Abstract
Recent metagenomics studies have begun to sample the genomic diversity among disparate habitats and relate this variation to features of the environment. Membrane proteins are an intuitive, but thus far overlooked, choice in this type of analysis as they directly interact with the environment, receiving signals from the outside and transporting nutrients. Using global ocean sampling (GOS) data, we found nearly ∼900,000 membrane proteins in large-scale metagenomic sequence, approximately a fifth of which are completely novel, suggesting a large space of hitherto unexplored protein diversity. Using GPS coordinates for the GOS sites, we extracted additional environmental features via interpolation from the World Ocean Database, the National Center for Ecological Analysis and Synthesis, and empirical models of dust occurrence. This allowed us to study membrane protein variation in terms of natural features, such as phosphate and nitrate concentrations, and also in terms of human impacts, such as pollution and climate change. We show that there is widespread variation in membrane protein content across marine sites, which is correlated with changes in both oceanographic variables and human factors. Furthermore, using these data, we developed an approach, protein families and environment features network (PEN), to quantify and visualize the correlations. PEN identifies small groups of covarying environmental features and membrane protein families, which we call “bimodules.” Using this approach, we find that the affinity of phosphate transporters is related to the concentration of phosphate and that the occurrence of iron transporters is connected to the amount of shipping, pollution, and iron-containing dust.
Integral membrane proteins play a fundamental role in sensing and interacting with the environment, allowing the influx and efflux of ions and molecules and relaying information about environmental conditions to the cell. Thus, the abundance and types of membrane protein families in a microbial community may give information about functional capabilities and nutritional requirements. In marine microorganisms, especially those inhabiting the oligotrophic (nutrient-poor) surface waters of the oceans, membrane protein content might provide insight into types of nutrients and conditions in the waters in which the organisms were isolated. For example, the recent discovery of spectral tuning of the light-driven proton pump proteorhodopsin reveals a relationship between a single amino acid mutation and dominant light wavelengths in the microbe's surroundings (Rusch et al. 2007).
Several recent studies have begun to relate functional attributes of microbial communities, such as central metabolism or broad functional classes (e.g., protein synthesis), to specific habitats (Tringe et al. 2005; Dinsdale et al. 2008) or environmental features (DeLong et al. 2006; Kunin et al. 2008; Gianoulis et al. 2009). In addition, new methods are allowing the integration of quantitative features of the environment alongside microbial function (DeLong et al. 2006; Gianoulis et al. 2009).
Given their important role in environmental sensing and transport, membrane proteins may serve as an even more sensitive barometer of environmental conditions than broad functional classes or central metabolism. In addition, integration of many different environmental conditions is needed to develop a comprehensive understanding of the complex interplay between environmental conditions and microbial communities. In particular, new techniques are needed to investigate the relationship between natural processes such as nutrient fluxes and the impact of humans on the environment (anthropogenic effects), such as pollution. Given the nutrient fluctuations and anthropogenic effects observed in the world's oceans, understanding the relationship between such factors and microbial adaptations is particularly timely. Indeed, Halpern et al. (2008) estimated that 40% of the world's oceans are substantially affected by human activity by computing indices for pollution, shipping, ultraviolet radiation, and climate change, among others.
To gain a better understanding of the relationship between environmental conditions and membrane protein content and abundance, we used 29 samples from the Global Ocean Sampling (GOS) Expedition (Rusch et al. 2007). This survey provided metagenomic sequence and environmental data (chlorophyll, water depth, sample depth, salinity, temperature), as well as GPS coordinates of the sampled sites. We used the GPS coordinates to extract additional environmental features from several disparate sources, providing both natural features, such as nutrient concentrations, and anthropogenic features, such as pollution. Integration of these quantitative measurements allowed us to investigate the relationship between microbial communities, nutrient dynamics, and anthropogenic effects, and in particular the relative importance of the various classes of membrane proteins in microbial adaptations.
Results
Integration of environmental features
The GPS coordinates provided for the sampled sites were essential to cross-reference different sources of information, mainly provided as annotated maps of the ocean. We integrated these data by interpolation of the map projections onto the GOS geographic coordinates (latitude and longitude information). To select the sites for further analysis, we used Google Earth to compare locations of GOS sites to locations of available data (some maps were sparse). We were able to extract an additional 11 environmental features for 29 sites: phosphate, nitrate, silicate, dissolved oxygen, and apparent oxygen utilization information from the World Ocean Database (Antonov et al. 2006; Garcia et al. 2006; Locarnini et al. 2006); pollution, shipping routes, ultraviolet radiation, ocean acidification, and climate change information from the National Center for Ecological Analysis and Synthesis (NCEAS) (Halpern et al. 2008); and dust levels, which serve as a proxy for oceanic iron concentrations, from Jickells et al. (2005) (see Supplemental material for additional information on environmental features). We have placed the features data for each of the sites on an interactive Google Earth map at http://metagenomics.gersteinlab.org/membrane/.
Membrane protein prediction/variation
Using PRODIV-TMHMM (Viklund and Elofsson 2004), we identified ∼1.3 million proteins of the 6 million proteins in the GOS protein data set (Yooseph et al. 2007) as having at least one membrane-spanning region. We filtered this set to include only high confidence peptides (see Supplemental material and Methods for more details on protein filtering), which resulted in 873,718 predicted membrane proteins. Due to the nature of the prediction, there is likely a bias against membrane proteins with a small number of transmembrane helices. Furthermore, as our selection method is quite stringent, we are likely underestimating total membrane protein content; however, the relative proportions between the sites should remain consistent. Membrane protein content ranged from 12.2% (Gulf of Maine) to 15.0% (Off Key West, FL and Roca Redonda) with an average of 14.2% (Supplemental Table 1). For comparison, in the known heterotrophic/photosynthetic microbial genomes, the predicted transmembrane helical protein content ranges from 21% (Acinetobacter baumannii) to 33% (Chloroflexus aurantiacus), with a median of 28%.
To examine functional differences across the sites, we homology-mapped 237,870 of the predicted membrane proteins to known annotation using clusters of orthologous groups (COG) (Tatusov et al. 2000). We filtered this set to the 151 membrane families involved in transport processes (transporters, channels, permeases) as these families should be particularly sensitive to environmental perturbations and, additionally, to strengthen the signal in our further analysis and prevent overfitting the data (Supplemental Table 4).
Standard methods
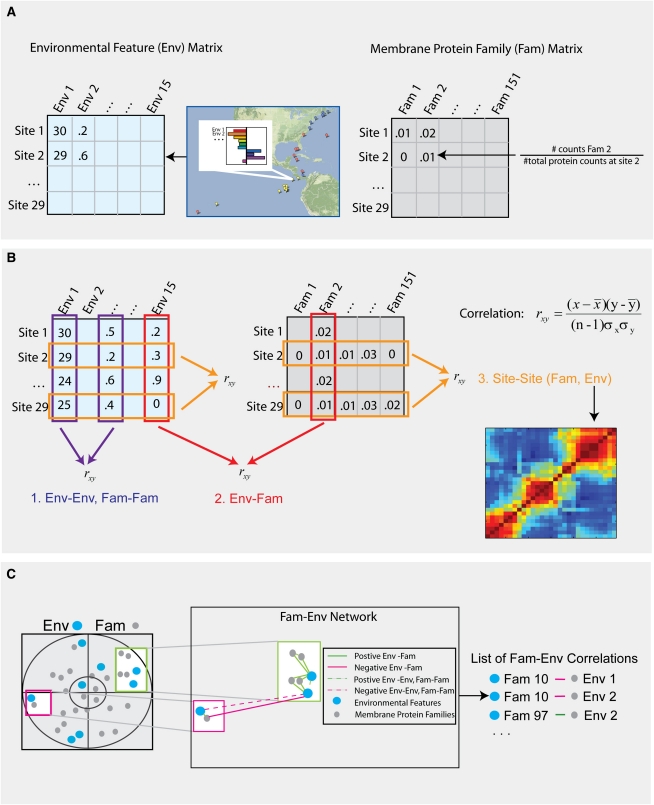
For the 29 sites, we computed the fraction of peptides belonging to each of the 151 families and created a membrane protein families matrix (the rows are the 29 sites, and the columns are the families) and, similarly, an environmental features matrix (the rows are the 29 sites, and the columns are the 15 environmental features) (Fig. 1A). Using these matrices, there are numerous straightforward correlations we can perform to investigate the relationship between and within the features and families across the sites (Fig. 1B). For example, one can compute the pairwise correlation across sites between different families, environmental features, or even between families and environmental features. In addition, one can transpose the above and correlate the sites on the basis of either environmental features or membrane protein families (resulting in a site-site correlation or similarity matrix) (see Supplemental Fig. 5). For simplicity, we refer to these site-site correlations (SS) as “SS-Env” or “SS-Fam,” for the environmental and membrane protein-based site-site correlations, respectively.
Figure 1.
(A) Environmental and membrane family matrix construction. (B) Pairwise correlations. Types of correlations that can be performed: (1) between either environmental features (Env-Env) or membrane protein families (Fam-Fam); (2) between an environmental feature and a membrane protein family (Env-Fam); or (3) between two sites (Site-Site) defined either through their membrane protein families (SS-FAM) or their environmental features (SS-Env) (see Fig. 2 for larger resulting heatmap and labels). (C) Membrane protein families and environmental features network (PEN) construction. Quantification of relationships between environmental features and membrane protein families by construction of Env-Fam networks from structural correlation coefficient plots.
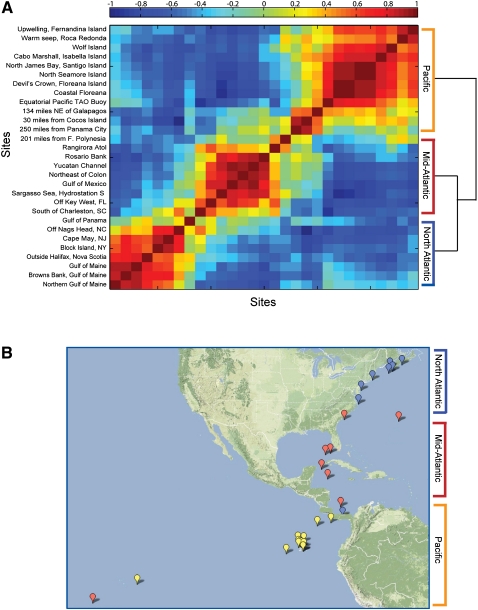
In particular, when calculating SS-Env, we observed significant variation between the sites as shown in Figure 2A, where site pairs are color-coded according to their similarity. Additionally, clustering the sites based on the similarity of the environmental features (see Methods) revealed a distinct latitudinal influence in the data, separating the sites into three groups (Fig. 2A,B): the North Atlantic, the Mid-Atlantic, and the Pacific. Such a finding is perhaps expected as the sites are not physically isolated from each other, and they were sampled from the North Atlantic through the Pacific over the course of 12 mo. Thus, adjacent samples were likely subjected to similar seasonal (temporal) effects, such as phytoplankton blooms, nutrient-carrying currents, and temperature; and similar spatial effects, such as nutrient gradients. In addition, specific environmental features appeared to have distinct patterns among the clusters. For example, phosphate concentrations were generally lower in the mid-Atlantic than the other two regions, while acidity was high, and pollution/shipping/climate changes were all relatively low in the Pacific.
Figure 2.
(A) Environmental features (Env) site correlations and clustering. Clustering of site-site correlations, where each site is defined by a vector of 15 environmental features (Site-Site Env heatmap). (B) Sites color-coded by environmental clustering; shows strong concordance with geographic location: North-Atlantic (blue), Mid-Atlantic (red), and Pacific (orange).
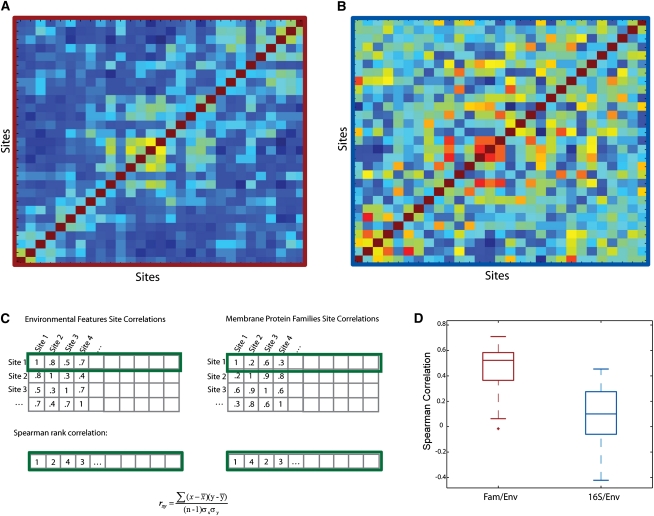
The SS-Fam matrix also showed variation across the sites (Fig. 3A; color bar reference Fig. 2A). Thus, even across these qualitatively similar ocean habitats, we are able to see differences in the abundance and types of membrane protein families in the genomes present. Interestingly, upon visual comparison with the site-site correlations of the environmental features matrix, we observe some concordance between regions of high and low correlations (cf. Figs. 2A and 3A; sites are ordered similarly). This suggests there is a relationship between sites such that sites with similar environmental features have similar membrane protein content and vice versa.
Figure 3.
Site-site correlations, where each site is defined by 151 membrane protein families (Site-Site-Fam, [SS-FAM]) (A) and 16S genes at the 20% divergence level (SS-16S) (B) (sites ordered as in Fig. 2A). (C) Method description: For each row of SS-FAM, we sort the correlation coefficients and convert them to rank-order. We then repeat this procedure for SS-16S and SS-Env (Fig. 2A); then, we compare the ranks of SS-FAM and SS-Env, as well as SS-16S and SS-Env. If the rank vectors are similar to one another, this implies that differences in one set of features are reflected in differences in a second set of features. For the FAM/Env, this is, indeed, the case; however, the low rank correlation between 16S/Env implies that 16S is not reflective of changes in environment as seen by the boxplot in D.
Environmental versus phylogenetic variation
A factor that could explain the observed variation across the sites is differences in species composition. The environmental differences would affect the types of species preferentially inhabiting these sites, and, in turn, this could explain the observed genomic variation. Thus, for comparison, we calculated the GOS SS-16S (20% 16S divergence groups) (Biers et al. 2009) to determine phylogenetic similarity of the sites (Fig. 3B). However, we were unable to find a significant relationship between the phylogenetic-based and environmental-based site-site similarity (for methods, see Supplemental Fig. 6). The average correlation between SS-16S and SS-Env was 0.2 (Fig. 3D); whereas, the average correlation between SS-Env and SS-Fam was 0.5 (Fig. 3D). This suggests that the observed membrane protein variation is more a function of the measured environmental features than of phylogenetic diversity. It is important to note, however, that we only had enough statistical power to look at the 20% divergence level of the 16S profiles, and we cannot rule out the possibility that a lower divergence level could result in a greater concordance between environmental site similarity and 16S profile similarity.
Variation in membrane protein families corresponds to environmentally distinct regions
Above, we show that the variation in membrane proteins is reflected in the variation in the environmental features; however, which families and features are contributing to the association remains unanswered. There are a host of multivariate statistical techniques for understanding these types of complex (many-to-many) relationships between data sets. Thus, we began our analysis by using a variety of standard and published techniques: (1) principal component analysis (PCA); (2) discriminative partition matching (DPM) (Gianoulis et al. 2009); and (3) regularized canonical correlation analysis (CCA) (Gonzalez et al. 2008). Furthermore, we developed a technique that we call “protein families and environmental features network” (PEN) to address limitations in the quantification of associations and visualization of the results of CCA.
Principal components analysis and discriminative partition matching
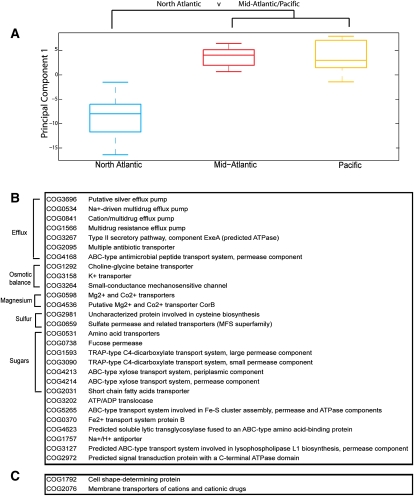
As demonstrated above, hierarchical clustering of the sites based on their environmental features revealed three distinct geographical regions (Fig. 2A,B). A similar pattern emerged after using a data reduction technique, PCA, of the sites and the proportion of membrane proteins at each site. In brief, each principal component is a weighted linear combination of features. These weights or scores can be used as new axes allowing the projection of the sites into a new lower dimensional space. We observed that sites deemed more similar in the environmental clustering also had a greater tendency to be closer together based on their membrane proteins. For example, in Figure 4A, the first component scores show that the occurrence of membrane proteins in the North Atlantic environmental cluster can be distinguished from the Mid-Atlantic and Pacific environmental clusters. As the clustering of the environmental features is done separately from finding variation in the membrane protein families, we independently show that the grouping of the sites based on environmental features is partially reflected in the membrane protein content.
Figure 4.
(A) Boxplot of PCA first component scores on the membrane protein family matrix. Separating sites by environmental clusters from Figure 2A shows that the North Atlantic scores are distinguishable from the Mid-Atlantic/Pacific. (B,C) Discriminate partition matching. Membrane protein families discriminating between site groups. Membrane protein families enriched in the North Atlantic (B) and Mid-Atlantic/Pacific (C).
As the PCA showed the Mid-Atlantic and Pacific to be similar, we grouped these sites into a “Mid-Atlantic/Pacific” cluster and used DPM to determine which specific families were discriminating between them and the North-Atlantic cluster. Briefly, DPM assesses whether the distribution of a specific protein family is significantly different and “discriminates” between the two partitions. Thirty families showed significant discrimination (q-value < 0.05) between the two site sets, and interestingly most were enriched in the North Atlantic (28/30) (Fig. 4B,C; Supplemental Table 5).
In the North Atlantic environmental cluster, there is enrichment in several proteins involved in inorganic ion transport. One such protein is a magnesium transporter, which is likely related to the higher chlorophyll content (P-value < 0.01) and thus bacterial abundance (Bird and Kalff 1984) in these regions. The North Atlantic sites also have higher pollution rates (P-value < 0.01) and possible nutrient availability due to coastal proximity; such features are also indicative of regions with increased cell growth and proliferation (Kirchman 2008). Magnesium is not only contained in the center of the chlorin ring, it is also a central player in the stabilization of DNA and RNA; thus one can presume that in dividing cells larger quantities of the ion would be required. Most interestingly, however, there is an increase in many families involved in efflux/secretion/antimicrobial processes. The enrichment in these proteins may reflect the microbes' need to expel antimicrobials, by-products of metabolism, or environmental toxins (Neyfakh 1997). In addition, there are a set of proteins related to protection from osmotic shock (glycine/betaine, K+, mechanosensitive channel), which may be acting to buffer against shifts in ocean solute concentrations (Poolman et al. 2002), again alluding to the increased pollutants, and possibly nutrient fluxes from land and rivers.
One interesting protein observed to increase in the North Atlantic is the ATP/ADP translocase. This protein is found in mitochondria, as well as obligate intracellular parasites of Chlamydiae and Rickettsiae, where they function to exchange host ATP for ADP, thereby sequestering host nutrients (Winkler and Neuhaus 1999). It is uncertain in what capacity they function. One intriguing possibility is that they originate from marine parasites, although a recent survey showed that several putative ATP/ADP translocases should have been annotated as more general nucleotide/H+ transporters (Tjaden et al. 1999); thus, simple misannotation of the class cannot be ruled out.
Canonical correlation analysis
Although the previous analysis is useful in finding discriminating families between environmentally distinct groups, it does not capitalize on the natural gradients in the environmental features we were able to extract. Consequently, we performed regularized CCA, which maximizes the correlation between linear combinations of the two sets of variables, to reveal a finer-grained picture of the relationship between environmental features and membrane protein families (Wichern and Johnson 2003). As our analysis of site-site correlations revealed concordance of the environmental features and membrane protein families in assessing the similarity of the sites, this analysis is justified and may be able to reveal more specific correlated features.
Similar to PCA, CCA gives us several principal directions that describe the greatest degree of covariation between features (Borga 1998). Interpretation of CCA is commonly performed by plotting the first two of these structural correlates, schematized in Figure 1C, left (Gonzalez et al. 2008). Those families and environmental features that are close in this structural correlation space are referred to as covarying.
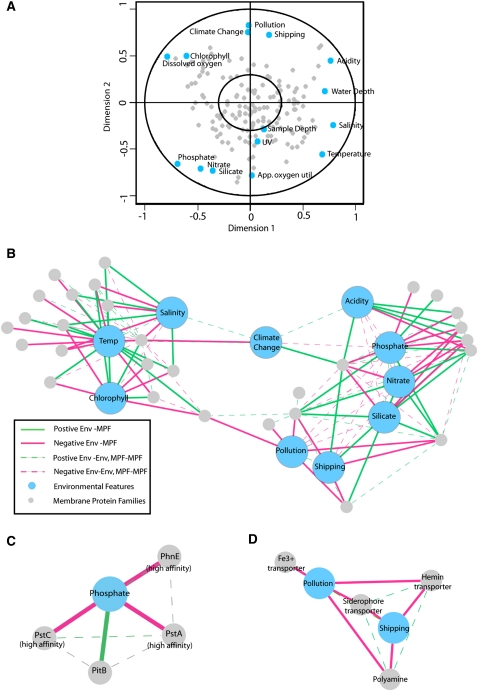
From the structural correlation plot (Fig. 5A), we observed all 15 environmental features and 107 out of 151 membrane protein families varying across the 29 sites. These points are outside the 0.3 circle and can thus be considered covarying with respect to the other set of features (Supplemental Tables 6, 7; Borga 1998; Wichern and Johnson 2003; Guo et al. 2006) (44/151 families were invariant, points inside the 0.3 circle). No single COG functional category was over-represented in either the variant or invariant set (P-value > 0.05). However, notably, 34 out of the 41 ABC transporters in the data set were shown to covary with the environmental features.
Figure 5.
(A) CCA structural correlations. Plot of first and second dimension of CCA with labeled environmental features (blue) and membrane protein families (gray). Within inner circle (0.3 circumference) features are invariant across the sites. (B) Membrane protein families and environmental features network. PEN construction from CCA structural correlations in the first and second dimension using a distance cutoff > |0.05| between all nodes (environmental features and membrane protein families). (Red edges) Negative associations; (green edges) positive associations. (C) Phosphate subnetwork. (D) Iron/polyamine subnetwork.
Between the environmental features, we observe many intuitive relationships. As an example, ocean-based pollution and shipping lanes are highly correlated as expected due to the overlap in measurement (same direction on plot) (Halpern et al. 2008). In addition, shipping itself is a contributor to ocean pollution given emissions from fuel burning and ballast water (which can bring invasive species) (Satir 2008). Predictably, dissolved oxygen shows a negative relationship with water temperature (as oxygen more readily dissolves in colder waters, opposite direction on plot) and also, as it is a by-product of primary production, a positive relationship with chlorophyll. In addition, the positive relationship between nitrate, phosphate, and silicate reflect similarities in the gradients of nutrients across the sites.
PEN
Solely using the structural correlations plot to analyze the results is problematical for several reasons. First, it is difficult to draw conclusions on the strength and directionality of a relationship between variables, especially negative relationships as they are not close in space, although such relationships can be identified by looking at the tabular form of the data. Second, the relative weight of the features' relationships can be difficult to visualize and compare. Third, there is no real means of quantifying covariation between specific sets of features, nor do standard visualization methods allow for comparisons in more than three dimensions. To better quantify and visualize the results of CCA, we developed a new approach we call “protein families and environmental features network” (PEN).
In brief, PEN creates a network from the CCA results, where each environmental feature and membrane protein is a node and the edges are weighted by taking the dot product between the structural correlations in the first and second dimensions (the procedure easily generalizes for the case of more than two dimensions).
We then use a simplified version of connected components analysis and prune all the edges with absolute value weights below 0.5 (see Methods). This simple metric provides an intuitive means of visualizing environmental/membrane protein clusters as it gives greater weight to features closer to the correlation circle (outer circle in Fig. 1C), as well as to features that have a small angle between them relative to the x-axis. Such features represent strongly covarying pairs or sets of features. We can use the topology of the network to identify these sets of tightly (negatively, red edges; positively, green edges) correlated environmental features and membrane proteins families that we term “bimodules” (Fig. 1C).
In the pruned network derived from the structural correlates, we observe two distinct bimodules (Fig. 5B), comprising families and environmental features that have both negative and positive relationships (see Supplemental Table 9 for edge weights). The first bimodule contains temperature, salinity, and chlorophyll with many shared connections between membrane families, and the second contains phosphate, nitrate, and silicate (which are themselves inversely related to acidity, shipping, and pollution). UV, dissolved oxygen, apparent oxygen utilization, sample depth, and water depth, although showing variation across the sites (outside the 0.3 circle in Fig. 5A), are not related to any specific membrane protein family and are thus not included in the graph. It is unlikely that these features are not affecting microbial diversity; it may be the case that limiting our genomic data to membrane proteins is not allowing us to highlight these influences.
From the network, we see both intuitive and nonintuitive relationships between the features and membrane protein families. For example, chlorophyll concentration and a magnesium ABC transporter (COG0598) are positively related likely due to the relationship between chlorophyll and bacterial abundance (and thus proliferation) (Bird and Kalff 1984) and to the fact that chlorophyll molecules contain a magnesium ion at the center of the ring structure. This was inferred from the DPM analysis as these transporters were enriched in the North Atlantic (area of high chlorophyll), but here we are able to explicitly see the relationship between the two variables.
A less intuitive relationship, but nonetheless interesting, is a negative relationship between an ABC transporter involved in polyamine (putrescine/spermidine) transport (COG1176) and ocean-based pollution/shipping (Fig. 5D). Polyamines are nitrogen-rich compounds found in all living matter, and they play an important role in the stabilization of DNA structure (Flink and Pettijohn 1975). Although their exact role is unknown, during cell growth in response to proliferative stimuli, both their uptake and biosynthesis are increased (Igarashi and Kashiwagi 2000). Possible sources of polyamines in ocean water are from the degradation of organic matter, amino acids, and proteins, where they are quickly taken up by bacteria (Lee and Jorgensen 1995). The negative relationship we observe might reflect the increased amount of polyamines in the environment in polluted, nutrient-rich waters, where fewer transporters would be needed for uptake. In these nutrient-rich areas, cell growth and death rates may be higher, leading to increased concentrations of polyamines.
Phosphate
The most pronounced negative relationship observed is that of phosphate concentrations and ABC-type phosphate transporters (COG0573 and COG0581) and phosphonate transporters (COG3639). These ABC transporters comprise pstA, pstC, and phnE of the phosphate (pho) regulon in Escherichia coli that have previously been shown to be involved in the active uptake of phosphate from the environment during phosphate limitation (Karp et al. 2002). Interestingly, we also observed the converse with the phosphate/sulfate permease pitB in E. coli (COG0306) (Fig. 5C). The relationship between pstA/C and phosphate starvation conditions has been well-characterized (Martiny et al. 2006, 2009); however, the positive relationship between the lower-affinity PitB and phosphate concentration suggests a more subtle influence of environmental parameters on modulating membrane content. That is, when phosphate concentration in the environment is low, more genes encoding high-affinity phosphate transporters (pstA/pstC/phnE) are present, and when phosphate concentration is high, more genes encoding a low-affinity transporter (pitB) are present.
Furthermore, we observe a positive relationship between an ABC transporter predicted to be involved in Lipophospholipase L1 biosynthesis (COG3127) and phosphate levels, suggesting increased cellular activities related to phospholipids with increased phosphate concentrations. Phosphate concentrations have been shown to modulate lipid content in marine bacteria, where organisms in low phosphate regions replace phospholipids with non-phosphorous-containing lipids (Van Mooy et al. 2009).
Iron
We observe a striking network of relationships between protein families involved in the active uptake of iron (COG0609: ABC Fe3+ siderophore transporter, COG1178: ABC Fe3+ transporter, and COG4558: ABC hemin transporter) and areas of high ocean-based pollution and shipping (Fig. 5D). Iron is a critical resource essential to microorganisms for a diverse array of enzymatic reactions and cellular processes such as respiration, photosynthesis, and nitrogen fixation. As such, its depletion has been shown to limit microbial growth even in the presence of other essential nutrients, such as phosphates and nitrates. Regions with such a limitation have been termed high nitrate/low chlorophyll (N/C) regions, an example of which is the Equatorial Pacific (Pacific) (Kirchman 2008). We hypothesize that the increase in gene content related to iron acquisition observed in low-pollution/shipping areas may reflect a greater difficulty in attaining this nutrient. Indeed, siderophores in particular are known to be produced by bacteria under iron-limited conditions and to actively sequester iron from the environment (Guan et al. 2001).
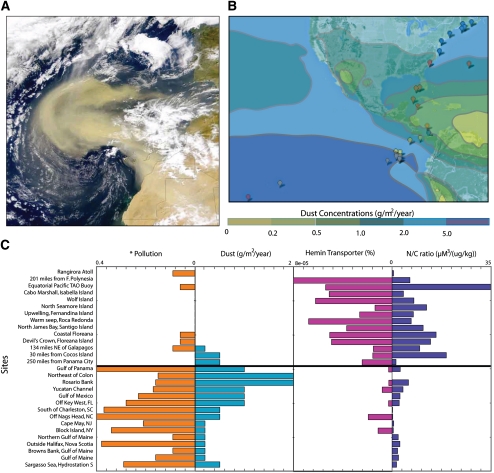
The main sources of iron in the ocean are aeolian dust from land (Fig. 6A), as well as terrestrial input near coastal regions, fluvial input, and upwelling from the ocean floor, all of which are lacking in these low-shipping/pollution sites. Interestingly, we observed that the areas of high ocean-based pollution/shipping (North Atlantic and Mid-Atlantic) parallel areas that may have higher iron concentration. Presently, there are no means to directly measure iron concentrations; however, oceanographers have shown that models of iron-containing dust (Fig. 6B; Jickells et al. 2005) can approximate iron concentrations. We found that iron values approximated from these dust models show significant negative correlation between COG4558 (P-value < 0.01) and COG0609 (P-value < 0. 01), as well as the N/C ratio across the sites (Fig. 6C). Such a trend is similar to our observation using shipping and pollution. In addition, searching the BRENDA database (Schomburg et al. 2002) for enzymes using iron as a cofactor revealed that an increase in these two families is negatively correlated to the amounts of enzyme present that require iron. Thus, similar to phosphate, it may be that in these low-pollution/shipping areas (open ocean, low aeolian dust input), microorganisms increase the production of siderophore and iron transporters to enable survival in a low iron environment.
Figure 6.
(A) Image of dust storm off Sahara desert (NASA). (B) Model of dust concentrations (color-coded) across GOS sites, adapted from Jickells et al. (2005). (C) Pollution levels (*, impact value; see Halpern et al. 2008), dust concentrations, percent of ABC-type hemin transport system proteins ([number of COG4558 proteins]/[number of total proteins at site]), and nitrate/chlorophyll ratio values across the 29 GOS sites. The black line shows separation of sites into two sets, one with high pollution and dust and low N/C and iron transporters and vice versa.
Unknown fraction
Intriguingly, of the 1.2 million unique proteins with at least one predicted membrane-spanning region, 15% had no homology with any protein currently in GenBank (E-value > 1 × 10−10), suggesting a large and hitherto unexplored space of membrane protein diversity. To further characterize this unknown fraction, we searched for known binding motifs by running each predicted membrane protein against PROSITE (Hulo et al. 2008), resulting in the functional characterization of 29,384 (15%) of this unknown fraction including previously unannotated ABC transporters, beta lactamases, G protein receptors, and lipocalins among others (data not shown).
Discussion
We presented the protein families and environmental features network (PEN) as a means of describing, quantifying, and exploring the relationships between and among sets of environmental features and occurrence of membrane protein families. Such graph theoretical approaches have been shown to be useful in the study of biological systems, particularly for understanding the complexity and global topology of relationships mediating protein and many other types of interactions (Barabasi and Oltvai 2004). PEN provides a simple flexible framework for exploring these complex relationships in the context of metagenomics data sets. Although complete characterization of an environment as complex and dynamic as the ocean is highly unlikely, through careful examination of the resulting bimodules, we demonstrate the usefulness of such studies even within these limitations. We are able to identify pertinent conditions affecting protein diversity and recapitulate potential explanations for the observed variation, illustrating the robustness of this type of analysis.
To date, most metagenomics studies integrating environment features have focused on the comparison of metabolic pathways or phylogenetic content among disparate habitats (Tringe et al. 2005; DeLong et al. 2006; Dinsdale et al. 2008; Gianoulis et al. 2009). Here, we focus on a specific set of membrane proteins sampled from sites with a high degree of environmental similarity (by removing outlying samples from the GOS data set, such as estuaries and lakes), and use quantitative environmental features to differentiate factors that are affecting the genomic content. By selecting only membrane proteins, we are able to see relationships between a microorganism's (or in this case, superorganism's) external barrier, mediating the transport of molecules in and out of the cell, and features of its environment. We show that, indeed, there is widespread variation in most membrane protein families, and these can be explicitly correlated to both nutrient availability and anthropogenic influences. In fact, the median structural correlation coefficient for the membrane proteins is 0.3, whereas for metabolic pathways it is 0.17 (Gianoulis et al. 2009), suggesting that membrane protein covariation is stronger with this set of environmental features (see Supplemental material).
Our results comparing membrane protein content to environmental features and species diversity add to the growing body of evidence suggesting that genome plasticity may be largely driven by environmental factors and less a result of species specificity. Given the large amount of horizontal gene transfer, observed intraribotype diversity, and the growing appreciation of the impact and prevalence of ocean viruses in surface waters (Williamson et al. 2008; Sharon et al. 2009), it might be expected that phylogenetic composition could play less of an role in determining membrane protein functions present in an organism. There are several instances in the literature suggesting that genome content differences even within species (“ecotypes”) reflect the environmental conditions in which they were extracted (Thompson et al. 2005; Martiny et al. 2006; Van Mooy et al. 2009). As an example, two ecotypes of the ocean-dominating Prochlorococcus, high-light (HL) and low-light (LL), are adapted to inhabit different levels of the water column, reflecting genomic adaptation to environmental characteristics (West and Scanlan 1999). In addition, in the GOS analysis of whole genomic content (Rusch et al. 2007), it was observed that there was a clear distinction between sites, and this was still evident upon limiting to or removing reads from dominant species, suggesting more global niche differences.
An advantage to our analysis is that it reveals not only the environmentally influenced fraction of the membrane proteins, but also provides a window into those membrane proteins that appear insensitive to this set of environmental features. For example, in our CCA analysis, we find 44 out of the 151 families to be invariant across the sites, including the ubiquitous chloride channel and type III secretion proteins involved in virulence, as noted previously to be abundant in marine bacteria (Persson et al. 2009). Within these invariant proteins there is a suggestion of functional importance, whether for essential cellular processes or processes intrinsic to their ocean habitats.
Across the variant set, we observed a significant proportion of ABC-type transporters (34/41) covarying with the environment, illustrating a possible case of streamlining for optimization and energy conservation. Responsible for the high-affinity transport of a wide array of substrates, and in some cases having broad specificity, these proteins provide an efficient means of transport in oligotrophic surface waters. As noted, these proteins had a strong tendency to be inversely correlated with the prevalence of their substrate, as in the case of phosphate, showing possible adaptation to phosphate-rich/poor conditions. Recently, Martiny et al. (2009) showed that the proteins surrounding the PhoB gene (the phosphate response regulator) in Prochlorococcus are enriched in GOS samples found in waters with low phosphate content. They limited sites selection to those sites with high Prochlorococcus hit counts (2.5 hits per 1000 bp), thus focusing on nutrient adaptation in this species (only 11 sites overlapped between studies). We observed the same trend in our results; however, we did not address any particular species, instead treating the environmental sites as a “superorganism.”
Through the GPS coordinates provided by the GOS project, we were able to tap into a wealth of available geospatial data from those measuring natural fluxes to those assessing human impact. It is important to note that due to the nature of collection, only five out of the 15 environmental features used in this study were collected at the same time as the metagenomics sampling was performed. The remainder of the environmental features were derived from historical information resulting in sometimes large differences in time and space resolution between the environmental feature data and the metagenomics survey (Supplemental Fig. 1). However, the characteristics of microbial communities are affected not just by the features at the time of sample collection but the history and flux of the features. We have only begun to skim the surface of the question of how much environmental history these communities carry, how much of a microbial footprint the environment reflects, and how much of our own footprint is reflected in both of them. The true test of these questions can only come through detailed examination of both microbial and environmental dynamics.
In addition, the analysis presented here is of the linear interactions between the environment and membrane proteins. Capturing the nonlinear interactions will require some modifications to existing techniques (e.g., kernel CCA), making it a particularly promising avenue for future research. We chose to first explore the linear interactions for the ease of their interpretability. We hope this work serves as a motivation for collecting additional oceanographic and metagenomics data sets and exploring higher-order relationships.
We have used metagenomics data to quantitatively investigate the relationship between gene content and abundance in differing habitats. The questions we have addressed here are certainly not new; however, metagenomics studies are beginning to reveal these relationships on much larger scales. Thus, the strength of metagenomics studies is not only in their ability to study uncultivable organisms, but also in their ability to integrate layers of data in the study of whole community dynamics, and to untangle the intricate web of dependencies within habitats.
Methods
Preprocessing GOS data
Sequences and metadata (salinity, chlorophyll, sample depth, water depth, temperature) from the GOS Expedition (Rusch et al. 2007) were downloaded from CAMERA (Seshadri et al. 2007). Sites were initially selected as in Gianoulis et al. (2009). All sites used a filter size of 0.1–0.8 μm. Peptides were mapped to sites as in Gianoulis et al. (2009). Briefly, each peptide was mapped to its open reading frame (ORF) and back to its read (which mapped to a site) through the scaffolds. If a peptide originated from two reads from different sites combined in a scaffold, they were placed in both sites. Cluster annotation in CAMERA was used to remove clusters of peptides that were labeled as spurious and that contained fewer than four sequences.
Environmental data integration
UV, shipping, pollution, climate change, and ocean acidification impact values for each of the sites were extracted using ArcGIS from maps from the National Center for Ecological Analysis and Synthesis (NCEAS) (Halpern et al. 2008). Each value represents the impact of the particular factor at the site based on the type of ecosystems present. The resolution of the data is 1 km2, and thus the value for the kilometer-square in which the site was contained was used. The other factors analyzed were not used due to the sparsity of the data at the GOS sites.
Phosphate, silicate, nitrate, dissolved oxygen, and apparent oxygen utilization annual values of the objectively analyzed mean for each site at surface levels were extracted from maps provided by the World Ocean Atlas 2005 (Antonov et al. 2006; Garcia et al. 2006; Locarnini et al. 2006). These environmental features are based on historical data regardless of year of observation, from various sources, with a resolution of 1° latitude/longitude.
Site selection
Sites were filtered for three main reasons: insufficient sample coverage, missing or nonrepresentative metadata, and metagenome composition outliers (see Supplemental Table 3 for a site-by-site breakdown; Supplemental Figs. 2–4). We selected the 29 sites based on availability of the environmental data as well as to measure subtle differences in genomic content across habitats. For example, Lake Gatan, a freshwater lake, and Punta Cormorant, a hypersaline lagoon, were removed as they were extreme environmental outliers with very different membrane protein (Supplemental Fig. 2) and genomic content (Rusch et al. 2007) with no representative metadata. While these features of the outlier sites in themselves are interesting, we wanted continuous differences in terms of environmental data and sequence data for further analysis.
Prediction of membrane proteins/mapping to COG
Each non-redundant sequence was run through PRODIV-TMHMM (Eddy 1998) to predict membrane-spanning regions and subsequently mapped to a clusters of orthologous group (COG) using BLASTP (E-value threshold 1e-10) (Altschul et al. 1990). Only for 0.2% of the sequences were the top two COG hits inconsistent; thus the top hit for each sequence was used. If >80% of the sequences in a COG were annotated as a membrane protein by PRODIV-TMHMM, the COG was labeled as a membrane COG (high-confidence membrane proteins). This threshold was chosen arbitrarily given the number of partial protein sequences in GOS and the error rate of PRODOV-TMHMM, and upon manual inspection of the COG descriptions. Membrane proteins that were not transporters, permeases, and channels (e.g., oxidative phosphorylation proteins) were manually removed to focus on transport and efflux processes. In addition, COGs that mapped to <1% of all sequences in the resulting sequence data set were removed. (Peptides mapping to viral sequences, 0.01% [Williamson et al. 2008], were included due to the insignificant number and prevalence of horizontal gene transfer.)
16S gene data
16S data were taken from Biers et al. (2009) at the 20% divergence level. Each site had an 18-element vector of counts for each “phylum” (as referred to in Biers et al. 2009).
Pairwise correlations/clustering
Matrices (rows are sites; columns are either percentage of membrane protein families, 16S diversity, or environmental features) were standardized prior to performing pairwise correlations (Pearson) of the sites (rows) and hierarchical clustering.
PEN
The membrane protein families and environmental features network was constructed using the structural correlations from regularized CCA. The dot product of the structural correlations in the first and second dimension between and within the membrane protein families and environmental features was calculated. The distance (dot product) threshold was set to >|0.5|, and between every satisfying pair (nodes) an edge was placed.
Acknowledgments
We thank Stacey Maples at the Yale University Map Department for help with data extraction. The instrumentation was supported by Yale University Biomedical High Performance Computing Center and NIH grant RR19895. M.G. acknowledges the support of the DOE.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102814.109.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Antonov J, Locarnini R, Boyer T, Mishonov A, Garcia H 2006. World Ocean Atlas 2005. In NOAA Atlas NESDIS 62 (ed. Levius S), pp. 182 U.S. Government Printing Office, Washington, DC [Google Scholar]
- Barabasi AL, Oltvai ZN 2004. Network biology: Understanding the cell's functional organization. Nat Rev Genet 5: 101–113 [DOI] [PubMed] [Google Scholar]
- Biers EJ, Sun S, Howard EC 2009. Prokaryotic genomes and diversity in surface ocean waters: Interrogating the global ocean sampling metagenome. Appl Environ Microbiol 75: 2221–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird D, Kalff J 1984. Empirical relationship between bacterial abundance and chlorophyll concentration in fresh and marine waters. Can J Fish Aquat Sci 41: 1015–1023 [Google Scholar]
- Borga M 1998. “Learning multidimensional signal processing.” PhD thesis, Linköping University, Linköping, Sweden [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503 [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. 2008. Functional metagenomic profiling of nine biomes. Nature 452: 629–632 [DOI] [PubMed] [Google Scholar]
- Eddy SR 1998. Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Flink I, Pettijohn DE 1975. Polyamines stabilise DNA folds. Nature 253: 62–63 [DOI] [PubMed] [Google Scholar]
- Garcia H, Locarnini R, Boyer T, Antonov J 2006. World Ocean Atlas 2005. In NOAA Atlas NESDIS 63 (ed. Levius S), pp. 342 U.S. Government Printing Office, Washington, DC [Google Scholar]
- Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, Yamada T, Paccanaro A, Jensen LJ, Snyder M, et al. 2009. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc Natl Acad Sci 106: 1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González I, Déjean S, Martin PGP, Baccini A 2008. CCA: An R package to extend canonical correlation analysis. J Stat Softw 23 http://www.jstatsoft.org/v23/i12 [Google Scholar]
- Guan LL, Kanoh K, Kamino K 2001. Effect of exogenous siderophores on iron uptake activity of marine bacteria under iron-limited conditions. Appl Environ Microbiol 67: 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Tatsuoka K, Liu R 2006. Histone acetylation and transcriptional regulation in the genome of Saccharomyces cerevisiae. Bioinformatics 22: 392–399 [DOI] [PubMed] [Google Scholar]
- Halpern BS, Walbridge S, Selkoe KA, Kappel CV, Micheli F, D'Agrosa C, Bruno JF, Casey KS, Ebert C, Fox HE, et al. 2008. A global map of human impact on marine ecosystems. Science 319: 948–952 [DOI] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ 2008. The 20 years of PROSITE. Nucleic Acids Res 36: D245–D249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Kashiwagi K 2000. Polyamines: Mysterious modulators of cellular functions. Biochem Biophys Res Commun 271: 559–564 [DOI] [PubMed] [Google Scholar]
- Jickells TD, An ZS, Andersen KK, Baker AR, Bergametti G, Brooks N, Cao JJ, Boyd PW, Duce RA, Hunter KA, et al. 2005. Global iron connections between desert dust, ocean biogeochemistry, and climate. Science 308: 67–71 [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Saier M, Paulsen IT, Collado-Vides J, Paley SM, Pellegrini-Toole A, Bonavides C, Gama-Castro S 2002. The EcoCyc Database. Nucleic Acids Res 30: 56–58 doi: 10.1093/nar/30.1.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL 2008. Microbial ecology of the ocean Wiley, New York [Google Scholar]
- Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P 2008. A bioinformatician's guide to metagenomics. Microbiol Mol Biol Rev 72: 557–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Jorgensen N 1995. Seasonal cycling of putrescine and amino acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry 29: 131–157 [Google Scholar]
- Locarnini R, Mishonov A, Antonov J, Boyer T, Garcia H 2006. World Ocean Atlas 2005. In NOAA Atlas NESDIS 61 (ed. Levitus S), pp. 182 U.S. Government Printing Office, Washington, DC [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci 103: 12552–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Huang Y, Li W 2009. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol 11: 1340–1347 [DOI] [PubMed] [Google Scholar]
- Neyfakh AA 1997. Natural functions of bacterial multidrug transporters. Trends Microbiol 5: 309–313 [DOI] [PubMed] [Google Scholar]
- Persson OP, Pinhassi J, Riemann L, Marklund BI, Rhen M, Normark S, Gonzalez JM, Hagstrom A 2009. High abundance of virulence gene homologues in marine bacteria. Environ Microbiol 11: 1348–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman B, Blount P, Folgering JH, Friesen RH, Moe PC, van der Heide T 2002. How do membrane proteins sense water stress? Mol Microbiol 44: 889–902 [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77 doi: 10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir T 2008. Ship's ballast water and marine pollution. In Integration of information for environmental security, pp. 467–477, 498 Springer, New York [Google Scholar]
- Schomburg I, Chang A, Hofmann O, Ebeling C, Ehrentreich F, Schomburg D 2002. BRENDA: A resource for enzyme data and metabolic information. Trends Biochem Sci 27: 54–56 [DOI] [PubMed] [Google Scholar]
- Seshadri R, Kravitz SA, Smarr L, Gilna P, Frazier M 2007. CAMERA: A community resource for metagenomics. PLoS Biol 5: e75 doi: 10.1371/journal.pbio.0050075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, Pinter RY, Partensky F, Koonin EV, Wolf YI, et al. 2009. Photosystem I gene cassettes are present in marine virus genomes. Nature 461: 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV 2000. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307: 1311–1313 [DOI] [PubMed] [Google Scholar]
- Tjaden J, Winkler HH, Schwoppe C, Van Der Laan M, Mohlmann T, Neuhaus HE 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J Bacteriol 181: 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. 2005. Comparative metagenomics of microbial communities. Science 308: 554–557 [DOI] [PubMed] [Google Scholar]
- Van Mooy BA, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblizek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, et al. 2009. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458: 69–72 [DOI] [PubMed] [Google Scholar]
- Viklund H, Elofsson A 2004. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci 13: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NJ, Scanlan DJ 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol 65: 2585–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichern R, Johnson D 2003. Applied mutlivariate statistical analysis Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
- Williamson SJ, Rusch DB, Yooseph S, Halpern AL, Heidelberg KB, Glass JI, Andrews-Pfannkoch C, Fadrosh D, Miller CS, Sutton G, et al. 2008. The Sorcerer II Global Ocean Sampling expedition: Metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3: e1456 doi: 10.1371/journal.pone.0001456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler HH, Neuhaus HE 1999. Non-mitochondrial ATP transport. Trends Biochem Sci 24: 64–68 [DOI] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol 5: e16 doi: 10.1371/journal.pbio.0050016 [DOI] [PMC free article] [PubMed] [Google Scholar]