Abstract
H2O2 is known as a signal molecule in plant cells, but its role in the regulation of aqbscisic acid (ABA) and gibberellic acid (GA) metabolism and hormonal balance is not yet clear. In this study it was found that H2O2 affected the regulation of ABA catabolism and GA biosynthesis during seed imbibition and thus exerted control over seed dormancy and germination. As seen by quantitative RT-PCR (QRT-PCR), H2O2 up-regulated ABA catabolism genes (e.g. CYP707A genes), resulting in a decreased ABA content during imbibition. This action required the participation of nitric oxide (NO), another signal molecule. At the same time, H2O2 also up-regulated GA biosynthesis, as shown by QRT-PCR. When an ABA catabolism mutant, cyp707a2, and an overexpressing plant, CYP707A2-OE, were tested, ABA content was negatively correlated with GA biosynthesis. Exogenously applied GA was able to over-ride the inhibition of germination at low concentrations of ABA, but had no obvious effect when ABA concentrations were high. It is concluded that H2O2 mediates the up-regulation of ABA catabolism, probably through an NO signal, and also promotes GA biosynthesis. High concentrations of ABA inhibit GA biosynthesis but a balance of these two hormones can jointly control the dormancy and germination of Arabidopsis seeds.
Keywords: ABA, ABA catabolism, Arabidopsis, GA, GA biosynthesis, hydrogen peroxide (H2O2), nitric oxide (NO), seed dormancy
Introduction
Seed germination is a complex process. Germination incorporates those events that commence with the uptake of water by the quiescent dry seed and terminate with the elongation of the embryonic axis (Bewley and Black, 1994; Holdsworth et al., 2008). Seeds of most angiosperms are dormant at maturity, and the dormancy must be lost before germination can occur (Bewley, 1997). Seed dormancy has been defined by Finch-Savage and Leubner-Metzger as the incapacity of a viable seed to germinate under favourable conditions (Finch-Savage and Leubner-Metzger, 2006). Many factors are involved in seed dormancy regulation, including some plant hormones, such as abscisic acid (ABA), gibberellic acid (GA), and ethylene (Bewley, 1997; Zhou et al., 1998; Ghassemian et al., 2000; Nakajima et al., 2006; Carrera et al., 2008; Holdsworth et al., 2008), some environmental factors, such as light intensity and low temperatures (Holdsworth et al., 2008), and several signalling molecules, such as nitric oxide (NO) and some reactive oxygen species (ROS) (Batak et al., 2002; Bethke et al., 2004, 2006; Sarath et al., 2007). However, the mechanisms of dormancy holding and breaking remain unclear because it is unknown how these factors are inter-related. The mechanisms of ABA catabolism and GA biosynthesis regulation are of particular interest.
H2O2 acts as a signalling molecule, participating in a series of processes including plant development, stress responses, and programmed cell death (Pei et al., 2000; Bethke and Jones, 2001; Apel and Hirt, 2004; Foyer and Noctor, 2005). In plants, H2O2 is generated in chloroplasts, mitochondria, and peroxisomes (Mittler et al., 2004). Plasma membrane NAD(P)H oxidase is reported to be the pivotal enzyme involved in H2O2 generation (Kauss and Jeblick, 1995, 1996; Mur et al., 1996; Shirasu et al., 1997). The effect of H2O2 on plant development, stress responses, and programmed cell death has been thoroughly investigated (Pei et al., 2000; Bethke and Jones, 2001; Apel and Hirt, 2004; Foyer and Noctor, 2005). The effect of H2O2 on seed germination has also been researched by some investigators. Fontaine et al. (1994) indicated that thioredoxin reduction by NADPH produced via the oxidative pentose phosphate pathway allows the mobilization of storage proteins of cereals, leading to germination. H2O2 is also regarded as having a function as a promoter of seed germination by oxidizing germination inhibitors in Zinnia elegans seeds (Ogawa and Iwabuchi, 2001). The sources of H2O2 during seed germination are not clear.
Bailly et al. (2008) indicated that in the dry state enzymes are probably not active and in this case ROS probably originate from non-enzymatic reactions such as lipid peroxidation or Amadori and Maillard reactions, and in hydrated seeds can be produced during the catabolism of lipids (glyoxysomes) and purines (peroxisomes), respiratory activity (mitochondria), electron transfer in photosystems (chloroplasts), or through the activity of NADPH oxidase (plasma membrane), amine oxidase, and peroxidase (cell wall) or cytochrome P450 (cytosol). They also indicated that accumulated H2O2 during imbibition is essential for seed dormancy breaking
ABA plays an important role in a number of physiological processes such as seed maturation, growth, and developmental regulation, seed dormancy, and adaptive responses to environmental stresses (Zeevaart and Creelman, 1988; Hoffmann-Benning and Kende, 1992; Kuwabara et al., 2003; Nambara and Marion-Poll, 2005). In addition, ABA has been shown to be an important positive regulator in both the induction of dormancy during seed maturation and the maintenance of the dormant state in imbibed seeds (Finkelstein et al., 2002; Himmelbach et al., 2003). ABA-deficient mutants in Arabidopsis, such as aba1, aba2, and aao3, show the absence of primary dormancy in mature seeds (Leon-Kloosterziel et al., 1996; Finkelstein et al., 2002; Himmelbach et al., 2003). Some ABA-insensitive mutants such as abi1, abi2, and abi3 also lack or have decreased primary dormancy in mature seeds (Raz et al., 2001; Finkelstein et al., 2002; Himmelbach et al., 2003; Kushiro et al., 2004; Nambara and Marion-Poll, 2005), whereas overexpression of some ABA biosynthesis genes increases seed ABA content and enhances seed dormancy or delays germination (Finkelstein et al., 2002; Kushiro et al., 2004; Nambara and Marion-Poll, 2005; Holdsworth et al., 2008).
Some investigations have shown that ABA catabolism also plays a major role in seed dormancy maintenance and dormancy break. Seeds of the mutant cyp707a2, lacking the key enzyme in ABA catabolism, (+)-abscisic acid 8'-hydroxylase, accumulate much more ABA and show stronger dormancy during imbibition than the wild type (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006). Earlier results (Liu et al., 2009) also indicated that CYP707A2 plays a major role in ABA catabolism during imbibition and regulates seed dormancy.
GA is a major plant hormone in a number of physiological processes, such as seed germination, stem elongation, leaf expansion, flowering, and seed development (Davies, 1993; Ogawa et al., 2003; Yamauchi et al., 2004). Together with ABA, GA is also involved in seed dormancy and germination control (Ogawa et al., 2003) and is found to promote seed germination in many species (Koornneef and van der Veen, 1980; White et al., 2000; Yamauchi et al., 2004). Inhibitors of GA biosynthesis, such as paclobutrazol (PAC) and uniconazole, reduce seed germination in Arabidopsis (Jacobsen and Olszewski, 1993; Leon-Kloosterziel et al., 1996; Toh et al., 2008). Several GA-deficient mutants, such as ga1-3 and ga2-1, have also delayed seed germination (Koornneef and van der Veen, 1980). It is proposed that GA plays two major roles in stimulating germination in Arabidopsis. The first role is in inducing radicle protrusion apparently by weakening the tissue that surrounds the embryo. The second role is in increasing the growth potential of the embryo, as indicated by the reduced growth rate of GA-deficient embryos (Groot and Karssen, 1987; Ogawa et al., 2003).
The roles of ABA and GA in seed germination control have been indicated to be antagonistic by some investigators (Razem et al., 2006; Weiss and Ori, 2007; Toh et al., 2008). For example, GA induces transcription of α-amylase in the aleurone layer of cereal seeds that is significantly suppressed by ABA (Rogers and Rogers, 1992; Gómez-Cadenas et al., 2001; Zentella et al., 2002). ABA is also reported to inhibit seed germination by inhibiting GA biosynthesis directly under high temperature (Toh et al., 2008). However, because of the complexity in their signalling pathways, the relationship of ABA and GA is not well understood in terms of their regulation. In this study, it was found that both GA and ABA are under the regulation of H2O2 in seed dormancy. Exogenous H2O2 increases ABA catabolism by enhancing the expression of CYP707A genes. At the same time, H2O2 enhances GA biosynthesis via enhancement of GA biosynthesis genes such as GA3ox and GAw20ox genes. The inhibition of seed germination by a low concentration of ABA is reversed by GA, but apparently GA cannot over-ride the effect of high concentration of ABA. The present results also suggest that the H2O2-enhanced ABA catabolism requires the participation of NO, another small signalling molecule.
Materials and methods
Plant materials
The plants were grown in a growth chamber with a 16 h photoperiod at a photon flux density of ∼200 μmol m−2 s−1 at a daytime temperature of 23 °C and a night-time temperature of 20 °C. In order to minimize the effect of seed maturation and storage conditions, plants of each genotype tested were grown in different sections of the same pot and seeds were harvested at the same time. Seeds were harvested in bulk 30 d after the petals appeared on the first flowers. These seeds maintained stronger dormancy. Only freshly harvested seeds were used in the experiments. The rest of the seeds were stored at –80 °C, at which temperature dormancy can be maintained for more than a year (Millar et al., 2006; Fujii et al., 2007).
T-DNA insertion line
The seeds of Arabidopsis thaliana cyp707a2 (SALK_083966) generated by the Salk Institute Genomic Analysis Laboratory (http://signal.sal.edu/) were obtained from the ABRC. The seeds were planted on agar plates containing kanamycin, and the kanamycin-resistant plants were transferred to soil. Seeds were harvested separately from individual plants. Subsequently, to confirm the mutant line as homozygous, PCR was performed with the genomic DNA of cyp707a2 using gene-specific oligonucleotides (LP, AATCCCAAATATGCCTTAGGC; and RP, TATGTGGGGACTTTGATGGAC).
Chemical treatments
Sodium nitroprusside (SNP) was used as the NO donor to release NO steadily, and 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (c-PTIO) was used as the NO scavenger (Bright et al., 2006). Diphenyliodonium (DPI) was used to inhibit NADPH oxidase to decrease production of H2O2 (Orozco-Cardenas et al., 2001). Diniconazole and PAC were used to inhibit ABA catabolism and GA biosynthesis, respectively (Han et al., 2004; Kitahata et al., 2005; Toh et al., 2008).
Germination assay
Fifty seeds were placed in 55 mm diameter Petri dishes with three Whatman No. 1 filter papers and 2.2 ml of sterile double-distilled water or treatment solutions. Plates were then placed in a 21 °C growth chamber under continuous light at 100 μM m−2 s−1 for 7 d. The seeds were regarded as germinated when the radicle emerged. Experiments were performed in quadruplicate for each treatment.
Determination of NO
NO was detected by the Nitric Oxide (total) Detection Kit (Assay Designs, USA). About 0.2 g of seeds were put into 1.5 ml tubes, then 200 μl of reaction buffer and 100 μl final dilution of NADH were added; 100 μl of water was added to a parallel tube as a control. Then 100 μl of nitrate reductase (NR) was added to the samples and 100 μl of reaction buffer was added to the control tubes. A 400 μl aliquot of reaction buffer without seeds, NADPH, and NR acted as a blank. The blank, control, and sample tubes were mixed well and incubated at 37 °C for 30 min. After incubation they were centrifuged at 3000 g for 1 min and 300 μl of supernatant was transformed to a new tube. A 100 μl aliquot of Griess reagent I was added to the control, sample, and blank, and after being well mixed, 100 μl of Griess reagent II was added. The tubes were mixed by shaking and then they were incubated at 25 °C for 10 min. The optical density (OD) of samples and controls was measured at 540 nm. The OD of each sample was labelled as ODs and that of each control as ODc. Then the average net OD was calculate and labelled as ODn. Each average ODn=average ODs–average ODc. Each ODn could be calculated from a standard curve. Sodium nitrate at 0–100 μM was used as a standard, and a standard curve was produced. The amount of NO released was equal to the amount of nitrate. The mechanism of this kit is transformation of NO to nitrite and its measurement.
Extraction and determination of ABA
For estimation of endogenous ABA levels of imbibed seeds, 0.2 g of seeds was homogenized in 1 ml of distilled water and then shaken at 4 °C overnight. The homogenates were centrifuged at 12 000 g for 10 min at 4 °C and the supernatant were directly used for ABA assay. ABA analysis was carried out using the radioimmumoassay (RIA) method as described by Quarrie et al. (1988). The 450 μl reaction mixture contained 200 μl of phosphate buffer (pH 6.0), 100 μl of diluted antibody (Mac 252) solution, 100 μl of [3H]ABA (∼8000 cpm) solution, and 50 μl of crude extract. The mixture was then incubated at 4 °C for 45 min and the bound radioactivity was measured in pellets precipitated with 50% saturated (NH4)2SO4– with a liquid scintillation counter.
Determination of H2O2
An Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen, Carlsbad, CA, USA) was used to measure H2O2 production in 2-week-old plants. Leaves were frozen in N2 and then ground. Then 500 μl of phosphate buffer (20 mM K2HPO4, pH 6.5) was added to 50 mg of ground frozen tissue. After centrifugation, 50 μl of the supernatant was incubated with 0.2 U ml−1 horseradish peroxidase and 100 μM Amplex Red reagent (10-acetyl-3,7-dihydrophenoxazine) at room temperature for 30 min in darkness. The fluorescence was quantified using FLUOStar Optima (excitation at 560 nm and emission at 590 nm) (Xing et al., 2008).
QRT-PCR analysis
Total RNA was isolated from seeds or leaves by an RNeasy kit (Invitrogen). DNA impurities in the isolated RNA were digested before synthesizing the cDNA by adding DNase (Invitrogen) and incubation for 30 min at 37 °C. DNase was then inactivated by incubation for 10 min at 65 °C. Then 2 μg of RNA was reverse transcribed to cDNA with SuperScriptIII RTS First-Strand cDNA Synthesis Kit (Invitrogen). After that, the cDNA was diluted 10 times, and 4 μl of cDNA was used to carry out the quantitative RT-PCR (QRT-PCR). IQ™ SYBR Green Supermix (Bio-Rad) was used for the QRT-PCR. Actin2 acted as the intramural standard. The QRT-PCR was executed with iCycle (Bio-Rad). The primers that were used in QRT-PCR are as follows: CYP707A1 (F, TTGGAAAGAGGAGACTAGAG; R, GTGAACCACAAAAGAGGAAC), CYP707A2 (F, AAATGGAGTGCACTCATGTC; R, CCTTCTTCATCTCCAATCAC), CYP707A3 (F, ATTCTTGTCCAGGCAATGAG; R, ATAGGCAATCCATTCTGAGG), CYP707A4 (F, GAAAGGAATACAGTACAGTC; R, GGATTAGATTTGGCTAACTAC), GA20ox1 (F, GCCTGTAAGAAGCACGGTTTCT; R, CTCGTGTATTCATGAGCGTCTGA), GA20ox2 (F, CCCAAGGCTTTCGTTGTCAA; R, CCGCTCTATGCAAACAGCTCT), GA20ox3 (F, TCGTGGACAACAAATGGCA; R, TGAAGGTGTCGCCTATGTTCAC), GA3ox1 (F, TCCGAAGGTTTCACCATCACT; R, TCGCAGTAGTTGAGGTGATGTTG), GA3ox2 (F, GTTTCACCGTTATTGGCTCTCC; R, TCACAGTATTTGAGGTGGTGGC), RD29A (F, TGCACCAGGCGTAACAGGTA; R, TAATCGGAAGACACGACAGGA), RD29B (F, GAGCATCCAAAGTGTTGAAGAAAGT; R, GGTCTTGCTCGTCATACTCATCAT), XTH5 (F, CACGTCGATGGATGTGAAGCT; R, CTTTCTGATCCCACCAACGTTT), EXP2 (F, CCTCCAAACTTTGCCTTAGCT; R, CGGCCAAGTCAAAGTGCTTAA), NCED6 (F, TGAGAGACGAAGAGAAAGAC; R, GTTCCTTCAACTGATTCTCG), NCED9 (F, GGAAAACGCCATGATCTCACA; R, AGGATCCGCCGTTTTAGGAT), GA2ox2 (F, CCCTCAAATTTTCCGTGAGT; R, CAGCATTTTACTCAGAGTGTC), CAT1 (F, ACACATACGTGTTTTGGTGTTGAGC; R, CACCCGAGTTTGTAGTGAAGAAAGG), CAT2 (F, CTCCAAGCTCTCTTCTCATCAAACCAT; R, GGAGCTCGGAGAAAGTCAGCACAA), CAT3 (F, GAGGGATATTCGTGGTTTTGCTGTC; R, TTTGTTTTCGGGTTAGGTTTCAACG), AT1G19230 (F, TCACTTTTACTGGGTCACAAGGGAG; R, AACTCCATGTTTGGCATG-GTTCA), AT4G11230 (F, GGTACCGCAAAACGGTATGGATGT; R, AATCATCTCCAGGGGAAGAAGTAATAGA), AT4G25090 (F, TTGGCAAAGAGTTTGGGTGATAGC; R, GGTAACAGAATTAGGGCCATGTTTAGC), Actin2 (F, TGTGCCAATCTACGAGGGT; R, GCTGGTCTTTGAGGTTTCC).
Generation of the CPY707A2-overexpressing line
Full-length Arabidopsis CPY707A2 cDNA was obtained by using reverse transcription-PCR and cloned into the pENTR-TOPO cloning vector (Invitrogen) and sequenced. After the LR reaction, CPY707A2 cDNA was inserted into the pGWB5 vector (a gift from Professor Liang, Yangzhou University) which had a 35S promoter; this vector was named pGWB5-CPY707A2. Transgenic Arabidopsis containing the cauliflower mosaic virus (CaMV) 35S promoter was generated using the floral dipping method (Clough and Bent, 1998) and transferred into Col-0 wild-type plants. Transformed plants were selected by growth on hygromycin-containing medium. Plants of the second generation after transformation were used for the experiments. The empty pGWB5 vector (the ccdb gene was substituted by a nonsense segment with a termination codon) which acted as control was also transferred into Col-0 wild-type plants.
Accession numbers
Sequence data from the article can be found in the GenBank data libraries or TIGR database (Arabidopsis thaliana Genome Project) under the following accession numbers: CYP707A1, At4g19230; CYP707A2, At2g29090; CYP707A3, At5g45340; CYP707A4, At3g19270; GA20ox1, At4g25420; GA20ox2, At5g51810; GA20ox3, At5g07200; GA3ox1, At1g15550; GA3ox2, At1g80340; XTH5, At5g13870; EXP2, At5g05290; RD29A, AT5g52310; RD29B, AT5g52300; GA2ox2, At1g30040; NCED6, At3g24220; NCED9, At1g78390; CAT1, At1g17680; CAT2, At4g35090; CAT3, At1g20620; and Actin2, At3g18780.
Results
H2O2 reduces dormancy of freshly harvested Arabidopsis seeds
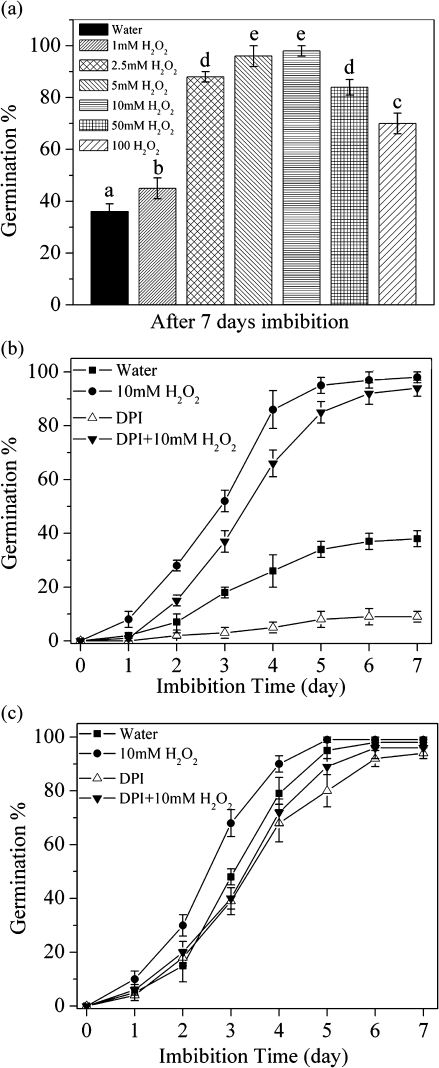
After 7 d of imbibition in 5 mM H2O2, the dormancy of freshly harvested Arabidopsis seeds was broken. Much higher H2O2 concentrations, such as 100 mM, had less effect (Fig. 1a). The H2O2 effect was further confirmed by supplying DPI, a H2O2 scavenger, to reduce the level of H2O2 (Levine et al., 1994; Alvarez et al., 1998; Lee et al., 1999). As shown in Fig. 1b, DPI enhanced seed dormancy significantly while exogenous H2O2 completely reversed the effect of DPI. When supplied to non-dormant seeds, H2O2 increased seed germination and DPI slightly slowed down the process (Fig. 1c).
Fig. 1.
The effect of H2O2 on seed germination and dormancy break in Arabidopsis. (a). Effect of different H2O2 concentrations on dormancy break of freshly harvested wild-type seed. Freshly harvested seeds were imbibed in water or different concentrations of H2O2 and the germination ratio was counted on the seventh day. (b) Effect of H2O2 and its production inhibitor DPI on freshly harvested seed dormancy and germination. (c) Effect of H2O2 and its production inhibitor DPI on non-dormant seed germination. Data represent the means±SE of four replicates, with 50 seeds each in a, b, and c. An ANOVA test followed by a rank test was performed. Different letters in (a) are used to indicate means that are significantly different (P <0.05).
H2O2 regulates genes involved in ABA catabolism and GA synthesis during imbibition
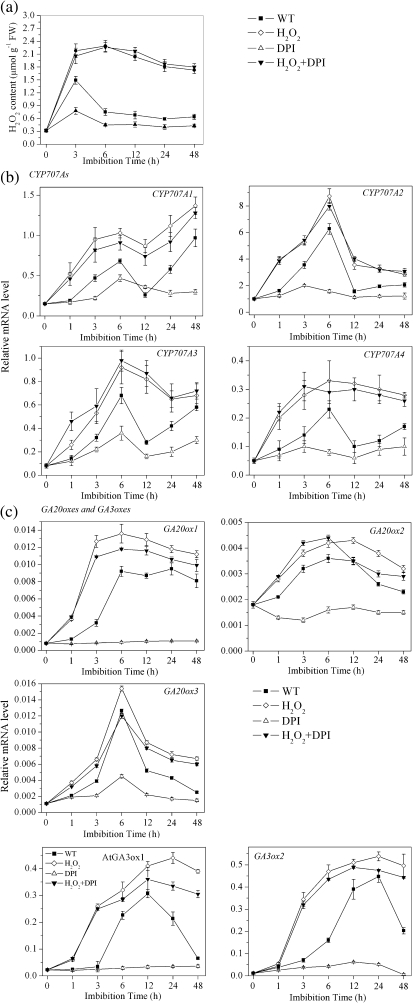
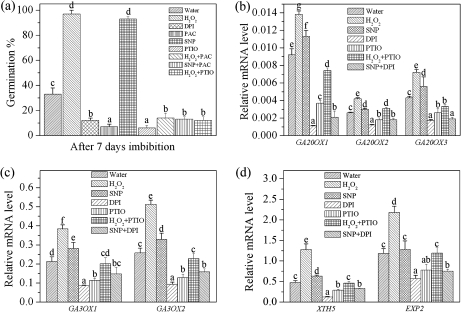
ABA and GA are known to regulate seed germination and dormancy but their signalling pathways have not yet been established. In the present experiments, H2O2 acted as a regulator of genes involved in both ABA and GA metabolism. As shown in Fig. 2a, the release of H2O2 from the imbibed seeds rapidly increased in the first few hours and reached a peak at 3 h, followed by a decrease after 6 h. DPI treatment decreased this H2O2 release significantly. When expression of ABA 8'-hydroxylase (CYP707A1, CYP707A2, CYP707A3, and CYP707A4), 9-cis epoxcartenoid dioxygenase (NECD6 and NCED9), GA 20-oxidase (GA20ox1, GA20ox2, and GA20ox3), GA 3-oxidase (GA3ox1 and GA3ox2), and GA 2-oxidase (GA2ox2) was investigated following different treatments during imbibition, the expression of four ABA catabolism genes (CYP707A genes) were induced within a few hours after imbibition of water, and then decreased after 6 h. When imbibed with H2O2, the transcription level of these four genes increased much more rapidly and was maintained at a high level during the entire imbibition period (Fig. 2a). When release of H2O2 was inhibited by DPI, there was no increase in transcription levels of these four genes compared with water. Further addition of exogenous H2O2 was able to reverse the inhibitory effect of DPI on transcription of the four genes (Fig. 2b). Transcription levels of the ABA biosynthetic genes NCED6 and NCED9 decreased in the water control within the first 6 h of imbibition and then increased. Exogenous H2O2 slightly enhanced the expression of these two genes (Supplementary Fig. S1 available at JXB online).
Fig. 2.
Effect of H2O2 on the expressions of ABA catabolism and GA biosynthesis genes during imbibition. (a) Change of H2O2 content during imbibition under different treatments in the first 48 h of imbibition. (b) Change in the transcript levels of ABA catabolism genes in the first 48 h of imbibition. All four CYP707A genes were determined by QRT-PCR. (c) Change in the transcript levels of GA biosynthesis genes in the first 48 h of imbibition. All three GA20ox genes and two GA3ox genes were determined by QRT-PCR. H2O2 (10 mM) and DPI (10 μM) were used for these experiments. Values are the means±SE (n=4 for a and n=3 for b and c). An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05).
Expression of three GA20ox and two GA3ox genes was also significantly induced in the water control in the initial hours of imbibition. As shown in Fig. 2c, transcription levels of all three GA20ox genes increased rapidly during the first 6 h. Levels of GA20ox1 remained high for the entire imbibition period, but GA20ox2 and GA20ox3 transcription levels decreased to a lower level after 6 h. Two GA3ox genes displayed a delayed initiation compared with the GA20ox genes. GA3ox1 reached its maximum at 12 h and decreased thereafter, while GA3ox2 peaked at 24 h. H2O2 enhanced the transcription of all five GA biosynthesis genes to a high level and these elevated levels were maintained throughout the remainder of the imbibition period. DPI significantly inhibited transcription of the five genes, but addition of exogenous H2O2 was able to reverse the DPI inhibition (Fig. 2c). The transcription level of the GA catabolic gene GA2ox2 decreased during the first 6 h of imbibition and increased thereafter. Exogenous H2O2 enhanced this gene expression slightly (Supplementary Fig. S1 at JXB online).
H2O2 requires NO for regulation of genes involved in ABA catabolism and breaking of seed dormancy
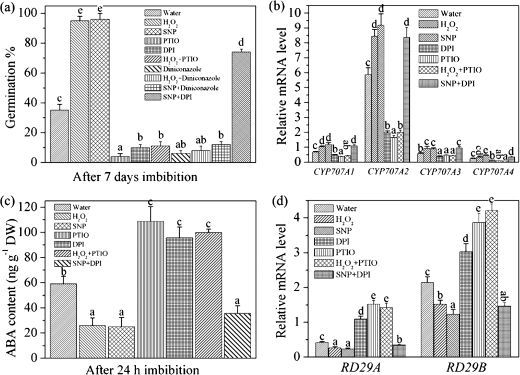
As shown in Fig. 3a, both NO and H2O2 caused dormancy break in freshly harvested seeds. Treatment with DPI or with the NO scavenger c-PTIO enhanced seed dormancy. Treatment with the NO donor SNP substantially reversed the inhibition caused by DPI, whereas exogenous H2O2 failed to reverse the inhibition caused by c-PTIO. The effect of H2O2 and SNP on seed dormancy break could be reversed by the ABA catabolism inhibitor diniconazole, which supposedly inhibits ABA 8'-hydroxylase activity (Fig. 3a).
Fig. 3.
The function of NO for H2O2-regulated ABA catabolism. (a) Effect of H2O2, NO, and their production inhibitor DPI or scavenger c-PTIO, or the ABA catabolism inhibitor diniconazole on freshly harvested seed dormancy and germination; the germination ratio was counted on the seventh day. (b) Change in the transcript levels of ABA catabolism genes under different treatments at 6 h imbibition. (c) Change in ABA content after 24 h imbibition. ABA contents were measured by RIA. (d) Change in the transcript levels of the ABA-regulated genes RD29A and RD29B under different treatments at 24 h imbibition. H2O2 (10 mM), DPI (10 μM), SNP (200 μM), c-PTIO (200 μM), and diniconazole (10 μM) were used for these experiments. Values are the means±SE (n=3 for a–d). An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05).
Earlier results indicated that CYP707A2 was much more abundant than the other CPY707A genes during germination (Saito et al., 2004; Okamoto et al., 2006), consistent with what was found in the present study (Fig. 3b). Both SNP and H2O2 enhanced CYP707A gene expression, especially that of CYP707A2 (Fig. 3b). Treatment with c-PTIO or DPI decreased CYP707A gene expression. Treatment with SNP reversed the effect of DPI, while exogenous H2O2 treatment was not able to reverse the effect of c-PTIO (Fig. 3b).
The changes in ABA content reflected the expression of ABA catabolic genes. SNP and exogenous H2O2 treatments enhanced ABA catabolic gene expression, while scavengers or inhibitors decreased expression of these ABA genes (Fig. 3c). SNP reversed the inhibitory effects of DPI, but exogenous H2O2 was largely ineffective at reversing the effects of c-PTIO on the expression of ABA catabolic genes.
Expression of the ABA response genes RD29A and RD29B (Yamaguchi et al., 2006; Fujii et al., 2007) showed a similar response (Fig. 3d) to that shown for ABA catabolic genes (Fig. 3c).
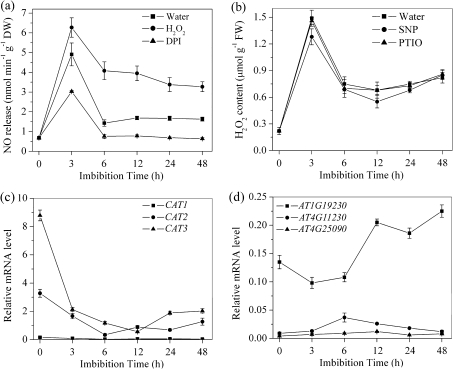
The apparent requirement for NO to elicit the H2O2-responsive expression of ABA catabolic genes and breaking of seed dormancy suggests that H2O2 may also regulate NO production. As shown in Fig. 4a, during imbibition, exogenous H2O2 increased NO production while DPI decreased it. However, SNP and its scavenger c-PTIO showed a slight effect on H2O2 production (Fig. 4b).
Fig. 4.
Effect of H2O2 and NO on each other's production and the transcription of CAT and NADPH genes during imbibition. (a) Changes in NO release during imbibition with water, H2O2 (10 mM), and its production inhibitor DPI (10 μM). (b) Changes in H2O2 release during imbibition with water, SNP (200 μM), and its scavenger c-PTIO (200 μM). (c) The transcript levels of CAT H2O2 catabolism genes during imbibition. (d). The transcript levels of H2O2 NADPH production genes during imbibition. Values are means±SE (n=4 for a and b, and n=3 for c and d). An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05).
Some genes involved in H2O2 production and catabolism were also measured during imbibition. Transcription of all three H2O2 catabolic genes, CAT1, CAT2, and CAT3, decreased rapidly and significantly during the first 3 h of imbibition (Fig. 4c). Transcription of the three NADPH oxidase genes, which are involved in H2O2 production, showed no change in the first 6 h of imbibition (Fig. 4d). Accumulation of H2O2 at the first stage of imbibition was apparently related primarily to decreasing H2O2 catabolism.
The expression of GA biosynthesis genes is enhanced by H2O2
Treatment with PAC, which inhibited GA biosynthesis (Kitahata et al., 2005; Toh et al., 2008), enhanced seed dormancy, and the enhancement of germination by H2O2 and NO could be reversed by PAC (Fig. 5a). When expression of GA biosynthesis genes was investigated after 24 h imbibition, the transcription of the GA20ox and GA3ox genes was significantly up-regulated by exogenous H2O2, slightly up-regulated by SNP, and down-regulated by DPI and c-PTIO (Fig. 5b, c). The down-regulation by c-PTIO was reversed significantly by addition of exogenous H2O2, but this enhanced value was still low compared with seeds imbibed in H2O2 (Fig. 5b, c). The inhibition by DPI was increased slightly by addition of SNP (Fig. 5b, c).
Fig. 5.
The function of NO in H2O2-regulated GA biosynthesis. (a) Effects of H2O2, NO, and their production inhibitor DPI or scavenger c-PTIO, or the GA biosynthesis inhibitor PAC on the dormancy and germination of freshly harvested seed; the germination ratio was counted on the seventh day. (b) Changes in the transcript levels of GA20ox GA biosynthesis genes under different treatments at 24 h imbibition. (c) Changes in the transcript levels of GA3ox GA biosynthesis genes under different treatments at 24 h imbibition. (d) Changes in the transcript levels of GA-regulated genes XTH5 and EXP2 under different treatments at 24 h imbibition. H2O2 (10 mM), DPI (10 μM), SNP (200 μM), c-PTIO (200 μM), and PAC (10 μM) were used for these experiments. Values are the means±SE (n=3 for a–d). An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05).
Expression of two GA-regulated genes, XTH5 and EXP2, was also measured after 24 h imbibition under different treatments (Rose et al., 2002; Yamauchi et al., 2004). Both XTH3 and EXP2 transcription levels were increased significantly by H2O2 and slightly by SNP, but were decreased by DPI and c-PTIO. Exogenous H2O2 reversed the effect of c-PTIO while SNP caused only a minimal reversal of the DPI effect (Fig. 5d).
H2O2 enhances and ABA suppresses the expression of GA biosynthesis genes
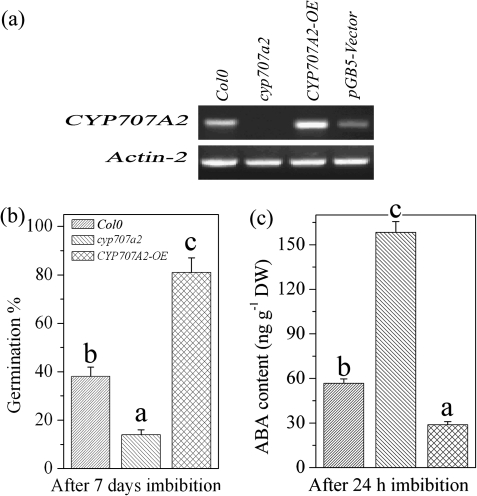
Treatment with H2O2 enhanced expression of GA biosynthesis genes during imbibition. It was therefore hypothesized that GA biosynthesis is up-regulated by H2O2 and suppressed by ABA. To examine this possibility, the ABA catabolism mutant cyp707a2 and its overexpression line CYP707A2-OE were used. As shown in Fig. 6a, the cyp707a2 mutant showed an absence of CYP707A2 gene expression while CYP707A2-OE had a high level of CYP707A2 gene expression. Freshly harvested cyp707a2 seeds showed strong dormancy, while CYP707A2-OE seeds had a much weaker dormancy response compared with wild-type seeds (Fig. 6b). After 24 h imbibition, cyp707a2 seeds retained a high ABA content whereas ABA levels in CYP707A2-OE seeds were much lower (Fig. 6c)
Fig. 6.
The CYP707A2 gene mediates seed dormancy and ABA catabolism. (a) Transcript levels of CYP707A2 in the wild type, cyp707a2, the CYP707A2 overexpression line, and pGB5 vector only in the wild type were analysed by RT-PCR. (b) The germination of the wild type, cyp707a2, and the CYP707A2 overexpression line in freshly harvested seeds. The germination rates were recorded after 7 d of imbibition. (c) ABA contents of the wild type, cyp707a2, and the CYP707A2 overexpression line after 24 h imbibition. Values are the means±SE (n=3 for b and c). An ANOVA test followed by a rank test was performed for b and c. Different letters are used to indicate means that are significantly different (P <0.05)
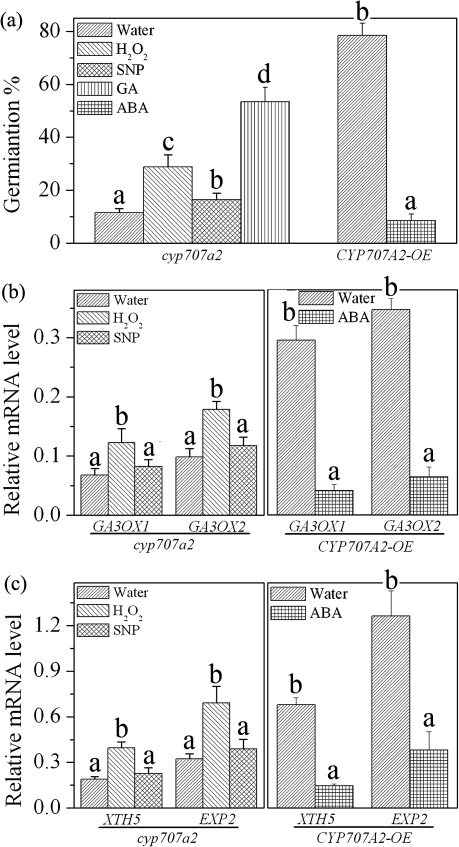
H2O2 and GA (10 μM) clearly enhanced germination of freshly harvested cyp707a2 seed, while NO did not (Fig. 7a). CYP707A2-OE germination was inhibited completely by treatment with 0.5 μM exogenous ABA (Fig. 7a). QRT-PCR analysis demonstrated that the transcription of GA3ox genes was enhanced by exogenous H2O2 rather than by SNP after 24 h imbibition in cyp707a2 (Fig. 7b). These results indicated that it was H2O2 rather than NO that exerts a regulatory effect on the expression of GA biosynthetic genes. QRT-PCR analysis also indicated that the transcription of GA3ox genes was high in CYP707A2-OE and was significantly decreased by treatment with 0.5 μM ABA (Fig. 7b). Expression of GA3ox genes was much lower in cyp707a2 compared with the wild type and CYP707A2-OE (Fig. 7b, c). Thus, ABA treatment appeared to suppress expression of GA biosynthesis genes.
Fig. 7.
H2O2 mediated GA biosynthesis and ABA suppressed GA biosynthesis. (a) The germination of cyp707a2 and the CYP707A2 overexpression line in freshly harvested seeds under different treatments. (b) Change in the transcript levels of GA3ox GA biosynthesis genes under different treatments in cyp707a2 and the CYP707A2 overexpression line after 24 h imbibition. (c) Change in the transcript levels of GA-regulated genes XTH5 and EXP2 under different treatments in cyp707a2 and the CYP707A2 overexpression line after 24 h imbibition. H2O2 (10 mM), SNP (200 μM), ABA (0.5 μM), and GA (10 μM) were used for these experiments. Values are the means±SE (n=3 for a–d). An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05)
The empty pGWB5 vector showed no effect on expression of any gene (data not shown). The transcription levels of the GA-regulated genes XTH5 and EXP2 were similar to those of GA3ox genes. In the cyp707a2 mutant, expression of these genes was enhanced by treatment with exogenous H2O2 but not by SNP after 24 h imbibition (Fig. 7c). Transcription of XTH5 and EXP2 was down-regulated by ABA in CYP707A2-OE after 24 h imbibition (Fig. 7c), suggesting that XTH5 and EXP2 transcription was much lower in cyp707a2 compared with the wild type and CYP707A2-OE (Figs 5d, 7c). All of these results suggest that H2O2 enhances GA biosynthesis while ABA suppresses GA biosynthesis.
Discussion
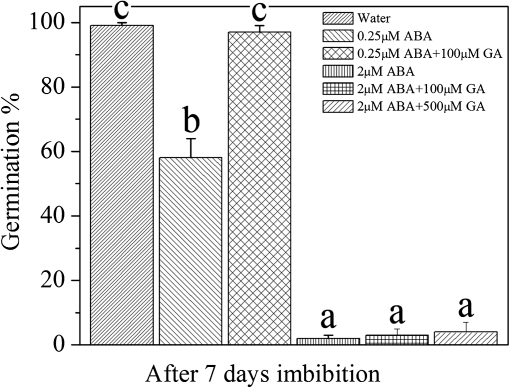
It is well known that ABA and GA play important roles in seed dormancy and germination (Finkelstein et al., 2002; Himmelbach et al., 2003; Ogawa et al., 2003; Razem et al., 2006; Weiss et al., 2007; Toh et al., 2008). Studies on numerous mutants have demonstrated that ABA catabolism and GA biosynthesis are required for seed germination (Koornneef and van der Veen, 1980; Groot and Karssen, 1987; Ogawa et al., 2003; Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006). The results from the present study may have clarified a signalling pathway for the mechanisms underlying these responses. A rapid NO-induced decrease in ABA is essential to break seed dormancy in Arabidopsis. Based on the present results, H2O2, acting as a signalling molecule, could regulate seed dormancy by triggering both ABA catabolism and GA biosynthesis. The up-regulation of ABA catabolism by H2O2 would be carried out through NO. Concomitantly, as long as a high concentration of ABA exists, it inhibits the expression of GA biosynthesis genes so that a balance of these two hormones jointly controls the dormancy and germination of Arabidopsis seeds (Fig. 8).
Fig. 8.
The relationship of ABA and GA in seed germination. The germination of non-dormant seeds under different treatments. An ANOVA test followed by a rank test was performed. Different letters are used to indicate means that are significantly different (P <0.05).
As shown in Fig. 1, exogenous H2O2 decreased dormancy in freshly harvested seed. Inhibiting H2O2 production in turn enhanced seed dormancy. The action of H2O2 was associated with expression of genes related to ABA and GA biosynthesis and catabolism. Exogenous H2O2 treatment clearly increased the expression of CYP707A, GA3ox, and GA20ox (Fig. 2b, c) and decreased seed dormancy (Fig. 1b). Using DPI to inhibit H2O2 decreased the expression of CYP707A, GA3ox and GA20ox genes (Fig. 2b, c) and enhanced seed dormancy (Fig. 1b). It was also observed that the inhibition by DPI of the expression of these genes and of seed germination was completely reversed by exogenous H2O2.
When ABA catabolism and GA biosynthesis were directly inhibited by diniconazole and PAC, the enhancement of seed germination by H2O2 disappeared (Figs 2, 4). H2O2 showed a slight effect on expression of genes involved in ABA biosynthesis and GA catabolism compared with its effects on expression of genes involved in ABA catabolism and GA biosynthesis (Fig. 2 and Supplementary Fig. S1). These results indicated that the effect of H2O2 on seed dormancy break might be connected with transcription of genes involved in both ABA catabolism and GA biosynthesis.
A role for NO, another widespread signalling molecule, in breaking of dormancy was also apparent. Up-regulation of genes responsible for ABA catabolism by H2O2 required the participation of NO. The enhancement of seed germination by H2O2 treatment was significantly reversed by the NO scavenger c-PTIO. DPI inhibited H2O2 generation and enhanced seed dormancy, but these effects were substantially reversed by addition of the NO donor SNP (Fig. 3a). Use of c-PTIO as an NO scavenger reversed the effect of H2O2 on the expression of CYP707A genes and of ABA catabolic genes (Fig. 3b–d). Treatment with H2O2 also modulated NO release during imbibition (Fig. 4a, b), and the accumulation of H2O2 at the first stage of imbibition, primarily by decreasing H2O2 catabolism (Fig. 4c, d).
The regulation by H2O2 of expression of GA biosynthetic genes is different from its effects on genes involved in ABA catabolism. Although SNP, similarly to H2O2, increased seed germination (Fig. 5a), SNP did not significantly increase transcription of GA biosynthetic genes when compared with H2O2 (Fig. 5b,c). SNP could not reverse the inhibition by DPI of transcription of GA biosynthesis genes, while exogenous H2O2 substantially restored the inhibition induced by c-PTIO (Fig. 5b, c). In particular, when the cyp707a2 mutant was used to measure the effect of H2O2 and NO on GA biosynthesis, it was found that SNP had no effect on transcription of GA biosynthesis genes, while H2O2 increased this transcription (Fig. 7b, c), It was also found that the expression of GA3ox genes was much lower in cyp707a2 than in the wild type imbibed with water or exogenous H2O2 (Figs 5c, 7b). The enhancement of seed germination induced by exogenous H2O2 and GA was also much lower in cyp707a2 compared with the effects on the wild type (Figs 3a, 7a). As shown in Fig. 6c, cyp707a2 had higher ABA levels compared with the wild type and CYP707A2-OE. The present results also support the conclusion that ABA down-regulated genes responsible for GA biosynthesis and ABA catabolism, both of which are functions that are necessary for breaking seed dormancy.
The transcription of GA biosynthesis genes was suppressed by exogenous ABA (Fig. 7b). From these results, it can be hypothesized that H2O2 could directly regulate GA biosynthesis and indirectly regulate ABA catabolism. The effect of SNP on GA biosynthesis may occur via regulation of ABA catabolism during imbibition. The results also suggest that GA reverses the inhibitory effect of a low concentration of ABA on seed germination but GA alone is insufficient to reverse germination inhibition when the ABA content is high (Fig. 8). Thus, ABA may regulate seed germination by two pathways, one that acts directly on some as yet uncharacterized factors and the second that acts through the regulation of GA biosynthesis. The data presented in Figs 3 and 5 indicate that even at low concentrations of ABA and under H2O2 and SNP treatments, the seeds failed to germinate if GA biosynthesis was inhibited by diniconozole or PAC. This indicates that both ABA catabolism and GA biosynthesis are absolutely necessary for seed dormancy break.
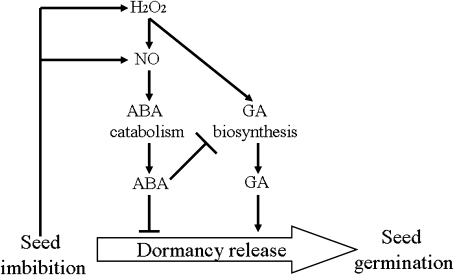
Figure 9 shows a hypothetical schematic model that could explain the results documented in the present paper. In this scheme, H2O2 may relieve dormancy of freshly harvested Arabidopsis seeds by two pathways. One pathway relies on enhancement of ABA catabolism and GA biosynthesis. The signal molecule NO does not regulate GA biosynthesis directly, but acts as an interim signalling molecule involved in H2O2 regulation of ABA catabolism. In the second pathway, ABA negatively regulates GA biosynthesis. In this way, both ABA and GA act in concert to regulate seed dormancy and germination.
Fig. 9.
Model showing how H2O2, NO, ABA, and GA regulate seed dormancy and germination. Seed imbibition leads to increases in H2O2 and NO. H2O2 up-regulates ABA catabolism through NO, and also GA biosynthesis. A high concentration of ABA also inhibits GA biosynthesis, but a balance of these two hormones jointly controls the dormancy and germination of Arabidopsis seeds.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Effects of H2O2 on the expression of ABA biosynthesis and GA catabolism genes during imbibition. The genes were analysed by QRT-PCR. H2O2 at 10 mM and DPI at 10 μM were used to manipulate H2O2 levels for these experiments. Values are the means with SE (n=4 for A and n=3 for B and C). (A) Changes in the transcript levels of NCED6 in the first 48 h during imbibition. (B) Changes in the transcript levels of NCED9 in the first 48 h during imbibition. (C) Changes in the transcript levels of GA2ox2 in the first 48 h during imbibition.
Supplementary Material
Acknowledgments
This work was supported by Hong Kong Research Grants Council (HKBU262708) and University Grants Committee of Hong Kong (AoE/B-07/99).
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F. From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. Comptes Rendu Biologies. 2008;331:806–814. doi: 10.1016/j.crvi.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Batak I, Devic M, Giba Z, Grubisic D, Poff KL, Konjevic R. The effects of potassium nitrate and NO-donors on phytochrome A and phytochrome B-specific induced germination of Arabidopsis thaliana seeds. Seed Science Research. 2002;12:253–259. [Google Scholar]
- Bethke PC, Gubler F, Jacobsen JV, Jones RL. Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta. 2004;219:847–855. doi: 10.1007/s00425-004-1282-x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Jones RL. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. The Plant Journal. 2001;25:19–29. doi: 10.1046/j.1365-313x.2001.00930.x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Libourei IG, Jones RL. Nitric oxide reduces seed dormancy in Arabidopsis. Journal of Experimental Botany. 2006;57:517–526. doi: 10.1093/jxb/erj060. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M. Seeds: physiology of development and germination. New York: Plenum Press; 1994. [Google Scholar]
- Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. The Plant Journal. 2006:113–122. doi: 10.1111/j.1365-313X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. The Plant Journal. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SL, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Davies E. Intercellular and intracellular signals in plants and their transduction via the membrane–cytoskeleton interface. Seminars in Cell Biology. 1993;4:139–147. doi: 10.1006/scel.1993.1017. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine O, Huault C, Pavis N, Billard JP. Dormancy breakage of Hordeum vulgare seeds: effects of hydrogen peroxide and scarification on glutathione level and glutathione reductase activity. Plant Physiology and Biochemistry. 1994;32:677–683. [Google Scholar]
- Foyer CH, Noctor G. Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant, Cell and Environment. 2005;28:1056–1071. [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. The Plant Cell. 2007:19,485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler E, Kawaide H, Kamiya Y, McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. The Plant Cell. 2000;12:1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentalla R, Walker-Simmons M, Ho THD. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. The Plant Cell. 2001;13:667–679. [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 1. 1987;71:525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Han SY, Kitahata N, Sekimata K, Saito T, Kobayashi M, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K, Yoshida S, Asami T. A novel inhibitor of 9- cis-epoxycarotenoid dioxygenase in abscisic acid biosynthesis in higher plants. Plant Physiology. 2004;135:1574–1582. doi: 10.1104/pp.104.039511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Current Opinion in Plant Biology. 2003;6:470–479. doi: 10.1016/s1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Hoffmann-Benning S, Kende H. On the role of abscisic acid and gibberellin in the regulation of growth in rice. Plant Physioogy. 1992;99:1156–1161. doi: 10.1104/pp.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, afterripening, dormancy and germination. New Phytologist. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Jeblick W. Pre-treatment of parsley suspension cultures with salicylic acid enhances spontaneous and elicited production of H2O2. Plant Physiology. 1995;108:1171–1178. doi: 10.1104/pp.108.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Jeblick W. Influence of salicylic acid on the induction of competence for H2O2 elicitation. Plant Physiology. 1996;111:755–763. doi: 10.1104/pp.111.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahat N, Saito S, Miyazawa Y, et al. Chemical regulation of abscisic acid catabolism in plants by cytochrome P450 inhibitors. Bioorganic and Medicinal Chemistry. 2005;13:4491–4498. doi: 10.1016/j.bmc.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theoretical and Applied Genetics. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO Journal. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara A, Ikegami K, Koshiba T, Nagata T. Effects of ethylene and abscisic acid upon heterophylly in Ludwigia arcuata (Onagraceae) Planta. 2003;217:880–887. doi: 10.1007/s00425-003-1062-z. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi H, Suh S, Doo IS, Oh KY, Choi EJ, Taylor SAT, Low PS, Lee Y. Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reaction oxygen species from guard cells of tomato and Commelina communis. Plant Physiology. 1999;121:147–152. doi: 10.1104/pp.121.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon RA, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Liu YG, Shi L, Ye NH, Liu R, Jia WS, Zhang JH. Nitric oxide-induced rapid decrease of ABA concentration is required in seed dormancy break in Arabidopsis. New Phytologist. 2009;183:1030–1042. doi: 10.1111/j.1469-8137.2009.02899.x. [DOI] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8'-hydroxylase. The Plant Journal. 2006;45:942–954. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van-Breusegem F. The reactive oxygen gene network in plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Naylor G, Warner SAJ, Sugars JM, White RF, Draper J. Salicylic acid potentiates defense gene expression in leaf tissue exhibiting acquired resistance to pathogen attack. The Plant Journal. 1996;9:559–571. [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, et al. Identification and characterization of Arabidopsis gibberellin receptors. The Plant Journal. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Iwabuchi M. A mechanism for promoting the germination of Zinnia elegans seeds by hydrogen peroxide. Plant and Cell Physiology. 2001;42:286–291. doi: 10.1093/pcp/pce032. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. CYP707A1 and CYP707A2, which encode abscisic acid 8'-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiology. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Narvaez-Vasquez J, Ryan CA. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. The Plant Cell. 2001;13:179–191. [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Quarrie SA, Whitford PN, Appleford NEJ, Wang TL, Cook SK, Henson IE, Loveys BR. A monoclonal antibody to (S)-abscisic acid: its characterization and use in a radioimmunoassay for measuring abscisic acid in crude extracts of cereal and lupin leaves. Planta. 1988;173:330–339. doi: 10.1007/BF00401020. [DOI] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M. Sequential steps for developmental arrest in Arabidopsis seeds. Development. 2001;128:243–252. doi: 10.1242/dev.128.2.243. [DOI] [PubMed] [Google Scholar]
- Razem FA, Baron K, Hill RD. Turning on gibberellin and abscisic acid signaling. Current Opinion in Plant Biology. 2006;9:454–459. doi: 10.1016/j.pbi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW. Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. The Plant Cell. 1992;4:1443–1451. doi: 10.1105/tpc.4.11.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiology. 2004;134:1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarath G, Hou G, Baird LM, Mitchell RB. Reactive oxygen species, ABA and nitric oxide interactions on the germination of warm-season C4-grasses. Planta. 2007;226:697–708. doi: 10.1007/s00425-007-0517-z. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. The Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Imamura A, Watanabe A, et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiology. 2008;146:1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiology. 2007;144:1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiology. 2000;122:1081–1088. doi: 10.1104/pp.122.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Jia WS, Zhang JH. AtMKK1 mediates ABA-induced CAT1 expression and H2O2produced via AtMPK6-coupled signaling in Arabidopsis. The Plant Journal. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. The Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annual Review of Plant Physiology and Plant Molecular Biology. 1988;39:439–473. [Google Scholar]
- Zentella R, Yamauchi D, Ho THD. Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. The Plant Cell. 2002;14:2289–2301. doi: 10.1105/tpc.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proceedings of the National Academy of Sciences, USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.