Abstract
Previous studies of highly branched mutants in pea (rms1–rms5), Arabidopsis thaliana (max1–max4), petunia (dad1–dad3), and rice (d3, d10, htd1/d17, d14, d27) identified strigolactones or their derivates (SLs), as shoot branching inhibitors. This recent discovery offers the possibility of using SLs to regulate branching commercially, for example, in chrysanthemum, an important cut flower crop. To investigate this option, SL physiology and molecular biology were studied in chrysanthemum (Dendranthema grandiflorum), focusing on the CCD8/MAX4/DAD1/RMS1/D10 gene. Our results suggest that, as has been proposed for Arabidopsis, the ability of SLs to inhibit bud activity depends on the presence of a competing auxin source. The chrysanthemum SL biosynthesis gene, CCD8 was cloned, and found to be regulated in a similar, but not identical way to known CCD8s. Expression analyses revealed that DgCCD8 is predominantly expressed in roots and stems, and is up-regulated by exogenous auxin. Exogenous SL can down-regulate DgCCD8 expression, but this effect can be overridden by apical auxin application. This study provides evidence that SLs are promising candidates to alter the shoot branching habit of chrysanthemum.
Keywords: Auxin, CCD8, chrysanthemum, shoot branching, strigolactone
Introduction
Chrysanthemum (Dendranthema grandiflorum) accounts for 30% of world cut flower production. Conventionally, the lateral shoots are controlled by manual decapitation or removing the axillary buds, which is labour-intensive and accounts for about one-third of total cultivation costs. Therefore, chrysanthemum producers are in need of novel approaches to control shoot branching.
Shoot branching depends on both the formation of axillary buds in the axils of leaves and their subsequent outgrowth. Outgrowth is a more interesting target for breeders, since it allows greater flexibility in branching. Axillary shoot apical meristems are established in the axils of the leaves produced by the primary shoot apical meristem and after forming only a few leaves, they can enter a dormant state. These dormant buds may be reactivated by endogenous or environmental signals, contributing to the enormous diversity of plant architectures observed in nature (Dun et al., 2006; Leyser, 2009).
Hormones play a central role in shoot branching control. Auxin inhibits bud outgrowth (Thimann and Skoog, 1933; Skoog and Thimann, 1934), whereas cytokinin promotes it (Sachs and Thimann, 1967). These hormones can efficiently regulate shoot branching, but their application also affects flowering, leaf development, and height, reducing their utility for the horticultural industry.
A series of branching mutants with fewer pleiotropic phenotypes has been identified. These include the more axillary branching (max) mutants of Arabidopsis, the ramosus (rms) mutants of pea (Pisum sativum), the decreased apical dominance (dad) mutants of petunia (Petunia hybrida), and the dwarf (d) or high tillering dwarf (htd) mutants of rice (Oryza sativa) (reviewed in Dun et al., 2006; Leyser, 2009). Reciprocal grafting and double mutant studies demonstrate that these genes are involved in the production and signalling of a graft-transmissible, upwardly moving branch inhibitor, which was recently shown to be strigolactone or a derivative (SL) (Beveridge et al., 1996, 2000; Napoli, 1996; Foo et al., 2001; Morris et al., 2001; Turnbull et al., 2002; Sorefan et al., 2003; Booker et al., 2004, 2005; Snowden et al., 2005; Simons et al., 2007; Gomez-Roldan et al., 2008; Umehara et al., 2008).
To date, four loci involved in SL biosynthesis and one locus involved in SL signalling have been molecularly identified (Stirnberg et al., 2002; Sorefan et al., 2003; Booker et al., 2004, 2005; Lin et al., 2009). A sixth gene in the pathway has been described, represented by rice D14, which acts either in a biosynthetic step downstream of SL or in SL signalling (Arite et al., 2009). Of the biosynthetic genes, two encode divergent plastidic carotenoid cleavage dioxygenases. MAX4/RMS1/DAD1/D10 encodes CAROTENOID CLEAVAGE DIOXYGENASE (CCD8) (Sorefan et al., 2003; Snowden et al., 2005; Arite et al., 2007), whereas MAX3/RMS5/HTD1/D17/SICCD7 encodes CCD7 (Booker et al., 2004; Johnson et al., 2006; Zou et al., 2006; Vogel et al., 2009). CCD8 and CCD7 sequentially cleave a variety of carotenoid substrates (Booker et al., 2004; Schwartz et al., 2004; Auldridge et al., 2006; Alder et al., 2008). D27 encodes a plastidic iron-containing protein, which possibly acts at an early point in the pathway (Lin et al., 2009). MAX1 encodes a cytP450 predicted to act downstream of CCD7 and CCD8 (Booker et al., 2005). MAX2/RMS4/D3 encodes a nuclear-localized F-box protein (Stirnberg et al., 2002; Ishikawa et al., 2005; Johnson et al., 2006), involved in SL signalling (Beveridge et al., 1996; Booker et al., 2005; Stirnberg et al., 2007).
The mode of action of SLs and their relationship with auxin is a matter of debate. One hypothesis is that SLs inhibit branching by reducing auxin transport canalization, preventing the establishment of polar auxin transport out of axillary buds into the main stem, and thus preventing their activation. According to this model, SLs act systemically to modulate auxin transport, and their ability to inhibit buds depends on the presence of competing auxin sources, such as the primary shoot apex or other active buds, for common auxin transport pathways in the main stem (Prusinkiewicz et al., 2009). However, it has also been proposed that SLs act directly in axillary buds to inhibit them by some unknown, but auxin-transport-independent mechanism (Brewer et al., 2009).
Another point of debate is the extent of negative feedback in the SL pathway and the role of auxin in the process. In most species, auxin positively regulates the expression of both CCD8 and CCD7 (Sorefan et al., 2003; Bainbridge et al., 2005; Foo et al., 2005; Johnson et al., 2006; Arite et al., 2007; Hayward et al., 2009). However in rice, CCD7/HTD1 is not stably up-regulated by auxin (Zou et al., 2006; Arite et al., 2007). There is also good evidence for widespread feedback regulation in the SL pathway (Foo et al., 2005; Snowden et al., 2005; Arite et al., 2007; Hayward et al., 2009). CCD7 and CCD8 transcripts generally accumulate above wild-type levels in SL mutants. This effect is much stronger in pea than in the other species. In rice, significant up-regulation was observed for CCD8/D10 but not for CCD7/HTD1 in the d3 mutant background (Arite et al., 2007). The relationship between auxin and feedback regulation of SL biosynthesis is unclear. SL mutants transport increased amounts of applied auxin compared to wild-type (Beveridge et al., 2000; Bennett et al., 2006; Lazar and Goodman, 2006; Lin et al., 2009), and in Arabidopsis this correlates with the over-accumulation of auxin in the polar transport streams of their stems (Prusinkiewicz et al., 2009). Furthermore, double mutants defective in both auxin signalling and SL signalling, feedback up-regulation of CCD7/MAX3 or CCD8/MAX4 is mostly abolished. This suggests that feedback signalling is substantially mediated by auxin (Hayward et al., 2009). Consistent with this idea, as described above, in rice there is a correlation between auxin-inducibility and feedback regulation (Arite et al., 2007). However, the situation in pea is different since auxin treatments cannot achieve the particularly high level of CCD8/RMS1 and CCD7/RMS5 up-regulation observed in rms mutants, suggesting an auxin-independent feedback (Foo et al., 2005; Johnson et al., 2006). It is therefore clear that comparative approaches in diverse species have contributed to our understanding of SL function, but also raised some interesting questions about the degree of conservation of the system between species.
Thus the analysis of the SL pathway in additional species has the potential to contribute to both the development of applications for SLs in agriculture and horticulture, and to resolving questions about the mechanism of action of SLs. To these ends, work aimed at the evaluation of SL physiology in chrysanthemum, as well as the identification of chrysanthemum genes required for SL synthesis, is described in this paper.
Materials and methods
Plant materials and growth condition
Chrysanthemum (Dendranthema grandiflorum) cv. Jinba plantlets were propagated under sterile conditions in jars containing MS agar medium (Murashige and Skoog, 1962) and grown in a tissue culture room at 21 °C, with 16/8 h light/dark photoperiods and a light intensity of 100–120 μmol m−2 s−2. Arabidopsis thaliana Columbia (WT) and max4-1 were grown in 4 cm square compartments containing F2 compost (Levington Horticulture, Ipswich, UK) and transferred to a greenhouse at 21 °C, with 16/8 h light/dark photoperiods.
Hormone stocks
Napthalene acetic acid (NAA) stock solution was dissolved in 70% ethanol, and GR24 (LeagGen Labs, Orange CT USA) was dissolved in acetone were injected into the apical or basal blocks, as required, to give a final concentration of 5 μM. Solvent treatments were used as controls.
Split plate assay
The hormone responses of buds on isolated stem segments were assessed using the method described by Chatfield et al. (2000).
Isolation of the DgCCD8 genes
Total RNA was extracted from the stem and roots of chrysanthemum plantlets using Trizol (Mylab, China). cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen). Degenerate primers were designed from the amino acid sequence of the Arabidopsis MAX4, petunia DAD1, and pea RMS1 gene and used to amplify a fragment of the DgCCD8 gene from cDNA, with forward primer (5′-GTSGTGAGRATGGAASCHGG-3′) and reverse primer (5′-CCATCATCYTCWTSGGTTGC-3′). The amplification products were used to design the primers for Rapid Amplification of cDNA Ends (RACE), which was used to obtain full-length DgCCD8 transcripts. For 3′ RACE, cDNA was synthesized with the adaptor oligo (dT) primer (5′-CTGATCTAGAAGGTACCGGATCCT(15)-3′) and amplified by PCR using the adaptor primer (5′-CTGATCTAGAAGGTACCGGATCCT-3′) and a specific primer 3P (5′-GGGTTGGGCGGTTTAGGATACCATTCG). For 5′ RACE, dC-tailed cDNA was synthesized using a specific primer 5P (5′-AGTCTATCTTAGTCAGAGTGTT-3′). The amplification was performed with primer AAP (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and the specific primer 5P. This PCR product was used as the template in an additional nested PCR using specific primer 5P1(5′-TGGCATCTCGGGCACAATCACA-3′) and primer AUAP (5′-GGCCACGCGTCGACTAGTAC-3′). Amplified fragments were cloned into the pMD18-T vector and sequenced. According to the sequences, a forward primer from the 5′-UTR region (5′-TAGCAAACCTCTTTATTACCGATGG-3′) and a reverse primer from 3′-UTR (5′-CTTTATTTCCGACATTTGCCCTTTC-3′) region of DgCCD8 were designed to isolate the complete DgCCD8 from both cDNA and genomic DNA. Genomic DNA was isolated from young leaves using the DNeasy Plant Mini Kit (Qiagen). The amplification products were used to determine the sequence of the chrysanthemum cDNAs and genomic clones, and the positions of introns in the genes.
Phylogenetic analysis
Orthologues of the known CCD8 genes were identified using InParanoid (Remm et al., 2001) to perform pairwise comparisons between each of the published or draft complete genome sequences: Arabidopsis thaliana (Rhee et al., 2003), Brachipodium distachyon (http://www.brachypodium.org/), Lotus japonicus (Sato et al., 2008), Medicago truncatula (http://www.medicago.org/), Oryza sativa (Ouyang et al., 2007), Physcomitrella patens (Rensing et al., 2008), Populus trichocarpa (Tuskan et al., 2006), Selaginella moellendorffii (http://genome.jgi-psf.org/Selmo1/Selmo1.home. html), Sorghum bicolor (Paterson et al., 2009), Vitis vinifera (Jaillon et al., 2007), and Zea mays (Schnable et al., 2009). The orthology of candidate genes was confirmed during phylogenetic reconstruction. Sequences were aligned using MUSCLE v3.6 (Edgar, 2004) using default settings. The maximum likelihood phylogeny was reconstructed under the Dayhoff substitution matrix using RA×ML (Stamatakis, 2006). Support for the maximum likelihood phylogenies was estimated from 100 rapid bootstrap resamplings (Stamatakis et al., 2008).
Generation of transgenic plants
For complementation experiments, the ORFs of DgCCD8a and DgCCD8b were cloned into pART7 fusing them with the 35S promoter, and then these fusions were transferred into the binary vector pART27 (Gleave, 1992). The constructs were transformed into Arabidopsis thaliana mutant max4-1 plants via Agrobacteriaum tumefaciens strain GV3101 using the floral dip method (Clough and Bent, 1998). The independent transformants were screened on Arabidopsis thaliana salts (ATS) agar medium containing 50 mg l−1 kanamycin. Independent, single insertion site, homozygous T3 lines were used to analyse the branching phenotype.
Decapitation assay
To quantify branching, a decapitation assay was used (Bainbridge et al., 2005). Seeds of max4-1, WT, [35S::DgCCD8a] max4-1 and [35S::DgCCD8b] max4-1 lines were sown in F2 compost and grown in a growth cabinet at 21 °C, 8/16 h light/dark photoperiods. After 28 d, plants were shifted to 16/8 h light/dark photoperiods to induce flowering. Primary bolts were decapitated once they reached 10–15 cm in height. Rosette branches were counted 10 d after decapitation.
Hormone treatments
For analysis of DgCCD8 expression after decapitation, auxin, or GR24 treatment, in vitro-grown chrysanthemum plantlet cuttings, 15 cm in height, were transferred into jars containing MS-agar medium with 5 μM GR24 or an equal volume of acetone. They were left intact or were decapitated, and Eppendorf tubes containing MS-agar medium with 5 μM NAA or an equal volume of ethanol were placed over the stumps. After 6 h treatment, the stem spanning the basal three nodes of six plantlets per treatment was excised and the tissue pooled to extract total mRNA, which was used to examine the accumulation of total DgCCD8 transcripts.
Semi-quantitative RT-PCR
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen) and reverse transcribed using Superscript II (Invitrogen) according to the manufacturer's manual. To investigate the total expression of DgCCD8 in chrysanthemum, the common primers RT1 (5′-GTTTGAATGGCATCCTGAGTCG-3′) and RT2 (5′-GTTGAGCACCACAAGCGTAAGC-3′) were used. Specific primers (5′-TTATTACCGATGGCTTCCTCCCTT-3′) and (5′-CCGGCACAATCACATAATGTTCG-3′) were used to analyse the expression of DgCCD8b. Amplification of ribosomal RNA (Yang et al., 2005) was performed using 25 cycles as a normalization control. To analyse the transcription from the transgenes in [35S::DgCCD8b] and [35s::DgCCD8a] max4 lines, RT1 (5′-GTTTGAATGGCATCCTGAGTCG-3′) and RT3 (5′-TGTTCACTGGGGTTAAGAGCGTC-3′) were used. UBQ was used as a normalization control with primers UF (5′-AACCCTTGAGGTTGAATCATC-3′) and UR (5′-GTCCTTCTTTCTGGTAAACGT-3′).
Results
The inhibition of chrysanthemum buds by SL and auxin
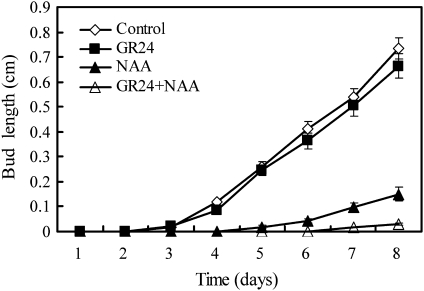
To assess whether SLs can inhibit chrysanthemum bud outgrowth, the hormone responses of buds on one-node stem segments held between two agar blocks in Petri dishes were tested (Chatfield et al., 2000). The segments were treated with basally supplied GR24, a synthetic strigolactone, apically supplied auxin (NAA) or both. Although NAA was sufficient to inhibit bud outgrowth, GR24 alone was surprisingly ineffective (Fig. 1). However, the combined application of GR24 and NAA completely inhibited bud outgrowth and was more effective than NAA applied alone.
Fig. 1.
Effect of GR24 and NAA on bud outgrowth on single-node isolated stem segments in chrysanthemum. One-node stem segments were excised from chrysanthemum plantlets and inserted between two agar blocks. The apical agar blocks contained either 5 μM NAA or 0 μM NAA (with an equal volume of ethanol as a control). The basal agar blocks contained either 5 μM GR24, or 0 μM GR24 (with an equal volume of acetone as a control). Bud lengths were measured every 24 h and the mean lengths are presented. Error bars represent the standard error of the means, n=19–20. The data presented are typical of three independent experiments.
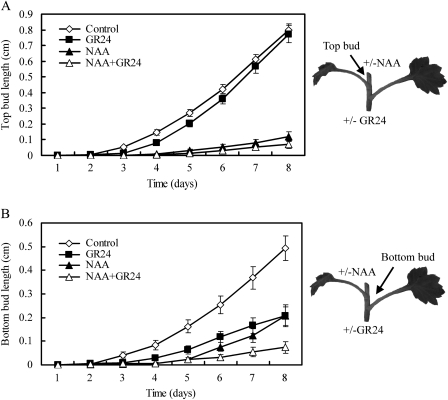
These results suggest that GR24 is only effective at inhibiting buds in the presence of an auxin source. To test this further, the activity of SLs supplied to two-node stem segments was determined, with the rationale that the second bud would provide a natural auxin source. In this configuration, the two buds respond in different ways to the treatments. The apical bud behaves similarly to buds in the one-node segment experiment (compare Fig. 1 with Fig. 2A). There is no effect of basally supplied GR24, but apical auxin has a strong inhibitory effect. By contrast, the basal bud was inhibited by GR24 alone to a similar degree as for apical NAA treatment alone (Fig. 2B). GR24 together with NAA was more effective at inhibiting both the apical and the basal buds than NAA and GR24 alone (Fig. 2A, B). NAA alone was less effective at inhibiting the basal bud compared with the apical bud (Fig. 2A, B). These results are consistent with the idea that GR24 is only effective at inhibiting a bud in the presence of at least one competing auxin source.
Fig. 2.
Effect of GR24 and NAA on bud outgrowth on two-node isolated stem segments in chrysanthemum. Two-node stem segments were excised from chrysanthemum plantlets and inserted between two agar blocks. The apical agar blocks contained either 5 μM NAA or 0 μM NAA (with an equal volume of ethanol as a control). The basal agar blocks contained either 5 μM GR24, or 0 μM GR24 (with an equal volume of acetone as a control). The length of the top bud (A) or bottom bud (B) was measured every 24 h and the mean lengths are presented. The arrows indicate the axil for which bud length was measured. Error bars represent the standard error of the mean, n=23–24. The data presented are typical of three independent experiments.
Isolation of DgCCD8
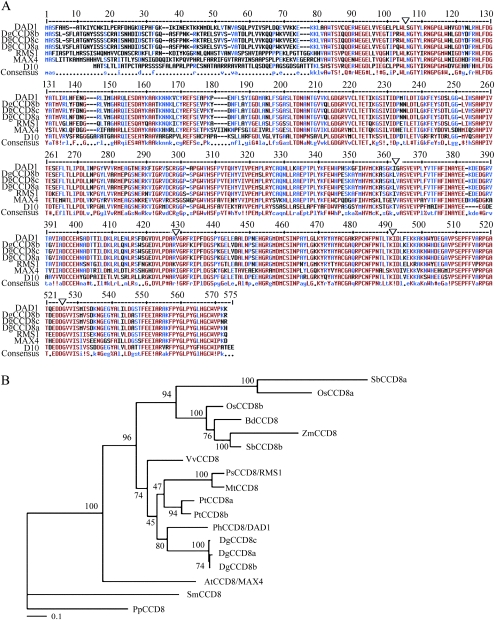
To study SL biosynthesis in chrysanthemum, its putative MAX4 orthologue, DgCCD8 was isolated. Amino acid sequence comparison of CCD8 from Arabidopsis thaliana (MAX4), pea (Pisum sativum, RMS1), petunia (Petunia hybrida, DAD1), and rice (Oryza sativa, D10) enabled the design of degenerate primers in the most conserved domains of these genes. A 750 bp fragment with strong sequence similarity to the CCD8 family was successfully amplified. A full-length cDNA of DgCCD8 was recovered by 5′-and 3′ Rapid Amplification of cDNA Ends (RACE) PCR. Subcloning and sequencing of 21 fragments resulted in the identification of three distinct DgCCD8 cDNAs, designated as DgCCD8a, DgCCD8b, and DgCCD8c. DgCCD8a cDNA comprises 1740 bp containing an open reading frame of 1671 bp encoding a predicted protein of 556 amino acids. DgCCD8b and DgCCD8c coding sequences are both 1668 bp in length and are predicted to encode proteins of 555 amino acids. Southern blot analysis showed that at least three copies of the DgCCD8 gene are present in the chrysanthemum genome (data not shown), consistent with its known hexaploidy (Nazeer and Khoshoo, 1983), so the three related sequences identified as DgCCD8a, DgCCD8b, and DgCCD8c may represent transcripts from these three loci, which are 98–99% identical. Alternatively, they could be alleles at two of those loci since, while the chrysanthemum line used is clonally propagated and thus of a single genotype, it is not inbred. Amino acid sequence comparisons between these three genes and their orthologues from Arabidopsis, pea, petunia, and rice show that the predicted DgCCD8s are 60–61% identical to AtMAX4, 72–73% identical to PsRMS1, 78% identical to PhDAD1, and 58% identical to OsD10 (Fig. 3A).
Fig. 3.
Isolation of DgCCD8. (A) Alignment of the predicted amino acid sequences of DgCCD8 compared with Arabidopsis (MAX4), pea (RMS1), petunia (DAD1), and rice (D10). Intron positions corresponding to the genomic DNA sequence are denoted by triangles. (B) Phylogenetic analysis. Maximum likelihood phylogeny of CCD8 orthologues reconstructed using RAxML (Stamatakis, 2006) under the Dayhoff substitution matrix. Node support values were estimated from 100 rapid bootstrap resamplings (Stamatakis et al., 2008). Proteins are labelled with a prefix that represents the species origin of the sequence: Arabidopsis thaliana AtCCD8 (At4g32810.1); Brachypodium distachyon BdCCD8 (Bd2g49670.1); Dendranthema grandiflorum DgCCD8a, DgCCD8b, and DgCCD8c; Medicago truncatula MtCCD8 (CR9563923.4); Oryza sativa OsCCD8a (Os01g38580.1), OsCCD8b (Os01g54270.1); Pisum sativum PsCCD8/RMS1 (AAS66906.1); Petunia hybrida PhCCD8/DAD1 (AAW33596.1); Populus trichocarpa PtCCD8a (eugene 300061708), PtCCD8b (gw1.XV111.1171.1); Physcomitrella patens PpCCD8 (e gw 1.14.69.1); Sorghum bicolor SbCCD8a (Sb05g00950), SbCCD8b (Sb03g034400); Selaginella moellendorffi SmCCD8 (egw1.86.30.1); Vitis vinifera VvCCD8 (GSVIVT0003 2423001); Zea mays ZmCCD8 (Zm2g147254).
To investigate further the relationship between the predicted CCD8 proteins, a phylogenetic tree of CCD8 proteins was constructed from a taxonomically diverse set of species (Fig. 3B). In line with taxonomy, DgCCD8s are placed in a well-supported clade with petunia.
Genomic fragments corresponding to DgCCD8 genes were isolated by PCR using a forward primer from the 5′-UTR regions and a reverse primer from the 3′-UTR regions of DgCCD8. Sequencing of clones led to the identification of five different CCD8-related genomic sequences. Comparison of the DgCCD8 cDNAs to the genomic sequence revealed that two genomic fragments correspond to the DgCCD8a and DgCCD8c cDNA, respectively. However, none corresponds to DgCCD8b. Thus, in total, six different DgCCD8 sequences, that are likely to represent two alleles at each of three DgCCD8 loci have been identified.
Alignment of all cDNAs with their corresponding genomic DNA revealed that all the DgCCD8 genes have the same intron–exon structure (Fig. 3A). Both the number of introns (five) and their positions in the genes are conserved with the previously reported gene structures.
Functional conservation of the DgCCD8s
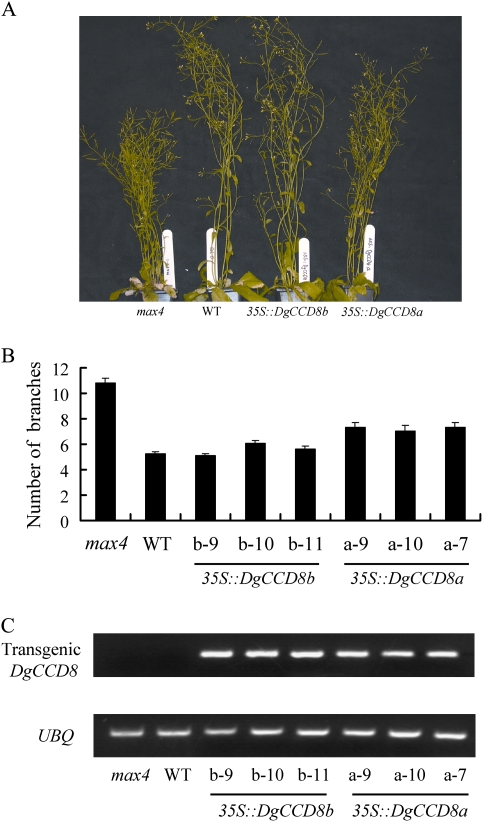
To determine whether the DgCCD8 genes are functionally conserved, the DgCCD8a and DgCCD8b ORFs from the Cauliflower Mosaic Virus 35S promoter (35S::DgCCD8a, 35S::DgCCD8b) were expressed in the Arabidopsis max4-1 mutant background. For each construct, multiple independent transgenic lines were generated, and those showing segregation patterns in the T2 consistent with a single insertion site (data not shown) were taken to homozygosity for detailed analysis. Branching was assessed using a decapitation assay previously shown to provide sensitive discrimination between wild-type and max4 mutant plants (Bainbridge et al., 2005). The mean number of rosette branches with a length of at least 2 cm was scored 10 d after decapitation to quantify the degree of rescue of the max4 mutant phenotype by the DgCCD8 genes. Figure 4B represents the three most strongly rescuing lines for each construct out of 11 35S::DgCCD8b and 15 35S::DgCCD8a max4 charaterized lines. Constitutive expression of DgCCD8b reduced the mean number of branches from 10.8 in max4 to 5–6, resulting in plants indistinguishable from wild-type (WT), which had a mean of 5.25 branches. However, constitutive expression of DgCCD8a was less effective, reducing the number of branches to 7.05–7.35 (Fig. 4A, B). This difference was not due to different levels of expression of the transgenes, because when RT-PCR was used to determine transcript abundance, the DgCCD8 transgenes were found to accumulate to similar levels between the 35S::DgCCD8b and 35S::DgCCD8a lines (Fig. 4C). Therefore, the increased effectiveness of DgCCD8b in rescuing the max4 phenotype compared to DgCCD8a is probably a post-transcriptional effect.
Fig. 4.
Complementation of Arabidopsis max4-1 mutant phenotype with DgCCD8. (A) Comparison of phenotypes of wild type, max4-1, and max4-1 transformed with the 35S::DgCCD8a and 35S::DgCCD8b constructs. (B) The number of secondary rosette branches produced by WT, max4-1, and the three most-strongly rescued independent homozygous lines transformed carrying either 35S::DgCCD8a or 35S::DgCCD8b. Branching was assessed using a decapitation assay. The mean number of rosette branches with a length of at least 2 cm 10 d after decapitation is shown (error bars =SEM, n=20). The data presented are typical of two independent experiments. (C) Analysis of DgCCD8 expression for the experiment presented in (B). Transcripts were assayed by reverse transcriptase PCR from total RNA from rosette leaves. One leaf from each of the 20 plants in each sample was collected and pooled after branching had been assessed. Detection of the UBQ transcript was used as a cDNA normalization control. The data presented are typical of two independent experiments. (This figure is available in colour at JXB online.)
Tissue specificity of DgCCD8 expression
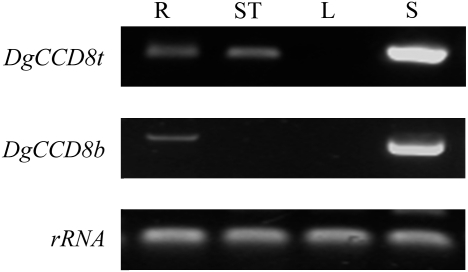
To gain an overall picture of the tissue specificity of DgCCD8 expression in chrysanthemum, the combined levels of all DgCCD8 transcripts in mRNA extracted from different tissues were examined by RT-PCR, using primers that do not discriminate between the different copies/alleles. DgCCD8 mRNA was detected in all tissues examined, except for the leaf. Expression in stems was higher than in roots and shoot apices (Fig. 5).
Fig. 5.
RT-PCR analysis of total DgCCD8 (DgCCD8t) and DgCCD8b expression in chrysanthemum. Total RNA was extracted from R, root; ST, shoot tips; L, leaf; and S, stem, with tissue samples taken from pools of six in vitro-grown plantlets. Detection of rRNA was used as a normalization control. The data presented are typical of three independent experiments.
To gain insight into whether there is any sub-functionalization within the family, the expression pattern of DgCCD8b was analysed further using specific primers. DgCCD8b was not detected in shoot apices, but only in stem and root tissues, with strongest expression in the stem (Fig. 5).
Induction of DgCCD8 expression by auxin
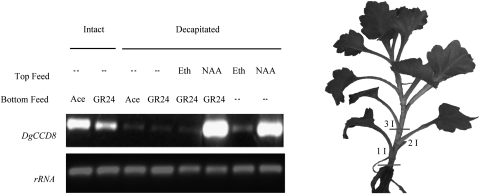
To investigate auxin regulation of DgCCD8, RT-PCR analysis was performed on basal stem segments following decapitation, with or without apical application of auxin (Fig. 6). Decapitation resulted in a drastic reduction in total DgCCD8 transcript abundance after 6 h but this reduction was successfully compensated by apical NAA application. These data indicate that auxin regulates DgCCD8 gene expression in a similar manner to that described in other species.
Fig. 6.
DgCCD8 transcript response to decapitation, auxin, and GR24. Top panel, DgCCD8 gene expression in the three basal internodes (1I–3I as represented by the lines in the scheme on the right). Pools of six plantlets were sampled either intact (left two lanes) or 6 h after decapitation (right six lanes). Plantlets were treated with 5 μM NAA (NAA) or 0 μM NAA [with an equal volume of ethanol as a control (Eth)] supplied to the decapitated stump, and/or with 5 μM GR24 (GR24) or 0 μM GR24 [with an equal volume of acetone as a control (Ace)] to the basal node. Bottom panel, rRNA gene expression was used as a cDNA normalization control. The data presented are typical of three independent experiments.
Feedback control of DgCCD8 expression
To investigate feedback control by SL on DgCCD8 expression, total DgCCD8 transcript abundance after 6 h treatment was compared with basal GR24 on intact and decapitated plants (Fig. 6). It was observed that, in intact plants, DgCCD8 expression is down-regulated by GR24 compared with untreated controls. However, this down-regulation is modest and is less than that observed upon decapitation. Furthermore, down-regulation by GR24 was not observed in decapitated plants, and DgCCD8 expression in decapitated plants treated with NAA was indistinguishable from the decapitated plants treated with both apical NAA and basal GR24.
Discussion
The mechanisms of bud inhibition by SL and auxin
Inhibition of bud outgrowth by an actively growing shoot apex is mediated by apically-derived auxin transported down the main stem (Thimann and Skoog, 1933), which acts indirectly without entering the bud (Morris, 1977; Booker et al., 2003). SLs have been proposed to act as second messengers for auxin, relaying the inhibitory signal from the main stem into the buds (Brewer et al., 2009). An alternative model is that apically-derived auxin moving in the main stem inhibits bud activity by reducing the sink strength of the stem for auxin, thus preventing canalization of auxin transport out of the bud (Prusinkiewicz et al., 2009). In this model, SLs inhibit bud activation by systemically dampening auxin transport canalization (Prusinkiewicz et al., 2009).
The effect of SL on the activity of solitary buds, in the absence of a competing auxin source has not previously been described, and has the potential to distinguish between these alternative models. When a solitary bud was treated with SL, it was activated with similar kinetics to an untreated bud. This argues against the idea that SLs act in a straightforward way by entering buds and inhibiting their activity. However, the result is entirely consistent with the idea that SLs act by dampening auxin transport canalization. Such a mechanism would only be effective in inhibiting bud activation in the presence of a competing auxin source, reducing the sink strength of the main stem for auxin. Accordingly, SL was effective at inhibiting solitary buds when apical auxin was simultaneously applied. Furthermore, SL was able to inhibit growth of the basal bud on an explant with two buds, while the apical bud was unaffected. The active apical bud presumably exports auxin into the stem, as has been demonstrated for Arabidopsis buds, providing a competing auxin source similar to apical auxin application (Prusinkiewicz et al., 2009).
Isolation of the chrysanthemum CCD8 genes
To investigate further the operation of the SL pathway in chrysanthemum, the chrysanthemum orthologues of CCD8 were isolated, and their orthology was confirmed by phylogenetic analysis and functional complementation of the Arabidopsis ccd8/max4-1 mutant. Our results suggest that there are at least three loci encoding CCD8 in chrysanthemum, consistent with its known hexaploidy. Furthermore, some evidence was found for sub-functionalization within the family. The two cDNAs tested were apparently not equally effective in rescuing max4-1, despite differing by only three non-conserved amino acids, and the more effective rescuing cDNA is expressed in only a sub-set of the tissues where the CCD8 transcripts were detected.
Despite the high functional conservation of the CCD8 genes between species, there are interesting differences in expression pattern. In Arabidopsis (Auldridge et al., 2006), petunia (Snowden et al., 2005), and pea (Foo et al., 2005), root expression of CCD8 is at least 10 times higher than shoot expression. By contrast, in rice (Arite et al., 2007) and chrysanthemum, shoot expression exceeds root expression. These differences may reflect different contributions of the root and shoot in the SL-regulation of shoot branching in the different species.
Feedback regulation of DgCCD8 expression
Previously negative feedback regulation has been observed for CCD8s in all species examined (Foo et al., 2005; Snowden et al., 2005; Arite et al., 2007; Hayward et al., 2009). However, the molecular basis for this feedback is unresolved. Three non-exclusive mechanisms have been proposed (Hayward et al., 2009). SL biosynthesis genes may be directly negatively regulated by SLs; feedback could be mediated indirectly by auxin since SL signalling results in a decreased auxin export from buds, and auxin is required for optimal CCD8 gene expression; or indirect feedback may be mediated by an unknown signal, which in pea is suggested to depend on the RMS2 gene (Foo et al., 2005; Dun et al., 2006).
In our studies, decapitation and exogenous application of auxin result in down- and up-regulation of DgCCD8 expression, respectively, suggesting positive regulation by auxin similar to other species. Furthermore, GR24 can down-regulate the expression of the DgCCD8 gene, demonstrating feedback control. However, application of GR24 to decapitated plants did not further repress DgCCD8 expression. Moreover, GR24 could not down-regulate DgCCD8 expression in the presence of exogenous apically applied auxin. In both these situations, GR24 would be expected to have only a modest effect on the amount of auxin in the stem, and thus these results are consistent with the hypothesis that GR24 acts mainly via auxin to mediate feedback regulation on DgCCD8.
Conclusion
It has been demonstrated that chrysanthemum buds can be inhibited by GR24 and thus there is potential to use SLs for the regulation of branching in chrysanthemum horticultural production. However, large-scale exogenous treatment techniques need to be developed and the cost of GR24 or analogues needs to be reduced for this to be practicable. In addition, chrysanthemum CCD8 genes have been isolated, providing the opportunity to use genetic modification to improve the chrysanthemum growth habit. Our results also contribute data from an additional species to ongoing debates about the mode of action of SLs in the inhibition of shoot branching, and its relationship with auxin. The effects of GR24 on bud activity and DgCCD8 expression support the idea that SLs inhibit bud activation by modulating auxin transport canalization, and that auxin is a major mediator of feedback by SL on SL synthesis.
Acknowledgments
This work was supported by the China 863 Program (2006AA100109) and 948 Project (2008-G3), the China Scholarship Council (CSC), and the UK Biotechnology and Biological Sciences Research Council as part of the European Research Area Network in Plant Genomics (BB/E024688/1). We thank Céline Mouchel for discussions on experimental design, and Céline Mouchel and Stephen Day for critical reading of the manuscript, Dan Jiang for the chrysanthemum material, and the University of York horticultural staff for plant care.
References
- Alder A, Holdermann I, Beyer P, Al-Babili S. Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochemical Journal. 2008;416:289–296. doi: 10.1042/BJ20080568. [DOI] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. The Plant Journal. 2007;51:1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant and Cell Physiology. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. The Plant Journal. 2006;45:982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. The Plant Journal. 2005;44:569–580. doi: 10.1111/j.1365-313X.2005.02548.x. [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology. 2006;16:553–563. doi: 10.1016/j.cub.2006.01.058. [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Ross JJ, Murfet IC. Branching in pea. Action of genes Rms3 and Rms4. Plant Physiology. 1996;110:859–865. doi: 10.1104/pp.110.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge CA, Symons GM, Turnbull CGN. Auxin inhibition of decapitation-induced branching is dependent on graft-transmissible signals regulated by genes Rms1 and Rms2. Plant Physiology. 2000;123:689–697. doi: 10.1104/pp.123.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signalling molecule. Current Biology. 2004;14:1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- Booker J, Chatfield S, Leyser O. Auxin acts in xylem-associated or medullary cells to mediate apical dominance. The Plant Cell. 2003;15:495–507. doi: 10.1105/tpc.007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P, Turnbull C, Srinivasan M, Goddard P, Leyser O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Developmental Cell. 2005;8:443–449. doi: 10.1016/j.devcel.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Dun EA, Ferguson BJ, Rameau C, Beveridge CA. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiology. 2009;150:482–493. doi: 10.1104/pp.108.134783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. The Plant Journal. 2000;24:159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dun EA, Ferguson BJ, Beveridge CA. Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiology. 2006;142:812–819. doi: 10.1104/pp.106.086868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA. The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. The Plant Cell. 2005;17:464–474. doi: 10.1105/tpc.104.026716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E, Turnbull CG, Beveridge CA. Long-distance signalling and the control of branching in the rms1 mutant of pea. Plant Physiology. 2001;126:203–209. doi: 10.1104/pp.126.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Hayward A, Stirnberg P, Beveridge C, Leyser O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiology. 2009;151:400–412. doi: 10.1104/pp.109.137646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiology. 2005;46:79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Johnson X, Brcich T, Dun EA, Goussot M, Haurogne K, Beveridge CA, Rameau C. Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals. Plant Physiology. 2006;142:1014–1026. doi: 10.1104/pp.106.087676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;10:472–476. doi: 10.1073/pnas.0509463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. The control of shoot branching: an example of plant information processing. Plant, Cell and Environment. 2009;32:694–703. doi: 10.1111/j.1365-3040.2009.01930.x. [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, et al. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell. 2009;21:1512–1525. doi: 10.1105/tpc.109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DA. Transport of exogenous auxin in two-branched dwarf pea seedlings (Pisum sativum L.) Planta. 1977;136:91–96. doi: 10.1007/BF00387930. [DOI] [PubMed] [Google Scholar]
- Morris SE, Turnbull CGN, Murfet IC, Beveridge CA. Mutational analysis of branching in pea. Evidence that rms1 and rms5 regulate the same novel signal. Plant Physiology. 2001;126:1205–1213. doi: 10.1104/pp.126.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Napoli C. Highly branched phenotype of the petunia dad1-1 mutant is reversed by grafting. Plant Physiology. 1996;111:27–37. doi: 10.1104/pp.111.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazeer MA, Khoshoo TN. Variation in the chromosome complement of Chrysanthemum morifolium complex. Nucleus. 1983;26:22–29. [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O. Control of bud activation by an auxin transport switch. Proceedings of the National Academy of Sciences, USA. 2009;106:17431–17436. doi: 10.1073/pnas.0906696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. Journal of Molecular Biology. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Research. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T, Thimann K. The role of auxins and cytokinins in the release of buds from dominance. American Journal of Botany. 1967;54:136–144. [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, et al. Genome structure of the legume, Lotus japonicus. DNA Research. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Loewen MC. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. Journal of Biological Chemistry. 2004;279:46940–46945. doi: 10.1074/jbc.M409004200. [DOI] [PubMed] [Google Scholar]
- Simons JL, Napoli CA, Janssen BJ, Plummer KM, Snowden KC. Analysis of the DECREASED APICAL DOMINANCE genes of petunia in the control of axillary branching. Plant Physiology. 2007;143:697–706. doi: 10.1104/pp.106.087957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Thimann KV. Further experiments on the inhibition of the development of lateral buds by growth hormone. Proceedings of the National Academy of Sciences, USA. 1934;20:480–485. doi: 10.1073/pnas.20.8.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. The Decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell. 2005;17:746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K, Booker J, Haurogne K, et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes and Development. 2003;17:1469–1474. doi: 10.1101/gad.256603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. The Plant Journal. 2007;50:80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HM. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129:1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- Thimann KV, Skoog F. Studies on the growth hormone of plants. III. The inhibiting action of the growth substance on bud development. Proceeding of the National Academy of Sciences, USA. 1933;19:714–716. doi: 10.1073/pnas.19.7.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO. Micrografting techniques for testing long-distance signalling in Arabidopsis. The Plant Journal. 2002;32:255–262. doi: 10.1046/j.1365-313x.2002.01419.x. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Walter MH, Giavalisco P, et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. The Plant Journal. 2009;61:300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- Yang DH, Yun PY, Park SY, et al. Cloning, characterization and expression of a Lateral suppressor-like gene from chrysanthemum (Dendranthema grandiflorum Kitamura) Plant Physiology and Biochemistry. 2005;43:1044–1051. doi: 10.1016/j.plaphy.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. The Plant Journal. 2006;48:687–696. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]