Abstract
Petal development and senescence entails a normally irreversible process. It starts with petal expansion and pigment production, and ends with nutrient remobilization and ultimately cell death. In many species this is accompanied by petal abscission. Post-harvest stress is an important factor in limiting petal longevity in cut flowers and accelerates some of the processes of senescence such as petal wilting and abscission. However, some of the effects of moderate stress in young flowers are reversible with appropriate treatments. Transcriptomic studies have shown that distinct gene sets are expressed during petal development and senescence. Despite this, the overlap in gene expression between developmental and stress-induced senescence in petals has not been fully investigated in any species. Here a custom-made cDNA microarray from Alstroemeria petals was used to investigate the overlap in gene expression between developmental changes (bud to first sign of senescence) and typical post-harvest stress treatments. Young flowers were stressed by cold or ambient temperatures without water followed by a recovery and rehydration period. Stressed flowers were still at the bud stage after stress treatments. Microarray analysis showed that ambient dehydration stress accelerates many of the changes in gene expression patterns that would normally occur during developmental senescence. However, a higher proportion of gene expression changes in response to cold stress were specific to this stimulus and not senescence related. The expression of 21 transcription factors was characterized, showing that overlapping sets of regulatory genes are activated during developmental senescence and by different stresses.
Keywords: Alstroemeria, cold stress, dehydration stress, flower, microarrays, petal, senescence, transcriptome
Introduction
Under optimal conditions floral lifespan is species specific and precisely controlled (Primack, 1985), senescence often being initiated following successful pollination. This is thought to be related to the resource investment of a flower, as well as other ecological factors (Rogers, 2006). While the flower is attached to the plant, environmental stresses such as temperature change can shorten floral lifespan. Once the flower is detached from the parent plant, one of the major factors affecting floral lifespan is supply of respiratory substrate. This is in part ameliorated by provision of a carbon source such as sucrose, or by removal of competing flowers on the same stem (Doi and Reid, 1995; Chanasut et al., 2003). Provision of sucrose can prevent or delay the up-regulation of senescence-associated genes (Hoeberichts et al., 2007), probably indirectly by delaying senescence. Petal senescence is typically characterized by a loss of fresh weight, protein content, and turgor, changes in pigmentation and, ultimately, in many species, programmed cell death and petal abscission (Wagstaff et al., 2003; van Doorn and Woltering, 2008). These changes are related to alterations in the patterns of gene expression which have been analysed in several species using microarrays (Channelière et al., 2002; van Doorn et al., 2003; Breeze et al., 2004; Price et al., 2008; Wagstaff et al., 2009). Many of the genes up-regulated during normal developmental petal senescence relate to remobilization of nutrients and include proteases, nucleases, lipases, and transporters (Hong et al., 2000; Wagstaff et al., 2002; Langston et al., 2005; Price et al., 2008). Genes encoding specific classes of transcription factors, NAC, MYB, MYC, MADS-box, WRKY, and zinc finger proteins, are also frequently up-regulated (Channelière et al., 2002; Breeze et al., 2004; Hoeberichts et al., 2007; Price et al., 2008; Wagstaff et al., 2009), indicating that they may play a regulatory role. Additionally, microarray screens and analyses of individual genes have revealed that a number of genes, generally considered to be stress related, are up-regulated during petal senescence. These include metallothioneins, abscisic acid (ABA)-responsive genes (Channelière et al., 2002; Breeze et al., 2004), and glutathione S-transferases (Meyer et al., 1991; Price et al., 2008).
The effects of stress on gene expression have mainly been studied in whole plants or leaves, and there is a body of evidence showing that gene expression changes in leaves are rapid, specific, and highly stress responsive (Buchanan-Wollaston et al., 2005; Gombert et al., 2006). A number of studies have concentrated on different forms of stress such as dark-induced senescence (Roca et al., 2004), cold stress (Khodakovskaya et al., 2005), and dehydration stress (Munne-Bosch and Alegre, 2004). These have identified sets of genes regulated specifically by each of these processes, and several of which are changed by a multitude of stress elicitors. Thus there is support for a model in which there is cross-talk between different stress-induced processes (Nakashima et al., 2009). Abscisic acid (ABA) is involved in coordinating responses to both drought and cold stress, and another common signalling mechanism may be via reactive oxygen species (ROS) (Takahashi et al., 2004). Members of the MYB, MYC, and NAC transcription factor classes are involved in ABA-regulated signal transduction relating to cold and drought responses (Shinozaki and Yamaguchi-Shinozaki, 2003) and cross-talk between other stress responses (Fujita et al., 2006). Mitogen-activated protein (MAP) kinases are also heavily implicated in coordinating stress responses. For example Arabidopsis MEKK1 is transcriptionally up-regulated by multiple stresses including drought and cold (Mizoguchi et al., 1996). The interaction between stress and developmental senescence has been studied in leaves (Buchanan-Wollaston et al., 2003; Lim et al., 2007), and many of the physiological changes induced by stress in leaves such as chlorophyll degradation, and remobilization of nutrients parallel those seen during natural senescence. This suggests that in leaves many stresses induce an early onset of developmental senescence. However, the sharing of gene expression is not complete, indicating that the situation is more complex.
In petals the overlap in gene expression between developmental senescence and stress-induced senescence in petals is poorly characterized in any species. Studies of the physiological effects of stress on cut flowers have focused on investigating ways to maximize floral longevity and determining ways to overcome the stresses experienced by flowers during their transport from grower to retailer (Ranwala and Miller, 1998; Andersen et al., 2004). Cold temperature storage at 4 °C is employed to delay flower development and causes little detrimental effect, in temperate species, if the stems are in solution, thereby retaining the maximum vase life. However, the majority of stems are transported without water, packed tightly in boxes. In these cases longevity may be reduced as a consequence of both drought and temperature stress (Rudnicki et al., 1991). In many species, excessive stress will induce temporary loss of petal turgor. This can be apparently reversed by a refreshment treatment; however, overall floral longevity is reduced.
Alstroemeria cv. Rebecca forms part of the Liliales and has large, colourful, relatively long-lived (10–15 d) flowers under optimal conditions. The time frame of developmental senescence has been characterized in previous studies (Wagstaff et al., 2001, 2003; Leverentz et al., 2002; Breeze et al., 2004) and has been shown to be almost completely ethylene independent (Wagstaff et al., 2005). Alstroemeria is an economically important cut flower. The nature of the commercial handling chain means that flowers are subjected to periods of many hours or even days stored, or transported, without water at ambient temperatures and/or several days stored in the dark at ∼4 °C (Reid, 2004). Thus young flowers may be subjected to cold and/or dehydration stress during this period.
In this study cDNA microarrays have been generated using RNA from petals of both young stress-treated flowers, following a recovery period, and naturally senescing petals. They have been used to investigate the overlap in gene expression between ambient dehydration stress, cold stress, and developmental senescence. Classes of genes specific to either or both stress treatments, specific to developmental senescence, or shared between stress and senescence have been identified. A significant finding is that the pattern of gene expression induced by ambient dehydration stress is similar to that seen during developmental senescence, whereas the pattern elicited by cold stress is different. Furthermore the patterns of gene expression of 21 transcription factors indicates a complex network of shared regulatory signals.
Materials and methods
Plant material
Alstroemeria inflorescences were harvested and transported to the laboratory in water. For developmental senescence studies individual cymes were detached and placed in glass vials of distilled water throughout the experiment in a growth room (21 °C, 16 h photoperiod, 12–14 μM m2 s−1, 60% relative humidity). For ambient stress experiments the inflorescences were trimmed to 60 cm, placed horizontally within a flower box in the dark, and left at ambient temperature (21 °C) for 6 h or 48 h prior to a 3 h refreshment period in distilled water in the growth room as above. For cold stress treatments the inflorescences were placed horizontally within a flower box in the dark and left at 4 °C for 3 d before refreshment in water in the growth room for 3 h or 24 h. Petals were then detached, frozen in liquid nitrogen, and used for RNA extraction (method as in Breeze et al., 2004). To determine longevity of stress-treated flowers, cymes were isolated and placed in vials of distilled water as described above.
cDNA libraries and EST analysis
A total of five unsubtracted cDNA libraries were constructed in addition to the SSH libraries detailed in Breeze et al. (2004). The developmental libraries contained material from (i) stages S0 and S1; (ii) stages S2 and S4; and (iii) stages S5 and S7. Stress libraries were made from (i) both ambient dry and (ii) both cold dry treatments detailed above. Dynal beads were used to isolate mRNA from 75 μg of total RNA for each stage or treatment of tissue. Equal amounts of mRNA from each stage/treatment totalling 5 μg were then used to construct each unsubtracted library using Stratagene's λ-zap cDNA synthesis and Gigapack III Gold cloning kit. Primary libraries of 1×106 pfu (plaque-forming units) were obtained. A mass excision of each library was performed and ∼106 clones were plated using blue/white selective media. Over 9600 clones were picked from each library and stored as glycerol stocks.
Inserts from selected clones were amplified by PCR using M13 forward and reverse primers either directly from glycerol stocks or from plasmid DNA isolated using a Qiaprep Miniprep kit (Qiagen, Crawley, UK). PCR products were purified using the Whatman vacuum manifold PCR cleanup system (Whatman, Maidstone, UK) or Qiaquick PCR cleanup kits (Qiagen). Sequencing was carried out using M13 forward and reverse primers with BigDye version 2 (Applied Biosystems, Foster City, CA, USA) and was analysed on an Applied Biosystems 3100 sequencer. All sequences determined are available on the NCBI expressed sequence tag (EST) database; accession numbers are detailed in Table S1 available at JXB online. Alignment of proteins and sequences was performed using BIOEDIT version 7.0.1 (Hall, 1999) and Seqman (Dnastar, Lasergene, Madison, WI, USA). Contig assembly was performed using Contig Express (Invitrogen, Paisley, UK). Sequences obtained were compared with database entries using the BLAST network service (NCBI). A match was assessed using a combination of low E value and the length of the homology in BLASTx. TAIR was used to provide extra details on the putative function of some of the genes.
Microarray construction
Each glycerol stock was used to inoculate a 1.5 ml LB culture containing 50 μg ml−1 ampicillin in a 96-well deep well culture plate. Cultures were grown overnight at 37 °C with shaking at 200 rpm. Plasmid DNA was extracted using the Whatman 96-well Plasmid Miniprep Kit, and ∼100 ng of DNA was used in a PCR to amplify the insert with M13 forward and reverse primers. PCRs were purified using the Whatman 96-well PCR Cleanup Kit protocol and the inserts were checked for size and for the presence of single clones on a 96-well gel (Amersham). Purified inserts were diluted by 50% with dimethylsulphoxide (DMSO) prior to printing onto Corning GAPS coated slides using a robotic Flexys array printer (Genomic Solutions, Ltd, Cambridgeshire, UK) with solid pins, having 120 μm tips delivering ∼340 pl. Slides were also spotted with Cy3/Cy5-labelled landmarks and DMSO blanks. The microarray contained: 960 probes derived from a cDNA library made from cold-stressed flowers, 960 probes derived from a cDNA library of ambient temperature dehydration-stressed flowers, 1269 ESTs from the subtractive libraries described in Breeze et al. (2004), a further 267 unsequenced probes from the same subtractive libraries, and 3053 probes picked randomly from the unsubtracted developmental libraries detailed above (S0–S1, S2, S4, and S5–S7). Each probe was printed in duplicate on each slide, giving a total of 13 018 probes per slide. Slide replication within the experiment gave a total of at least eight replicates per probe.
Hybridization and washing
Probes were made using 20 μg of total RNA from a TRI reagent (Sigma) extraction (see Breeze et al., 2004 for the method) that was subsequently purified using an Rneasy column (Qiagen) according to the manufacturer's instructions. Indirect post-labelling with Cy3 or Cy5 dye was performed with the Amersham Cyscribe kits according to the manufacturer's instructions. Unlabelled cDNA was purified by ethanol precipitation, and unincorporated Cy dye was removed at the final step using a Nucleospin column (Macheray Nagel). Samples were combined and dried down using a vacuum centrifuge and then resuspended in sterile distilled water. The final steps of probe manufacture and denaturing were performed as detailed in the Amersham Cyscribe handbook and included blocking poly(T)+ tails with an oligo(dA)80 primer with a final volume of 60 μl. All probes were replicated at least twice with reciprocal Cy dye labelling, and hybridization was performed using back to back manual hybridization with two array slides, so the minimum number of technical replicates for any sample was four. Slides were blocked in a solution of 5× SSC, 0.1% SDS, and 1% bovine serum albumin (BSA) for 45 min at 42 °C and then washed in four changes of milliQ water and twice in isopropanol. Hybridization was performed at the same temperature in a humid chamber. After hybridization for ∼16 h, slides were washed in 1× SSC/0.2% SDS for 10 min; 0.1× SSC/0.2% SDS for 2×10 min. Slides were scanned on an Affymetrix 428 scanner at a resolution of 10 μm per pixel.
Statistical analysis
Images were first assigned spot identity and quantified using Imagene (BioDiscovery). Genes that were absent in all samples of any experiment were filtered out using Excel (Microsoft) and the data were entered into Genespring (Silicon Genetics) for subsequent analysis. Where possible, two-colour analysis was performed; probes were normalized per chip and per gene using a Lowess algorithm with a 40% cut-off. Genes showing >2-fold changes between conditions and with a spread of <1.4 standard deviations from the mean were entered into a Welch t-test or Welch analysis of variance (ANOVA) as appropriate. In experiments such as the developmental study where four stages were compared, a circular design was employed and one-colour analysis was therefore used. Genes were corrected to a baseline of zero and then normalized on a per-chip and per-gene basis. A Welch ANOVA was subsequently used to identify genes showing significant changes between conditions (Supplementary Table S2 at JXB online). Clones containing genes of interest were grown and plasmid DNA extracted as described above prior to sequencing on an ABI 3700 capillary sequencing machine. Contingency χ2 test was performed using Minitab15 (Minitab Inc., PA, USA).
Quantitative RT-PCR (qRT-PCR)
RT-PCR was carried out using material from two different harvest times with similar results. Primers suitable for use in real-time PCR were designed using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), checked for primer pair and primer self-complementarity using Operon's oligo toolkit (http://www.operon.com/oligos/toolkit.php?), and the secondary structure of the amplicon analysed using Mfold (http://bibiserv.techfak.uni-bielefeld.de/cgi-bin/mfold_submit) to ensure no significant folding at the primer binding sites. A list of the primer sequences is provided in Supplementary Table S3 at JXB online.
Total RNA from Alstroemeria petals at the various stages of development or from the different treatments was treated with RNase-free DNase (Ambion TURBO DNase) for 60 min at 37 °C followed by inactivation. cDNA synthesis was performed on 2 μg of RNA using Euroscript reverse transcriptase (Eurogentec) with both oligo d(T) primer and random nonamers, according to the manufacturer's instructions.
For the qRT-PCR 1 μl of cDNA was used in each reaction consisting of 12.5 μl of 2× qPCR mastermix for SYBR green No ROX (Eurogentec), 5 pmol each of forward and reverse primer, and 0.75 μl of SYBR green (1/2000 dilution) (Eurogentec) in a final volume of 25 μl. A standard curve was generated by 10-fold serially diluting a standard cDNA mix made up of equal aliquots of cDNA from all developmental stages in order to span the likely range of gene expression. All sample and standard reactions were performed in triplicate. The real-time PCR was carried out using the iCycler iQ Real Time Detection system (Bio-Rad). The reactions were incubated at 50 °C for 2 min for the uracil-N-glycosylase reaction, then heated to 95 °C for 10 min followed by 50 cycles of 15 s at 95 °C and 30 s at 60 °C. Melt curve analysis from 45 °C to 95 °C was performed on the end-products of the PCR to demonstrate that only a single product was being amplified. Each reaction was optimized so that the PCR efficiency was between 90% and 110%. The relative gene expression was calculated from the threshold cycle (Ct value) and was normalized using 18S rRNA expression as an internal control.
Results
Stress treatments induced early petal abscission
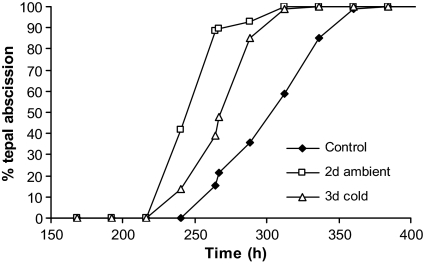
Two stress treatments were employed in this study, chosen to mirror post-harvest conditions frequently experienced by cut flowers during the transport chain. First dehydration stress was imposed by storing flowers dry at ambient temperature for a short period (6 h) or a longer period (48 h) and then refreshing them in water for 3 h (Fig. 1A–C). The second stress was a cold treatment: in a separate experiment flowers were cold stressed dry at 4 °C for 72 h (Fig. 1D), and then refreshed in water for a short (3 h) or longer (24 h) period. Cold-stressed flowers were slightly wilted when removed from the cold. However, even a short period of refreshment in water appeared to restore turgidity such that inflorescences were indistinguishable from freshly harvested material. Dehydration-stressed flowers, especially when treated for 48 h, showed considerable wilting when first examined, but again a short refreshment period appeared to restore turgidity. The treatments were both started when the flowers were at stage S0 (freshly harvested tight buds, Fig. 1A), and cold treatment arrested any further development. However, the ambient dehydration treatment allowed development as far as stage S1 (loose bud) over the 48 h period (stages previously defined in Breeze et al., 2004) such that each bud was opening and showing colour (Fig. 1C). This is less than a flower would develop over the same time period if held in water. Other differences only became apparent several days later when the flowers were fully open; for example, anthocyanin pigment development was much reduced in the cold-stressed flowers (Fig. 1E). The longevity of ambient dehydration or cold dehydration-stressed flowers, once replaced in water, was measured as the time taken to tepal abscission (Fig. 2). Cold dehydration stress (72 h) reduced time to 50% abscission by nearly 2 d. Ambient dehydration stress (48 h) reduced the time to 50% abscission by 3 d, with the flowers only lasting 12 d following the refreshment treatment, compared with 15 d shown by the controls. In general, however, the appearance and development of flowers following stress treatment were similar to those of the controls. Therefore it seems that phenotypically stressed flowers followed a normal pattern of development over a more rapid time frame.
Fig. 1.
Appearance of flowers (A) at harvest prior to any stress treatment (control), (B) after removal from 6 h dehydration stress at 21 °C, (C) after removal from 48 h dehydration stress at 21 °C, (D) after 72 h cold dry stress at 4 °C, and (E) opening of cold-stressed flower compared with control at stage S5, the latter showing much more intense pigmentation. Inflorescences showed some wilting following stress treatments, but this was completely ameliorated by a 3 h refreshment treatment in water at ambient temperature.
Fig. 2.
Effect of stress treatments on rate of tepal abscission. Ambient dehydration stress (48 h followed by 3 h refreshment period) accelerated time to 50% tepal abscission by ∼2 d, whereas 3 d cold stress (followed by a 3 h refreshment period) accelerated time to 50% tepal abscission by ∼1 d. n=20 flowers, each of which had six tepals.
ESTs from developmental and stress-treated petal cDNA libraries
ESTs (1849) were derived from four sources. These were previously published subtracted libraries (Breeze et al., 2004) randomly picked from unsubtracted cDNA libraries representing stages from S0 to S7, from an ambient temperature dehydration-treated library, and from a cold-treated cDNA library. The sequences yielded 270 contigs and 553 singletons. The stress treatments were applied as described in the previous section and were followed by a refreshment period. Using database comparison, contigs and sequences were putatively assigned to 566 independent genes (Supplementary Table S1 at JXB online). As previously described in Breeze et al. (2004), sequences showing the closest homology to metallothioneins were heavily represented in both the developmental and stress-treated libraries. They comprised 358 sequences in 10 contigs and six singletons which represented 19% of the total sequences analysed (Table 1). A putative cell wall protein and a ubiquitin carrier-like protein as well as a genes related to anthocyanin and epicuticular wax biosynthesis were also well represented in the developmental libraries. The ESTs from the stress-treated libraries also included a CXE carboxyl esterase gene and a glutathione S-transferase.
Table 1.
Most abundant classes of ESTs from developmental senescence and stress-treated libraries
| Contig | No. of clones | Source library | Putative function | Nearest match | e-value |
| Developmental libraries | |||||
| Contig 175 | 102 | S0S1 | Metallothionein-like protein | AY833008 | 7E-24 |
| Contig 172 | 49 | S0S1 | Large subunit rRNA | DQ008809 | 1E-73 |
| Contig 180 | 40 | S2S4 | Metallothionein type II | AF039003 | 9E-27 |
| Contig 19 | 30 | S0S1 | Metallothionein type II | AF039003 | 9E-27 |
| Contig 239 | 19 | S0S1 | Metallothionein type II | AF147786 | 4E-23 |
| Contig 252 | 19 | S2S4 | Metallothionein type II | AF039003 | 2E-28 |
| Contig 35 | 19 | S2S4 | Metallothionein-like protein | AF279655 | 3E-30 |
| Contig 158 | 17 | SSH0-2 | Putative cell wall P8 protein | AC146631 | 0.0009 |
| Contig 203 | 16 | S0S1 | Cytochrome b6/f complex subunit VIII | BAF64867 | 0.001 |
| Contig 198 | 15 | S0S1 | Ubiquitin carrier-like protein | DQ294271 | 1E-101 |
| Contig 150 | 13 | SSH0-2 | GSDL-motif lipase | AY491975 | 6E-62 |
| Contig 242 | 13 | S2S4 | Metallothionein type I | AF039002 | 1E-22 |
| Contig 214 | 11 | S0S1 | 40S ribosomal protein S17 (RPS17D) | AT5G04800 | 3E-64 |
| Contig 128 | 10 | SSH2-45 | Dihydroflavonol-4-reductase | AY374471 | 1E-54 |
| Contig 221 | 10 | S2S4 | Large subunit rRNA gene | DQ008808 | 0 |
| Contig 45 | 10 | SSH2-0 | Aldehyde decarbonylase (epicuticular wax associated) | ATHCER1 | 2E-56 |
| Stress libraries | |||||
| Contig 175 | 48 | COLD | Metallothionein-like protein | AY833008 | 7E-24 |
| Contig 172 | 22 | COLD | Large subunit rRNA | DQ008809 | 1E-73 |
| Contig 19 | 15 | COLD | Metallothionein type II | AF039003 | 9E-27 |
| Contig 242 | 12 | COLD | Metallothionein type I | AF039002 | 1E-22 |
| Contig 263 | 12 | COLD | Arabidopsis unknown protein/CXE carboxylesterase | AT1G47480 | 0 |
| Contig 180 | 9 | COLD | Metallothionein type II | AF039003 | 9E-27 |
| Contig 186 | 5 | COLD | No significant BLAST match | ||
| Contig 187 | 4 | COLD | Elongation factor-1 alpha 2 | ||
| Contig 239 | 4 | COLD | Metallothionein-like protein (ML2) | AF147786 | 4E-23 |
| Contig 252 | 4 | COLD | Metallothionein type II | AF039003 | 2E-28 |
| Contig 154 | 3 | COLD | Ubiquitin precursor-like | EU249995 | E-116 |
| Contig 193 | 3 | COLD | Glutathione transferase | AJ441055 | 4E-72 |
| Contig 35 | 3 | COLD | Metallothionein-like protein | AF279655 | 3E-30 |
Petal libraries: S0S1, S2S4 are non-subtracted libraries from combined stages S0 and S2, and S2 and S4 respectively; SSH0-2 and SSH2-45 are subtracted libraries of stage S2 from S0 and combined stages S4 and S5 from S2, respectively; COLD library combined a 3 d cold treatment followed by a 3 h or 24 h refreshment period (see Materials and methods for further details).
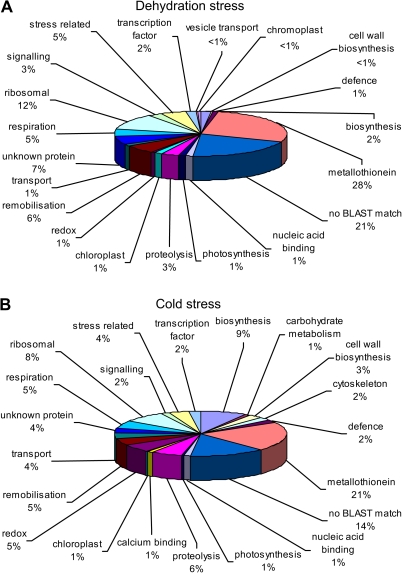
Of particular interest were ESTs from the stressed libraries. The 187 ESTs from the cold-stressed library and the 216 ESTs from the dehydration-stressed library were grouped according to function (Fig 3A, B). The distribution of functional classes differed significantly between the cold-treated and dehydration-treated libraries (analysed by contingency χ2). A higher proportion of genes from the cold-treated library were involved in biosynthesis, and genes related to redox processes were also overrepresented compared with the ambient dehydration stress library.
Fig. 3.
Functional analysis of all ESTs derived from (A) dehydration-stressed (6 h or 48 h followed by 3 h refreshment) and (B) cold-stressed (3 d cold treatment followed by a 3 h or 24 h refreshment period) cDNA libraries.
Gene expression during developmental senescence and following stress treatments
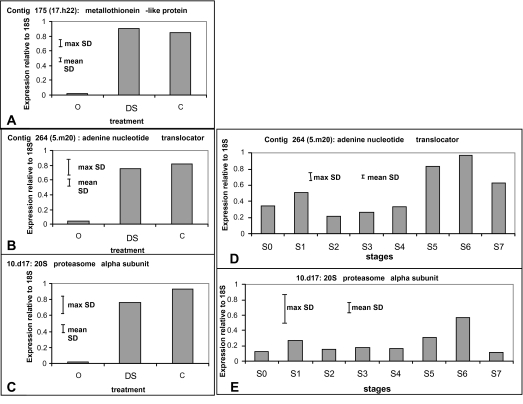
Three genes were selected to compare gene expression following the stress treatments and during developmental senescence using qRT-PCR. These genes had putative functions as a metallothionein-like gene, an adenine-nucleotide translocator, and the α-subunit of the 20S proteasome, and were selected to represent different functional classes. Expression of all three genes was up-regulated >20-fold with either the cold (3 d followed by 3 h refreshment) or ambient dehydration (48 h followed by 3 h refreshment) treatments (Fig. 4A–C) applied as described in previous sections. Breeze et al. (2004) showed by northern analysis that metallothionen gene expression was first detectable at stage S3 and increased with petal age thereafter. The expression patterns for the other two selected genes followed a similar pattern (Fig. 4D, E). In the case of both the adenine nucleotide translocator and the 20S proteasome α subunit, expression was low in young petals, with a minor peak around the time of petal opening (stage S1) followed by a steady increase from stage S4 (early senescence) to stage S6 (mid to late senescence). In both cases, however, expression was reduced in the latest stage of senescence (stage S7).
Fig. 4.
Real-time qPCR of three genes to analyse their expression following stress treatments (A, B, and C) and throughout development and senescence from stage S0 (closed bud) to stage S7 (petal abscision) (D and E). (A) Metallothionein, (B) and (D) adenine nucleotide translocator, (C) and (E) 20S proteasome α subunit. O is unstressed flowers at stage S0, C is 3 d cold treatment followed by a 3 h refreshment period, and WD is ambient dehydration stress of 48 h followed by a 3 h refreshment period. Each experiment was repeated on a second biological replicate with similar results. Maximum and minimum SDs for each experiment are shown.
Dehydration stress and developmental senescence elicited gene expression changes in a common group of genes.
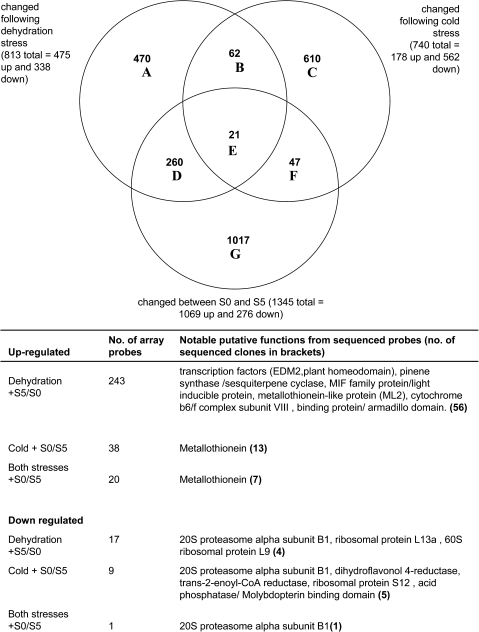
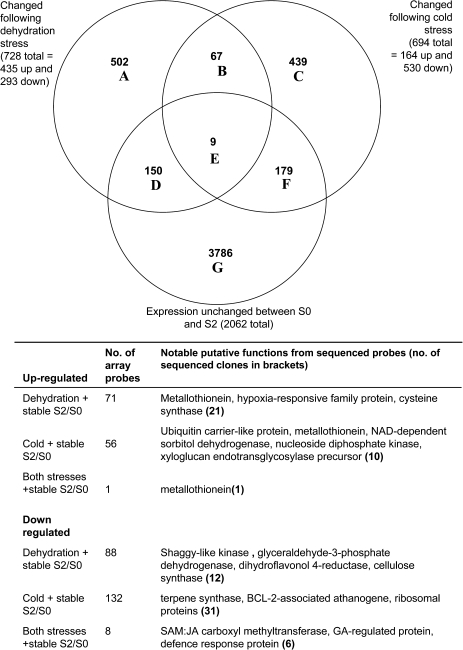
To compare expression patterns across more genes, microarrays were constructed using the 1849 sequenced clones, and a further 4660 unsequenced clones from the same cDNA libraries. Changes in expression following the two stress treatments were compared with changes between stage S0 (young buds) and stage S5 (early senescence). These stages were chosen to represent genes whose expression changes with the onset of developmental senescence (Fig. 5, Supplementary Tables S2 and S4 at JXB online). The most striking feature was the greater sharing of gene expression between developmental senescence and ambient dehydration stress (281/1345) compared with developmental senescence and cold stress (68/1345). The similarity in gene expression patterns between ambient dehydration stress and developmental senescence was more marked in the up-regulated genes. Of the 1069 probes up-regulated between stages S0 and S5, 25% were also up-regulated following the ambient stress treatments while only 5% of them were up-regulated with the cold stress treatments. However, of the 276 probes down-regulated between stages S0 and S5, 7% were also down-regulated following the ambient stress treatments and 4% of them were down-regulated following cold treatments, thus the levels were more similar. Probes whose expression changed following dehydration stress and between stages S0 and S5 included several transcription factors as well as metallothioneins and an armadillo domain protein. Amongst those whose expression changed following cold stress and during developmental senescence, genes included a dihydroflavonol-4-reductase and metallothionein-related genes. Another feature of these results was the large number of probes whose expression pattern was unique to the specific treatment or developmental senescence. Expression of only 21 array probes was changed by both stress treatments and developmental senescence. All of the sequenced up-regulated probes were metallothionein related (or rRNA) whereas the single down-regulated probe was homologous to the 20S proteasome α subunit B1.
Fig. 5.
Venn diagram and details of microarray probes whose signal changed following cold stress (3 d cold treatment followed by a 3 h or 24 h refreshment period) or ambient dehydration stress (6 h or 24 h followed by a 3 h refreshment period) compared with developmental changes between stage S0 and stage S5 (stage S0 petals were from closed buds and stage S5 represents the first visible signs of senescence). The two subtreatments associated with each stress were grouped together. Details of main gene functions associated with some of the Venn diagram intersections are listed below the diagram.
Some gene expression changes were exclusive to stress, and were not shared with developmental changes
Microarrays were used to assess to what extent changes in gene expression at the end of the stress treatment resembled changes seen in petals of the same chronological age held under optimal conditions. Probes whose expression changed post-stress were compared with probes showing little developmental change in non-stressed conditions between stages 0 and 2 (tight bud to open flower) (Fig. 6, Supplementary Tables S2 and S4 at JXB online). Similar numbers of probes changed in expression following either stress treatment but were stable in expression between stages S0 and S2. Of those whose expression changed following dehydration stress, putative functions included respiration, cellulose biosynthesis, and a signalling-related kinase. Those that changed in response to cold included ubiquitin-mediated proteolysis, terpene and anthocyanin biosynthesis, ribosomal proteins, and a BCL-2-associated athanogene. Only a small number of probes changed in expression following both treatments but were also stable in expression from stages S0 to S2. Only one probe, a metallothionein, was up-regulated by both treatments but not from stages S0 to S2, and the eight probes whose expression was down-regulated included a gibberellin (GA)-regulated protein, a defence response protein, and a S-adenosyl-L-methionine:jasmonic acid (SAM:JA) carboxyl methyltransferase.
Fig. 6.
Venn diagram and details of microarray probes whose signal changed following cold stress (3 d cold treatment followed by a 3 h or 24 h refreshment period) or ambient dehydration stress (6 h or 24 h followed by 3 h refreshment period) compared with probes whose signal did not change during early unstressed floral development (from stage S0 to stage S2) The two subtreatments associated with each stress were grouped together. Details of the main gene functions associated with some of the Venn diagram intersections are listed below the diagram.
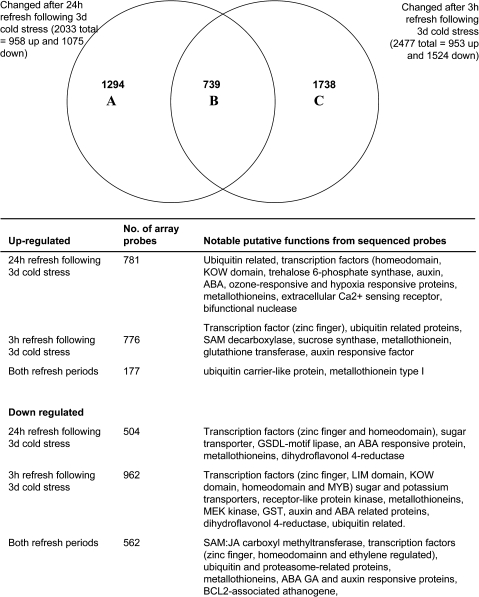
The length of stress treatment and the rehydration period affected gene expression patterns
Microarray analysis was used to investigate the effects of a longer (24 h) or shorter (3 h) refreshment period on gene expression (Fig. 7, Supplementary Tables S2 and S4 at JXB online). This experiment was designed to identify which genes show transitory changes in expression following the cold treatment and which show more stable changes in expression. Expression of less than half of the probes was changed following both refreshment periods, indicating that the length of the refreshment period has an important effect on gene expression profiles and that transcript half-life is relatively short. However, of those genes whose expression was down-regulated after the 3 h recovery period 37% were still down-regulated after 24 h recovery relative to unstressed controls. In contrast, only 19% of probes whose expression was up-regulated following 3 h of refreshment were still up-regulated after 24 h of refreshment. The expression of a number of transcription factors of various classes was differentially affected by the two refreshment periods, as well as genes related to fatty acid biosynthesis, a dihydroflavonol-4-reductase, and metallothionein-related genes.
Fig. 7.
Venn diagram and details of microarray probes whose signal changed following 3 d cold stress followed by a period of 3 h or 24 h refreshment. Details of the main gene functions associated with the Venn diagram classes are listed below the diagram.
Many more genes showed altered expression following 48 h of treatment than after just 6 h of treatment (Fig. 8, Supplementary Tables S2 and S4 at JXB online). However, changes in the expression of substantial numbers of probes were induced by either treatment. Again expression of several transcription factors was changed differentially between the two treatments, as well as a serine/threonine protein kinase, a cysteine protease, a GA-regulated protein, and an ABA-regulated protein.
Fig. 8.
Venn diagram and details of microarray probes whose signal changed following ambient dehydration stress for 6 h or 48 h, followed by a 3 h refreshment. Details of the main gene functions associated with the Venn diagram classes are listed below the diagram.
Stress treatments elicit changes in the expression of genes related to plant growth regulator (PGR) signalling
EST sequencing identified 24 genes related to PGR signalling. Of these, a large number were altered in expression in response to the stress treatments (Table 2). Of the six auxin-related genes whose expression changed as a result of the stress treatments, four were down-regulated by the cold treatments, while expression of only two was up-regulated. In contrast, cold treatment appeared to up-regulate expression of four out of the six ABA-related genes. Expression of ethylene-, JA-, and GA-related genes was down-regulated by cold, whereas expression of the cytokinin-regulated gene was up-regulated. The dehydration treatments generally induced more down-regulation of PGR-related genes compared with up-regulation (Table 2). Expression of only two PGR-related genes was up-regulated: an auxin-regulated protein and an ABA-regulated protein.
Table 2.
Expression patterns of genes related to plant growth regulator signalling, in response to stress treatments
| PGR | ID/contig | Function | Cold treatments | Dehydration treatments |
| Auxin | 12.m15 | ATAPP1 (aminopeptidase P1), auxin polar transport | D | |
| Contig 94 | Auxin binding/ubiquitin-protein ligase (AFB2) | D | ||
| 4.b13 | Auxin response factor 4 | U | ||
| 15.a10 | Auxin-induced protein | D | ||
| 14.c13 | Auxin-induced protein | D | D | |
| Contig 229 | Auxin-regulated protein | U | U | |
| ABA | Contig 226 | ABA ripening protein-like protein | U | |
| 15.c18 | ABA-responsive protein (GRAM domain) | D | ||
| Contig 1 | ABA-responsive protein-related (GRAM domain) | D | D | |
| 10.b17 | AtHVA22a protein, ABA- and stress-inducible | U | ||
| 11.l15 | Supersensitive to ABA and drought | D | ||
| 1.j9 | Transcription factor, ABA and drought responsive | U | D | |
| 8.p19 | Abscisic stress ripening protein-like protein | U | U | |
| Ethylene | 15.h15 | EIL2, ethylene transcriptional factor | D | D |
| JA | Contig 57 | SAM:JA carboxyl methyltransferase | D | D |
| Cytokinin | 10.j15 | Proline dehydrogenase/oxygenase, cytokinin inducible | U | |
| GA | Contig 122 | Gibberellin-regulated family protein | D | |
| Contig 119 | Gibberellin-regulated protein GASA2 precursor | D | D |
Cold refers to either of the two cold treatments: 3 d cold followed by 3 h or 24h refreshment, and dehydration to either of the two ambient dehydration treatments: 6 h or 48 h followed by a 3 h refreshment.
D, down-regulated; U, up-regulated. where no response is recorded, array values did not pass the statistical tests; for contigs, a change is noted if the signals from one or more of the probes from that contig on the arrays changed significantly and consistently.
Transcription factor-like genes change in expression in response to stress treatments and developmental senescence, suggesting complex patterns of regulation
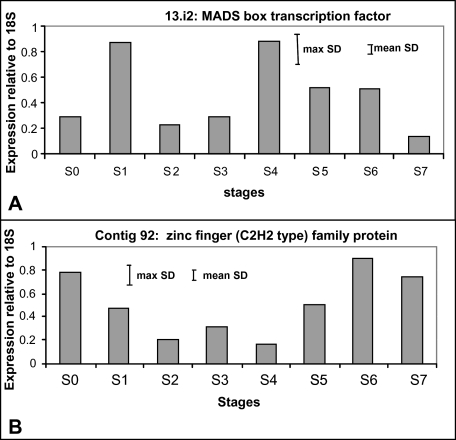
Thirty-six EST sequences were identified as transcriptional regulators representing at least 24 different genes (Table 3). The largest group (eight genes) was represented by zinc finger proteins, but transcription factors of the Myb, Lim, Hap5B, and MADS box gene families were also present. Expression patterns for 21 different contigs and singletons could be analysed from the arrays (Table 3), revealing different expression patterns. Expression of only four of the transcriptional control genes changed during development from S0 to S5. Real time RT-PCR confirmed this result for two of the transcription factors: a MADS box and a C2H2-zinc finger-type transcription factor (Fig. 9), also revealing a more complex expression pattern during development. Transcript levels of the C2H2-zinc finger transcription factor peaked at stage S0 (tightly closed bud) and again at stage S6 (mid-senescent petals) (Fig. 9A), whereas the MADS box gene peaked at stage S1 (young bud) and stage S4 (open flower) (Fig. 9B). Expression of greater numbers of the transcriptional regulators was affected by the stress treatments. Fourteen changed in response to one or other of the two cold treatments, and 17 in response to one or other of the two ambient dehydration treatments. Expression of the same number of transcription factors was changed after a 3 h and 24 h refreshment period following the 3 d cold treatment, but expression of only six was changed at both time points, and expression of four was altered after a 3 h but not after a 24 h refreshment period. Cold treatment appeared to induce more down-regulation than up-regulation of transcription factor expression, and transcript levels of five regulators were reduced as a result of both cold treatments, whereas none was up-regulated. However, expression of only two transcriptional regulators was up- or down-regulated by both the ambient dehydration stress treatments. Each transcriptional control gene showed a unique pattern of expression on the array, indicating a complex network of transcriptional regulation.
Table 3.
Expression profiles of transcriptional regulators from microarray analysis
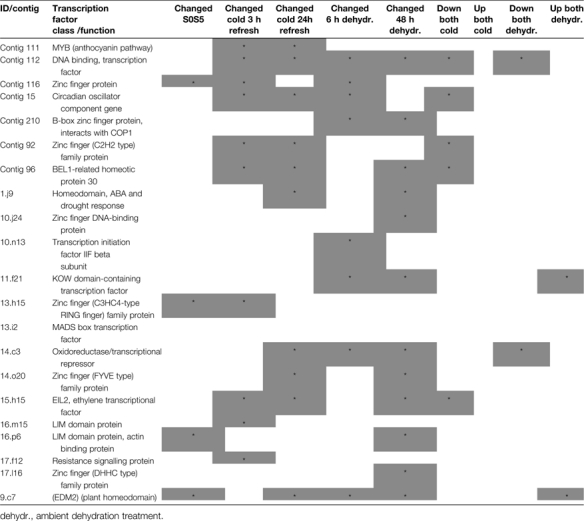 |
For contigs, a change is noted if the signals from one or more of the probes from that contig on the arrays changed significantly. Treatments are detailed in text.
dehydr., ambient dehydration treatment.
Fig. 9.
Real-time qPCR of two transcription factors to analyse their expression throughout development and senescence from stage S0 (closed bud) to stage S7 (petal abscission) (A) MADS box transcription factor and (B) zinc finger (C2H2 type) family protein. Each experiment was repeated on a second biological replicate with similar results. Maximum and minimum SDs for each experiment are shown.
Discussion
Both leaves and flowers show a remarkable recovery from moderate stress treatments (Hsiao, 1973) and, in the case of Alstroemeria flowers, their appearance following a recovery period was almost indistinguishable from that of untreated controls. However, the acceleration of leaf and flower abscission following dehydration stress is also well established (Hsiao, 1973). In Alstroemeria, following both cold and ambient dehydration stress, floral lifespan was substantially reduced from the 15 d achievable under optimal conditions (Chanasut et al., 2003). However, there were differences between the two stress treatments. Cold treatment arrested development which did not proceed appreciably during the 72 h of the treatment; however, during the ambient dehydration stress the flowers continued to develop, albeit more slowly than under optimal conditions. The effects of cold treatment are consistent with those seen in other flowers, for example rose (Faragher et al., 1986) and petunia (Ferrante et al., 2006), and in other lilies where prolonged cold storage also has an adverse effect on longevity (Ranwala and Miller, 2005). The differential physiological response of the flowers to the two kinds of stress treatment indicated that different sets of genes were being activated, as has been found in other systems (Nakashima et al., 2009) where complex networks of regulatory signals activate partially overlapping pathways.
Over 800 new Alstroemeria petal ESTs were added to the EST collection in this work, almost doubling the number of ESTs reported in previously published work (Breeze et al., 2004) and providing additional ESTs from stressed material. ESTs derived from stressed material comprised a high proportion of metallothionein genes (>20% for both cold- and dehydration-stressed material) though the proportion was not as high as the 83% reported for Alstroemeria petal libraries enriched for genes expressed in later stages of senescence (Breeze et al., 2004). This abundance of metallothionein genes has been previously reported for libraries derived from senescent leaves (Gibbings et al., 2003) petals (Channèliere et al., 2002), and ripening fruit (Aharoni et al., 2000; Moyle et al., 2004). Likewise, in studies surveying the stressed transcriptome, metallothioneins are highly represented, for example in drought-stressed rice leaves (Reddy et al., 2002) and cold-stressed Arabidopsis leaves (Jung et al., 2003). The function of such high levels of metallothioneins is not clear; however, they may be involved in sequestering metal ions to avert increases in ROS levels during stress (Reddy et al., 2002). The EST collection also included genes relating to other important cellular processes that have been previously associated with leaf and petal senescence in other systems such as remobilization of nutrients. A comparison of the functional classes of the ESTs between those derived from the cold-stressed and ambient dehydration-stressed cDNA libraries indicates a difference in the representation of different biochemical processes. Genes related to biosynthetic processes were under-represented in the library derived from dehydration-stressed material compared with the cold-stressed material. Biosynthetic process-related genes were down-regulated during senescence in Alstroemeria (Breeze et al., 2004), suggesting a possible closer similarity between dehydration stress and senescence that between cold stress and senescence.
Since stress treatments appear to accelerate processes associated with developmental senescence, it was predicted that genes whose expression was up-regulated by the stress treatments would also increase in expression during developmental senescence. This was confirmed in the case of three genes: a metallothionein and two genes related to remobilization and proteolysis, respectively. In the latter two genes, expression was reduced by the latest stage of senescence, suggesting that at this stage remobilization processes were being down-regulated. This indicates that some processes such as remobilization and ubiquitin-mediated proteolysis, associated in Alstroemeria and in other systems with floral senescence (e.g. Courtney et al., 1994), are being activated by the stress treatment. Gene expression was assessed here after a refreshment period which restored the appearance of the flowers to that of pre-senescent open flowers. Thus senescence-associated transcripts normally only evident after stage S4 are switched on, at least in some cells, in petals that have the appearance of stage S2–S3 flowers. This appears to indicate a change in the coordination of developmental senescence induced by the stress.
Microarray experiments were used to investigate to what extent stress treatments simply accelerated normal developmental/senescence processes or to what extent genes specific to the stress were up- or down-regulated. In general the microarray results were confirmed by the RT-PCR and previously published northern analysis (Breeze et al., 2004). Metallothionein expression was up-regulated on the arrays by a mean of 21-fold (±3.37 SEM over 140 array probes) between stages S0 and S5, and this is supported by a strong up-regulation on the northern (Breeze et al., 2004). Likewise, metallothionein expression was up-regulated by 2.1-fold (±0.11 SEM over 150 array probes) following cold stress (3 d followed by 3 h refreshment) and by 3.6-fold (±0.28 SEM over 150 array probes) following ambient dehydration stress (48 h followed by 3 h refreshment), in agreement with the strong up-regulation seen with the real-time RT-PCR (Fig. 4A). For the zinc finger (C2H2 type) family transcription factor, the RT-PCR and array trends were again in general agreement. On the array, stage S5 expression was 0.4-fold of stage S0 expression (±0.19 SEM over three probes), while on the RT-PCR it was 0.6 fold (Fig 8B).
In common with several other senescence-related experiments in other tissue types, far more genes were up-regulated by both development and stress than were down-regulated (Andersson et al., 2004; Lin and Wu, 2004). However, a key finding from the work presented here is that changes in gene expression induced by ambient dehydration stress were more similar to the pattern induced during developmental senescence than that induced by cold stress. Thus the data suggest that more senescence-related metabolic changes might be anticipated in flowers treated with dry ambient conditions than those treated with cold, supported by evidence that developmental events such as flower opening continue to some extent during dry ambient stress (Fig. 1). The fact that fewer genes are down-regulated compared with up-regulated may be due to fewer genes being expressed at stage S0 relative to later developmental stages. The comparison between the selected stages (stage S0 and S5) will also have missed genes whose expression patterns are more complex and only change in the intervening stages (stages S1–S4).
Many of the individual probes up-regulated by stress and development encode metallothionein-like proteins (Breeze et al., 2004); however, other genes encoding other proteins were also up-regulated, including ribosomal proteins, regulators, and enzymes related to volatile production. The latter, characterized by a terpene synthase/cyclase, was unexpected given that this variety of Alstroemeria is not scented. However, several of its wild ancestors are scented (HJ Rogers et al. unpublished data) so part of the terpenoid biosynthesis pathway (Dudareva et al., 2003) may still be present in this species. A gene putatively encoding dihydroflavonol-4-reductase was down-regulated in response to cold treatment as well as during developmental senescence. This enzyme is the first step in anthocyanin biosynthesis (Holton and Cornish, 1995), and its down-regulation following cold treatment may be related to the reduced pigmentation seen in cold-treated flowers compared with controls. Expression of proteasome subunits was also noted in senescing Arabidopsis leaves (Guo et al., 2004), and some groups have reported an increase in proteasome and ubiquitin activity during senescence (Roberts et al., 2002). However, this is believed to be due to post-transcriptional events (Basset et al., 2002). Proteasome subunit genes were repressed during Alstroemeria petal senescence and in Alstroemeria petals that had undergone stress treatments.
In Arabidopsis, expression of the majority of drought-responsive genes returned to control levels following 3 h of re-watering (Huang et al., 2008). Here results from the two refreshment periods after cold stress suggested that there was significant recovery. However, for a substantial number of genes, 3 h refreshment was insufficient to restore control transcriptional levels. This effect was more marked for down-regulated compared with up-regulated gene expression, suggesting that up-regulation of gene expression was more transient. The transient nature of gene expression induced by cold treatment may reflect the activation of transcriptional cascades known to play an important part in cold acclimation (Zhu et al., 2007). Of the 21 transcription factors examined here on the Alstroemeria microarrays, some expression was shared between the shorter or longer stress or refreshment periods. However, expression of some transcriptional regulators was only changed at one of the two time points. This suggests the possibility of transcriptional cascades also occurring during the refreshment period. Transcriptomic studies have investigated gene expression in Arabidopsis following recovery from cold stress (Oono et al., 2006) and found that expression of a substantial number of genes was up-regulated during cold acclimation but subsequently down-regulated during a 24 h recovery period, and vice versa. Likewise new sets of genes were induced during recovery from dehydration stress (Oono et al., 2003), indicating that different gene sets were required for acclimation and de-acclimation to stress treatments.
Expression of a number of genes related to plant growth regulator signalling was altered in response to the cold stress treatments in Alstroemeria petals, as was found with cold stress in Arabidopsis seedlings (Lee et al., 2005). The greater number of auxin-related ESTs from Alstroemeria petals that were down-regulated by the cold treatment is in agreement with Lee et al. (2005). The down-regulation of the genes related to methyl-jasmonate biosynthesis and ethylene response in the Alstroemeria petals following both cold treatments is also in accordance with results from Arabidopsis (Lee et al., 2005). The majority of the ABA-related genes were up-regulated following cold treatment, again in agreement with data from Arabidopsis seedlings (Lee et al., 2005), although in rice, less cross-talk was found between ABA and cold signalling than between drought and ABA (Rabbani et al., 2003).
Dehydration stress also appeared mainly to down-regulate expression of PGR-related genes, although expression of far fewer auxin-responsive genes was affected by dehydration stress compared with cold stress. Down-regulation of ABA-responsive genes was unexpected as dehydration is generally associated with up-regulation of ABA signalling (e.g. Rabbani et al., 2003). Note, however, that gene expression here was assessed after a refreshment period, so it may be that ABA signalling was only up-regulated during the period of stress, and that there is overcompensated down-regulation during refreshment. This seems likely since in other flowers such as rose (Le Page-Degivry et al., 1991) and daylily (Panavas et al., 1998) endogenous ABA levels rise in response to stress treatments when expression is measured immediately following the stress with no refreshment period.
A number of different transcription factor types were identified from the ESTs, although no WRKY type transcription factors were found. This was unexpected as several WRKY transcription factors are important in leaf senescence in both dicotyledonous (Eulgem et al., 2000) and monocotyledonous plants (Ross et al., 2007). Different patterns of expression were revealed by the microarray analysis for 21 putative transcription factors, indicating some sharing of transcriptional regulation between the stress and developmental senescence (Table 3). This overlap is well documented in leaves both at the level of genes involved in effecting these processes (Weaver et al., 1998) and at the level of transcription factors (Singh et al., 2002). Less progress has been made in understanding the transcriptional control of floral senescence, although some of the characterized transcriptional regulators have also shown an overlap in expression between petal senescence and stress (van der Krol et al., 1999). Relatively few of the transcription factors identified from the Alstroemeria ESTs changed in expression during senescence (from stage S0 to S5) on the microarrays; however, analysis of a zinc finger (C2H2 type) family protein and a MADS box protein by RT-PCR reveals a bimodal expression pattern. Expression was high in buds at stage S0 and S1, respectively, and then again from stage S5 and S4, respectively; hence both these transcription factors may well be involved in senescence regulation. Transcription factors associated with petal senescence have been identified in a few systems such as rose (Channelière et al., 2002), petunia (van der Krol, 1999), and wallflower (Price et al., 2008). However the detailed expression during petal senescence of multiple transcription factors has not been studied in many species. MADS box transcription factors may well be generally important in petal senescence since overexpression of a MADS box transcription factor in Arabidopsis delayed petal senescence and abscission (Fang and Fernandez, 2002). Several transcription factors showed patterns of regulation in one or other of the stress treatments that mimicked developmental patterns, but none responded in the same way to all stresses and development. This may indicate the means by which specific responses are elicited to particular stresses. Further analysis of the roles of the different Alstroemeria petal transcription factors may provide further insights into the regulation patterns of stress responses and developmental senescence.
Supplementary data
Supplementary data are available at JXB online.
Table S1. EST data for sequenced clones including systematic name, EMBL code, contig assignment, putative function, and accession number of nearest match.
Table S2. Normalized microarray data for all probes on the array and all experiments comparing expression of stages S0 versus S2, S0 versus S5, and the four stress treatments versus their respective controls.
Table S3. Primers used for RT-PCR analysis.
Table S4. Gene lists associated with Venn diagrams (Figs 5–8). Statistical analysis of data is detailed in the Materials and methods. Systematic names are listed for each category on the Venn diagrams.
Supplementary Material
Acknowledgments
The authors would like to thank Rowena Naylor and Rachel Edwards (Warwick HRI) and Gareth Lewis and Ezra Linley (Cardiff) for sequencing. Plant material was supplied by Adrian Franke (Oak Tree Nurseries, Egham, UK) and David Cross (New Cross Nurseries, Barnham Lane, Walberton, UK). The work was funded by a Defra grant No. HH2605.
References
- Aharoni A, Keizer LCP, Bouwmeester HJ, et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. The Plant Cell. 2000;12:647–661. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen L, Williams MH, Serek M. Reduced water availability improves drought tolerance of potted minature roses: is the ethylene pathway involved? Journal of Horticultural Science and Biotechnology. 2004;79:1–13. [Google Scholar]
- Andersson A, Keskitalo J, Sjodin A, et al. A transcriptional timetable of autumn senescence. Genome Biology. 2004;5:R24. doi: 10.1186/gb-2004-5-4-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset G, Raymond P, Malek L, Brouquisse R. Changes in the expression and the enzymic properties of the 20S proteasome in sugar-starved maize roots. Evidence for an in vivo oxidation of the proteasome. Plant Physiology. 2002;128:1149–1162. doi: 10.1104/pp.010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Wagstaff C, Harrison E, Bramke I, Rogers HJ, Stead AD, Thomas B, Buchanan-Wollaston V. Gene expression patterns to define stages of post-harvest senescence in Alstroemeria petals. Plant Biotechnology Journal. 2004;2:155–168. doi: 10.1111/j.1467-7652.2004.00059.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D. The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnology Journal. 2003;1:3–22. doi: 10.1046/j.1467-7652.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- Chanasut U, Rogers HJ, Leverentz MK, Griffiths G, Thomas B, Wagstaff C, Stead AD. Increasing flower longevity in Alstroemeria. Postharvest Biology and Technology. 2003;29:325–333. [Google Scholar]
- Channelière S, Riviere S, Scalliet G, et al. Analysis of gene expression in rose petals using expressed sequence tags. FEBS Letters. 2002;515:35–38. doi: 10.1016/s0014-5793(02)02413-4. [DOI] [PubMed] [Google Scholar]
- Courtney SE, Rider CC, Stead AD. Changes in protein ubiquitination and the expression of ubiquitin-encoding transcripts in daylily petals during floral development and senescence. Physiologia Plantarum. 1994;91:196–204. [Google Scholar]
- Doi M, Reid MS. Sucrose improves the postharvest life of cut flowers of a hybrid Limonium. HortScience. 1995;30:1058. [Google Scholar]
- Dudareva N, Martin D, Kish KM, Kolosova N, Gorenstein N, Fäldt J, Miller B, Bohlmann J. (E)-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: function and expression of three terpene synthase genes of a new terpene synthase subfamily. The Plant Cell. 2003;15:1227–1241. doi: 10.1105/tpc.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Fang S-C, Fernandez DE. Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiology. 2002;130:78–89. doi: 10.1104/pp.004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragher JD, Mayak S, Tirosh T. Physiological response of cut rose flowers to cold storage. Physiologia Plantarum. 1986;67:205–210. [Google Scholar]
- Ferrante A, Vernieri P, Tognoni F, Serra G. Changes in abscisic acid and flower pigments during floral senescence of petunia. Biologia Plantarum. 2006;50:581–585. [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gibbings JG, Cook BP, Dufault MR, Madden SL, Khuri S, Turnbull CJ, Dunwell JM. Global transcript analysis of rice leaf and seed using SAGE technology. Plant Biotechnology Journal. 2003;1:271–285. doi: 10.1046/j.1467-7652.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- Gombert J, Etienne P, Ourry A, Le Dily F. The expression patterns of SAG12/Cab genes reveal the spatial and temporal progression of leaf senescence in Brassica napus L. with sensitivity to the environment. Journal of Experimental Botany. 2006;57:1949–1956. doi: 10.1093/jxb/erj142. [DOI] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant, Cell and Environment. 2004;27:521–549. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Research Symposium Series. 1999;41:95–98. [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF. Sucrose prevents up-regulation of senescence-associated genes in carnation petals. Journal of Experimental Botany. 2007;58:2873–2885. doi: 10.1093/jxb/erm076. [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Wang T-W, Hudak KA, Schade F, Froese CD, Thompson JE. An ethylene-induced cDNA encoding a lipase expressed at the onset of senescence. Proceedings of the National Academy of Sciences, USA. 2000;97:8717–8722. doi: 10.1073/pnas.140213697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao TC. Plant responses to water stress. Annual Review of Plant Physiology. 1973;24:519–570. [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S-H, Lee J-Y, Lee D-H. Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Molecular Biology. 2003;52:553–567. doi: 10.1023/a:1024866716987. [DOI] [PubMed] [Google Scholar]
- Khodakovskaya M, Li Y, Li JS, Vankova R, Malbeck J, McAvoy R. Effects of cor15a-IPT gene expression on leaf senescence in transgenic Petunia×hybrida and Dendranthema×grandiflorum. Journal of Experimental Botany. 2005;56:1165–1175. doi: 10.1093/jxb/eri109. [DOI] [PubMed] [Google Scholar]
- Langston BJ, Bai S, Jones ML. Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene-insensitive (etr1-1) transgenic petunias. Journal of Experimental Botany. 2005;56:15–23. doi: 10.1093/jxb/eri002. [DOI] [PubMed] [Google Scholar]
- Le Page-Degivry MT, Orlandini M, Garello G, Barthe P, Gudin SJ. Regulation of ABA levels in senescing petals of rose flowers. Plant Growth Regulation. 1991;10:6–72. [Google Scholar]
- Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold responsive transcriptome and its regulation by ICE1. The Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverentz MK, Wagstaff C, Rogers HJ, Stead AD, Chanasut U, Silkowski H, Thomas B, Weichert H, Feussner I, Griffiths G. Characterization of a novel lipoxygenase-independent senescence mechanism in Alstroemeria peruviana floral tissue. Plant Physiology. 2002;130:273–283. doi: 10.1104/pp.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annual Review of Plant Biology. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lin J-F, Wu S-H. Molecular events in senescing Arabidopsis leaves. The Plant Journal. 2004;39:612–628. doi: 10.1111/j.1365-313X.2004.02160.x. [DOI] [PubMed] [Google Scholar]
- Meyer RC, Jr., Goldsbrough PB, Woodson WR. An ethylene-responsive flower senescence-related gene from carnation encodes a protein homologous to glutathione S transferases. Plant Molecular Biology. 1991;17:277–281. doi: 10.1007/BF00039505. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Irie K, Hirayama T, Hayashida N, Yamaguchi- Shinozaki K, Matsumoto K, Shinozaki K. A gene encoding a mitogen-activated protein kinase kinase kinase is induced simultaneously with genes for a mitogen-activated protein kinase and an S6 ribosomal protein kinase by touch, cold, and water stress in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1996;93:765–769. doi: 10.1073/pnas.93.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR. Developing pineapple fruit has a small transcriptome dominated by metallothionein. Journal of Experimental Botany. 2004;56:101–112. doi: 10.1093/jxb/eri015. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch S, Alegre L. Die and let live: leaf senescence contributes to plant survival under drought stress. Functional Plant Biology. 2004;31:203–216. doi: 10.1071/FP03236. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiology. 2009;149:88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Seki M, Satou M, Iida K, Akiyama K, Sakurai T, Fujita M, Yamaguchi-Shinozaki K, Shinozaki K. Monitoring expression profiles of Arabidopsis gene expression during rehydration process after dehydration using ca. 7000 full-length cDNA microarray. The Plant Journal. 2003;34:868–887. doi: 10.1046/j.1365-313x.2003.01774.x. [DOI] [PubMed] [Google Scholar]
- Oono Y, Seki M, Satou M, Iida K, Akiyama K, Sakurai T, Fujita M, Yamaguchi-Shinozaki K, Shinozaki K. Monitoring expression profiles of Arabidopsis genes during cold acclimation and deacclimation using DNA microarrays. Functional and Integrative Genomics. 2006;3:212–234. doi: 10.1007/s10142-005-0014-z. [DOI] [PubMed] [Google Scholar]
- Panavas T, Walker EL, Rubinstein B. Possible involvement of abscisic acid in senescence of daylily petals. Journal of Experimental Botany. 1998;49:1987–1997. [Google Scholar]
- Price AM, Aros Orellana DF, Stevens R, Acock R, Buchanan-Wollaston V, Stead AD, Rogers HJ. A comparison of leaf and petal senescence in wallflowers (Erysimum linifolium) reveals common and distinct patterns of gene expression and physiology. Plant Physiology. 2008;147:1898–1912. doi: 10.1104/pp.108.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primack RB. Longevity of individual flowers. Annual Review of Ecology and Systematics. 1985;16:15–37. [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwala AP, Miller WB. Gibberellin(4+7), benzyladenine, and supplemental light improve postharvest leaf and flower quality of cold-stored ‘Stargazer’ hybrid lilies. Journal of the American Society of Horticultural Science. 1998;123:563–568. [Google Scholar]
- Ranwala AP, Miller WB. Effects of cold storage on post harvest leaf and flower quality of potted Oriental-, Asiatic- and LA-hybrid lily cultivars. Scientia Horticulturae. 2005;105:383–392. [Google Scholar]
- Reddy AR, Ramakrishna W, Sekhar CA, Nagabhushana I, Babu PR, Bonaldo MF, Soares MB, Bennetzen JL. Novel genes are enriched in normalized cDNA libraries from drought stressed seedlings of rice (Oryza sativa L. subsp. Indica cv. Nagina 22) Genome. 2002;45:204–211. doi: 10.1139/g01-114. [DOI] [PubMed] [Google Scholar]
- Reid M. Produce facts Alstroemeria, Peruvian lily recommendations for maintaining postharvest quality. University of California, Davis: Postharvest Technology Research & Information Center; 2004. [Google Scholar]
- Roberts I, Murray PF, Passeron S, Barneix AJ. The activity of the 20S proteasome is maintained in detached wheat leaves during senescence in darkness. Plant Physiology and Biochemistry. 2002;40:161–166. [Google Scholar]
- Roca M, James C, Pruzinska A, Hortensteiner S, Thomas H, Ougham H. Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of. Lolium temulentum. Phytochemistry. 2004;65:1231–1238. doi: 10.1016/j.phytochem.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Rogers HJ. Programmed cell death in floral organs: how and why do flowers die? Annals of Botany. 2006;97:309–315. doi: 10.1093/aob/mcj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2007;49:827–842. [Google Scholar]
- Rudnicki RM, Nowak J, Goszczynska DM. Cold storage and transportation conditions for cut flowers, cuttings and potted plants. Acta Horticulturae. 1991;298:225–236. [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, et al. Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Molecular Biology. 2004;56:29–55. doi: 10.1007/s11103-004-2200-0. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, van Poecke RMP, Vorst OFJ, Voogt C, van Leeuwen W, Borst-Vrensen TWM, Takatsuji H, van der Plas LHW. Developmental and wound-, cold-, desiccation-, ultraviolet-B-stress-induced modulations in the expression of the Petunia zinc finger transcription factor gene. ZPT2-21. Plant Physiology. 1999;121:1153–1162. doi: 10.1104/pp.121.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Balk PA, van Houwelingen AM, Hoeberichts FA, Hall RD, Vorst O, van der Schoot C, van Wordragen MF. Gene expression during anthesis and senescence in Iris flowers. Plant Molecular Biology. 2003;53:845–863. doi: 10.1023/B:PLAN.0000023670.61059.1d. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Woltering EJ. Physiology and molecular biology of petal senescence. Journal of Experimental Botany. 2008;59:453–480. doi: 10.1093/jxb/erm356. [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Chanasut U, Harren FJM, L–J Laarhoven, Thomas B, Rogers HJ Stead AD. Ethylene and flower longevity in Alstroemeria: relationship between tepal senescence, abscission and ethylene biosynthesis. Journal of Experimental Botany. 2005;56:1007–1016. doi: 10.1093/jxb/eri094. [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Leverentz MK, Griffiths G, Thomas B, Chanasut U, Stead AD, Rogers HJ. Protein degradation during senescence of Alstroemeria petals. Journal of Experimental Botany. 2002;53:233–240. doi: 10.1093/jexbot/53.367.233. [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Malcolm P, Rafiq A, Leverentz M, Griffiths G, Thomas B, Stead A, Rogers HJ. Programmed cell death (PCD) processes begin extremely early in Alstroemeria petal senescence. New Phytologist. 2003;160:49–59. doi: 10.1046/j.1469-8137.2003.00853.x. [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Rogers HJ, Leverentz MK, Griffiths G, Thomas B, Chanasut U, Stead AD. Characterisation of Alstroemeria flower vase life. Acta Horticulturae. 2001;543:161–175. [Google Scholar]
- Wagstaff C, Yang TJW, Stead AD, Buchanan-Wollaston V, Roberts JA. A molecular and structural characterisation of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. The Plant Journal. 2009;57:690–705. doi: 10.1111/j.1365-313X.2008.03722.x. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM. A comparison of the expression patterns of several senescence associated genes in response to stress and hormone treatment. Plant Molecular Biology. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Zhu J, Dong C-H, Zhu J-K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.