494) were perfectly aligned to genomic clone R13H8, and the four other probes to genomic clone F55A3. The four probes that aligned to genomic clone F55A3 showed possible cross-hybridization and were discarded from eQTL mapping. Furthermore, one of the probes (17494) had a mean log2 intensity value of <7.5 and was also discarded. The two remaining probes (3998 and 7870) showed a similar eQTL profile and had a colocating eQTL on chromosome IV. This eQTL had in both cases a –log10 P significance of ≥4. For clarity, we took the mean log2 intensity of these two probes per RIL as a measure for daf-16 expression and subsequently calculated the eQTL profile as shown in Figure 3.
Threshold determination
We used a permutation approach to determine the thresholds for the different mapping strategies (as in the method of Li et al. 2006). For each of the used models for eQTL mapping, we used 23,000 permutations. For each permutation, we randomly picked a spot; each spot could only be picked once. The gene expression and relative lifespan values were than randomly distributed over the RILs (and time points) and used for mapping. In this way, we obtained a threshold for each of the explaining factors. For the single time points, we used a FDR of 0.01 to adjust for multiple testing (Li et al. 2006). The genome-wide threshold for this FDR is –log10 P = 3.8 for each of the three time points. For the combined models (t1 to t2 and t2 to t3), we used a genome-wide threshold of –log10 P = 4, which resembles an FDR of 0.006, 0.001, and 0.006 for marker, age, and the interaction between marker and age, respectively.
To determine the threshold for the single gene examples, we used 1000 permutations as in the genome-wide threshold. The difference is that we use the gene under study in all of the permutations. The P-values for the gene specific thresholds were determined at FDR = 0.05.
Genome-wide eQTL mapping
Prior to eQTL mapping, we made sure that the absolute correlation of all marker and the dyes was less than 0.35. Furthermore, the absolute correlation between markers on different chromosomes was less than 0.55 for all markers. Also, before calculating the eQTL profiles, the outliers (> mean + 2 SD or < mean − 2 SD) per spot per stage were removed, to prevent the signals from dust, etc.
First, we used a linear model to calculate the linkage of each marker with the measured expression levels for each of the time points separately (to make Fig. 1). We used the log2 single channel normalized intensities as a measure for gene expression. We did not use the ratios because they can be notoriously problematic when it comes to inflation of variance. The model used for each of the three age groups was as follows: gene expression = marker(effect) + error. In this way, we obtained the genome-wide eQTL profiles for all genes for the three age groups.
Second, to quantify the heritable differences in gene expression that are age dependent, we extended our linear model by analyzing two age groups at once and including the relative age of the RILs as an explanatory factor. The model used for both combinations, t1 (juvenile) and t2 (reproductive), and t2 and t3 (old worms) was as follows: gene expression = marker(effect) + relative age + interaction (marker × relative age) + error. The age groups t1 and t2 in one model are referred to as “developing worms”; the age groups t2 and t3 in one model are referred to as “aging worms.” The significance and effect of each marker, relative age (time), and the interaction between the two were obtained for all genes on the array.
Determination of temporal expression patterns
The significance (P ≤ 0.0001; see Threshold Determination, above) of the relative age (time) was used to determine if a gene was differentially expressed between the three age (time) groups. The effect of this factor explaining gene expression differences was used to determine if the expression went up or down during the two age/time periods (t1–t2 and t2–t3), so we could assign each gene affected by age to one of the eight possible classes (see Supplemental Text, Fig. 1).
Determination of variance differences
We determined for all genes if a difference in variance exists between the juvenile and reproducing worms and between reproducing an old worms. A two-sided F-test was used to determine the significance of the difference in variance. As a threshold, we used a P < 0.0001. Genes that had a significant difference in transcript variance where then assigned to increasing or decreasing in developing and aging worms. Overrepresentation (or possible underrepresentation) of genes with an eQTL in the different significantly increased or decreased transcript level variance groups was determined by a hypergeometric test. As example, the group of 1289 genes with significantly increased variance in developing worms contained 113 genes with a gxaeQTL. We asked what the likelihood is to find this by chance when the total number of genes (18,893) contains 280 genes with a gxaeQTL.
We used three methods to investigate if a possible relation between mean expression and an increase or decrease in variance influenced our results. First, we calculated the correlation (Pearson) between the variance and mean expression level. For all three stages, these were low but positive (0.30, 0.32, and 0.40, respectively); therefore, a possible increase in variation of the low expressed genes by normalization is very unlikely to have influenced our results. Second, when we divided the expression levels per stage by the mean expression level per stage, we found an even larger difference between genes with increased variance (3403) and decreased variance (641), both at P < 0.0001, which confirms and strengthens our initial result. Third, when we used a permutation approach, we only found a small difference between genes that have an increased or decreased variance when comparing stages 2 and 3 (979 and 743, respectively). In this permutation approach, we made two random groups per gene from the expression levels of stage 2 and 3; then, we calculated the variances, means, and P-values. Again, this result shows that there is a large difference between the number of genes that have and increased variance and those that have a decreased variance when stages 2 and 3 are compared.
Acknowledgments
We thank Mike Herman for stimulating discussions. This work was supported by the Ecogenomics Consortium of the Netherlands Genomics Initiative (NGI) to A.V. and L.B.S.
Footnotes
[Supplemental material is available online at http://www.genome.org. The gene expression data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE17071.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102160.109.
References
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé MET, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. 2006. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441: 1011–1014 [DOI] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Collins JJ, Huang C, Hughes S, Kornfeld K 2008. The measurement and analysis of age-related changes in Caenorhabditis elegans. In WormBook (ed. The C. elegans Research Community). doi: 10.1895/wormbook.1.137.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Allikmets R 1995. Evolution of ATP-binding cassette transporter genes. Curr Opin Genet Dev 5: 779–785 [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Curado J, Church GM 2009. Meta-analysis of age-related gene expression profiles identities common signatures of aging. Bioinformatics 25: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C 2002. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834 [DOI] [PubMed] [Google Scholar]
- Doroszuk A, Snoek LB, Fradin E, Riksen J, Kammenga J 2009. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res 37: e110 doi: 10.1093/nar/gkp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR, Aten JE, Farber EA, de Vera V, Gularte A, Islas-Trejo A, Wen P, Horvath S, Lucero M, Lusis AJ, et al. 2009. Genetic dissection of a major mouse obesity QTL (Carfhg2): Integration of gene expression and causality modeling. Physiol Genomics 37: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigl JB, Langer S, Barwisch S, Pfleghaar K, Lederer G, Speicher MR 2004. Analysis of gene expression patterns and chromosomal changes associate with aging. Cancer Res 64: 8550–8557 [DOI] [PubMed] [Google Scholar]
- Gerrits A, Li Y, Tesson BM, Bystrykh LV, Weersing E, Ausema A, Dontje B, Wang X, Breitling R, Jansen RC, de Haan G 2009. Expression quantitative trait loci are highly sensitive to cellular differentiation state. PLoS Genet 5: e1000692 doi: 10.1371/journal.pgen.1000692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteling EW, Riksen JAG, Bakker J, Kammenga JE 2007. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity 98: 28–37 [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJM, Marra MA, Brooks-Wilson AR, et al. 2005. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15: 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AA, Hunter CP, Tsung BT, Tucker-Kellogg G, Brown EL 2000. Genomic analysis of gene expression in C. elegans. Science 290: 809–812 [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap J-P 2001. Genetical genomics: The added value from segregation. Trends Genet 17: 388–391 [DOI] [PubMed] [Google Scholar]
- Jiang M, Ryu J, Kiraly M, Duke K, Reinke V, Kim SK 2001. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc Nat Acad Sci 98: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga JE, Doroszuk A, Riksen JAG, Hazendonk E, Spiridon L, Petrescu AJ, Tijsterman M, Plasterk RH, Bakker J 2007. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet 3: e34 doi: 10.1371/journal.pgen.0030034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, Garcia JM, Van Den Ackerveken G, Snoek LB, Peeters AJM, Vreugdenhil D, Koornneef M, Jansen RC, et al. 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Nat Acad Sci 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz CL, Shapira M, Chen K, Baillie DL, Tan M-W 2007. Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem Biophys Res Commun 363: 438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JA, Hazendonk E, Prins P, Plasterk RH, Jansen RC, et al. 2006. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet 2: e222 doi: 10.1371/journal.pgen.0020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891 [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE 2002. Transcriptional profile of aging in C. elegans. Curr Biol 12: 1566–1573 [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan M-W, Rahme LG, Ausubel FM 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 96: 47–56 [DOI] [PubMed] [Google Scholar]
- Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG 2007. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative hybridization. Genome Res 17: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H 2004. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet 36: 197–204 [DOI] [PubMed] [Google Scholar]
- Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L 2008. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, et al. 2006. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet 2: e172 doi: 10.1371/journal.pgen.0020172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoni MF, Sebastiani P, Kohane IS 2002. Cluster analysis of gene expression dynamics. Proc Nat Acad Sci 99: 9121–9126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L 2006. Genetics of global gene expression. Nat Rev Genet 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302 [DOI] [PubMed] [Google Scholar]
- Schadt E, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6: e107 doi: 10.1371/journal.pbio.0060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L 2008. Gene–environment interaction in yeast gene expression. PLoS Biol 6: e83 doi: 10.1371/journal.pbio.0060083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Khaitovich P, Bahn S, Pääbo S, Lachmann M 2006. Gene expression becomes heterogeneous with age. Curr Biol 16: R359–R360 [DOI] [PubMed] [Google Scholar]
- Southworth LK, Owen AB, Kim SK 2009. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet 5: e1000776 doi: 10.1371/journal.pgen.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan M, Yamaza H, Sun Y, Sinclair J, Li H, and Zou S 2007. Temporal and spatial transcriptional profiles of aging in Drosophila melanogaster. Genome Res 17: 1236–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fang LL, Johnsen R, Baillie DL 2004. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem Biophys Res Commun 323: 104–111 [DOI] [PubMed] [Google Scholar]