Abstract
One of the key advantages of using Drosophila melanogaster as a genetic model organism is the ability to conduct saturation mutagenesis screens to identify genes and pathways underlying a given phenotype. Despite the large number of genetic tools developed to facilitate downstream cloning of mutations obtained from such screens, the current procedure remains labor intensive, time consuming, and costly. To address this issue, we designed an efficient strategy for rapid identification of heterozygous mutations in the fly genome by combining rough genetic mapping, targeted DNA capture, and second generation sequencing technology. We first tested this method on heterozygous flies carrying either a previously characterized dac5 or sensE2 mutation. Targeted amplification of genomic regions near these two loci was used to enrich DNA for sequencing, and both point mutations were successfully identified. When this method was applied to uncharacterized twr mutant flies, the underlying mutation was identified as a single-base mutation in the gene Spase18-21. This targeted-genome-sequencing method reduces time and effort required for mutation cloning by up to 80% compared with the current approach and lowers the cost to <$1000 for each mutant. Introduction of this and other sequencing-based methods for mutation cloning will enable broader usage of forward genetics screens and have significant impacts in the field of model organisms such as Drosophila.
The availability of genetic tools in Drosophila to generate, screen, and characterize mutations with phenotypes of interest makes it one of the most powerful genetic model organisms. One of the most commonly used mutagens, ethyl methane sulfonate (EMS), has been widely used in genetic screens, and mutations identified in more than 3000 genes have been recorded in FlyBase (http://flybase.org/). In order to clone an EMS or other chemical mutagen-induced novel mutation, it is often necessary to reduce the candidate locus to a small interval, typically of ∼20 kb or less in size, by fine genetic mapping (H Bellen, pers. comm.). However, due to the large number of flies that must be scored in order to achieve such resolution, the fine mapping step is quite labor intensive and costly. Additionally, for some genomic regions, it is not always possible to map a mutation to a small interval due to limitations such as low recombination rates and lack of mapping stocks.
Several molecular methods have been proposed for cloning EMS mutations without the need for genetic mapping. One method, named TILLING, is based on Cel-I-mediated heteroduplex DNA cleavage and can be used to identify changes in genes of interest between the wild-type and mutant flies (Winkler et al. 2005). It is ideal for isolating alleles of given genes from a large collection of EMS-induced mutants; however, this method is not well suited for identifying mutations obtained from forward genetic screens, since candidate genes need to be predefined for testing using TILLING. Another approach of mutation cloning is to take advantage of the recent development of second-generation sequencing technology. Rapid progress in sequencing technologies has dramatically increased the throughput and reduced the cost of DNA sequencing (Mardis 2008a,b; Shendure and Ji 2008; Ansorge 2009). Recently, identification of homozygous mutations by direct whole-genome sequencing has been reported (Sarin et al. 2008; Smith et al. 2008; Srivatsan et al. 2008; Blumenstiel et al. 2009). Mutations in genes that are important for development, however, are often homozygous lethal, and it is necessary to detect heterozygous mutations. The detection of heterozygous mutations requires much more sequencing coverage (Bentley et al. 2008; Ley et al. 2008; Wheeler et al. 2008). We estimate that about 30× sequencing coverage is necessary to detect heterozygous mutations with high sensitivity (>95%) and accuracy (error rate <10−6) (Supplemental Fig. 1). At the current cost of about $1500 for every 10× sequencing coverage of the Drosophila genome, the direct whole-genome sequencing approach is still too expensive for broad usage. This approach is further limited, as parental fly strains and multiple alleles often need to be sequenced in parallel to distinguish single nucleotide polymorphisms (SNPs) and other changes from causative mutations.
When a cloning by sequencing approach is used, rough genetic mapping of the mutation is often highly valuable, as it can greatly reduce downstream data analysis efforts. Since approximately one base mutation is induced per 400 kb when flies are treated with 25 mM EMS, a commonly used experimental condition, a total of about 400 base changes exist in each mutant fly (Cooper et al. 2008; Blumenstiel et al. 2009). Mapping of the mutation can greatly reduce the number of candidate changes that need to be analyzed. Although fine genetic mapping is quite labor intensive, intermediate levels of genetic mapping where a mutation is mapped to within a few megabases (Mb) can be achieved efficiently using methods such as P element or SNP mapping in Drosophila (Zhai et al. 2003; Chen et al. 2008). Once a mutation is mapped to an interval of several megabases or less, it is then necessary to sequence only the candidate region (often <1% of the genome) instead of the entire genome, resulting in significant cost reduction. Recently, several DNA enrichment methods have been developed that enable enrichment of DNA from targeted regions (Albert et al. 2007; Dahl et al. 2007; Porreca et al. 2007; Gnirke et al. 2009; Ng et al. 2009; Okou et al. 2009). With optimized probe design and capture conditions, the vast majority of a targeted region (>95%) can be efficiently enriched (H Wang and R Chen, unpubl.). Among the numerous methods for targeted DNA enrichment, padlock capture, which relies on a combination of oligonucleotide hybridization and enzymatic activity, shows the greatest specificity (Bau et al. 2009; Li et al. 2009b). As shown in Figure 1A, a pool of probes that contain target-specific capturing arms can be annealed to the target regions. DNA synthesis using the target regions as templates will generate circular DNA, which is selectively amplified by PCR using common sequences flanking the capture arms as primers. A key feature of padlock probes is that they can be regenerated by PCR, thereby greatly reducing costs when multiple enrichments are needed, as in the case of sequencing complementing mutations of the same locus.
Figure 1.
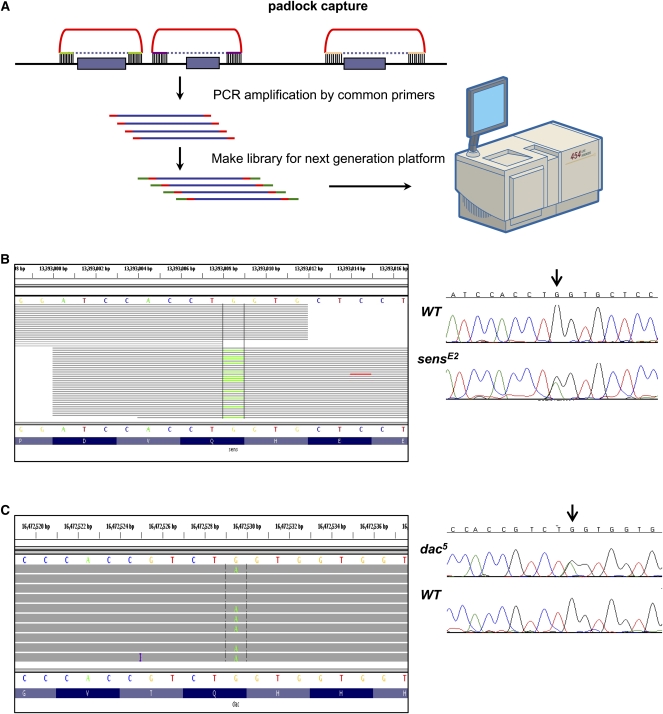
Two known mutations were detected using padlock capture sequencing. (A) Flowchart of mutation detection by padlock capture and next-generation sequencing technologies. Padlock capture technology requires probes that contain two target-specific capturing arms (green) connected by a common linker (red). The unique targeting arms of individual targeting oligonucleotides are designed to hybridize immediately upstream and downstream from each exon (purple bars) of interest. Hybridization to genomic DNA is followed by an enzymatic gap filling and ligation step, such that a copy of the sequence of interest is incorporated into a circle (purple dashed line). Then the enriched DNA is PCR amplified and is used to prepare libraries for the next-generation platform sequencing. (B) sensE2 and (C) dac5 mutation detection. Reads alignment at the mutation is shown between the vertical lines. Each read is drawn as a gray line, and bases that are different from the reference are colored. Sanger sequencing is conducted to confirm the mutation with the heterozygous mutated base indicated by the arrow.
To test the feasibility of identifying heterozygous mutations by targeted genome sequencing, we first applied the proposed method to two known mutations, dac5 and sensE2. Exons within 0.5 Mb flanking the two known mutations were enriched and sequenced using both the 454 Life Sciences (Roche) and the Illumina GAII systems. We found that the causative point mutations could be reliably identified by either platform using only a small fraction of each platform's capacity. Next, we applied this method to uncover the mutation in a previously uncharacterized mutant, twr1, and identified a missense mutation in the Spase18-21 gene. Two lines of evidence support that the identified mutation in Spase18-21 is indeed the causative mutation for the twr phenotype. First, we identified mutations in Spase18-21 in two additional twr mutant alleles, twr2 and twr11. Second, a P-element insertion that fails to complement the twr1 mutant allele maps to the 5′ end of the Spase18-21 gene. In summary, we conclude that combining rough genetic mapping, DNA targeted capture, and second generation sequencing is highly cost effective and immediately applicable to mutation cloning in Drosophila, which will enable broader usage of the forward genetic screen.
Results
We to identify a strategy for cloning of these lethal mutations in the Drosophila genome. As an alternate approach to whole-genome sequencing, targeted genomic sequencing offers the potential of significant cost reduction by reducing the portion of the genome sequenced (Li et al. 2009b). This is particularly suitable for model organisms in which genetic mapping can rapidly reduce candidate loci to a reasonable genomic interval. In Drosophila, the combination of genetics markers, P-element insertions, and chromosomal deficiency and duplication stocks, make it possible to map a lethal mutation within a few hundred kilobase interval efficiently. As a result, instead of the entire genome, <0.1% of the genome needs to be sequenced to identify an underlying mutation. Therefore, significant cost reduction can be potentially achieved if the small interval can be enriched from the genome and sequenced.
Capture sequencing as a cost-effective strategy for detecting heterozygous mutations
To test this approach, we performed targeted sequencing on two previously characterized mutant alleles, one in the gene senseless (sens), sensE2, and the other in dachshund (dac), dac5 (Fig. 1B,C). Using the padlock method, DNA oligo probes covering all exons within the 0.5-Mb region centered on each of these two mutations were designed and used to enrich DNA from adult heterozygous mutant flies (Fig. 1A). As shown in Figure 1B, sensE2 is a nonsense mutation caused by a G to A transition in the sens gene at 13,393,009 bp on chromosome 3L (version 5.1). Flies homozygous for the sensE2 mutation are embryonic lethal. To enrich DNA for the annotated exons in the 0.5-Mb region surrounding sens, a total of 257 amplicons covering 86,435 exon bases were designed (Supplemental File 1). Using this probe set, DNA from sensE2 heterozygous flies was enriched and about 37,000 sequencing reads were obtained using the 454 platform. These reads were mapped to the targeted region, giving an average sequencing coverage of 44× with median coverage of 38×. At least 1× coverage was observed for 92.6% of the targeted bases, and 75.2% of bases had at least 10× coverage. In total, 62 SNPs were detected, including the known SNP. As shown in Figure 1B, a total of 33 reads covering the mutated base in sensE2 were obtained, with 21 reads representing the wild-type allele and 12 representing the mutant allele, resulting in a SNP with a quality score of 84. This finding was confirmed by direct Sanger sequencing of a PCR product containing the mutated base (Fig. 1B, arrow). Similarly, for the 0.5-Mb genomic locus around dac, a total of 249 amplicons covering 112,284 annotated exon bases were designed. Capture sequencing was performed on dac5 heterozygous DNA, which contains a G-to-A transition at 16,472,530 bp on chromosome 2L (version 5.1). A total of 24,530 454 sequencing reads were obtained and mapped to the targeted region, which was equivalent to an average coverage of 22× with median coverage of 11×. A total of 97,917 (87%) bases were covered at least once while 64,580 (57.5%) of all targeted bases had 10× coverage or higher and 11 SNPs were detected in total. A total of 10 reads covering the known dac5 mutant allele were obtained, with four reads representing the wild-type allele and six reads representing the mutant allele, resulting in a SNP with a quality score of 87 (Fig. 1C). Consistent with this result, the heterozygous mutation was further confirmed by directly sequencing the PCR product (Fig. 1C, arrow). Taken together, these results demonstrate that previously known mutant alleles in heterozygous flies can be identified by capture sequencing.
As shown above in our capture sequencing experiment for the sensE2 mutant flies, although the mean coverage is 44×, ∼25% of targeted bases have a coverage of 10× or lower, and 7.4% of all targeted bases are not sequenced at all. Therefore, we asked whether higher sequencing coverage would increase the portion of bases with sufficient sequencing coverage. DNA from two independent capture experiments on the sensE2 heterozygous flies was sequenced on the Illumina GAII platform at 90× and 140× coverage. As expected, an increase in sequencing depth increases the percentage of bases with 10× coverage (Supplemental Fig. 1C). For example, at 140× sequencing depth, 85.6% of the bases have at least 10× coverage, while only 3.9% of the bases remain uncovered. In addition, the coverage profile is very similar between these two capture experiments (data not shown), suggesting that the method is quite robust and reproducible. As the capture region of sensE2 represents just 0.05% of the genome, the total amount of sequence data generated at 140× coverage of this region is equivalent to only ∼0.07× coverage of the whole genome.
Identification of novel mutations using the capture sequencing approach
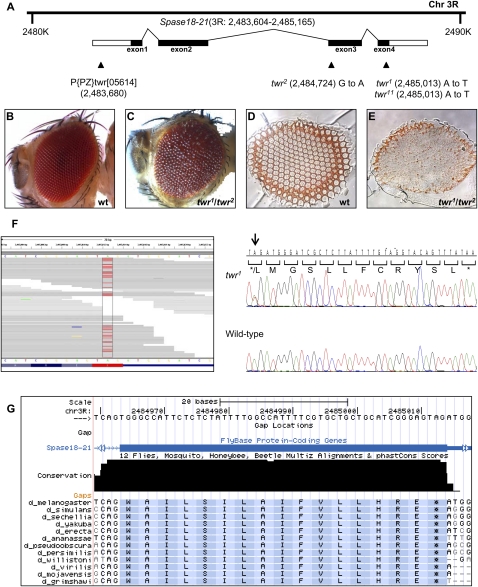
To determine whether the capture sequencing method is applicable to identification of novel mutations, we tested our approach on twr mutant flies. twr mutants were first identified through an EMS mutagenesis screen of the ANTP-C region (Lewis et al. 1980; Hazelrigg and Kaufman 1983). In this screen, multiple twr alleles were recovered with three stocks that are available from the Bloomington stock center including twr1, twr2, and twr11 (Fig. 2A). Although twr mutant flies are homozygous lethal, trans heterozygous twr mutant flies exhibit disorganization of ommatidia with degenerative photoreceptors (Fig. 2C,E). The twr mutant has been mapped to cytological position 84A, but the molecular nature of these mutations remains unknown. To identify the underlying mutations in twr, we designed a capture probe set spanning the 84A region, starting from 2.27 to 2.73 Mb on chromosome 3R (Supplemental File 1). This probe set amplifies a total of 285 amplicons containing 74,001 bp of exon sequences in the targeted region. DNA enriched by this probe set was sequenced using the GAII platform and a total of 206,729 32-bp reads were generated and mapped to the targeted region, resulting in an average of 74× coverage of the targeted region. As a result, only 4.8% of the targeted region was missing, while 78% of the targeted region had >10× sequence coverage. Interestingly, a large number of changes were identified, including 105 heterozygous changes and 21 homozygous changes, most of which probably reflect the prevalence of SNPs in the Drosophila genome. A large portion of these potential variants were synonymous changes, while only 26 resulted in amino acid changes (Supplemental File 2). To distinguish potential mutations from SNPs, it would be best to obtain sequences from the parental strain. Unfortunately, the twr1 allele was generated more than 20 yr ago, and the parental strain used for mutagenesis is no longer available. To solve this problem, capture sequencing was performed on heterozygous flies carrying an independently derived twr mutant allele, twr2. As both twr1 and twr2 alleles were induced from the same parental strain, these two mutant fly strains should share parental SNPs. Indeed, among the 26 heterozygous variants identified in the heterozygous twr1 flies, 24 are also identified in the twr2 flies, indicating that these variants are likely to be SNPs inherited from their parental strain (Supplemental File 2). Only two single-base variants appear to be twr1 specific and were likely induced during mutagenesis, one at base position 2,485,103 A → T and the second one at 2,518,875 G → A. Variant at base position 2,518,875 results in amino acid change from Gly to Ser in gene Ccp84Ad (chitin cuticular protein at 84Ad). Another mutation at base 2,485,103 results in a significant change in gene Spase18-21. Among the 221 reads covering this position, 135 reads contains the wild-type base (A) and the remaining 86 were mutated to base T (Fig. 2F). As a result, the normal stop codon is replaced by Leu, causing an addition of 12 amino acids to the end of the Twr protein (Fig. 2F). Interestingly, the amino acid sequence of the last coding exon and the position of the stop codon of Spase18-21 are completely conserved in 12 Drosophila species (Fig. 2G). Further support that the variant in Spase18-21 is the causative mutation for the twr phenotype comes from sequencing of the two additional twr mutant alleles, twr2 and twr11. An identical mutation was observed in the twr11 allele, while a single base pair change that disrupts the splicing acceptor site of exon 3 of the Spase18-21 gene was identified in twr2 mutant flies (Fig. 2A; data not shown). These data suggested that twr flies indeed carry mutations in the Spase18-21 gene. This conclusion is further supported by characterization of a homozygous lethal P-element insertion, twr05614 (Fig. 2A). twr05614 is an allele of twr that fails to complement twr1 mutant. Inverse PCR confirms the insertion site at position 3R: 2,483,680 bp, just 76 bp 5′ of the Spase18-21 gene as recorded in FlyBase (Fig. 2A; data not shown). Furthermore, when the P-element is mobilized, wild-type reverted flies are recovered, indicating that the phenotype observed in twr1/twr05614 is caused by the P-element insertion. Taken together, we conclude that twr mutant flies carry mutations in Spase18-21.
Figure 2.
Padlock capture was successfully used to identify novel mutations. (A) The genomic locus of twr and its alleles are shown. Mutations were detected within the Spase18-21 gene in three different twr alleles, twr1, twr2, and twr11, using capture sequencing. Alleles twr1 and twr11 shared the same mutation of an A-to-T transition at position 3R:2,485,014. twr2 was found to have a different transition of G to A at position 3R: 2,484,724. A P-element twr[05614] that failed to complement twr1 was found by inverse PCR to be located 76 bp downstream from the Spase18-21 start site at position 3R:2,483,680. Compared with the wild type (B,D), twr1/twr2 transheterozygous adult flies (C,E) show rough eye phenotype and missing photoreceptors. (F) Alignment of reads at the position of the twr1 mutation is shown. Alignment of capture sequencing reads covering the heterozygous mutation in the twr1 is shown on the left. The mutated base pair is highlighted in red. Direct Sanger sequencing was performed to confirm the mutation in twr1, with the heterozygous mutated base indicated by the arrow. The addition of 12 amino acids that were caused by the mutation are also shown. (G) Alignment of the SPASE18-21 amino acid sequence within several Drosophila species is shown, indicating a high degree of identity.
Discussion
Forward genetic approaches are widely used from bacteria to mice and play an essential role in modern biology by identifying genes with interesting phenotypes. In model organisms, the forward genetic screen remains one of the most powerful tools to study biological pathways. The major hurdle in forward genetic screens is the cloning step. It can take months of effort to identify the molecular lesion in higher eukaryotes, even with the extensive genetic tools that are available in Drosophila. In our study, we demonstrate that the combination of rough genetic mapping, DNA capture, and next-generation sequencing is a viable method for rapid and cost-effective mutation identification. First, the ability of next-generation sequencing to generate billions of bases at low cost makes it efficient to sequence a megabase region. As a result, only intermediate levels of genetic mapping are needed to narrow down a mutation to within a few megabases, thereby eliminating the labor-intensive, time-consuming genetic fine-mapping step. As a result, the time for cloning a mutation can be shortened dramatically from 6 mo to ∼2 mo along with significantly less effort. Second, the DNA capture method makes it possible to enrich specific genomic regions for sequencing. To achieve high sensitivity (>95%) and specificity, we have estimated that 30× sequencing coverage is needed to identify heterozygous mutations with high accuracy (less than one error per megabase) and sensitivity (>95%) for 90% of the genome (Supplemental Fig. 1). Furthermore, to increase the likelihood of identifying changes and distinguishing pre-existing SNPs from true mutations, it is best to sequence multiple alleles within the same complementation group, along with their respective controls. Therefore, the choice of sequencing approach is primarily driven by the sequencing cost. Even at the currently low sequencing cost of $750 per gigabase, detection of mutations by direct whole-genome sequencing is quite costly. Together with sequencing library construction cost, $4500 per stain at 30× sequence coverage, a total of $14,400 is needed to perform whole-genome sequencing for two alleles with their parental strain as control (Table 1). In contrast, the DNA capture technology used in our study offers greatly reduced sequencing costs while increasing the sequencing coverage at targeted regions. Exons within a 0.5-Mb region surrounding two known mutations, dac5 and sensE2, were captured and sequenced. Even with high coverage of 140× for the targeted region, the total amount of sequence needed is still <1% that of whole-genome sequencing. As a result, the current sequencing cost for captured DNA is <$400 per strain, bringing the total cost below $2000 for characterizing two alleles plus the parental strain (Table 1). Furthermore, even with a 10-fold reduction of the current sequencing cost, the capture sequencing approach will still offer significant savings over the whole-genome shotgun approach (Table 1, projected cost columns). Third, the high coverage obtained from capture sequencing ensures sensitivity and accuracy in detecting mutations. At 40× sequencing coverage, 57.5% and 75.2% of bases enriched in dac5 and sensE2 sequencing showed >10× coverage, respectively. The portion of highly covered bases can be further increased by simply performing additional sequencing. For example, 85.6% of exon bases at the sens locus were sequenced at least 10 times when the sequence coverage was increased to 140×. This high coverage is essential for the accurate identification of mutations.
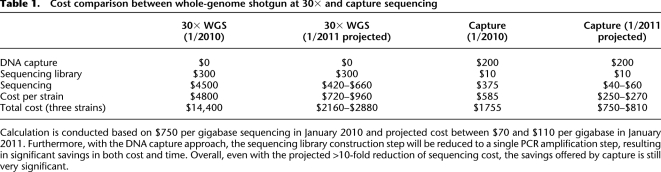
Table 1.
Cost comparison between whole-genome shotgun at 30× and capture sequencing
Calculation is conducted based on $750 per gigabase sequencing in January 2010 and projected cost between $70 and $110 per gigabase in January 2011. Furthermore, with the DNA capture approach, the sequencing library construction step will be reduced to a single PCR amplification step, resulting in significant savings in both cost and time. Overall, even with the projected >10-fold reduction of sequencing cost, the savings offered by capture is still very significant.
Regardless of the sequencing strategy, rough genetic mapping data will be highly desired during data analysis and should be included as a key part of mutation cloning. There is approximately one SNP per 200 bp in each Drosophila strain (Platts et al. 2009). Indeed, a large number of SNPs have been observed when mutant fly sequences are compared with that of the reference genome. For example, in the twr1 region we found 126 nucleotide changes, of which 26 caused changes in the amino acid sequence. Therefore, to distinguish SNPs from mutations, it is crucial to have the parental strain sequenced as a control. A polymorphism should exist in both the parent and the mutant, while EMS-induced mutations are mutant specific. To minimize complications such as spontaneous mutations accumulating in the parental stocks, it is recommended to obtain sufficient DNA from parental flies at the time of mutagenesis for reference. In cases where the parental strain is no longer available, parallel sequencing of multiple alleles can also serve this purpose. Another issue during data analysis is the fact that several hundred base changes can be induced by EMS across the genome. Several measures can be implemented to facilitate identification of the causative mutation. First, a large number of EMS-induced changes can be excluded by genetic mapping data. Only changes within the targeted region are relevant. Second, data obtained from multiple independent alleles can be compared to identify shared mutant genes. As EMS-induced mutations are random, this will greatly reduce false positives. Third, a large number of changes will be benign. A base change should be excluded if it fails to affect an amino acid or an mRNA splice site. In addition, as more Drosophila sequencing is carried out, the establishment of a database cataloging common SNPs will be a critical step to facilitate downstream analysis.
Through capture sequencing, we were able to identify mutations in both dac5 and sensE2 heterozygous flies. In addition, we have identified that the novel underlying mutation in twr1 lies within the Spase18-21 gene. The molecular nature of the twr1 mutation is a base mutation at the stop codon, resulting in an addition of 12 amino acids to the C terminus of the protein. It is interesting that such a small change in the SPASE18-21 protein in twr1 has such a dramatic impact on its normal function. SPASE18-21 is a subunit of a signal peptidase that catalyzes the cleavage of signal peptides within the endoplasmic reticulum. The amino acid sequence of SPASE18-21 is nearly identical across all 12 sequenced Drosophila species. Therefore, it is conceivable that small changes in the protein might lead to an alteration in the protein's tertiary structure that could affect its interaction with other subunits in the multisubunit signal peptidase complex, thereby dramatically reducing the enzymatic activity of this complex.
We found that typically 10%–15% of the bases are not well covered by capture sequencing using padlock probes. The efficiency can probably be improved by several methods. First, the probe design can be further optimized. A combination of changing probe design, adding blocking oligos at overrepresented regions, and adding additional capture probes for underrepresented regions, has been shown to improve capture results (Li et al. 2009b). Second, a set of padlock probes specific for under-represented regions can be designed and used in a separate experiment in addition to the original kit. Without competition from overenriched regions, under-represented regions are likely to be captured more efficiently. Third, other enrichment methods such as DNA hybridization liquid capture can be used either in conjunction with the padlock approach or independently. Near complete capture can be achieved with DNA hybridization methods when the probe design is optimized (H Wang and R Chen, unpubl.). Finally, to improve the sensitivity of detecting mutations, capture sequencing can be conducted for all alleles of the same complementation group when available. This is feasible, as the cost of capture sequencing is quite low. As independent alleles usually harbor mutations at different positions in a given gene, the problem of missing small portions of a gene sequence is minimized.
In summary, with the development of capture probe sets across the entire genome, establishment of a SNP database, and further development of sequencing technologies and data analysis tools, mutation cloning in Drosophila will become straightforward and cost efficient. Compared with the current genetic fine-mapping plus Sanger sequencing approach, which typically requires at least 6 mo and costs at least $1500 in reagent alone, our proposed method can be completed within 2 mo with partial effort and costs <$1000. As a result, we expect that the use of chemical mutagenesis in Drosophila research will be further broadened, including the generation of a complete collection of EMS-induced mutations. Genetic mapping can be conducted in most model organisms efficiently, and, hence, this approach should also be readily applicable to other model organisms such as mice.
Methods
Fly genetics and DNA preparation
All flies used in this study were maintained on standard Drosophila medium in a 25°C room with a light cycle of 12 h light and 12 h dark. Genomic DNA from flies was prepared using Buffer A (100 mM Tris-HCl at pH 7.5, 100 mM EDTA, 100 mM NaCl, 0.5% SDS), followed by LiCl/KAc incubation and ethanol precipitation.
Padlock probe design
Targets were defined as protein-coding sequences of three candidate regions in the Drosophila genome (US National Center for Biotechnology Information [NCBI], April 2006). We developed a probe design algorithm to search for an optimal set of padlock probes covering an arbitrary set of nonrepetitive genomic targets (Porreca et al. 2007). This algorithm weights candidate probes based on several sequence features that were previously not considered in eMIP probe design, including the melting temperature, size of the capturing arms, and gap sizes (AJ Gore and K Zhang, unpubl., in prep.). The average size of the target regions is 140 bp, with a standard deviation of 14.6. Sequences for the designed probes can be found in Supplemental File 1. These Agilent oligos were released and converted to padlock probes using the protocol described previously (Deng et al. 2009).
Capture of targeted sequences
We hybridized targeting oligos to genomic DNA in 20 μL of 1× Ampligase buffer (Epicentre) with 200 ng of genomic DNA and 2 ng of targeting oligos, incubating the reactions at 95°C for 2 min and 60°C for 20 h. Then, we added 1 μL of gap-filling mix (0.5 uM dNTPs, 0.5 U of Taq Stoffel Fragment [Applied Biosystems], and 0.5 U of Ampligase in 1× Ampligase buffer), and incubated the reaction at 60°C for 20 h. To degrade linear species, we added 2 μL of exonuclease mix (containing 50 U of exonuclease I and 500 U of exonuclease III; New England BioLabs), and incubated the reaction at 37°C for 2 h and then at 95°C for 2 min.
Amplification of enriched DNA
Enriched DNA was amplified by PCR reaction. A total of 1 μL of captured DNA was used as template in a 12.5-μL reaction. PCR conditions were: 95°C for 15min, 30 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and, finally, 72°C for 5 min. PCR products were separated on a 2% agarose gel. We recovered amplicons corresponding to the expected size range (170–210 bp), purified them, and resuspended the products in 20 μL of TE (pH 8.0).
Library preparation
The purified PCR amplicons were directly used as the DNA template for the 454 Life Sciences (Roche) library. The 454 GS FLX Titanium libraries were prepared according to the manufacturer's protocol. To prepare Illumina libraries, purified PCR amplicons were first digested with MmeI: 16 units of MmeI (2 U/μL; New England BioLabs), 100 μM SAM in 1× New England BioLabs Buffer 4 at 37°C for 1 h. Digestions were again column purified and digested with 3 U of USER enzyme (1 U/μL; New England BioLabs) at 37°C for 2 h, then with 10 U of S1 nuclease (10 U/μL; Invitrogen) in 1× S1 nuclease buffer at 37°C for 10 min. The fragmented DNA was column purified and used as DNA template for the Illumina library. Illumina libraries were generated by following the Illumina pair-end library preparation protocol.
Mutation calling
To identify candidate mutations, custom ruby scripts developed for Atlas-SNP were used to combine BLAT and cross_match alignments with SAMtools SNP calling for a custom capture SNP-calling pipeline (http://nebc.nox.ac.uk/bioinformatics/docs/cross_match.html; Kent 2002; Li et al. 2009a). 454 Life Sciences (Roche) and Illumina GAII sequencing reads were anchored to the Drosophila genome (dm3) using BLAT parameters appropriate to each platform's read length. Reads with a unique or “best” hit were then locally aligned by cross_match and the output from both platforms combined and converted to SAM/BAM format. SAMtools was used to generate pileup files with the “pileup –cf” option and the samtools.pl script was used to identify candidate SNPs with these paramaeters: “varFilter –D 500.” Increasing the maximum read depth is necessary due to the enrichment of captured regions. As an example, parameters for Illumina GAII BLAT mapping were: “–repMatch = 128 –minIdentity = 90 –stepSize = 6 –minScore = 18,” followed by cross_match alignment with flags: “−minscore = 24 –bandwidth = 6 –gap_init = −2 –penalty = −1 –gap_ext = −1 –raw –masklevel = 0.”
Mutation validation
To confirm mutations identified by the 454 Life Sciences (Roche) and Illumina GAII parallel sequencing, a direct PCR sequencing approach was used. Specific PCR primers were designed surrounding the SNPs, and target SNPs were amplified. PCR products were purified with ExoSAP-IT (USB Corp.). Sequencing was performed using an ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit v3.1 according to the manufacturer's recommendations. The ABI 3700 capillary electrophoresis system was used to carry out the electrophoretic separations, and sequencer software was used to analyze the data.
Acknowledgments
We thank Graeme Mardon for providing dac5 and sensE2 flies, for scientific discussion, and for review of the manuscript. We thank Fuli Yu and Yong Wang for insights on mutation detection. We also thank Hugo Bellen for critical reading of the manuscript. Finally, we thank the staff of the Human Genome Sequencing Center who performed the sequencing of genomic libraries. B.D. is supported by training grant T32 EYO7102-16. H.W. is supported by postdoc fellowship EY19430-01. This work is supported by the Retinal Research Foundation and NEI/NIH grant R01EY016853 to R.C.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA012301.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.102921.109.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 [DOI] [PubMed] [Google Scholar]
- Ansorge WJ 2009. Next-generation DNA sequencing techniques. New Biotechnol 25: 195–203 [DOI] [PubMed] [Google Scholar]
- Bau S, Schracke N, Kranzle M, Wu H, Stahler PF, Hoheisel JD, Beier M, Summerer D 2009. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal Bioanal Chem 393: 171–175 [DOI] [PubMed] [Google Scholar]
- Bentley DRS, Balasubramanian HP, Swerdlow GP, Smith J, Milton CG, Brown KP, Hall DJ, Evers CL, Barnes HR, Bignell JM, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel JP, Noll AC, Griffiths JA, Perera AG, Walton KN, Gilliland WD, Hawley RS, Staehling-Hampton K 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ahlford A, Schnorrer F, Kalchhauser I, Fellner M, Viragh E, Kiss I, Syvanen AC, Dickson BJ 2008. High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat Methods 5: 323–329 [DOI] [PubMed] [Google Scholar]
- Cooper JL, Greene EA, Till BJ, Codomo CA, Wakimoto BT, Henikoff S 2008. Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics 180: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H 2007. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci 104: 9387–9392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T, Kaufman TC 1983. Revertants of dominant mutations associated with the Antennapedia gene complex of Drosophila melanogaster: Cytology and genetics. Genetics 105: 581–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Kaufman TC, Denell RE, Tallerico P 1980. Genetic analysis of the Antennapedia gene complex (Ant-C) and adjacent chromosomal regions of Drosophila melanogaster. I. Polytene chromosome segments 84b-D. Genetics 95: 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009a. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gao Y, Aach J, Zhang K, Kryukov G, Xie B, Ahlford A, Yoon JK, Rosenbaum AM, Zaranek AW, et al. 2009b. Multiplex padlock capture and sequencing reveal human hypermutable CpG variations. Genome Res 19: 1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2008a. The impact of next-generation sequencing technology on genetics. Trends Genet 24: 133–141 [DOI] [PubMed] [Google Scholar]
- Mardis ER 2008b. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9: 387–402 [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Locke AE, Steinberg KM, Hagen K, Athri P, Shetty AC, Patel V, Zwick ME 2009. Combining microarray-based genomic selection (MGS) with the Illumina Genome Analyzer platform to sequence diploid target regions. Ann Hum Genet 73: 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts AE, Land SJ, Chen L, Page GP, Rasouli P, Wang L, Lu X, Ruden DM 2009. Massively parallel resequencing of the isogenic Drosophila melanogaster strain w(1118); iso-2; iso-3 identifies hotspots for mutations in sensory perception genes. Fly 3: 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. 2007. Multiplex amplification of large sets of human exons. Nat Methods 4: 931–936 [DOI] [PubMed] [Google Scholar]
- Sarin S, Prabhu S, O'Meara MM, Pe'er I, Hobert O 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods 5: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H 2008. Next-generation DNA sequencing. Nat Biotechnol 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B, Shen L, Donahue WF, Tusneem N, Stromberg MP, et al. 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res 18: 1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4: e1000139 doi: 10.1371.journal.pgen.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452: 872–876 [DOI] [PubMed] [Google Scholar]
- Winkler S, Schwabedissen A, Backasch D, Bokel C, Seidel C, Bonisch S, Furthauer M, Kuhrs A, Cobreros L, Brand M, et al. 2005. Target-selected mutant screen by TILLING in Drosophila. Genome Res 15: 718–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, et al. 2003. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci 100: 10860–10865 [DOI] [PMC free article] [PubMed] [Google Scholar]