Abstract
In a recent issue of Molecular Cell, Feige et al. (2009) utilize the murine immunoglobulin system to shed light on a long-standing puzzle: how do cells coordinate folding of different polypeptides that ultimately form a complex?
Some cells are virtual protein factories; antigen-activated B-lymphocytes, for example, produce several thousand antibody molecules per second per cell. The protein-folding research community should tip its hat at the feat accomplished by these cellular factories, particularly when the product immunoglobulins are heterodimeric complexes of heavy and light chains, specifically stitched together by both intra- and interchain disulfides and held in the archetypal final Y structure by homotypic and heterotypic protein interfaces. Obtaining this correctly assembled final fold is crucial, not only to produce the requisite yield of antibodies, but also to avoid the formation of off-pathway amyloidogenic aggregated species—a process to which immunoglobulins are definitely vulnerable.
The busy folding world of the B cell endoplasmic reticulum (ER) pointed the way to the chaperone revolution more than two decades ago when the “heavy chain binding protein,” BiP, was first described (Haas and Wabl, 1983) and found to be homologous to the 70 kDa heat-shock proteins (Munro and Pelham, 1986), all now lumped into the ubiquitous Hsp70 molecular chaperone family (Bukau and Horwich, 1998). The hypothesis was then articulated that BiP held the newly synthesized heavy chains in a state that remained competent for immunoglobulin maturation until they could be matched up with light chains seeking their compatible partners.
But the devil is in the details in the pathway of antibody biosynthesis. How are the steps in heterodimeric complex assembly optimally timed? When do the disulfides form? How is chaperone recognition mediated so as not to compete with folding and oxidation? What features of antibody sequences encode the information for this complex interplay within the cell? To answer all these questions requires a detailed energy landscape for the folding of immunoglobulin heavy and light chains, complete with connections to their productive complex formation and linkages to other required components in the ER-folding cocktail, such as BiP, protein disulfide isomerase, and peptidyl-prolyl isomerase.
Feige et al. (2009) now provide tantalizing first steps in this complete description by using complementary in vitro biophysical studies of murine IgG heavy chain domain CH1 and in vivo analysis of the secretion efficiency and BiP interactions of heavy chains containing wild-type and mutant versions of this domain. They report compelling evidence that sequence-encoded properties of the isolated domain enable it to serve as a controlling element for in vivo folding and assembly of the intact immunoglobulin.
The first exciting hint was the discovery that the CH1 domain is unable to fold on its own. It lacks the well-established hallmarks of intrinsically unfolded proteins (Dunker et al., 2008)—many neutral polar residues, glycines, few hydrophobic residues—and so its secret to folding ineptitude must be of a different nature. The next provocative in vitro observation was that native CH1 domain secondary, tertiary, and quaternary structures all formed at the same rate upon mixing the unfolded CH1 domain with its partner CL domain, arguing for binding-induced folding limited by a single rate-determining step. Indeed, in vitro refolding of other immunoglobulin domains is rate limited by isomerization of specific trans prolyl bonds (favored in the absence of structure) to cis (Isenman et al., 1979; Thies et al., 1999), and CH1 domain folding upon interaction with its partner CL domain also proved to be rate limited by a trans-to-cis prolyl bond rotation. Substitution of each of the prolines involved in a cis peptide bond in native CH1 individually to alanine, which essentially eliminates possible isomerization to cis, showed clearly that native folding required the Phe31-Pro32 bond to achieve its cis arrangement but was unaffected by the state of isomerization of the other native cis X-Pro bonds. Moreover, the slow step in CL-assisted folding of the CH1 domain was catalyzed in vitro by addition of the ER peptidyl-prolyl isomerase, cyclophilin B. Inspection of the native structure of the CH1 domain shows that the Phe31-Pro32 bond is at the core of a well-packed region, which could not organize itself if this bond were in the trans configuration.
But these data are only one part of the life story of immunoglobulin folding and assembly: an intrachain disulfide must form prior to CL-induced folding. Moreover, the newly synthesized CH1 domain must have an “on-deck” circle where it can safely await the availability of its partner as well as avoid premature secretion or break down by ER quality control. Our old friend BiP emerges as the earliest helper in this cascade of steps: reduced, intrinsically unfolded CH1 domain forms a stable complex with BiP in vitro, consistent with several studies implicating this domain in BiP binding in cells. In vitro, oxidized CH1 also can bind BiP, with only slightly lower affinity. This finding leaves open the possibility that the CH1 intrachain disulfide forms in vivo while it is bound to BiP. The authors have not addressed the timing or catalyst assistance of intrachain disulfide formation in CH1. Intriguingly, a predicted site for BiP binding (Blond-Elguindi et al., 1993) within CH1 is proximal to both Cys25, which participates in the intradomain disulfide, and Phe31-Pro32, which requires isomerization to cis for native folding.
These exciting in vitro results do not necessarily tell us about antibody folding in vivo. Hence, a capstone aspect of this felicitous collaboration between the Buchner and Hendershot labs is the demonstration that secretion of folded antibodies required the presence of the CH1 domain, either wild-type or with Pro32 preserved (and either of the other cis-bond-forming prolines substituted to alanine), and the wild-type CL domain. Introduction of the folding-incompetent CH1 domain into the light chain in place of its CL domain abrogated its ability to be secreted and instead caused it to be retained in the ER, in complex with BiP. This crippled light chain was also incapable of successful complex formation and secretion with its normal partner heavy chain. These data strongly support the relevance of the in vitro data to the biosynthetic folding and assembly pathway of antibodies.
Although this “how I met my perfect partner” story significantly enhances our understanding of antibody biosynthesis (Figure 1), many fascinating questions remain, including the timing and involvement of peptidyl-prolyl isomerase in catalysis of the Phe31-Pro32 peptide bond rearrangement, the timing of disulfide formation and extent to which a protein disulfide isomerase family member catalyzes it, the sequence origin of the folding deficiency of CH1, the localization of all of these folding events in the ER, and the extent to which the functions of all of the ER-folding assistants are coordinated by their participation in a multifunctional “foldosome” machine (Meunier et al., 2002). Nonetheless, this elegant study using complementary in vitro and in vivo approaches points the protein-folding community in the right direction and shows that seemingly daunting, complex folding questions in the cell are ripe for creative experimental strategies. Moreover, there is widespread interest in developing better systems for production of properly folded antibodies for therapeutic uses. The insights provided by studies like that from Feige et al. (2009) will greatly enhance our ability to engineer antibody production systems.
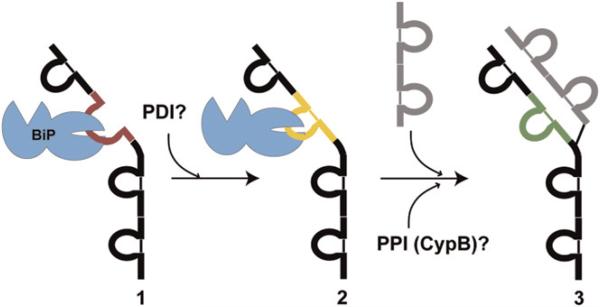
Figure 1. The Sequence of Events in Cellular Folding and Assembly of IgG Antibodies.
(1) The unfolded CH1 domain (red) of a newly synthesized IgG heavy chain is bound by BiP. (2) The intramolecular disulfide bond in the CH1 domain (yellow) forms, possibly while the domain is bound to BiP, or upon its release, and most likely catalyzed by a PDI family member. (3) The CH1 domain folds to its native structure (green) upon interaction with the complementary CL domain within its partner light chain (gray structure). Peptidyl-proline isomerization must accompany native structure formation and most likely is catalyzed by an ER PPIase such as cyclophilin B. In this cartoon, we show only one of the two heavy and two light chains in the final assembled antibody.
REFERENCES
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething M-JH. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. Mol. Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas IG, Wabl M. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Isenman DE, Lancet D, Pecht I. Biochemistry. 1979;18:3327–3336. doi: 10.1021/bi00582a020. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood Y-K, Chung KT, Hendershot LM. Mol. Biol. Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Pelham HRB. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Thies MJ, Mayer J, Augustine JG, Frederick CA, Lilie H, Buchner J. J. Mol. Biol. 1999;293:67–79. doi: 10.1006/jmbi.1999.3128. [DOI] [PubMed] [Google Scholar]