Abstract
Fucoxanthin chlorophyll proteins (Fcps), the light-harvesting antennas of heterokont algae, are encoded by a multigene family and are highly similar with respect to their molecular masses as well as to their pigmentation, making it difficult to purify single Fcps. In this study, a hexa-histidine tag was genetically added to the C-terminus of the FcpA protein of the pennate diatom Phaeodactylum tricornutum. A transgenic strain expressing the recombinant His-tagged FcpA protein in addition to the endogenous wild type Fcps was created. This strategy allowed, for the first time, the purification of a specific, stable trimeric Fcp complex. In addition, a pool of various trimeric Fcps was also purified from the wild-type cells using sucrose density gradient ultracentrifugation and gel filtration. In both the His-tagged and the wild-type Fcps, excitation energy coupling between fucoxanthin and chlorophyll a was intact and the existence of a chlorophyll a/fucoxanthin excitonic dimer was demonstrated using circular dichroism spectroscopy. Mass spectrometric analyses of the trimeric His-tagged complex indicated that it is composed of FcpA and FcpE polypeptides. It is confirmed here that a trimer is the basic organizational unit of Fcps in P. tricornutum. From circular dichroism spectra, it is proposed that the organization of the pigments on the polypeptide backbone of Fcps is a conserved feature in the case of chlorophyll a/c containing algae.
Keywords: Diatoms, FcpA, His-tag, thylakoid, photosynthesis
Introduction
Diatoms (Bacillariophyceae) are unicellular, eukaryotic algae which belong to the Heterokont group of algae. This group of algae was derived by a secondary endosymbiosis event between a red alga and a heterotrophic eukaryote (Bhattacharya and Medlin, 1998; Archibald and Keeling, 2002; Yoon et al., 2002). Diatoms are major players in the biochemical cycles of carbon, nitrogen, silica, and phosphorus. By participating in these vital element cycles they have a profound effect on the global climate both in marine and freshwater environments (Wilhelm et al., 2006). Despite the fact that the general organization of the photosynthetic apparatus of diatoms, plants, and red algae is very similar, there are significant differences in membrane topology, and polypeptide and pigment composition. The thylakoid membranes are not differentiated into distinct granal and stromal regions but organized into bands consisting of three thylakoids each (Gibbs, 1962). Furthermore, unlike in higher plants, no specific localization of photosystem (PS) I and PSII within the diatom thylakoid membrane system is known; they seem to be rather homogeneously distributed (Pyszniak and Gibbs, 1992).
In contrast to higher plants and green algae, the light-harvesting complex (LHC) of diatoms contains chlorophyll (Chl) c instead of Chl b as the accessory Chl and fucoxanthin (Fx) instead of lutein as the major carotenoid. Since they are the major components of the LHC, the antennae are referred to as Fcps (Fucoxanthin chlorophyll a/c binding proteins). The major xanthophyll cycle pigments in diatoms are diadinoxanthin (Ddx) and its de-epoxidized form diatoxanthin (Dtx). The localization of these pigments within the Fcp antennae has been demonstrated and diatoxanthin in particular has been shown to play a critical role in the protective process of non-photochemical quenching (NPQ) under conditions of excess illumination (Lavaud et al., 2003; Guglielmi et al., 2005; Gundermann and Büchel, 2008; Goss and Jakob, 2010).
Fcps in diatoms are membrane intrinsic light-harvesting antenna complexes which are members of the LHC superfamily encoded by a conserved multigene family (Bhaya and Grossman, 1993; Green and Pichersky, 1994; Green and Durnford, 1996). They function either in light harvesting and/or photoprotection. Fcps share significant similarity in sequence and in predicted structure with plant LHCII. Fcps, like LHC polypeptides in plants, are inserted in thylakoid membranes by three membrane spanning α-helices. The similarity between these is especially strong in helix 1 and 3 (Green and Pichersky, 1994). However, mature Fcps have been found to be smaller in size than the plant LHCs due to shorter loops and termini, which, in turn, makes them more hydrophobic than plant LHCs.
All known Fcps have a molecular weight between 17–23 kDa (Fawley and Grossman, 1986; Caron and Brown, 1987) and are classified into three major groups (termed I, II, and III) based on sequence analyses. Group I represents the major Fcps in diatoms and it includes the genes fcp1-3 and fcp5 from Cyclotella cryptica and fcpA-F from Phaeodactylum tricornutum, which are hypothesized to serve in the function of light harvesting (Durnford et al., 1996; Eppard et al., 2000; Green, 2003; Bowler et al., 2008). Group II is represented by the genes fcp4 of C. cryptica and its homologues lhca and lhcr in Thalassiosira pseudonana and P. tricornutum, respectively (Armbrust et al., 2004). It was shown recently that in Cyclotella meneghiniana fcp4 encodes PSI specific antenna proteins (Veith et al., 2009). LI818 is a light-inducible protein found in the green alga Chlamydomonas reinhardtii. The respective homologue genes, i.e. fcp6, 7, and 12 in C. cryptica belong to Group III of the Fcps, which are proposed to function in light protection (Richard et al., 2000; Beer et al., 2006; Zhu and Green, 2008; Peers et al., 2009). The genome of P. tricornutum also contains sequences which represent light-inducible proteins similar to LI818, which are denoted as Lhcx proteins (Bowler et al., 2008).
Biochemical investigations have shown that the major antenna proteins of PSII in plants are trimerically arranged Lhcb1–b3 proteins, while the minor antennae consist of Lhcb4–b6 proteins (also termed as CP24, CP26, and CP29 proteins). It has also been confirmed at a structural level by high resolution X-ray diffraction studies that the LHCII in higher plants is a stable trimer (Liu et al., 2004). Unfortunately, such a wide array of information regarding the biochemical identities and structural organization of Fcps is not yet available for diatoms; however, this has started to change in recent years. Guglielmi et al. (2005) reported the isolation of an Fcp trimer from P. tricornutum by applying a rapid sequence of thylakoid solubilization and subsequent gel filtration. Büchel (2003) reported the presence of two different Fcp fractions in C. meneghiniana; which were different in molecular weight and oligomeric status. Furthermore, Beer et al. (2006) identified the subunits of the higher oligomer FCPb and the trimer FCPa in C. meneghiniana. Lepetit et al. (2007) recently showed that, in P. tricornutum, the basic stable unit of organization of Fcps is a trimer. It was speculated that the co-existence of higher oligomers and trimers of Fcps in P. tricornutum was unlikely. In addition, FcpA–F were identified in the main antenna complex fraction by mass spectrometry with FcpC, D, and E having the strongest signals, while no subpopulation of trimers could be isolated and characterized. The high degree of similarity in molecular weights and possibly also in pigmentation among all Fcps is so far the biggest hurdle for the purification of different Fcp complexes or single Fcps. The availability of the genome (Bowler et al., 2008) and genetic tools for stable genetic transformation of P. tricornutum (Apt et al., 1996; Zaslavskaia et al., 2000; Kroth, 2007; Siaut et al., 2007; Maheswari et al., 2008) makes this diatom a very suitable model organism to study the organization of the photosynthetic apparatus. Moreover, such tools allow adding affinity tags to any chosen Fcp protein, enabling their purification via affinity chromatography.
In order to elucidate possible differences in function between different Fcps and to examine the molecular organization of Fcp complexes, specific Fcps, either as single polypeptides or as a complex in a highly pure and native state, have to be obtained. Here, the aim was to study FcpA in more detail and thus the gene of P. tricornutum was genetically manipulated by tagging the 3′ terminus with a nucleotide sequence encoding a hexa histidine (His) tag. The transgenic strain expressing the His-tagged FcpA protein could then be used to isolate an Fcp complex containing a specific fcp gene product, FcpAHis, for the first time. Data are presented here concerning the purification and biochemical, as well as spectroscopic, characterization of the pure His-tagged FcpA complex in comparison with the wild-type (WT) Fcp pool in P. tricornutum.
Materials and methods
Cloning of a construct for expression of recombinant FcpAHis and transformation of P. tricornutum
The complete nucleotide sequence for fcpA was available at the EMBL–EBI database with gene ID number Z24768 as deposited by Bhaya and Grossman (1993) and can also be found in the US Department of Energy's Joint Genome Institute P. tricornutum genome database (http://genome.jgi-psf.org/Phatr2/Phatr2.home.html (Bowler et al., 2008) with protein ID 18049. Genomic DNA was isolated from wild-type cells (UTEX culture collection, strain 646). Using the following oligonucleotide primers, the coding region of the fcpA gene was amplified by PCR: (F) 5′-GA(GATATC)ATTCAAGATGAAATTTGC-3′ and (R) 5′-A(GTCGAC)TTAATGGTGATGGTGATGGTGAGGAAGGATAG-3′. Primer (F) inherently contained an EcoRV restriction site whereas the reverse primer (R) was designed to introduce a SalI restriction site and simultaneously to generate a fragment encoding a His-tag at the 3′ terminus of the gene. The restriction sites within the primers are indicated in parentheses while the underlined sequence codes for six histidine residues. This insert was introduced in the multiple cloning site of the P. tricornutum transformation vector pPha-T1 (Genbank accession number AF219942) as developed by Zaslavskaia et al. (2000), by digesting the vector and the PCR fragment with EcoRV and SalI and subsequently ligating the construct into the vector. The resulting plasmid pPha-T1FcpAHis, carries a His-tagged fcpA gene under the control of the native fcpA promoter (Fig. 1). It was allowed to amplify in E. coli under ampicillin pressure. Plasmid DNA was isolated and used for the transformation of WT P. tricornutum cells.
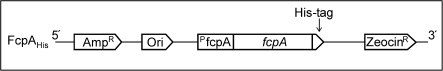
Fig. 1.
Schematic representation of the fcpA gene under the control of the fcpA promoter (pfcpA) and position of the histidine-tag in the constructed plasmid pPha-T1FcpAHis. The sequence encoding a six histidine residue was inserted at the 3′ end of the coding region of the fcpA gene. The transformation vector pPha-T1 carries a gene for Ampicillin (Amp) resistance upstream of the fcpA gene and for Zeocin resistance downstream of the fcpA gene. The origin of replication (Ori) for amplification in E. coli is also found upstream of the fcpA gene.
Transformation of P. tricornutum with pPha-T1FcpAHis was carried out according to Apt et al. (1996) and Zaslavskaia et al. (2000) with minor modifications. WT cells plated on Artificial Seawater medium (ASP) agar (Provasoli et al., 1957) were bombarded using the Bio-Rad Biolistic PDS-1000/He Particle delivery system (Bio-Rad laboratories, Hercules, CA) fitted with a 1350 psi rupture disc. 1 μg plasmid DNA was coated onto M17 tungsten particles (Bio-Rad) and used for bombardment. Bombarded cells were illuminated continuously for 24 h before plating them on ASP agar plates supplemented with 75 μg ml−1 zeocin to select possible transformants. Clones which could survive zeocin selection were screened for expression of the His-tagged recombinant protein at the mRNA level (data not shown). The recombinant clones now contained one or more copies of the His-tagged fcpA gene along with a background of all other WT fcp genes. The clone FcpA1.2 was observed to have the highest expression level and was chosen for further work. The presence of the His-tagged gene within this clone was again confirmed by PCR and by sequence analyses (data not shown). It was cultured and subcultured regularly in ASP medium containing zeocin under low light conditions for a period of 6 months before His-tagged FcpA was isolated.
Culture conditions
P. tricornutum Böhlin WT (UTEX culture collection, strain 646) and the mutant, FcpA1.2, were grown in ASP medium for 10 d at 18 °C under constant aeration by bubbling sterile air, and at a light intensity of 40 μE m−2 s−1 and a 16/8 h light/dark cycle.
Thylakoid preparation
Cells were harvested from 10-d-old cultures at a Chl a concentration of approximately 10–15 mg l−1 within 1–2 h after the onset of light, and thylakoids were prepared as described in Veith and Büchel (2007) with a few minor changes. Harvested cells were resuspended in homogenization buffer (10 mM HEPES, 2 mM KCl, 5 mM EDTA, 1 M sorbitol, pH 7.4) and broken by repeated French Press cycles (Polytec/Thermo) for six times at a pressure of 20 000 psi. The cell lysate was spun for 2 min at 4 °C at 3000 g to avoid contamination with unbroken cells. The pellet was resuspended in homogenization buffer and centrifuged again at 3000 g for 2 min at 4 °C. The supernatants from both centrifugation steps were collected and ultracentrifuged for 40 min at 206 000 g. The pelleted thylakoid membranes were resuspended in washing buffer A (10 mM HEPES, 2 mM KCl, pH 7.4) and ultracentrifuged once again for 40 min at 163 000 g. The pellet of thylakoid membranes was resuspended in washing buffer A and the chlorophyll content was determined according to Jeffrey and Humphrey (1975).
Purification of Fcps
Sucrose density gradient ultracentrifugation:
Thylakoids of the WT and the mutant were solubilized with 15 mM β-dodecyl maltoside (β-DDM) at a concentration of 0.25 mg ml−1 Chl a (detergent: Chl a, 30:1) in a 2 ml end volume for 20 min under constant shaking on ice and loaded onto a discontinuous sucrose density gradient and ultracentrifuged at 141 000 g (Rotor: Beckmann SW28) for 22 h at 4 °C. The sucrose gradient consisted of seven steps of sucrose solutions made in washing buffer A supplemented with 0.03% β-DDM and were layered in the order of 25%, 17.5%, 15%, 12.5%, 10%, 7.5%, and 5%. The brown-coloured Fcp band localized between 10% and 12.5% sucrose. It was harvested carefully to avoid any contamination with the green photosystem fraction which was localized at the interface of the 15% sucrose and 17.5% sucrose density bands. Fcp fractions from the WT and mutant were then concentrated in concentration devices with 30 kDa cut-off (Amicon) and were frozen under liquid nitrogen and stored at –80 °C until further purification.
Gel filtration:
For further purification of Fcps in the WT, the Fcp band obtained from sucrose density gradient ultracentrifugation was applied to a self-packed gel filtration column (Superdex 200, GE Healthcare) connected to an ÄKTA purifier P-900 (Amersham Biosciences). Flow-rate was maintained at 0.5 ml min−1 during the run and absorbances of the eluates were analysed at 437 nm, 280 nm, and 675 nm. The column was calibrated with FCPa and FCPb complexes from C. meneghiniana since their oligomeric status and molecular weights had previously been established allowing their use as molecular weight markers (Büchel, 2003; Beer et al., 2006). The major peak fractions were collected and concentrated in 30 kDa cut-off concentration devices. This pure WT Fcp pool was immediately analysed by spectroscopy and then frozen in liquid nitrogen and stored at –80 °C for further analyses.
Nickel-NitriloTriacetate (Ni-NTA) IMAC:
For the transgenic strain, the concentrated Fcp band from the sucrose density gradient was applied to a self-packed Ni-NTA column (Chelating Sepharose Fast Flow, Amersham Biosciences) pre-equilibrated with binding buffer (25 mM phosphate, 150 mM KCl, 5 mM imidazole, 0.6 mM β-DDM, pH 7.4) and allowed to incubate for 1 h at 4 °C for binding of the His-tagged proteins to the column material. The column was washed with washing buffer B (25 mM phosphate, 50 mM KCl, 0.6 mM β-DDM, pH 7.4) at 0.5 ml min−1 flow-rate until the absorbance at 672 nm was less than 0.005 absorbance units. His-tagged FcpA (FcpAHis) was subsequently eluted using elution buffer (25 mM phosphate, 15 mM KCl, 400 mM imidazole, 0.6 mM β-DDM, pH 7.4) in 0.5 ml aliquots, which were concentrated using 30 kDa microcon concentration devices (Amicon).
Characterization of FcpA
Gel filtration:
To analyse the oligomeric state, FcpAHis corresponding to ∼2.5 μg Chl a was applied to a self-packed column (Superdex 200, GE Healthcare) connected to an ÄKTA purifier P-900 (Amersham Biosciences). Run conditions were identical to those used for the WT.
Electrophoresis:
15% TRIS-Tricine denaturing gels were made according to Schägger and von Jagow (1987). Samples were denatured with Rotiload (Roth) for 15 min and gels were stained with Coomassie Blue G 250 or were silver stained.
Western blot:
Unstained gels were rinsed briefly with double-distilled water and blotted onto PVDF membranes (Roth) according to Veith and Büchel (2007). Immunodetection was carried out using ECL detection solutions (Amersham Biosciences) after decorating either with antibodies made against all Fcp polypeptides from C. meneghiniana isolated as described in Büchel (2003); α-Fcp, or with a monoclonal His antibody; α-His (Sigma).
Mass spectrometry (MS):
The band of interest was cut out from a silver-stained gel and freeze-dried. A tryptic digest and nano LC-ESI-MS/MS analysis of the peptides were carried out according to Veith et al. (2009) without changes.
HPLC:
Cells were pelleted at 5000 rpm and lyophilized. They were then suspended in extraction solution (1:2; cell volume:extraction volume) of 90% methanol buffered with 1 mM TRIS-HCl (pH 7.5), and disrupted mechanically in the presence of glass powder by an homogenization pistil followed by sonication for 30 s. Cells were then allowed to stand on ice for 1 min. This process was repeated 10 times after which the solution was spun at 13 000 rpm for 15 min.
In the case of Fcps isolated from the WT and mutant, samples were precipitated directly in 90% final concentration of methanol and spun briefly. The supernatants were analysed for pigment content by HPLC and the amounts of pigments were quantified according to Papagiannakis et al. (2005).
Spectroscopic analyses:
Absorbance spectra were recorded with a spectrophotometer (Jasco V550) using a bandpass of 1 nm and 1 cm optical path length at room temperature. Samples were adjusted to an absorbance of 0.03 at the QY band of Chl a, which corresponded to a Chl a concentration of ∼0.3 μg ml−1. Fluorescence spectra were recorded at room temperature in a fluorescence spectrometer (Jasco FP6500) with a bandpass of 3 nm both on the excitation and emission side and at 0.1 nm data pitch. Emission spectra were recorded between 600–800 nm when the samples were illuminated at λex=465 nm to preferentially excite Chl c. Excitation spectra were recorded at λem=675 nm upon excitation from λex=400–600 nm. A rhodamine-B spectrum was used as a reference for the correction at the excitation side and the photomultiplier was corrected using a calibrated lamp spectrum. Circular dichroism (CD) spectra were measured at room temperature using 1 cm optical pathlength, 2 nm bandwidth, and a 2 s response time in the range of 370–750 nm in a CD spectrometer (Jasco 810). Concentration of the samples was adjusted to ∼5 μg ml−1 of Chl a. Five spectra were recorded sequentially in each case to improve the signal-to-noise ratio and the data presented herewith are an average of these sequential spectra.
Results and discussion
Purification of a single Fcp and its biochemical characterization was the primary goal of this study. The availability of the fcpA gene sequence in P. tricornutum, a suitable vector (pPha-T1), and a method for its stable transformation, made it possible to create a transgenic strain of P. tricornutum in which the fcpA gene carries a fragment encoding a hexa-histidine tag on the 3′ terminus of the gene. Expression of the modified gene is under the control of the native promoter of the gene (FcpA promoter; Zaslavskaia et al., 2000) in the transformed cell lines, thereby avoiding artificial overexpression of FcpAHis. The modified gene also encodes the complete FcpA plastid targeting presequence with a conserved sequence at the predicted signal peptide cleavage site (Kilian and Kroth, 2005; Gruber et al., 2007). It has been shown previously, with the presequence of the γ subunit of the P. tricornutum plastid ATP synthetase, that preproteins are also targeted correctly to the plastid when a His-tag has been engineered to their C-terminus (Apt et al., 2002). Targeting of the FcpAHis gene product to the thylakoid membranes should therefore function analogously to the WT FcpA gene product. The Fcp complex, isolated by the use of this His-tag, is compared here with the pool of WT Fcp complexes.
Purification of Fcps
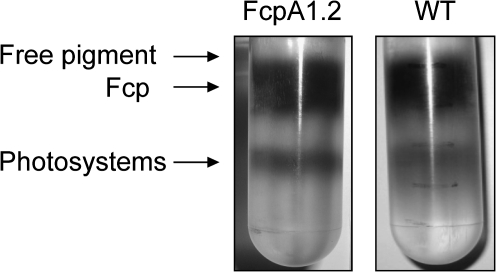
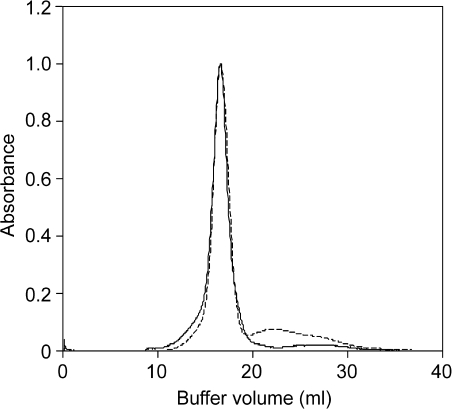
Discontinuous sucrose density gradient ultracentrifugation was carried out for both the WT and the transgenic strain as a prepurification step to separate the photosystem fractions from the Fcp pool. A fractionation pattern similar to that reported by Guglielmi et al. (2005) and Lepetit et al. (2007) was observed within the gradients in both cases (Fig. 2). A golden-olive coloured fraction was localized at the 7.5% sucrose step. It was mainly found to contain free pigment based on spectroscopic and SDS-PAGE analyses and was thus discarded. A green-coloured fraction containing photosystem(s) was fractionated at the interface of the 17.5% and the 25% sucrose step and was also not considered further. The brown-coloured fraction which occurred between the 12.5% and 15% sucrose steps was found to be enriched in Fcp and was harvested for further purification. This Fcp fraction contained about 65–70% Chl a of that found in thylakoids, both in WT and the mutant. The WT Fcp fraction from sucrose density gradients was further purified by gel filtration. Figure 3 shows the profile of the WT Fcp band from the sucrose density gradient applied to a gel filtration column. A major peak was observed at 16.6 ml followed by a tail peak (20–29 ml). Fractions were collected from the major peak (16.6 ml) and used as the WT Fcp pool after spectroscopic confirmation that the energy transfer was intact (see below).
Fig. 2.
Fractionation of thylakoids, on a sucrose density gradient after solubilization with 15 mM β-DDM (detergent:Chl a, 30:1). The left tube shows the results of fractionation of the transgenic strain, FcpA1.2, whereas the fractionation of the WT thylakoids under identical conditions is shown on the right.
Fig. 3.
Elution profile of gel filtration of FcpAHis (solid line) and of WT Fcp (dashed line) fractions obtained from sucrose density gradient centrifugation. Absorbance was measured at 437 nm.
For the transgenic strain, the His-tagged FcpA was further purified from the Fcp fraction collected after sucrose density gradient centrifugation using IMAC.
Polypeptide analysis
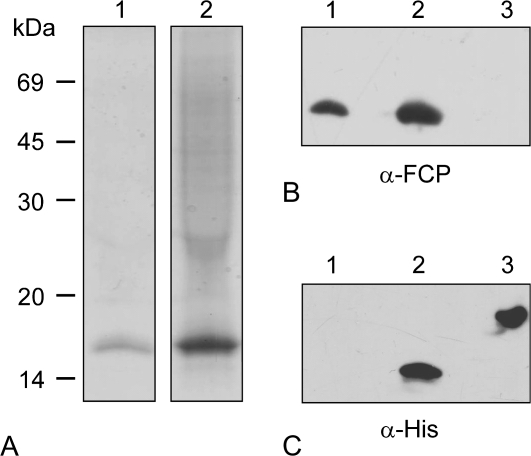
Denaturing SDS-PAGE was carried out to analyse the purity of the FcpAHis and to estimate the relative molecular mass of its polypeptide(s). The predicted molecular mass of FcpAHis is 18.9 kDa. Figure 4A presents a Coomassie-stained gel loaded with WT Fcp (lane 1) and FcpAHis (lane 2). A single prominent band around 19 kDa proved that contaminants like photosystem fractions were absent in the FcpAHis fraction. WT Fcp had been loaded at the same concentration of Chl a as the FcpAHis, however, signals were stronger in the case of the mutant Fcp. Since this turned out to be reproducible, independent of the batches of preparation used, it was concluded that WT Fcp contained lower amounts of polypeptides than the FcpAHis, i.e. the FcpAHis fraction contains fewer pigments per polypeptide (see also the HPLC analysis described below).
Fig. 4.
Coomassie-stained SDS-PAGE (A) and Western blots (B, C) of WT Fcps (lanes 1) and FcpAHis containing fractions isolated using IMAC (lanes 2). In (A), samples corresponding to 0.5 μg Chl a were loaded. Adjacent on the left the molecular weights in kDa of marker proteins are given. The Western blot in (B) was developed with an α-Fcp antibody, whereby His-tagged PsbS was loaded in lane 3 as a negative control. The Western blot in (C) was developed with an α-His antibody, whereby His-tagged PsbS in lane 3 now served as a positive control.
Samples were also analysed by immunoblots with antibodies either against the His-tag (α-His) or against all Fcps of C. meneghiniana (α-Fcp). His-tagged PsbS protein from higher plants expressed in E. coli with a predicted molecular mass of 24 kDa was used as a positive control for the anti-His antibody. In cases where the blot was developed with α-Fcp (Fig. 4B), signals were detected in the fractions from WT Fcp and also in the FcpAHis fraction, confirming that the purified product consisted of Fcp polypeptides in each case. As expected, only a single sharp signal was seen at 19 kDa in the FcpAHis sample when developed with α-His (Fig. 4C) and none in the WT Fcp, confirming the presence of the His-tagged protein.
Oligomeric state
FcpAHis was subjected to analytical gel filtration to investigate the oligomeric state as well as the homogeneity. Figure 3 shows the gel filtration profile of FcpAHis where a single sharp peak was observed at 16.6 ml. An additional broad but minor peak was observed between 24–27 ml and spectroscopic investigations showed that the former peak was composed solely of Fcps and the latter was devoid of them. After spectroscopic investigations, the latter peak was assumed to be free pigment and was not considered further. It has been shown for Fcps from C. meneghiniana that the xanthophyll cycle pigments are lost more easily when they are purified via ion exchange chromatography as compared to those purified by sucrose gradient (Beer et al., 2006). It is therefore assumed that a part of the latter (free pigment) peak could have been pigments originally associated with the FcpAHis complex. However, the loosely bound pigments decoupled from the complex during the washing conditions on the IMAC as well as during gel filtration. The WT Fcp fraction of P. tricornutum was also eluted from the same column under identical conditions at 16.6 ml (see above). Peak fractions in both WT Fcp and FcpAHis were analysed for energy transfer and it was confirmed that the Fcps were intact (see below). Trimeric FCPa complex from C. meneghiniana also eluted at 16.5 ml under identical conditions (K. Gundermann, personal communication). The similarity in the elution volume thus related the oligomeric state of the trimeric FCPa complex from C. meneghiniana (Büchel, 2003; Beer et al., 2006) and the isolated FcpAHis complex along with the WT Fcp from P. tricornutum. Neither higher oligomers nor monomers were observed in the samples. Thus, it was concluded that FcpAHis in P. tricornutum is part of a trimeric complex. Furthermore, we also confirmed that all Fcps in the WT sample were also trimeric under this particular condition of solubilization of thylakoids.
To date FCP complexes in different oligomeric states have been isolated both from the centric diatom C. meneghiniana and from the pennate diatom P. tricornutum. Co-existing trimeric FCPa complexes and higher oligomeric FCPb complexes were isolated and characterized from C. meneghiniana (Büchel, 2003; Beer et al., 2006). In P. tricornutum, it was shown that there exists an antenna complex FCPo in a higher oligomeric state and a trimeric FCP complex, comparable to FCPb and FCPa in C. meneghiniana, respectively. The major difference as compared to the antenna complexes from C. meneghiniana was that at least in vitro the FCPo and FCP complexes were never co-existent (Lepetit et al., 2007). FCPo complexes were obtained by mild solubilization of thylakoids (detergent:Chl, 5:1), while the trimeric FCP complexes were purified using relatively harsher conditions (detergent: Chl, 20:1 and 40:1). In our case the detergent:Chl a ratio was set to 30:1, explaining that both the complexes of FcpAHis and the WT Fcp pool were trimeric. From our observations, we can support the hypothesis that a trimer is the basic stable unit of organization of all Fcps in P. tricornutum.
Polypeptide composition of trimers
MS was used to identify the polypeptides which built up the FcpAHis-containing complex. Trypsin digestion of the 19 kDa band from a silver-stained gel followed by nano LC-ESI-MS/MS led to the identification of several peptide sequences. These were used to identify proteins within a combined diatom database as described in Veith et al. (2009). Two Fcp proteins were unambiguously identified (Table 1). These were Lhcf1 (JGI protein ID: 18049) and Lhcf5 (JGI protein ID: 30648) from P. tricornutum, which are also annotated as FcpA and FcpE, respectively. The presence of just these two gene products is in contrast to the WT fraction, in which Lepetit et al. (2007) demonstrated the presence of FcpA, B, C, D, E, and F using mass spectrometry as well. Thus, the FcpAHis complex is a specific trimer made up of FcpA and FcpE subunits. Since the applied LC-ESI-MS/MS technique is not a direct quantitative approach, further work needs to be done to elucidate the quantitative contributions of these two polypeptides in the trimer.
Table 1.
Nano LC-ESI-MS/MS analysis of a FcpAHis containing complex
| Protein ID (JGI-Phatr2) | Name | Identified peptides | z | Xcorr |
| 18049 | Lhcf1 (FcpA) | NNYLDFGWDTFSEDK | 2 | 4.55 |
| NNYLDFGWDTFSEDKK | 2 | 3.99 | ||
| DITGGEFVGDFR* | 2 | 3.93 | ||
| 30648 | Lhcf5 (FcpE) | NDFIDFGWDSFDEETK | 2 | 4.04 |
| DITGGEFVGDFR* | 2 | 3.93 | ||
| ISMoLAVAGYLVQENGIR | 2 | 3.24 |
JGI-Phatr2, Joint Genome Institute-vs2 of the P. tricornutum genome. z, charge. Xcorr, cross-correlation factor. Mo, oxidized Met. * The peptide is present in both models.
Spectroscopic characterization and pigment analysis
The pigment composition of the trimeric His-tagged complex and the WT Fcp pool was compared by HPLC analyses, which resulted in some differences. In both cases the Chl c content was similar with about 0.36–0.38 mol per mol Chl a (Table 2). However, the Fx content in the case of the His-tagged complex was about 1.2 mol mol−1 of Chl a, while in WT Fcp the content was about 1.5 times higher. To our knowledge, there is no literature report of such high values for Fx in Fcps of P. tricornutum, except for one of the fractions obtained by Guglielmi et al. (2005) using gel filtration. In order to obtain large amounts of protein, the cultures used here were grown under low light by bubbling sterile air (see the Materials and methods), allowing the cells to multiply faster than in shaking cultures (data not shown), and in addition to grow to a high density (10–15 mg Chl a l−1). At this Chl a concentration, the cultures appeared dark brown in colour, indicating that the amount of fucoxanthin, the major carotenoid inside the FCPs, was up-regulated. To evaluate this hypothesis, whole cells were measured as well and an unusually high content of Fx was found in both the WT and the transgenic strain (0.78–0.80 mol Fx mol−1 Chl a) compared with the value of 0.68 published for cells not grown to such densities (Lavaud et al., 2003). In addition, pigmentation of Fcps in P. tricornutum was usually studied using Fcp complexes purified either by gel filtration or by sucrose density centrifugation (Guglielmi et al., 2005; Lepetit et al., 2007), whereas a combined approach was used in this study for the WT Fcp pool, i.e. a second purification step that should reduce contamination. However, it cannot be ruled out that some contamination with Fx from the free pigment fraction took place when using these two, still very gentle, preparation methods sequentially.
Table 2.
HPLC analysis of whole cells and isolated Fcp complexes
| mol mol−1 Chl a | Fx | Chl c | Ddx+Dtx |
| WT cells | 0.782±0.136 | 0.219±0.013 | 0.124±0.021 |
| FcpA1.2 cells | 0.806±0.052 | 0.232±0.030 | 0.120±0.008 |
| WT Fcp | 1.710±0.137 | 0.369±0.027 | 0.207±0.020 |
| FcpAHis | 1.199±0.090 | 0.376±0.027 | 0.055±0.040 |
Pigment composition is indicated on mol mol−1 Chl a basis and values represent average and standard deviations of three to five independent measurements.
FcpA and FcpE belong to group I of Fcps which are thought to be the classical light-harvesting Fcps. It was therefore unexpected to find lower amounts of fucoxanthin in the FcpAHis complexes compared with the WT Fcp pool. It is known that the xanthophyll cycle pigments can be lost from the antenna complexes more easily than Fx, Chl c and Chl a due to their probable peripheral binding site (Beer et al., 2006). It was therefore not surprising that the concentrations of Ddx+Dtx were higher in the WT Fcp fraction than in the FcpAHis complex (Table 2), because WT Fcp was not subjected to the slightly harsher condition of IMAC during purification. In the case of the trimeric FcpAHis complex, the pigment data include only the pool of pigments tightly associated with the polypeptides, which seem to be the ‘core’ pigments quite similar in all FCP complexes isolated so far. Thus, a characterization of more individual Fcps is needed to prove that the different Fcps can bind different pigments in different stoichiometries.
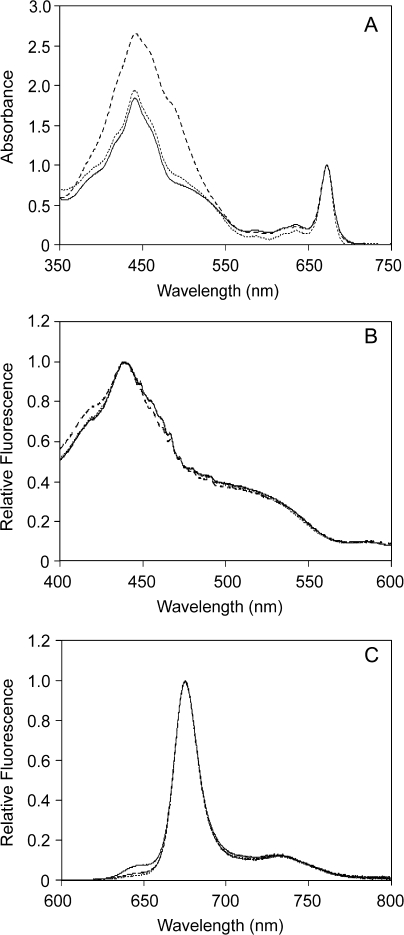
Figure 5A shows a comparison between room temperature absorbance spectra of WT Fcp, the FcpAHis complex, and FcpAHis after subsequent gel filtration. These spectra are very similar to those reported for Fcps from other diatoms (Büchel, 2003; Lepetit et al., 2007). In all cases, the absorption maxima at the QY region of Chl a were consistently found at 672 nm. A small peak at 636 nm represented the QY band of Chl c in each case. In the Soret region, the absorption maximum was observed at 441.5 nm for the WT Fcp whereas the FcpAHis complex showed a slight blue shift of 1.5 nm. This minor hypochromic shift was attributed to the conditions of the IMAC and the subsequent gel filtration, and therefore to a loss of loosely bound pigments. This became more obvious by a significant decrease in the absorbance in the Soret region in FcpAHis complexes compared with WT Fcp by ∼25%. The loss of pigments was even more pronounced in the case of a shoulder at 486 nm, which is present in WT Fcp and absent in the FcpAHis trimer. This shoulder probably reflects on the higher amount of xanthophyll cycle pigment pool and/or fucoxanthin in the WT, owing to the fact that it has a pool of different Fcps. This observation was supported by the pigment analysis that the WT Fcp and the pure FcpAHis complex have different amounts of associated pigments (see above).
Fig. 5.
Absorption spectra (A), fluorescence excitation spectra measured at 675 nm (B) and fluorescence emission spectra (C) recorded upon excitation at 465 nm were recorded using WT Fcp purified on gel filtration (dashed line), FcpAHis after IMAC (solid line) and FcpAHis further purified on gel filtration (dotted line) at a Chl concentration of ∼0.3 μg ml−1, and normalized to the QY absorption (A), the maximum in the Soret (B) and the maximal Chl a emission (C).
Fluorescence excitation spectra (Fig. 5B) for both WT Fcp and the trimeric FcpAHis complex demonstrated that the isolated complexes were intact with respect to energy transfer from Chl c and Fx to Chl a. It also showed that the additional pigments seen in absorbance spectra of the WT Fcp compared with FcpAHis (Fig. 5A) were not involved in the transfer of excitation energy to Chl a. Thus, the additional pigments found by HPLC in the WT complexes, i.e. fucoxanthin, diadinoxanthin, and diatoxanthin, most probably represent pigments not properly working in excitation energy transfer. Excitation at 465 nm (Soret band of Chl c) to record the fluorescence emission resulted in one sharp peak at 675.1 nm and at 675.7 nm for the FcpAHis complex and the WT Fcp, respectively (Fig. 5C). A very small shoulder at 636 nm was observed in the case of FcpAHis purified only via IMAC suggesting that a small amount of Chl c was decoupled from the complex or that this shoulder arose due to the presence of free Chl c on the IMAC column. This small Chl c emission disappeared after gel filtration and indirectly proved that whatever Chl c emission was observed after IMAC was only due to free Chl c independent of the FcpAHis complex. The additional gel filtration in FcpAHis was helpful to determine the oligomeric state and to confirm that the complex was extremely stable and functional even after multiple series of purification processes.
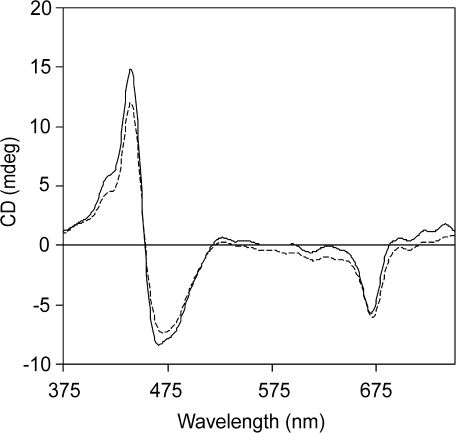
The CD spectra of WT Fcps as well as FcpAHis complex were observed to be very similar to each other (Fig. 6). They also resembled the previously reported CD spectra from P. tricornutum (Lepetit et al., 2007) and also from C. meneghiniana (Büchel, 2003) concerning the spectral structure. A prominent single negative band was observed at (–)669 nm in the case of FcpAHis and at (–)670 nm for the WT Fcps which was attributed to Chl a. This peak showed a 3 nm blue shift as compared to the QY band of Chl a in the absorbance spectrum which might suggest the existence of a heterogeneous pool of Chl a in Fcps (Mimuro et al., 1990). As this was a single band, it was concluded that no Chl a–Chl a excitonic interactions exist in Fcps of P. tricornutum. Minor negative bands were observed at (–)612 nm and (–)615 nm for FcpAHis and WT, respectively, corresponding either to the Qx absorbance of Chl a or to the QY absorbance of Chl c. A huge split signal was observed in the Soret region at (–)467 nm and (+)440 nm for FcpAHis, resembling the spectral fingerprint observed earlier for Fcps in P. tricornutum (Lepetit et al., 2007), clearly indicated the existence of a Chl a-carotenoid excitonic dimer. This again established the intactness of the isolated complex. Lepetit et al. (2007) could not find any differences in the CD spectra of Fcps in P. tricornutum in higher oligomeric or in the trimeric state, like those observed in the case of C. meneghiniana earlier (Büchel, 2003). The similarity in the spectral shape of FcpAHis and WT Fcps in our case to that of the FCPo and FCP complex suggests that the arrangement of pigments on the polypeptide backbone in all Fcps is similar in P. tricornutum. Furthermore, the common split signal in FCP complexes of C. meneghiniana and P. tricornutum indicate that the Chl a-fucoxanthin excitonic dimer is probably a conserved feature of the Fcps in diatoms.
Fig. 6.
Circular dichroism spectra of WT Fcp (dashed line) and of FcpAHis (solid line). Concentration of both samples was adjusted to 5 μg ml−1 Chl a.
In summary, it was possible to isolate for the first time a trimeric FCP complex containing a specific gene product, FcpA. The pigments bound to this complex were functional in excitation energy transfer. The complex was found to be composed of FcpA as well as FcpE, which indirectly demonstrated that the other well expressed polypeptides in P. tricornutum, mainly FcpB, FcpC, and FcpD, have to be localized in different trimers. Thus, this is the first report on different subpopulations of Fcp complexes in P. tricornutum.
Acknowledgments
We would like to thank Kathi Gundermann for her excellent scientific advice through all stages of this project. Kerstin Pieper is gratefully acknowledged for supplying isolated, His-tagged PsbS. We thank Professor W. Kühlbrandt, MPI of Biophysics, Frankfurt; for allowing us to use the CD spectrometer in his laboratory. We acknowledge the funding for this project from DFG to CB and MS (Bu 812/4-1), to MM (MI 373/11-1), and PK (KR 1661/4-2).
Glossary
Abbreviations
- ASP
artificial seawater medium
- β-DDM
β-dodecyl maltoside
- Chl
chlorophyll
- CD
circular dichroism
- Ddx
diadinoxanthin
- Dtx
diatoxanthin
- Fx
fucoxanthin
- Fcp
fucoxanthin chlorophyll protein
- FCP
fucoxanthin chlorophyll protein complex
- FcpAHis
His-tagged FcpA
- HPLC
high pressure liquid chromatography
- IMAC
immobilized metal affinity chromatography
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- LHC
light-harvesting complex
- Ni-NTA
nickel nitrilo triacetate
- NPQ
non-photochemical quenching
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- WT
wild type
References
- Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Molecular and General Genetics. 1996;252:572–579. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG. In vivo characterization of diatom multipartite plastid targeting signals. Journal of Cell Science. 2002;115:4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ. Recycled plastids: a green movement in eukaryotic evolution. Trends in Genetics. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Beer A, Gundermann K, Beckmann J, Büchel C. Subunit composition and pigmentation of fucoxanthin-chlorophyll proteins in diatoms: evidence for a subunit involved in diadinoxanthin and diatoxanthin binding. Biochemistry. 2006;45:13046–13053. doi: 10.1021/bi061249h. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Medlin L. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. Journal of Phycology. 1998;31:489–498. [Google Scholar]
- Bhaya D, Grossman AR. Characterization of gene clusters encoding the fucoxanthin chlorophyll proteins of the diatom Phaeodactylum tricornutum. Nucleic Acids Research. 1993;21:4458–4466. doi: 10.1093/nar/21.19.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Büchel C. Fucoxanthin-chlorophyll proteins in diatoms: 18 and 19 kDa subunits assemble into different oligomeric states. Biochemistry. 2003;42:13027–13034. doi: 10.1021/bi0349468. [DOI] [PubMed] [Google Scholar]
- Caron L, Brown J. Chlorophyll–carotenoid protein complexes from the diatom Phaeodactylum tricornutum: spectrophotometric, pigment, and polypeptide analysis. Plant and Cell Physiology. 1987;28:775–785. [Google Scholar]
- Durnford DG, Aebersold R, Green BR. The fucoxanthin-chlorophyll proteins from a chromophyte alga are part of a large multigene family: structural and evolutionary relationships to other light-harvesting antennae. Molecular and General Genetics. 1996;253:377–386. doi: 10.1007/s004380050334. [DOI] [PubMed] [Google Scholar]
- Eppard M, Krumbein WE, von Haesler A, Rhiel E. Characterization of fcp4 and fcp12, two additional genes encoding light harvesting proteins of Cylotella cryptica (Bacillariophyceae) and phylogenetic analysis of this complex gene family. Plant Biology. 2000;2:283–289. [Google Scholar]
- Fawley MW, Grossman AR. Polypeptides of a light-harvesting complex of the diatom Phaeodactylum tricornutum are synthesized in the cytoplasm of the cell as precursors. Plant Physiology. 1986;81:149–155. doi: 10.1104/pp.81.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs SP. The ultrastructure of the chloroplasts of algae. Journal of Ultrastructure Research. 1962;7:418–435. doi: 10.1016/s0022-5320(62)90038-2. [DOI] [PubMed] [Google Scholar]
- Goss R, Jakob T. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynthesis Research. 2010 doi: 10.1007/s11120-010-9536-x. DOI 10.1007/s11120-010-9536-x. [DOI] [PubMed] [Google Scholar]
- Green BR. The evolution of light-harvesting antennas. In: Green BR, Parson WW, editors. Light-harvesting antennas in photosynthesis. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. pp. 129–168. [Google Scholar]
- Green BR, Durnford DG. The chlorophyll–carotenoid proteins of oxygenic photosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:685–714. doi: 10.1146/annurev.arplant.47.1.685. [DOI] [PubMed] [Google Scholar]
- Green BR, Pichersky E. Hypothesis for the evolution of three-helix Chl a/b and Chl a/c light-harvesting antenna proteins from two-helix and four-helix ancestors. Photosynthesis Research. 1994;39:149–162. doi: 10.1007/BF00029382. [DOI] [PubMed] [Google Scholar]
- Gruber A, Vugrinec S, Hempel F, Gould SV, Maier UG, Kroth PG. Protein targeting into complex diatom plastids: functional characterization of a specific targeting motif. Plant Molecular Biology. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- Guglielmi G, Lavaud J, Rousseau B, Etienne AL, Houmard J, Ruban AV. The light-harvesting antenna in the diatom Phaeodactylum tricornutum. Evidence for a diadinoxanthin-binding subcomplex. The FEBS Journal. 2005;272:4339–4348. doi: 10.1111/j.1742-4658.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- Gundermann K, Büchel C. The fluorescence yield of the trimeric fucoxanthin-chlorophyll protein FCPa in the diatom Cyclotella meneghiniana is dependent on the amount of bound diatoxanthin. Photosynthesis Research. 2008;95:229–235. doi: 10.1007/s11120-007-9262-1. [DOI] [PubMed] [Google Scholar]
- Jeffrey SW, Humphrey GF. New spectrometric equations for determining chlorophyll a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochemie und Physiologie der Pflanzen. 1975;167:191–194. [Google Scholar]
- Kilian O, Kroth PG. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. The Plant Journal. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- Kroth PG. A tool to study protein targeting in diatoms. In: van der Giezen M, editor. Methods in molecular biology, protein targeting protocols. Vol. 390. Totowa, NJ: Humana Press Inc.; 2007. pp. 257–267. [PubMed] [Google Scholar]
- Lavaud J, Rousseau B, Etienne AL. Enrichment of the light-harvesting complex in diadinoxanthin and implications for the nonphotochemical fluorescence quenching in diatoms. Biochemistry. 2003;42:5802–5808. doi: 10.1021/bi027112i. [DOI] [PubMed] [Google Scholar]
- Lepetit B, Volke D, Szabo M, Hoffmann R, Garab G, Wilhelm C, Goss R. Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry. 2007;46:9813–9822. doi: 10.1021/bi7008344. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An Z, Chang W. Crystal structure of spinach major light-harvesting complex at 2.72Å resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- Maheswari U, Mock T, Armbrust EV, Bowler C. Update of the Diatom EST database: a new tool for digital transcriptomics. Nucleic Acids Research. 2008;37:D1001–D1005. doi: 10.1093/nar/gkn905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimuro M, Katoh T, Kawai H. Spatial arrangement of pigments and their interaction in the fucoxanthin-chlorophyll a/c protein assembly (FCPA) isolated from the brown alga Dictyota dichotoma. Analysis by means of polarized spectroscopy. Biochimica et Biophysica Acta - Bioenergetics. 1990;1015:450–456. [Google Scholar]
- Papagiannakis E, van Stokkum IHM, Fey H, Büchel C, van Grondelle R. Spectroscopic characterization of the excitation energy transfer in the fucoxanthin–chlorophyll protein of diatoms. Photosynthesis Research. 2005;86:241–250. doi: 10.1007/s11120-005-1003-8. [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- Provasoli L, McLaughlin JJA, Droop MR. The development of artificial media for marine algae. Archives of Microbiology. 1957;25:392–428. doi: 10.1007/BF00446694. [DOI] [PubMed] [Google Scholar]
- Pyszniak AM, Gibbs SP. Immunocytochemical localization of photosystem I and the fucoxanthin-chlorophyll a/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma. 1992;166:208–217. [Google Scholar]
- Richard C, Ouellet H, Guertin M. Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Molecular Biology. 2000;42:303–316. doi: 10.1023/a:1006340308077. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406:23–35. doi: 10.1016/j.gene.2007.05.022. [DOI] [PubMed] [Google Scholar]
- VeithT Brauns J, Weisheit W, Mittag M, Büchel C. Identification of a specific fucoxanthin–chlorophyll protein in the light harvesting complex of photosystem I in the diatom Cyclotella meneghiniana. Biochimica et Biophysica Acta - Bioenergetics. 2009;1787:905–912. doi: 10.1016/j.bbabio.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Veith T, Büchel C. The monomeric photosystem I-complex of the diatom Phaeodactylum tricornutum binds specific fucoxanthin chlorophyll proteins (FCPs) as light-harvesting complexes. Biochimica et Biophysica Acta - Bioenergetics. 2007;1767:1428–1435. doi: 10.1016/j.bbabio.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Wilhelm C, Büchel C, Fisahn J, et al. The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae, a putative consequence of secondary endosymbiosis. Protist. 2006;157:91–124. doi: 10.1016/j.protis.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proceedings of the National Academy of Sciences, USA. 2002;26:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossmann AR, Apt KE. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. Journal of Phycology. 2000;36:379–386. [Google Scholar]
- Zhu SH, Green BR. Light-harvesting and photoprotection in diatoms: identification and expression of LI818-like proteins. In: Allen JF, Gantt E, Golbeck JH, Osmond B, editors. Photosynthesis. Energy from the sun. 2008. 14th International Congress on Photosynthesis: Heidelberg: Springer, 261–264. [Google Scholar]