Abstract
A genome-wide, whole brain approach to investigate genetic effects on neuroimaging phenotypes for identifying quantitative trait loci is described. The Alzheimer's Disease Neuroimaging Initiative 1.5 T MRI and genetic dataset was investigated using voxel-based morphometry (VBM) and FreeSurfer parcellation followed by genome-wide association studies (GWAS). One hundred forty-two measures of grey matter (GM) density, volume, and cortical thickness were extracted from baseline scans. GWAS, using PLINK, were performed on each phenotype using quality-controlled genotype and scan data including 530,992 of 620,903 single nucleotide polymorphisms (SNPs) and 733 of 818 participants (175 AD, 354 amnestic mild cognitive impairment, MCI, and 204 healthy controls, HC). Hierarchical clustering and heat maps were used to analyze the GWAS results and associations are reported at two significance thresholds (p<10−7 and p<10−6). As expected, SNPs in the APOE and TOMM40 genes were confirmed as markers strongly associated with multiple brain regions. Other top SNPs were proximal to the EPHA4, TP63 and NXPH1 genes. Detailed image analyses of rs6463843 (flanking NXPH1) revealed reduced global and regional GM density across diagnostic groups in TT relative to GG homozygotes. Interaction analysis indicated that AD patients homozygous for the T allele showed differential vulnerability to right hippocampal GM density loss. NXPH1 codes for a protein implicated in promotion of adhesion between dendrites and axons, a key factor in synaptic integrity, the loss of which is a hallmark of AD. A genome-wide, whole brain search strategy has the potential to reveal novel candidate genes and loci warranting further investigation and replication.
Introduction
Recent advances in brain imaging and high throughput genotyping techniques enable new approaches to study the influence of genetic variation on brain structure and function (Bearden et al., 2007; Cannon et al., 2006; Glahn et al., 2007a; Meyer-Lindenberg and Weinberger, 2006; Potkin et al., 2009a). The NIH Alzheimer's Disease Neuroimaging Initiative (ADNI) is an ongoing 5-year public–private partnership to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), genetic factors such as single nucleotide polymorphisms (SNPs), other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). Given the availability of genome-wide SNP data and repeat structural and functional neuroimaging data as part of this initiative, ADNI provides a suitable data set for a large scale imaging genetics study. Using the ADNI baseline MRI data set, we present an imaging genetics framework that employs a whole genome and whole brain strategy to systematically evaluate genetic effects on brain imaging phenotypes for discovery of quantitative trait loci (QTLs).
Imaging genetics is an emergent transdisciplinary research field where the association between genetic variation and imaging measures as quantitative traits (QTs) or continuous phenotypes is evaluated. Imaging genetics studies have certain advantages over traditional case control studies. QT association studies have been shown to have increased statistical power and thus decreased sample size requirements (Potkin et al., 2009b). In addition, imaging phenotypes may be closer to the underlying biological etiology of the disease making it easier to identify underlying genes (e.g., Potkin et al., 2009a). Given these observations, the method proposed in this paper focuses on identifying strong associations between regional imaging phenotypes as QTs and SNP genotypes as QTLs and aims to provide guidance for refined statistical modeling and follow-up studies of candidate genes or loci.
SNPs and other types of polymorphisms in single genes such as APOE have been related to neuroimaging measures in both healthy controls and participants with brain disorders such as MCI and AD (e.g., Lind et al., 2006; Wishart et al., 2006). However, the analytic tools that relate a single gene to a few imaging measures are insufficient to provide insight into the multiple mechanisms and imaging manifestations of these complex diseases. Genome-wide association studies (GWAS) are increasingly performed (Balding, 2006; Hirschhorn and Daly, 2005; Purcell et al., 2007; Zondervan and Cardon, 2007), but effectively relating high throughput SNP data to large scale image data remains a challenging task. As pointed out by Glahn et al. (2007b), in imaging genetics, prior studies typically make significant reduction in one or both data types in order to complete analyses. For example, whole brain studies usually focus on a small number of genetic variables (e.g., Ahmad et al., 2006; Brun et al., in press; Filippinia et al., 2009; Nichols and Inkster, 2009; Pezawas et al., 2004; Shen et al., 2007), while whole genome studies typically examine a limited number of imaging variables (e.g., Baranzini et al., 2009; Potkin et al., 2009a; Seshadri et al., 2007). This restriction of target genotypes and/or phenotypes greatly limits our capacity to identify important relationships.
To overcome this limitation, we present a whole genome and whole brain search strategy for discovering imaging genetics associations to guide further detailed analyses. In addition, we present the results from implementation of this technique, including the identification of new genetic loci potentially involved in hippocampal and global brain atrophy associated with MCI and AD. In the present study, a detailed set of regions of interest (ROIs) extracted using voxel-based morphometry (VBM) and FreeSurfer automated parcellation defined 142 imaging phenotypes from across the brain (Risacher et al., 2009). A separate GWAS analysis using PLINK software (Purcell et al., 2007) was completed for each of these 142 imaging phenotypes. Hierarchical clustering and heat maps (Eisen et al., 1998) were used to display and evaluate the association patterns between top SNPs and top imaging phenotypes for multiple statistical thresholds. Subsequent pattern analysis of these heat maps not only confirmed prior findings (e.g., APOE and TOMM40 SNPs were among the top ranked list) but also revealed novel QTLs which warranted further analyses. Two types of refined imaging genetics analysis were performed for one of the top SNPs (NXPH1, rs6463843), including a VBM analysis assessing global grey matter (GM) density and a regional analysis of target phenotypes. These focused analyses resulted in interesting imaging genetics findings about the target SNP, including an overall and regional decrease in GM density associated with TT genotype relative to the GG genotype with an increased vulnerability to this effect in AD participants.
Materials and methods
Sample
Data used in the preparation of this article were obtained from the ADNI database (http://www.loni.ucla.edu/ADNI). The following data from 818 ADNI participants were downloaded from the ADNI database: all baseline 1.5 T MRI scans, the Illumina SNP genotyping data, demographic information, APOE genotype, and baseline diagnosis information. Two participants had genotypic data but no baseline MRI scans and were excluded from all analyses.
The ADNI was launched in 2004 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public–private partnership. The Principle Investigator of this initiative is Michael W. Weiner, M.D., VA Medical Center and University of California-San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations. Presently, more than 800 participants, aged 55 to 90 years, have been recruited from over 50 sites across the United States and Canada, including approximately 200 cognitively normal older individuals (i.e., healthy controls or HCs) to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. Baseline and longitudinal imaging, including structural MRI scans collected on the full sample and PIB and FDG PET imaging on a subset are collected every 6–12 months. Additional baseline and longitudinal data including other biological measures (i.e. cerebrospinal fluid (CSF) markers, APOE and full-genome genotyping via blood sample) and clinical assessments including neuropsychological testing and clinical examinations are also collected as part of this study. Written informed consent was obtained from all participants and the study was conducted with prior institutional review board's approval. Further information about ADNI can be found in the study of Jack et al. (2008) and Mueller et al. (2005a,b) and at www.adni-info.org.
DNA isolation and SNP genotyping
Single nucleotide polymorphism (SNP) genotyping for more than 620,000 target SNPs as was completed on all ADNI participants using the following protocol. Seven milliliters of blood was taken in EDTA containing vacutainer tubes from all participants and genomic DNA was extracted using the QIAamp DNA Blood Maxi Kit (Qiagen, Inc., Valencia, CA) following the manufacturer's protocol. Lymphoblastoid cell lines were established by transforming B lymphocytes with Epstein-Barr virus as described by Neitzel (1986). Genomic DNA samples were analyzed on the Human610-Quad BeadChip (Illumina, Inc. San Diego, CA) according to the manufacturer's protocols (Infinium HD Assay; Super Protocol Guide; Rev. A, May 2008). Before initiation of the assay, 50 ng of genomic DNA from each sample was examined qualitatively on a 1% Tris–acetate–EDTA agarose gel to check for degradation. Degraded DNA samples were excluded from further analysis. Samples were quantitated in triplicate with PicoGreen® reagent (Invitrogen, Carlsbad, CA) and diluted to 50 ng/μl in Tris–EDTA buffer (10 mM Tris,1 mM EDTA, pH 8.0). DNA (200 ng) was then denatured, neutralized, and amplified for 22 h at 37 °C (this is termed the MSA1 plate). The MSA1 plate was fragmented with FMS reagent (Illumina) at 37 °C for 1 h, precipitated with 2-propanol, and incubated at 4 °C for 30 min. The resulting blue precipitate was resuspended in RA1 reagent (Illumina) at 48 °C for 1 h. Samples were then denatured (95 °C for 20 min) and immediately hybridized onto the BeadChips at 48 °C for 20 h. The BeadChips were washed and subjected to single base extension and staining. Finally, the BeadChips were coated with XC4 reagent (Illumina), dessicated, and imaged on the BeadArray Reader (Illumina). The Illumina BeadStudio 3.2 software was used to generate SNP genotypes from bead intensity data. All SNP genotypes are publicly available for download at the ADNI website (http://www.loni.ucla.edu/ADNI).
MRI analysis and extraction of imaging phenotypes
Two widely employed automated MRI analysis techniques were used to process and extract brain-wide target MRI imaging phenotypes from all baseline scans of ADNI participants as previously described (Risacher et al., 2009). First, voxel-based morphometry (VBM; Ashburner and Friston, 2000; Good et al., 2001; Mechelli et al., 2005) was performed to define global grey matter (GM) density maps and extract local GM density values for 86 target regions (Table 1). Second, automated parcellation via FreeSurfer V4 (http://surfer.nmr.mgh.harvard.edu/) was conducted to define 56 volumetric and cortical thickness values (Table 2). All included ADNI participants had a minimum of two 1.5 T MP-RAGE scans at baseline following the ADNI MRI protocol (Jack et al., 2008). Each raw scan was independently processed using FreeSurfer and VBM.
Table 1.
VBM phenotypes defined as mean GM densities of various regions of interest (ROIs). SPM5 was applied for computing voxel-wise GM density values, while the MarsBaR ROI toolbox was used to define ROIs in the MNI space. A total number of 43 × 2 = 86 phenotypes were calculated. Each of the 43 IDs shown in the table corresponds to two phenotypes: one for the left side and the other for the right side. For example, “LAmygdala” indicates the mean GM density of the left amygdala. Each region marked with * in the table is a combined set of more than one MarsBaR ROI. For example, “RMeanLatTemporal” indicates the mean GM density of the right lateral temporal region defined by a set of MarsBaR ROIs, including right inferior temporal gyrus, right middle temporal gyrus, and right superior temporal gyrus.
| Phenotype ID | Region of interest (Phenotype is defined as the mean GM density of the ROI) | Phenotype ID | Region of interest (Phenotype phenotype is defined as the mean GM density of the ROI) |
|---|---|---|---|
| Amygdala | Amygdala | MidTempPole | Middle temporal pole |
| Angular | Angular gyrus | MidTemporal | Middle temporal gyrus |
| AntCingulate | Anterior cingulate | Olfactory | Olfactory gyrus |
| Fusiform | Fusiform gyrus | Parahipp | Parahippocampal gyrus |
| Heschl | Heschl's gyrus | PostCingulate | Posterior cingulate |
| Hippocampus | Hippocampus | Postcentral | Postcentral gyrus |
| InfFrontal_Oper | Inferior frontal operculum | Precentral | Precentral gyrus |
| InfFrontal_Triang | Inferior frontal triangularis | Precuneus | Precuneus |
| InfOrbFrontal | Inferior orbital frontal gyrus | Rectus | Rectus gyrus |
| InfParietal | Inferior parietal gyrus | Rolandic_Oper | Rolandic operculum |
| InfTemporal | Inferior temporal gyrus | Supfrontal | Superior frontal gyrus |
| Insula | Insula | SupOrbfrontal | Superior orbital frontal gyrus |
| Lingual | Lingual gyrus | SupParietal | Superior parietal gyrus |
| MedOrbFrontal | Medial orbital frontal gyrus | SupTempPole | Superior temporal pole |
| MedSupFrontal | Medial superior frontal gyrus | SupTemporal | Superior temporal gyrus |
| MidCingulate | Middle cingulate | SuppMotorArea | Supplementary motor area |
| MidFrontal | Middle frontal gyrus | Supramarg | Supramarginal gyrus |
| MidOrbFrontal | Middle orbital frontal gyrus | Thalamus | Thalamus |
| Phenotype ID | Regions of interest (phenotype is defined as the average GM density of multiple MarsBaR ROIs) | ||
| MeanCing* | Anterior cingulate, middle cingulate, and posterior cingulate | ||
| MeanFrontal* | Inferior frontal operculum, inferior orbital frontal gyrus, inferior frontal triangularis, medial orbital frontal gyrus, middle frontal gyrus, middle orbital frontal gyrus, superior frontal gyrus, medial superior frontal gyrus, superior orbital frontal gyrus, rectus gyrus, rolandic operculum, and supplementary motor area | ||
| MeanLatTemporal* | Inferior temporal gyrus, middle temporal gyrus, and superior temporal gyrus | ||
| MeanMedTemporal* | Amygdala, fusiform gyrus, Heschl's gyrus, hippocampus, lingual gyrus, olfactory gyrus, parahippocampal gyrus, middle temporal pole, and superior temporal pole | ||
| MeanOccipital* | Calcarine gyrus, cuneus, inferior occipital gyrus, middle occipital gyrus, and superior occipital gyrus | ||
| MeanParietal* | Angular gyrus, inferior parietal gyrus, superior parietal gyrus, supramarginal gyrus, and precuneus | ||
| MeanTemporal* | Amygdala, fusiform gyrus, Heschl's gyrus, hippocampus, lingual gyrus, olfactory gyrus, parahippocampal gyrus, inferior temporal gyrus, middle temporal gyrus, middle temporal pole, superior temporal pole, and superior temporal gyrus | ||
Table 2.
FreeSurfer phenotypes defined as volumetric or cortical thickness measures of various regions of interest (ROIs). FreeSurfer was applied for automated parcellation to extract volume and cortical thickness values for a total number of 28×2=56 ROIs. Each of the 28 IDs shown in the table corresponds to two phenotypes: one for the left side and the other for the right side. For example, “LAmygVol” indicates the volume of the left amygdala, while “RSupTemporal” indicates the (mean) thickness of the right superior temporal gyrus.
| Phenotype ID | Phenotype description |
|---|---|
| AmygVol | Volume of amygdala |
| CerebCtx | Volume of cerebral cortex |
| CerebWM | Volume of cerebral white matter |
| HippVol | Volume of hippocampus |
| InfLatVent | Volume of inferior lateral ventricle |
| LatVent | Volume of lateral ventricle |
| EntCtx | Thickness of entorhinal cortex |
| Fusiform | Thickness of fusiform gyrus |
| InfParietal | Thickness of inferior parietal gyrus |
| InfTemporal | Thickness of inferior temporal gyrus |
| MidTemporal | Thickness of middle temporal gyrus |
| Parahipp | Thickness of parahippocampal gyrus |
| PostCing | Thickness of posterior cingulate |
| Postcentral | Thickness of postcentral gyrus |
| Precentral | Thickness of precentral gyurs |
| Precuneus | Thickness of precuneus |
| SupFrontal | Thickness of superior frontal gyrus |
| SupParietal | Thickness of superior parietal gyurs |
| SupTemporal | Thickness of superior temporal gyrus |
| Supramarg | Thickness of supramarginal gyrus |
| TemporalPole | Thickness of temporal pole |
| MeanCing | Mean thickness of caudal anterior cingulate, isthmus cingulate, posterior cingulate, and rostral anterior cingulate |
| MeanFront | Mean thickness of caudal midfrontal, rostral midfrontal, superior frontal, lateral orbitofrontal, and medial orbitofrontal gyri and frontal pole |
| MeanLatTemp | Mean thickness of inferior temporal, middle temporal, and superior temporal gyri |
| MeanMedTemp | Mean thickness of fusiform, parahippocampal, and lingual gyri, temporal pole and transverse temporal pole |
| MeanPar | Mean thickness of inferior and superior parietal gyri, supramarginal gyrus, and precuneus |
| MeanSensMotor | Mean thickness of precentral and postcentral gyri |
| MeanTemp | Mean thickness of inferior temporal, middle temporal, superior temporal, fusiform, parahippocampal, and lingual gyri, temporal pole and transverse temporal pole |
For VBM analysis, SPM5 (http://www.fil.ion.ucl.ac.uk/spm/) was used to create an unmodulated normalized GM density map (1×1×1 mm voxel size, 10 mm FWHM Gaussian kernel for smoothing) in the MNI space for each scan as previously described (Risacher et al., 2009). A mean GM density map was created as an average of two independent smoothed, unmodulated normalized GM density maps for each participant using SPM5. The MarsBaR region of interest (ROI) toolbox (Brett et al., 2002; Tzourio-Mazoyer et al., 2002) as implemented in SPM5 was then used to extract a single mean GM density value for 86 target regions in MNI space (Table 1) to be used as target QTs for the imaging genetic analyses. In addition to the individual MarsBaR ROIs, larger target regions defined by combining the mean GM density value from a set of MarsBaR ROIs were used as imaging phenotypes. All individual and combined mean GM density values are referred to as VBM phenotypes; see Table 1 for a total list and explanation of the 86 VBM phenotypes.
For automated segmentation and parcellation, FreeSurfer V4 was employed to automatically label cortical and subcortical tissue classes using an atlas-based Bayesian segmentation procedure (Dale et al., 1999; Fischl and Dale, 2000; Fischl et al., 2002, 1999) and to extract target region volume and cortical thickness, as well as to extract total intracranial volume (ICV) for all participants. Extracted FreeSurfer values for two independently processed MP-RAGE images of the same participant were averaged to create a mean value for volumetric and cortical thickness measures for all target regions. Mean volumetric and cortical thickness measures extracted using automated parcellation are referred to as FreeSurfer phenotypes; see Table 2 for a total list of the 56 FreeSurfer phenotypes defined for selected target regions.
Genome-wide association analysis of imaging phenotypes
APOE genotype
The APOE gene is an important target gene in AD research (Farrer et al., 1997). However, the two previously identified APOE SNPs important in AD susceptibility (rs429358, rs7412) were not available on the Illumina array. Therefore, we determined the genotypes of the two APOE SNPs (rs429358, rs7412) using the APOE ε2/ε3/ε4 status information from the ADNI clinical database for each participant.
Quality control
The original genotype data contained 620,903 markers, including 620,901 genomic markers on the Illumina chip plus 2 APOE SNPs whose values were obtained from the APOE status data. Only SNP markers were analyzed in this study. The following quality control (QC) steps were performed on these genotype data using the PLINK software package (http://pngu.mgh.harvard.edu/∼purcell/plink/), release v1.06. SNPs were excluded from the imaging genetics analysis if they could notmeet any of the following criteria: (1) call rate per SNP ≥90%, (2) minor allele frequency (MAF) ≥5%, and (3) Hardy–Weinberg equilibrium test of p≤10−6 using healthy control (HC) subjects only. Participants were excluded from the analysis if any of the following criteria was not satisfied: (1) call rate per participant ≥90% (1 participant was excluded); (2) gender check (2 participants were excluded); and (3) identity check (3 sibling pairs were identified with PI_HAT over 0.5; one participant from each pair was randomly selected and excluded). Population stratification analysis suggested the advisability of restricting analyses to non-Hispanic Caucasians (79 participants were excluded from this report). After the QC procedure, 733 out of 818 participants and 530,992 out of 620,903 markers remained in the analysis and the overall genotyping rate for the remaining dataset was over 99.5%.
GWAS analyses
One hundred forty-two separate GWAS analyses on 142 selected imaging phenotypes (86 VBM phenotypes and 56 FreeSurfer phenotypes) were completed using the quality-controlled SNP data. All the imaging phenotypes were adjusted for the baseline age, gender, education, handedness, and baseline intracranial volume (ICV) using the regression weights derived from the HC participants, prior to any of the GWAS analyses (Risacher et al., 2009). Using the PLINK software package (v1.06) with the quantitative trait association option, each GWAS analysis calculated the main effects of all SNPs on the target quantitative imaging phenotype. An additive SNP effect was assumed and the empirical p-values were based on the Wald statistic (Purcell et al., 2007). Right hippocampal GM density was selected for a detailed sample analysis of a target QC because it had the largest number of associations at p<10−6. A Manhattan plot and a quantile-quantile (Q–Q) plot were used to visualize GWAS results for the right hippocampal GM density. All association results surviving the significance threshold of p<10−6 were saved and prepared for additional pattern analysis.
Sample definition and demographics
The sample employed in the GWAS analyses of FreeSurfer phenotypes included participants that passed the genotype QC procedure and FreeSurfer processing. The sample used in the GWAS analyses of VBM phenotypes included participants that passed the genotype QC procedure, FreeSurfer processing, and VBM processing. Demographic information, including baseline age, years of education, gender distribution, and handedness distribution, was compared between baseline diagnostic groups for each sample separately using one-way ANOVAs and chi-squared analyses as applicable in SPSS (version 16.0.1).
Pattern analyses of GWAS results
To expedite the review of GWAS results and data reduction for subsequent analyses, we employed heat map and hierarchical clustering approaches (Eisen et al., 1998; Levenstien et al., 2003; Sloan et al., submitted for publication) for visualizing associations between identified SNPs and their associated imaging phenotypes at various significance levels. Heat maps are colored images mapping given values (in this study, −log10(p) of the corresponding association) to coded colors. Generally, heat maps have dendrograms, representing hierarchical clustering results along both the x-axis and y-axis (in this study, x: imaging phenotypes, y: SNPs). R (v.2.9.0) (http://www.r-project.org/), an open source statistical computing package, was employed to create the heat maps. Hierarchical clustering was completed using Euclidean distance methods to define dissimilarity between two nodes and average of distances between all pairs of objects in two clusters to measure the distance between two clusters. On each heat map, significant associations between imaging phenotypes and SNPs were marked with an “x” to facilitate visual evaluation of the results. The color bar on the left side of the heat map encodes the chromosome IDs for the corresponding SNPs. In addition to the heat maps, a summary statistic detailing the number of significant associations at the p<10−6 level for each imaging phenotype and SNP was evaluated to help guide the refined analyses. In the present study, all imaging GWAS results are presented and analyzed using heat maps and summary statistics.
Detailed analysis of a target SNP identified by cluster analysis
An in-depth analysis was performed for one of the top SNPs selected by inspecting the heat maps and summary statistics. The refined analysis included two steps: (1) a global voxel-based analysis on the entire brain using VBM and (2) regional analyses of identified target phenotypes. We included both types of analyses as they provide complementary information relevant to assessing risk for AD or disease progression (Risacher et al., 2009; Saykin et al., 2006).
For global analyses, VBM was performed on a voxel-by-voxel basis using a general linear model (GLM) approach as implemented in SPM5. After identifying the SNP of interest, a two-way ANOVA assessing the effects of baseline diagnostic group and SNP genotype value was performed to compare the smoothed, unmodulated normalized GM maps to determine any significant effects of diagnosis, SNP genotype, and SNP-by-diagnosis interactions on global GM density between and within groups. Contrasts between genotypes were displayed with a significance threshold of p<0.01 corrected for multiple comparisons using a false discovery rate (FDR) technique when including the entire sample. For contrasts within a single diagnostic group, the p<0.01 (FDR) threshold was too stringent given the reduced power and no significant voxels were observed. Therefore, we used a slightly less stringent significance threshold of p<0.001 (uncorrected for multiple comparisons) when examining SNP effects with in a diagnostic group, in order to evaluate the pattern of GM density associated with genotype. A minimum cluster size (k) of 27 voxels was required for significance in all comparisons and an explicit GM mask was used to restrict analyses to GM regions. Age, gender, education, handedness and baseline ICV were included as covariates in all analyses.
For ROI analyses, a two-way multivariate ANOVA in SPSS (version 16.0.1) was completed to determine the effect of baseline diagnosis and genotype on bilateral hippocampal and mean medial temporal lobar GM density. Similar to the VBM analysis, age, gender, education, handedness, and baseline ICV were included as covariates in all comparisons. Independent effects of baseline diagnosis and genotype, as well as the interaction effect of baseline diagnosis×genotype for each SNP, were assessed for selected imaging variables. All graphs were created using SigmaPlot (version 10.0).
Results
Sample characteristics after QC
After quality control of the genotyping data including the exclusion of 79 participants to avoid potential population stratification confounds, 733 out of 818 ADNI participants remained in the present study. Among these 733 participants, 729 sets of scans were successful in FreeSurfer segmentation and parcellation and were included in GWAS analyses of FreeSurfer phenotypes (56 volumetric and cortical thickness values described in Table 2). Seven hundred fifteen participants had successful VBM processing and were used in GWAS analyses of VBM phenotypes (86 GM density values described in Table 1). Table 3 shows the demographics information of the sample analyzed for both FreeSurfer and VBM studies. In both samples, gender and education are significantly different (overall p<0.05) among baseline diagnostic groups (HC, MCI, AD). In the subsequent GWAS analyses, baseline age and gender, as well as education, handedness, and baseline ICV are included as covariates.
Table 3.
Demographic information and total number of participants involved in each analysis. Of 818 ADNI participants, 733 remained after quality control of the genotyping data and consideration of population stratification. Among these 733 participants, 729 subjects succeeded in FreeSurfer segmentation and parcellation and were involved in the GWAS analysis of FreeSurfer phenotypes. Of these, 715 subjects had successful VBM processing and were involved in the GWAS analysis of VBM phenotypes. Basic demographics information is shown for both groups of participants.
| Category | FreeSurfer phenotypes (729 subjects) | VBM phenotypes (715 subjects) | ||||||
|---|---|---|---|---|---|---|---|---|
| HC | MCI | AD | p-value | HC | MCI | AD | p-value | |
| Number of subjects | 203 | 351 | 175 | – | 203 | 346 | 166 | – |
| Gender (M/F) | 111/92 | 229/122 | 97/78 | 0.019 | 111/92 | 225/121 | 90/76 | 0.017 |
| Baseline age (years; mean±SD) | 76.1 ± 5.0 | 75.1 ± 7.3 | 75.5 ± 7.6 | 0.283 | 76.1 ± 5.0 | 75.1 ± 7.4 | 75.5 ± 7.6 | 0.285 |
| Education (years; mean±SD) | 16.1 ± 2.7 | 15.7 ± 3.0 | 14.9 ± 3.0 | 0.0004 | 16.1 ± 2.7 | 15.7 ± 3.0 | 14.9 ± 3.0 | 0.0003 |
| Handedness (R/L) | 188/15 | 318/33 | 163/12 | 0.53 | 188/15 | 314/32 | 157/9 | 0.31 |
GWAS of imaging phenotypes
For convenience, in this paper, an SNP is described by its rs number together with its respective gene (i.e., the closest gene, as annotated in Illumina's Human610-Quad SNP list). Shown in Fig. 1 are all the imaging genetics associations at a significance threshold of p<10−7 (a typical threshold for genome-wide significance), which are discovered by GWAS analysis of 142 imaging phenotypes (i.e., quantitative traits, or QTs).
Fig. 1.
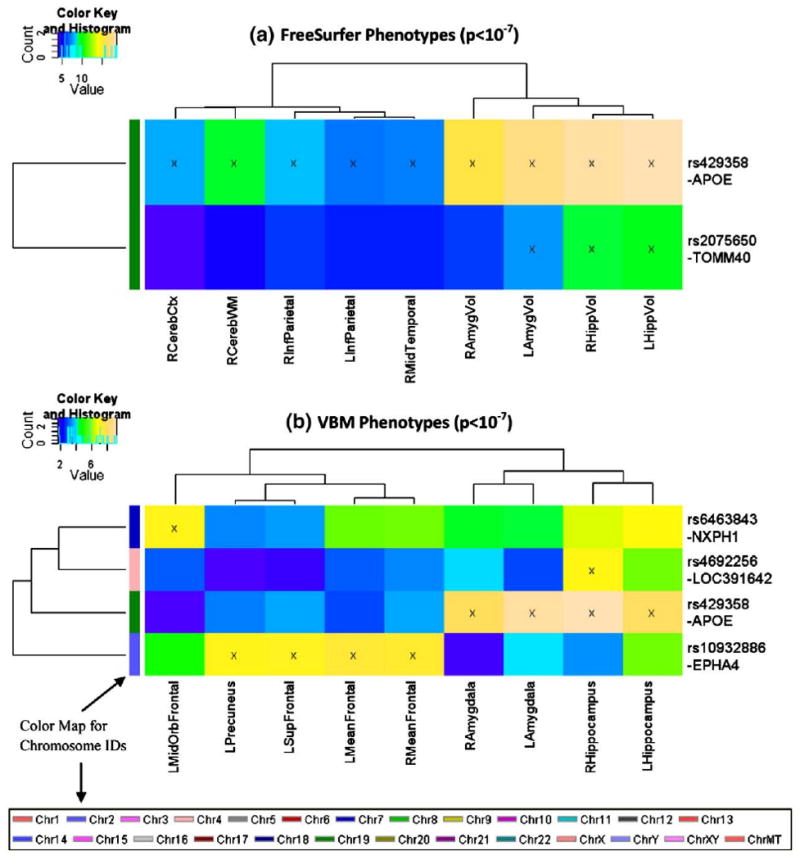
Heat maps of SNP associations with quantitative traits (QTs) at the significance level of p<10−7. GWAS results at a statistical threshold of p<10−7 using QTs derived from FreeSurfer (top) and VBM/MarSBaR (bottom) are shown. −log10(p-values) from each GWAS are color-mapped and displayed in the heat maps. Heat map blocks labeled with “x” reach the significance level of p<10−7. Only top SNPs and QTs are included in the heat maps, and so each row (SNP) and column (QT) has at least one “x” block. Dendrograms derived from hierarchical clustering are plotted for both SNPs and QTs. The color bar on the left side of the heat map codes the chromosome IDs for the corresponding SNPs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
At the p<10−7 significance level, 22 strong SNP-QT associations (see blocks labeled with “x” in Fig. 1) were identified in the GWAS analyses, and five SNPs were involved in these associations. As a well-established AD risk factor (Farrer et al., 1997), the APOE SNP rs429358 confirmed to have multiple associations with both FreeSurfer QTs and VBM QTs, showing as the most prominent imaging genetics pattern at the significance level of p<10−7. In addition, associations with multiple FreeSurfer QTs were identified for rs2075650 (TOMM40), supporting the recent finding of TOMM40 as a gene adjacent to APOE and an additional contributor to AD (Osherovich, 2009; Potkin et al., 2009a). Three additional SNPs were found to have strong associations with one or more VBM QTs: rs6463843 (NXPH1), rs4692256 (LOC391642), and rs10932886 (EPHA4). Further information about these SNPs is available in Table 4.
Table 4.
Top quantitative trait (QT) loci ranked by the total number of associations at the significance level of p<10−6. Relevant information about top ranked SNPs and their respective genes (i.e., the closet gene, as annotated in Illumina's Human610-Quad SNP list (except APOE information extracted from dbSNP)) is shown in this table, including SNP, chromosome (CHR), coordinate (Build 36.2), gene, location, and position. In addition, the number of QTs that are associated with each SNP at the significance level of p<10−6 is also shown. The SNPs are ordered according to the last column.
| SNP | CHR | Coordinate | Gene | Location | Position | Number of QT associations | ||
|---|---|---|---|---|---|---|---|---|
| VBM | FreeSurfer | Total | ||||||
| rs10932886 | 2 | 221428332 | EPHA4 | Flanking_3UTR | −562,659 | 27 | 0 | 27 |
| rs429358 | 19 | 50103781 | APOE | Coding | Exon 4 | 4 | 15 | 19 |
| rs7610017 | 3 | 190826118 | TP63 | Flanking_5UTR | −5792 | 19 | 0 | 19 |
| rs6463843 | 7 | 8805242 | NXPH1 | Flanking_3UTR | −46124 | 9 | 0 | 9 |
| rs2075650 | 19 | 50087459 | TOMM40 | Intron | −31 | 0 | 5 | 5 |
| rs16912145 | 10 | 59752674 | UBE2D1 | Flanking_5UTR | −12071 | 4 | 0 | 4 |
| rs12531488 | 7 | 144523019 | LOC643308 | Flanking_5UTR | −154052 | 3 | 0 | 3 |
| rs7526034 | 1 | 63359561 | LOC199897 | Flanking_5UTR | −103696 | 0 | 2 | 2 |
| rs7647307 | 3 | 69705878 | LOC642487 | Flanking_5UTR | −31337 | 0 | 2 | 2 |
| rs4692256 | 4 | 27353816 | LOC391642 | Flanking_3UTR | −156945 | 1 | 0 | 1 |
A number of imaging phenotypes were identified to have strong associations with target SNPs in the GWAS analyses, suggesting that these values may be sensitive QTs to imaging genetics studies of AD. As expected, both the left and right amygdalar and hippocampal regions were found to be strongly associated with rs429358 (APOE) using volumetric and GM density measures. In addition, rs2075650 (TOMM40) was significantly associated with bilateral hippocampal volume and left amygdalar volume. Additional imaging phenotypes found to be sensitive QTs, include (a) volume measures from the right cerebral cortex and cerebral white matter, (b) cortical thickness measures from left and right inferior parietal gyri, and right middle temporal gyrus, and (c) GM density measures from the left middle orbital frontal gyrus, left precuneus, left superior frontal gyrus, and left and right mean frontal lobe regions (see MeanFrontal definition in Table 1).
Heat maps of clustered associations at a somewhat less stringent significance level (p<10−6) are shown in Fig. 2. As expected, more SNPs and QTs are involved. The top 10 SNPs and their respective genes ranked by the total number of significant QT associations at p<10−6 are shown in Table 4. With more SNPs and QTs available in the heat maps, interesting clustering patterns in both the imaging and genetics dimensions were revealed by examining the corresponding dendrograms (i.e., hierarchical clustering results). In the imaging dimension (x-axis), many pairs of left and right measures of the same structure were clustered together, supporting the symmetric relationship between these phenotypes and genetic variation. In addition, regional similarity was also detected including a prominent pattern of multiple orbital frontal measures clustered together in Fig. 2b. In the genomic dimension (y-axis), three SNPs from LOC391642 were grouped together in Fig. 2b, suggesting an increased likelihood of linkage disequilibrium (LD) effects.
Fig. 2.
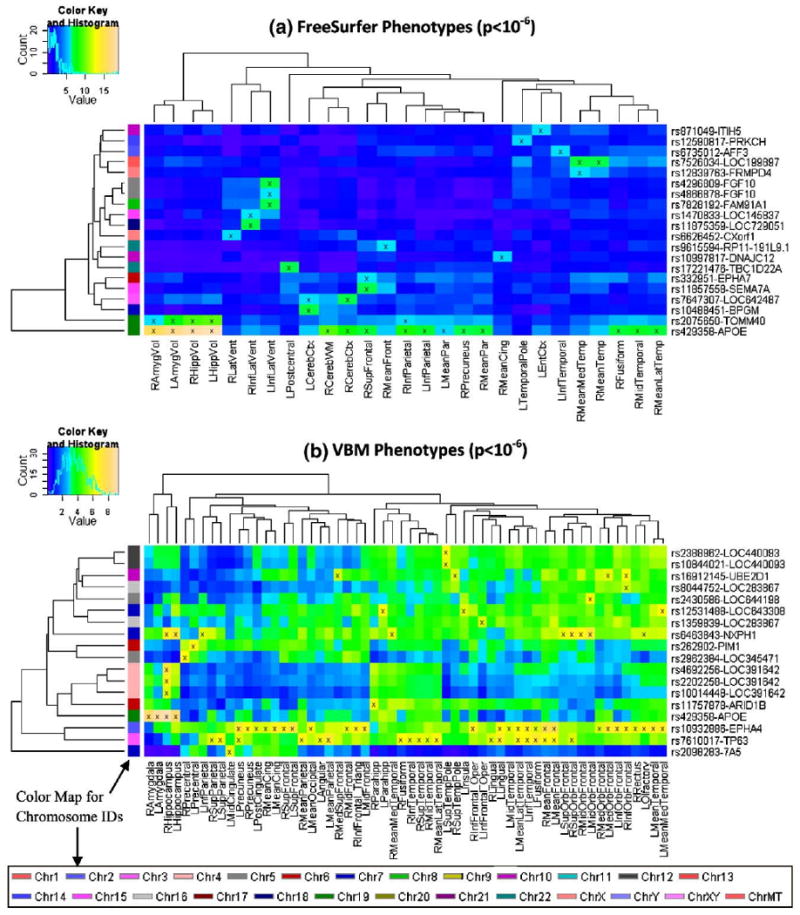
Heat maps of SNP associations with quantitative traits (QTs) at the significance level of p<10−6. GWAS results at a statistical threshold of p<10−6 using QTs derived from FreeSurfer (top) and VBM/MarSBaR (bottom) are shown. −log10(p-values) from each GWAS are color-mapped and displayed in the heat maps. Heat map blocks labeled with “x” reach the significance level of p<10−6. Only top SNPs and QTs are included in the heat maps, and so each row (SNP) and column (QT) has at least one “x” block. Dendrograms derived from hierarchical clustering are plotted for both SNPs and QTs. The color bar on the left side of the heat map codes the chromosome IDs for the corresponding SNPs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Refined analysis for a sample target QT
Subsequent analyses focused on a target QT and a target SNP selected from heat maps in Fig. 2. Shown in Fig. 3 are the Manhattan and Q–Q plots of the GWAS for the target QT, right hippocampal GM density (RHippocampus in Fig. 2b). In the Q–Q plot, for most of the p-values, the observed p-values from GWAS are almost the same as the expected p-values from the null hypothesis. There was little or no evidence of systematic bias, which could be caused by factors such as a strong population substructure and genotyping artifacts. The p-values in the upper tail of the distribution do show a significant deviation suggesting strong associations between these SNPs and the QT.
Fig. 3.
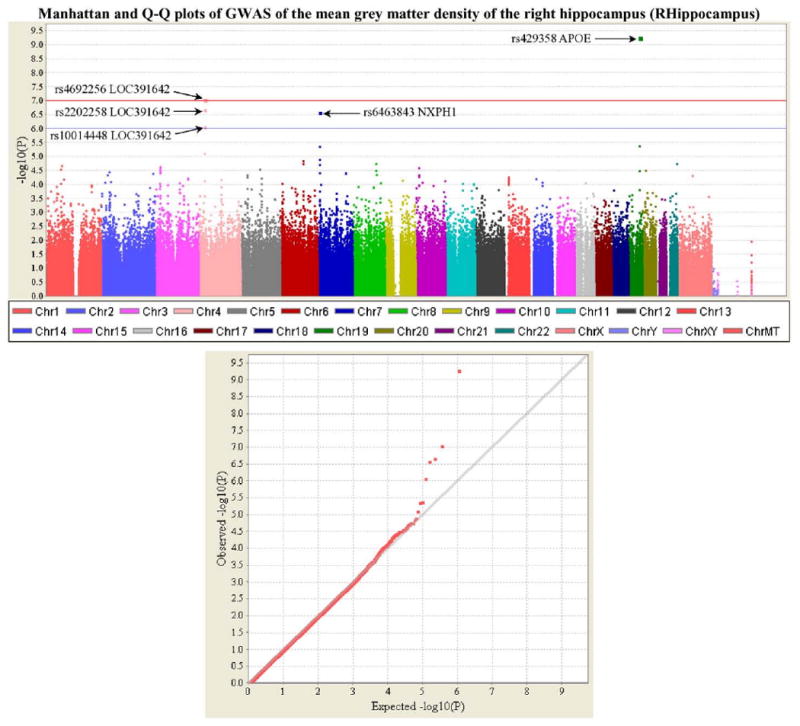
Manhattan and Q–Q plots of genome-wide association study (GWAS) of an example quantitative trait (QT). The QT examined in this analysis is the mean GM density of the right hippocampus (i.e., VBM phenotype RHippocampus, see Table 1) which was calculated using VBM/MarsBaR and adjusted for age, gender, education, handedness and ICV. Shown on the top panel is the Manhattan plot of the p-values (−log10(observed p-value)) from GWAS analysis of the QT. The horizontal lines display the cutoffs for two significant levels: blue line for p<10−6, and red line for p<10−7. Shown on the bottom panel is the quantile–quantile (Q–Q) plot of the distribution of the observed p-values (−log10(observed p-value)) in this sample versus the expected p-values (−log10(expected p-value)) under the null hypothesis of no association. Genomic inflation factor (based on median chi-squared) is 1.01667. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Refined analysis for a sample target SNP
A target SNP, rs6463843 (NXPH1), was selected for detailed imaging analyses since it was the only SNP strongly associated with both left and right hippocampi other than rs429358 (APOE) and rs2075650 (TOMM40). The results of a two-way ANOVA using VBM to compare the effects of baseline diagnostics group and rs6463843 (NXPH1) genotype on global GM density are shown in Fig. 4. After evaluating hippocampal GM density group means for each diagnosis-genotype group, we chose to contrast GG vs. TT (GG>TT) using all participants (n=715; 166 AD (44 TT, 78 GT, 44 GG); 346 MCI (82 TT, 170 GT, 94 GG); 203 HC (35 TT, 105 GT, 63 GG)). As shown in Fig. 4a, TT participants had significantly reduced global GM density throughout the brain relative to GG participants (p<0.01 (FDR), k=27). Maximal differences between groups were found in a number of regions known to be associated with AD, including the medial temporal lobe (−36, −30, −17; T=5.20) and frontal (19, 56, −15; T=5.56), parietal (26, −59, 67; T=5.71) and temporal (−59, 2, −30; T=4.81) lobe cortical surfaces. In order to determine whether a particular diagnostic group was responsible for the effects seen in the full sample contrast of GG>TT, we evaluated the same comparison within each baseline diagnostic group (Fig. 4b; AD, MCI, HC). The pattern of significant voxels for GG>TT was largest in the AD group, with highly significant clusters in the right hippocampus (31, −26, −15; T=5.34), left medial temporal lobe (−25, −32, −7; T=4.37), and frontal lobe (−35, 49, −13; T=4.33). MCI and HC groups also showed significant voxels in the contrast of GG>TT, with maximum voxels found in the inferior frontal lobe (45, 25, −13; T=3.82) and middle frontal lobe (−25, 6, 62; T=4.58), respectively. The AD panel in Fig. 4b showed more prominent patterns, while the MCI and HC panels appeared less structured. This suggested a possible SNP-by-diagnosis interaction effect on brain structure, which is examined below at a more detailed level for several candidate imaging phenotypes. Furthermore, the inclusion of APOE genotype as a covariate did not significantly alter these effects (data not shown).
Fig. 4.
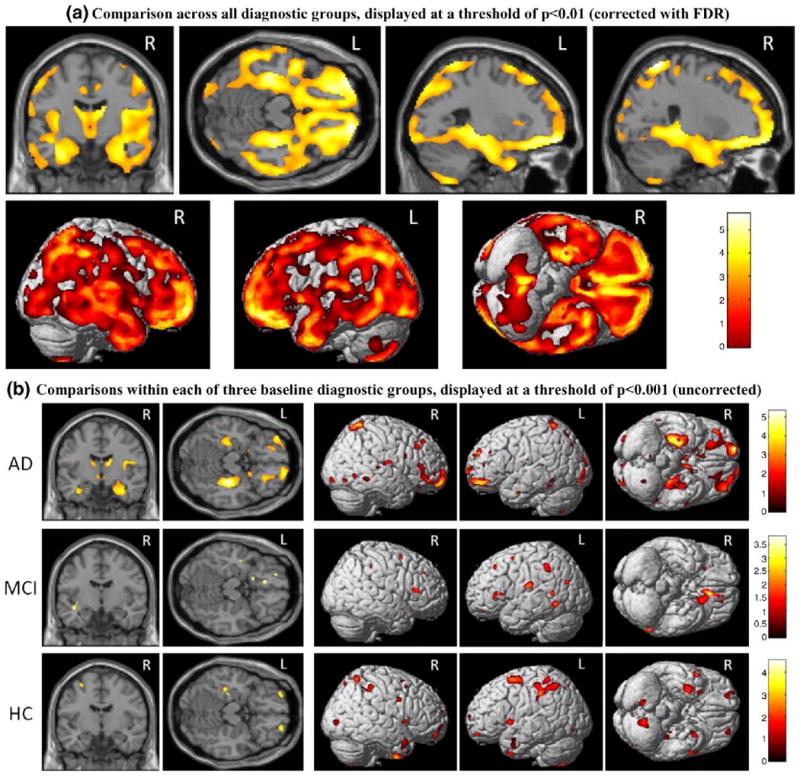
VBM genetics analysis for rs6463843 (NXPH1). A two-way ANOVA was performed on mean GM density maps to compare rs6463843 SNP genotype and baseline diagnostic group within the ADNI cohort. Analysis of the contrast of two genotype groups, GG>TT, is shown (n=715; 166AD(44 TT, 78 GT, 44 GG); 346 MCI (82 TT, 170 GT, 94 GG); 203 HC (35 TT, 105 GT, 63 GG)). Age, gender, education, handedness, and baseline ICV are included as covariates in all comparisons. Shown in the top panel (a) are the results of comparison involving all 715 subjects (i.e., across all the diagnostic groups), which are displayed at a threshold of p<0.01 (corrected with FDR) with minimum cluster size (k)=27. Shown in the bottom panel (b) are the results of comparisons within each of the three baseline diagnostic groups (AD, MCI, and HC), which are displayed at a threshold of p<0.001 (uncorrected), with minimum cluster size (k)=27.
Based on the heat map and VBM results, four GM density measures were further evaluated as phenotypes for additional associations with rs6463843 (NXPH1). As shown in Fig. 5, expected baseline diagnostic differences in left (Fig. 5a; F(7,708)=79.4, p<0.001) and right (Fig. 5b; F(7,708)=78.4, p<0.001) hippocampal GM density, as well as left (Fig. 5c; F(7,708)=60.3, p<0.001) and right (Fig. 5d; F(7,708)=59.4, p<0.001) mean medial temporal lobe GM density were found. Pairwise comparisons indicated that AD participants had significantly reduced hippocampal and mean medial temporal lobe GM density relative to both MCI and HC participants (all p<0.001). MCI participants also showed a significantly reduced GM density in all these regions relative to HCs (p<0.001). The main effect of genotype across all participants was also significant for left and right hippocampal GM density (left, F (7,708)=10.4; right, F(7,708)=9.9, both p<0.001) and left and right mean medial temporal lobe GM density (left, F(7,708)=7.9; right, F(7,708)=9.0, both p<0.001). Paired comparisons indicated significantly reduced left and right hippocampal and mean medial temporal lobar GM density in participants with a TT genotype relative to those with a GG genotype in the rs6463843 (NXPH1) SNP (p<0.01). In addition, participants with the TT genotype had significantly reduced left and right mean medial temporal lobe GM density relative to TG heterozygotes (p<0.01). The interaction effect of baseline diagnosis and rs6463843 genotype was also significant for right hippocampal GM density (p<0.05), but not for the other three regions, which suggested that AD patients with TT genotype were particularly vulnerable to increased GM density loss in right hippocampus.
Fig. 5.
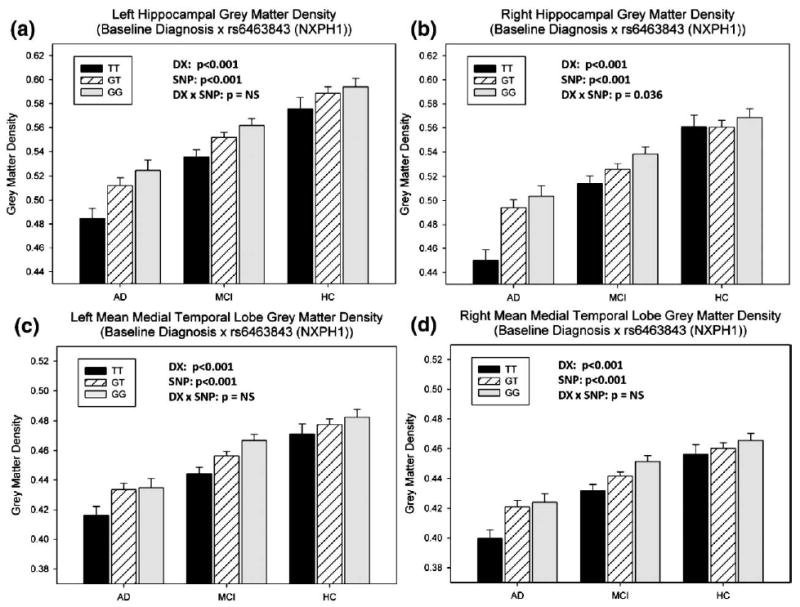
Refined analysis of sample imaging phenotypes in relation to rs6463843 (NXPH1) and baseline diagnosis. Two-way ANOVAs were applied to examine the effects of rs6463843 (NXPH1) and baseline diagnosis on four target GM density measures: (a–b) left and right hippocampal GMDs, and (c–d) left and right mean medial temporal lobe GMDs. All the analyses included age, gender, education, handedness and baseline ICV as covariates. n=715 subjects were involved: 166 AD (44 TT, 78 GT, 44 GG); 346 MCI (82 TT, 170 GT, 94 GG); 203 HC (35 TT, 105 GT, 63 GG). The p-values for the main effect of diagnosis (DX), the main effect of SNP (SNP), and the interaction effect of SNP-by-diagnosis (DX×SNP) were shown in each plot.
Discussion
Methodological overview
Employing a whole genome and entire brain strategy, we presented an imaging genetics methodological framework for systematically identifying associations between genotypes and imaging phenotypes, and demonstrated the utility of this method using the ADNI cohort. Our imaging genetics method can be broadly summarized as the following four steps after quality control and preprocessing: (1) imaging phenotype definition, (2) GWAS of image phenotypes, (3) cluster and heat map analysis of imaging GWAS results, and (4) refined statistical modeling.
Imaging phenotype definition
Eight-six GM density ROI measures and 56 volume and cortical thickness ROI measures were extracted, using VBM and FreeSurfer methods respectively, and analyzed as image phenotypes in independent GWAS analyses. This approach is complementary to another recently proposed imaging genetics analysis method, voxel-wise GWAS (vGWAS) (Stein et al., submitted for publication). The vGWAS technique explores SNP associations with all voxels in the image space. Our study is ROI-based, analyzing fewer but anatomically meaningful imaging phenotypes and thus, requires less computational resources. In addition, we used multiple techniques to define imaging phenotypes. Among the top 5 SNPs identified as part of the present study (Table 4), rs10932886 (EPHA4), rs7610017 (TP63) and rs6463843 (NXPH1) are primarily associated with VBM QTs, rs2075650 (TOMM40) is associated with FreeSurfer QTs, and rs429358 (APOE) is associated with ROIs extracted using both techniques. These results suggest that the VBM and FreeSurfer QTs are not equally sensitive to the same genetic markers and consequently may provide complementary information. The VBM measures we employed are not modulated (Good et al., 2001) and therefore measure GM densities (Ashburner and Friston, 2000), which are different from the volume and thickness measures that FreeSurfer generates for analysis. The complementary nature of GM density, volumetric, and cortical thickness ROIs in assessing of early AD, MCI, and pre-MCI samples is consistent with our recent findings examining ADNI baseline MRI data (Risacher et al., 2009) as well as an independent cohort (Saykin et al., 2006).
GWAS of image phenotypes
Following quality control of the genotyping data, genome-wide association studies were conducted on each of the 142 imaging phenotypes. The entire set of the GWAS analyses was performed and completed on a 112-node parallel computing environment within 20 min, suggesting an excellent potential for larger scale future extensions. One extension could be to investigate more sophisticated statistical models (e.g., exploring SNP-by-SNP or SNP-by-diagnosis interactions). Another extension could be to involve more imaging phenotypes from other imaging modalities or longitudinal data.
Cluster and heat map analysis of imaging GWAS results
Heat maps and hierarchical clustering have been used frequently for grouping results in gene expression analysis for pattern discovery (Eisen et al., 1998; Levenstien et al., 2003). In imaging genetics, heat maps can be equally useful for performing relevant pattern analysis tasks thanks to the rich information contained within the maps and their effective mechanism to organize and visualize complicated imaging GWAS results. A straightforward use of a heat map is to select target QTs, SNPs, or associations for further analyses. Due to its intuitive representation, some obvious patterns (e.g., the APOE SNP in Fig. 1) can be easily identified. For less obvious cases, other criteria could be used, for example, the selection of rs6463843 (NXPH1) because of its associations with multiple candidate phenotypic regions (i.e., hippocampus) affected by AD (Fig. 2b). In addition, a heat map can also be used to discover new patterns or structures. All the QTs and SNPs are hierarchically clustered as dendrograms on the x-axis and y-axis, respectively. In the genomic domain, for those SNP clusters that do not match the existing LD relationships, the dendrogram provides the ability to identify novel inter-SNP structures (e.g., Sloan et al., submitted for publication). In the imaging domain, for those phenotype clusters that do not follow a regional or bilaterally symmetric pattern, there might be an opportunity to identify an underlying brain connectivity pattern associated with a genetic variation.
Refined statistical modeling
In this paper, each heat map includes all the strong associations at a given significance threshold level, and can be used to guide further analyses using refined statistical models (e.g., involving diagnosis and other biomarkers, addressing interaction effects, etc.). These analyses can be performed using different strategies as follows: (1) select a target phenotype from the heat map and examine its whole genome mapping (e.g., Fig. 3); (2) pick a target SNP from a heat map and perform detailed image analysis (e.g., Fig. 4); and (3) choose a target SNP-QT association based on a heat map and/or an imaging analysis results, and perform a refined statistical modeling (e.g., Fig. 5). In this study, we conducted sample analyses for each of the above cases. The ultimate goal of these types of analyses is to identify genetic markers affecting brain structure and function, how these imaging and genetic markers interact with each other, as well as with diagnosis and/or other clinically and biologically relevant measures, and to gain a better understanding of disease risk and pathophysiology.
Imaging and genetics findings
The APOE SNP rs429358 and TOMM40 SNP rs2075650 were confirmed to be top markers affecting multiple brain structures in a mixed population of HC, MCI and AD (Farrer et al., 1997; Osherovich, 2009; Potkin et al., 2009a). Other SNPs, including rs10932886 (EPHA4), rs7610017 (TP63) and rs6463843 (NXPH1), were also among the top markers influencing brain structures in our analysis (Table 4). These SNPs and the genes in which they are found or flank have a number of important functions and potential pathways through which they may influence the pathophysiological processes underlying AD.
The EPHA4 [EPH receptor A4] gene belongs to the ephrin receptor subfamily of the protein-tyrosine kinase family (Fox et al., 1995). The interaction between neuronal EphA4 and glial ephrin-A3 was found to bidirectionally control synapse morphology and glial glutamate transport, which may ultimately regulate hippocampal function (Carmona et al., 2009). In addition, EphA4 and EphB2 receptors were reported to be reduced in the hippocampus before the development of impaired object recognition and spatial memory in transgenic mouse models of AD (Simon et al., 2009). The TP63 [Tumor protein 63] gene encodes a member of the p53 family of transcription factors (Yang et al., 1998). A literature search did not locate any articles associating TP63 with AD, cognitive impairment or neurode-generation. Additional imaging genetics analyses on both rs10932886 (EPHA4) and rs7610017 (TP63) appear warranted for future study.
The NXPH1 [Neurexophilin 1] gene is a member of the neurexophilin family and encodes a secreted protein which features a variable N-terminal domain, a highly conserved, N-glycosylated central domain, a short linker region, and a cysteine-rich C-terminal domain. This protein forms a very tight complex with alpha neurexins, a group of proteins that promote adhesion between dendrites and axons (Missler and Sudhof, 1998). This gene has previously been implicated as a candidate gene for neuroticism (van den Oord et al., 2008). In the present study, a VBM analysis of rs6463843 (NXPH1) revealed significantly reduced global and regional GM density in participants with the TT genotype relative to those with the GG genotype. Additional analyses indicated an interaction between rs6463843 (NXPH1) and baseline diagnostic group in which AD patients homozygous for the T allele were differentially vulnerable to decreased GM density in the right hippocampus, a finding presumably reflecting greater atrophy associated with this genotype in patients with AD.
Heat maps of imaging genetics associations at two significance threshold levels (p<10−7 and p<10−6) were also reported. At the conventional p<10−7 significance threshold, measures of hippocampal and amygdalar GM density and volume were strongly associated with the APOE and TOMM40 SNPs. Ten additional imaging phenotypes were strongly associated with at least one of the top SNPs (Fig. 1). We also examined a somewhat less stringent threshold (p<10−6) in order to identify additional SNP and imaging QT associations, as well as to examine patterns of genotype and phenotype clustering. SNPs associated with multiple unrelated or loosely related imaging phenotypes may represent an interesting genetic marker affecting overall brain structure or neurodegeneration. In addition, imaging variables associated with a number of SNPs from multiple genes may be particularly sensitive phenotypic markers for examining disease associated genetic variation. Therefore, heat maps at multiple statistical thresholds are useful in identifying candidate SNPs and imaging phenotypes warranting further investigation.
Limitations and future directions
The majority of analyses presented in this study focused on the extraction and evaluation of imaging phenotypes and the relationship of genetic variation to these phenotypes. However, we also included a limited assessment of the effects of baseline diagnostic group and the interaction effect of SNP and diagnosis in the analysis of candidate SNPs and phenotypes. Future studies could incorporate additional variables (e.g., clinical measures, other types of imaging and biomarkers) in the GWAS design to examine their effects and interactions with SNPs and/or target imaging phenotypes. The present analysis did not address epistasis or gene–gene interactions, a potentially very important topic. Future analyses should include models that incorporate epistatic interactions which are likely to be important for understanding susceptibility and protective factors in AD and other complex diseases.
Although we employed reasonably stringent thresholds for assessing genome-wide significance, a large number of ROIs represent a multiple comparison problem. The issue of determining the proper statistical threshold for a whole genome and whole brain search for associations is a challenging area for investigation (Nichols and Holmes, 2002; Nichols and Inkster, 2009; Stein et al., submitted for publication). The issue is complicated by the fact that variables within both the genomic and neuroimaging dimensions are non-independent due to LD and spatial autocorrelation, respectively. The determination of the effective number of independent statistical tests under these conditions is an area of investigation. Models for the joint distribution of both dimensions under the null hypothesis require development and validation.
Replication of current and future GWAS results in independent samples will remain of critical importance for confirmation. Although our follow-up analyses examine additional statistics at a more detailed level for yielding additional insights, these statistics are non-independent of the statistics used to select candidate ROIs and candidate associations. Given the recent interest in the non-independent analysis issue (e.g., Kriegeskorte et al., 2009), independent datasets for replication will be important for future studies to confirm the findings. For the current ADNI sample, given its modest size, we were unable to use one half of the data for hypothesis generation and the other half for confirmation, since one half of the data (i.e., n=367 in this study) cannot provide sufficient power to detect moderate/small genetic effects (Potkin et al., 2009b). With additional replication and extension opportunities under development, we anticipate that there will be ample statistical power and the ability to replicate potentially important findings in multiple independent data sets in the future.
At present there are few opportunities for replication of imaging genetics results such as those emerging from ADNI given the unique nature of this multi-dimensional data set. Fortunately, a worldwide ADNI consortium is actively being developed and large scale international data sets are likely to become available in the next few years that can provide adequate replication samples. In addition, the new NIH sponsored AD Genetics Consortium (ADGC) is assembling large meta-analytic databases of GWAS results that can provide confirmation of novel findings. Finally, the AlzGene meta-analytic database (www.alzgene.org) of candidate genes for AD, curated by Lars Bertram and colleagues (Bertram et al., 2007), provides a regularly updated source for determining the replication and validation status of AD genes.
The AAL atlas (Tzourio-Mazoyer et al., 2002) used to create the ROIs for the VBM analysis in this study is based on a single individual. To take anatomical variability into account, an important future direction will be to employ a probabilistic atlas, e.g., the Harvard-Oxford atlas (distributed with the FSL software package; http://fsl.fmrib.ox.ac.uk/fsl/), or the LONI probabilistic brain atlas (Shattuck et al., 2008). The most appropriate method to derive a GM-based summary statistic (e.g., density or volume) for a probabilistic ROI is a topic warranting investigation.
Despite the limitations and challenges, the encouraging experimental results obtained using the proposed analytic framework appear to have substantial potential for enabling the discovery of imaging genetics associations and for localizing candidate imaging and genomic regions for refined statistical modeling and further characterization. Ultimately, imaging genetics holds the promise of providing important clues to pathophysiology that could inform development of methods for earlier detection and therapeutic intervention.
Acknowledgments
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; principal investigator: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contributions from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, Alzheimer's Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
Data analysis was supported in part by the following grants from the National Institutes of Health: NIA R01 AG19771 to A.J.S. and P30 AG10133 to Bernardino Ghetti, MD and NIBIB R03 EB008674 to L.S., by the Indiana Economic Development Corporation (IEDC 87884 to AJS), by Foundation for the NIH to A.J.S., and by an Indiana CTSI award to L.S.
The FreeSurfer and PLINK analyses were performed on a 112-node parallel computing environment, called Quarry, at Indiana University. We thank the University Information Technology Services at Indiana University for their support.
We thank the following people for their contributions to the ADNI genotyping project: (1) genotyping at the Translational Genomics Institute, Phoenix AZ: Jennifer Webster, Jill D. Gerber, April N. Allen, and Jason J. Corneveaux; and (2) sample processing, storage and distribution at the NIA-sponsored National Cell Repository for Alzheimer's Disease: Kelley Faber.
References
- Ahmad RH, Emily MD, Daniel RW. Imaging genetics: perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry. 2006;59:888–897. doi: 10.1016/j.biopsych.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry–the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, Angelakopoulou A, Hall L, Richardson JC, Prinjha RK, Gass A, Geurts JJ, Kragt J, Sombekke M, Vrenken H, Qualley P, Lincoln RR, Gomez R, Caillier SJ, George MF, Mousavi H, Guerrero R, Okuda DT, Cree BA, Green AJ, Waubant E, Goodin DS, Pelletier D, Matthews PM, Hauser SL, Kappos L, Polman CH, Oksenberg JR. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, van Erp TG, Dutton RA, Tran H, Zimmermann L, Sun D, Geaga JA, Simon TJ, Glahn DC, Cannon TD, Emanuel BS, Toga AW, Thompson PM. Mapping cortical thickness in children with 22q 11.2 deletions. Cereb Cortex. 2007;17:1889–1898. doi: 10.1093/cercor/bhl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Brett MA, Jean-Luc, Valabregue Romain, Poline Jean-Baptiste. Region of interest analysis using an SPM toolbox [Abstract]. Presented at the 8th International Conference on Functional Mapping of the Human Brain; Sendai, Japan. 2002. [Google Scholar]
- Brun CC, Lepore N, Pennec X, Lee AD, Barysheva M, Madsen SK, Avedissian C, Chou YY, deZubicaray GI, McMahon K, Wright MJ, Toga AW, Thompson PM. Mapping the regional influence of genetics on brain structure variability–a tensor-based morphometry study. NeuroImage. doi: 10.1016/j.neuroimage.2009.05.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Thompson PM, van Erp TG, Huttunen M, Lonnqvist J, Kaprio J, Toga AW. Mapping heritability and molecular genetic associations with cortical features using probabilistic brain atlases: methods and applications to schizophrenia. Neuroinformatics. 2006;4:5–19. doi: 10.1385/NI:4:1:5. [DOI] [PubMed] [Google Scholar]
- Carmona MA, Murai KK, Wang L, Roberts AJ, Pasquale EB. Glial ephrin-A3 regulates hippocampal dendritic spine morphology and glutamate transport. Proc Natl Acad Sci U S A. 2009;106:12524–12529. doi: 10.1073/pnas.0903328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale A, Fischl B, Sereno M. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer L, Cupples L, Haines J, Hyman B, Kukull W, Mayeux R. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis, APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Filippinia N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, Whitcher B, Matthews PM. Anatomically-distinct genetic associations of APOE ε4 allele load with regional cortical atrophy in Alzheimer's disease. NeuroImage. 2009;44:724–728. doi: 10.1016/j.neuroimage.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno M, Dale A. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fox GM, Holst PL, Chute HT, Lindberg RA, Janssen AM, Basu R, Welcher AA. cDNA cloning and tissue distribution of five human EPH-like receptor protein-tyrosine kinases. Oncogene. 1995;10:897–905. [PubMed] [Google Scholar]
- Glahn DC, Paus T, Thompson PM. Imaging genomics: mapping the influence of genetics on brain structure and function. Hum Brain Mapp. 2007a;28:461–463. doi: 10.1002/hbm.20416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007b;28:488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, J LW, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci. 2009;12:535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenstien MA, Yang Y, Ott J. Statistical significance for hierarchical clustering in genetic association and microarray expression studies. BMC Bioinformatics. 2003;4:62. doi: 10.1186/1471-2105-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind J, Larsson A, Persson J, Ingvar M, Nilsson LG, Bäckman L, Adolfsson R, Cruts M, Sleegers K, Van Broeckhoven C, Nyberg L. Reduced hippocampal volume in non-demented carriers of the apolipoprotein E epsilon4: relation to chronological age and recognition memory. Neurosci Lett. 2006;396:23–27. doi: 10.1016/j.neulet.2005.11.070. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Price CJ, Friston KJ, Ashburner J. Voxel-based morphometry of the human brain: methods and applications. Curr Med Imaging Rev I. 2005:1–9. [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexophilins form a conserved family of neuropeptide-like glycoproteins. J Neurosci. 1998;18:3630–3638. doi: 10.1523/JNEUROSCI.18-10-03630.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005a;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005b;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols TE, Inkster B. Comparison of whole brain multiloci association methods. OHBM'09: 15th Annual Meeting of Organization for Human Brain Mapping; San Francisco, CA. 2009. [Google Scholar]
- Osherovich L. TOMMorrow's AD marker. SciBX. 2009;2(30) doi: 10.1038/scibx.2009.1165. [DOI] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E, Weiner M, Macciardi F. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 2009a;4:e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn Neuropsychiatry. 2009b;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ, Wishart HA, Rabin LA, Santulli RB, Flashman LA, West JD, McHugh TL, Mamourian AC. Older adults with cognitive complaints show brain atrophy similar to that of amnestic MCI. Neurology. 2006;67:834–842. doi: 10.1212/01.wnl.0000234032.77541.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri S, DeStefano A, Au R, Massaro J, Beiser A, Kelly-Hayes M, Kase C, D'Agostino R, DeCarli C, Atwood L, Wolf P. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham study. BMC Med Genet. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Saykin AJ, Chung MK, Huang H. IEEE BIBE. 2007. Morphometric analysis of hippocampal shape in mild cognitive impairment: an imaging genetics study; pp. 211–217. [Google Scholar]
- Simon AM, de Maturana RL, Ricobaraza A, Escribano L, Schiapparelli L, Cuadrado-Tejedor M, Perez-Mediavilla A, Avila J, Del Rio J, Frechilla D. Early changes in hippocampal EPH receptors precede the onset of memory decline in mouse models of Alzheimer's disease. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1096. [DOI] [PubMed] [Google Scholar]
- Sloan C, Shen L, West J, Wishart H, Flashman L, Rabin L, Santulli R, Guerin S, Rhodes C, Tsongalis G, McAllister T, Ahles T, Lee S, Moore J, Saykin A. Genetic pathway-based hierarchical clustering analysis of older adults with cognitive complaints and amnestic mild cognitive impairment using clinical and neuroimaging phenotypes. doi: 10.1002/ajmg.b.31078. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga A, Saykin AJ, Shen L, Foroud T, Pankratz N, Huentelman MJ, Craig DW, Gerber JD, Allen A, Corneveaux J, DeChairo BM, Potkin SG, Jack C, Weiner M, Thompson P. Voxelwise genome-wide association study (vGWAS) submitted for publication. [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van den Oord EJ, Kuo PH, Hartmann AM, Webb BT, Moller HJ, Hettema JM, Giegling I, Bukszar J, Rujescu D. Genomewide association analysis followed by a replication study implicates a novel candidate gene for neuroticism. Arch Gen Psychiatry. 2008;65:1062–1071. doi: 10.1001/archpsyc.65.9.1062. [DOI] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, Rabin LA, Santulli RB, Flashman LA, Guerin S, Mamourian AC, Belloni D, Rhodes CH, McAllister TW. Increased prefrontal activation during working memory in cognitively intact APOE ε4 carriers. Am J Psychiatry. 2006;163:1603–1610. doi: 10.1176/ajp.2006.163.9.1603. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2:2492–2501. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]


