SUMMARY
Human tumors are characterized by widespread reduction in microRNA (miRNA) expression [1], although it is unclear how such changes come about and whether they have an etiological role in the disease. Importantly, miRNA-knockdown has been shown to enhance the tumorigenic potential of human lung adenocarcinoma cells [2]. A defect in miRNA-processing is one possible mechanism for the global down-regulation. To explore this possibility in more detail in vivo we have manipulated Dicer1 gene dosage in a mouse model of retinoblastoma. We show that while monoallelic loss of Dicer1 does not affect normal retinal development it dramatically accelerates tumor formation on a retinoblastoma-sensitized background. Importantly, these tumors retain one wild-type Dicer1 allele and exhibit only partial decrease in miRNA-processing. Accordingly, in silico analysis of human cancer genome data reveals frequent hemizygous, but not homozygous, deletions of DICER1. Strikingly, complete loss of Dicer1 function in mice did not accelerate retinoblastoma formation. miRNA profiling of these tumors identified members of the let-7 and miR-34 families as candidate tumor suppressors in retinoblastoma. We conclude that Dicer1 functions as a haploinsufficient tumor suppressor. This finding has implications for cancer aetiology and cancer therapy.
Keywords: Dicer, microRNA, retinoblastoma, tumor suppressor, haploinsufficiency
Introduction
A large body of evidence indicates that alterations in the expression of miRNAs contribute to cancer pathologies [3]. miRNAs act as agents of the RNA interference pathway to silence their cognate coding target genes either by cleaving mRNA molecules or inhibiting their translation [4]. By silencing tumor suppressive and oncogenic mRNAs, miRNAs themselves can function as oncogenes or tumor suppressors, respectively [5]. The let-7 family, for instance, limit lung tumorigenesis through inhibition of several oncogenes including members of the Ras family or HMGA2 [6, 7]
Initial efforts to link miRNAs with cancer were based on expression analyses in which tumors were compared with normal tissues. Such expression profiling analyses revealed characteristic miRNA signatures of human cancers [8]. Surprisingly, they also highlighted an overall down-regulation of mature miRNAs in several types of mouse and human cancer [1]. This observation raised two important questions: (i) one of causality - is there a causative link between global down-regulation of miRNAs and cancer progression or is the down-regulation a byproduct of tumor development? (ii) and one of mechanism - what are the molecular mechanisms and the genetic events underlying such widespread changes in tumors? The first question was addressed by RNAi targeting of factors involved in miRNA maturation, such as DICER1 and DROSHA, demonstrating that global down-regulation of miRNA-processing increased the transforming properties of a lung adenocarcinoma cell line in in vitro culture assays and in xenograft experiments [2]. Widespread silencing of miR expression was proposed to be, at least partly, a consequence of Myc-mediated transcriptional repression [9], however, the data also raised the possibility that DICER1 might be a target of genetic disruption in human cancers. Surprisingly, however, although reduced levels of DICER1 in tumors have been reported [10, 11], no loss-of-function mutations in DICER1 have been reported to date. There have however been reports of truncating mutations in TARBP2, encoding an integral component of a DICER1-containing complex, in sporadic and hereditary carcinomas with microsatellite instability [12]. Frameshift mutations in TARBP2 diminished TRBP protein expression and cause a partial defect in the processing of miRNAs. Importantly, the TRBP impairment is associated with a partial destabilization of the DICER1 protein. These data raised the possibility that Dicer down-regulation rather than its complete loss of function is selected for during tumorigenesis.
In order to directly address this question, we manipulated Dicer1 (referred thereafter as Dic) gene dosage in a mouse model of retinoblastoma. As germline inactivation of Dic in mice causes an early embryonic lethal phenotype [13], we specifically inactivated Dic in retinoblasts by combining a conditional floxed allele of Dic [14] with the retinal Chx10Cre transgenic line [15]. We chose this genetic model system for two main reasons. First, Chx10Cre-mediated inactivation of one Dic allele decreases the production of mature miRNAs without affecting retinogenesis [16]. Second, the Chx10Cre mice have been used to create the first preclinical mouse model of retinoblastoma [17, 18]. Chx10Cre-mediated inactivation of the Retinoblastoma (Rb) gene leads to inappropriate exit from the cell cycle of retinal progenitor cells and a block in rod maturation [19]. These mice do not develop retinoblastoma as expression of another Rb family member, p107, increases in a compensatory manner [18]. However, on a p107-null background, retinal Rb-inactivation (Chx10Cre; Rblox/lox; p107−/−) leads to the formation of early hyperproliferative lesions [17], which are often referred to as retinomas [20]. Importantly, these lesions rarely go on to become aggressive and invasive tumors and only do so in older animals suggesting that additional oncogenic lesions are required for full-blown tumorigenesis. For example, conditional inactivation of p53 in mice is one mechanism through which these lesions can progress into aggressive retinoblastoma [17]. Accordingly, amplification and overexpression of MDMX, a key negative regulator of p53, is a frequent event that is selected for during human retinoblastoma formation [21, 22].
Results
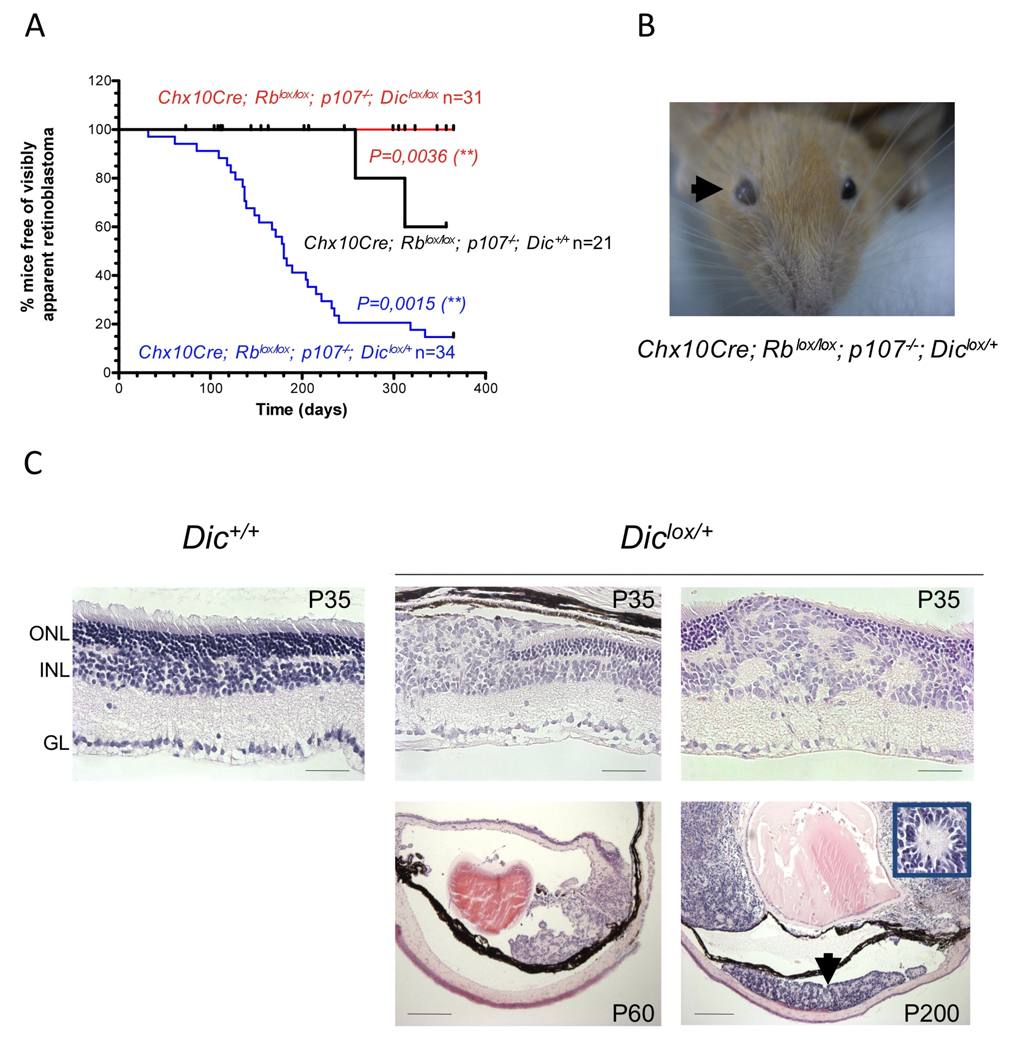
We first confirmed that specific loss of one Dic allele in retinal progenitor cells does not affect normal retinal development. As expected, histological analysis of retinae of several Chx10Cre; Diclox/+ mice at various stages of postnatal development did not reveal any morphological abnormalities (Supplemental Figure S1). Strikingly, however, on the retinoblastoma-sensitized background, loss of one Dic allele dramatically accelerated tumor formation (Figure 1A). Virtually all Chx10Cre; Rblox/lox; p107−/−; Diclox/+ mice develop aggressive and invasive intraocular retinoblastoma (Figure 1 B and 1C), with an average time to visible tumors of 180 days (Figure 1A). Half of these mice had anterior chamber invasion that was clearly visible by 10 weeks of age. By contrast, Chx10Cre; Rblox/lox; p107−/−; Dic+/+ developed retinoblastoma only with slow and inconsistent kinetics (Figure 1A). Moreover, while more than 30% of Chx10Cre; Rblox/lox; p107−/−; Diclox/+ mice had developed bilateral retinoblastoma with clear evidence of anterior chamber invasion Chx10Cre; Rblox/lox; p107−/−; Dic+/+ mice only ever developed unilateral tumors (data not shown). Finally, metastatic tumors that had invaded local tissues outside of the eye through the optic nerve were only observed in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ mice (data not shown).
Figure 1. Dicer1 (Dic) heterozygosity enhances tumorigenesis on a retinoblastoma-sensitized background.
(A) Kaplan-Meier curve showing the time to first observation of externally visible retinoblastoma. This time was respectively markedly decrease in Chx10Cre;Rblox/lox;p107−/−;Diclox/+ mice and increased in Chx10Cre;Rblox/lox;p107−/−;Diclox/lox mice (log rank test, P=0,0015 and P=0,0036) relative to Chx10Cre;Rblox/lox;p107−/−;Dic+/+ littermates. (B) A 3 months-old Chx10Cre;Rblox/lox;p107−/−;Diclox/+ mouse with aggressive retinoblastoma. (C) Hematoxylin and eosin stain with the three retinal nuclear layers (GL: ganglion layer; INL: inner nuclear layer; ONL: outer nuclear layer) indicated. Early retinoblastoma lesions at P35 and invasive tumors seeding the vitreous at P60 are found in Chx10Cre;Rblox/lox;p107−/−;Diclox/+ mice. P200 shows late stage retinoblastoma that had filled the vitreous and the anterior chamber (arrow). inset: a representative Homer-Wright rosette found in Chx10Cre;Rblox/lox;p107−/−;Diclox/+ tumors. Scale bars in the top panels = 40µm and = 200µm in the lower panels.
We next examined retinae histologically at postnatal days P35 and onwards. As previously described [18], the retinal cytoarchitecture of Chx10Cre; Rblox/lox; p107−/−; Dic+/+ at P35 is slightly disrupted due to focal expansion of immature cells from the inner nuclear layer (INL) and their protrusion through the outer plexiform layer (OPL) (Figure 1C). Additionally, defects in the maturation of the rod photoreceptors lead to a hypocellular outer nuclear layer (ONL) although the three nuclear layers are still detected in these mice. In contrast, the three nuclear layers can no longer be distinguished in the Dic heterozygous mutants (Figure 1C). The laminar organization in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ retinae was focally severely disrupted, with immature cells from the INL invading the OPL and extending up to the apical surface of the retina. This resulted in the disruption of the interaction between photoreceptor outer segments and the retinal pigment epithelium (RPE) and focally to dramatic ONL hypocellularity, presumably as a consequence of photoreceptor cell death. The focal nature of the phenotype is consistent with the previously reported mosaic expression pattern of Cre in Chx10Cre transgenic mice [15, 23]. Beyond P35, larger dysplastic lesions, found mainly at the periphery (6/7 eyes analyzed), seeded the vitrous (Figure 1c, P60) and eventually invaded the anterior chamber of the eye (Figure 1C, P200). The lesions contained Homer-Wright rosettes (Figure 1C), which consist of a radial arrangement of cells around a central tangle of neuronal processes. Interestingly, these histological structures are often found in a subset of human retinoblastoma [24].
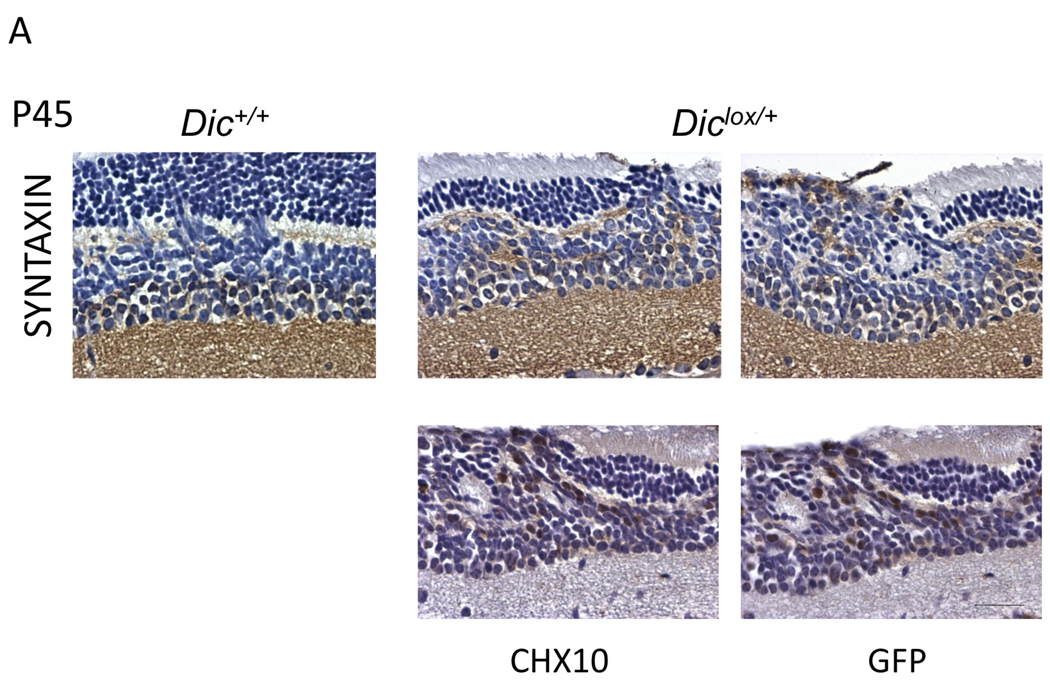
This histopathological analysis indicated that the phenotype observed in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ retinae is similar in nature but significantly more severe than that observed in Chx10Cre; Rblox/lox; p107−/− retinae. We therefore hypothesized that the dysplasia and the partial degeneration observed in Dic heterozygous retinae might result from severe expansion of a pool of retinal progenitor cells that normally reside in the INL. Accordingly, immunostaining showed that the cells that disrupted synaptogenesis in the OPL and extended all the way to the apical surface of the retinae expressed the progenitor cell markers Syntaxin and Chx10 [25–27] (Figure 2A, P45). The early tumors also stained for Calretinin, which labels a subset of amacrine and ganglion cells. However, Calretinin expression was less abundant than Syntaxin, more scattered and variable from animal to animal (data not shown). Calbindin, which labels horizontal cells and a subset of amacrine cells weakly, was either expressed at very low levels or undetectable in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ early lesions (data not shown). Together these data indicate that the lesions are composed of retinal progenitor cells biased towards the amacrine cell fate. Importantly, these immature cells were GFP-positive and therefore Cre-positive (Figure 2B). Cre was fused to GFP in the Chx10Cre transgenic mice [15] so that Cre-positive cells can be identified using anti-GFP-antibodies.
Figure 2. Immunostaining of the Chx10Cre;Rblox/lox;p107−/−;Diclox/+ retinoblastoma lesions.
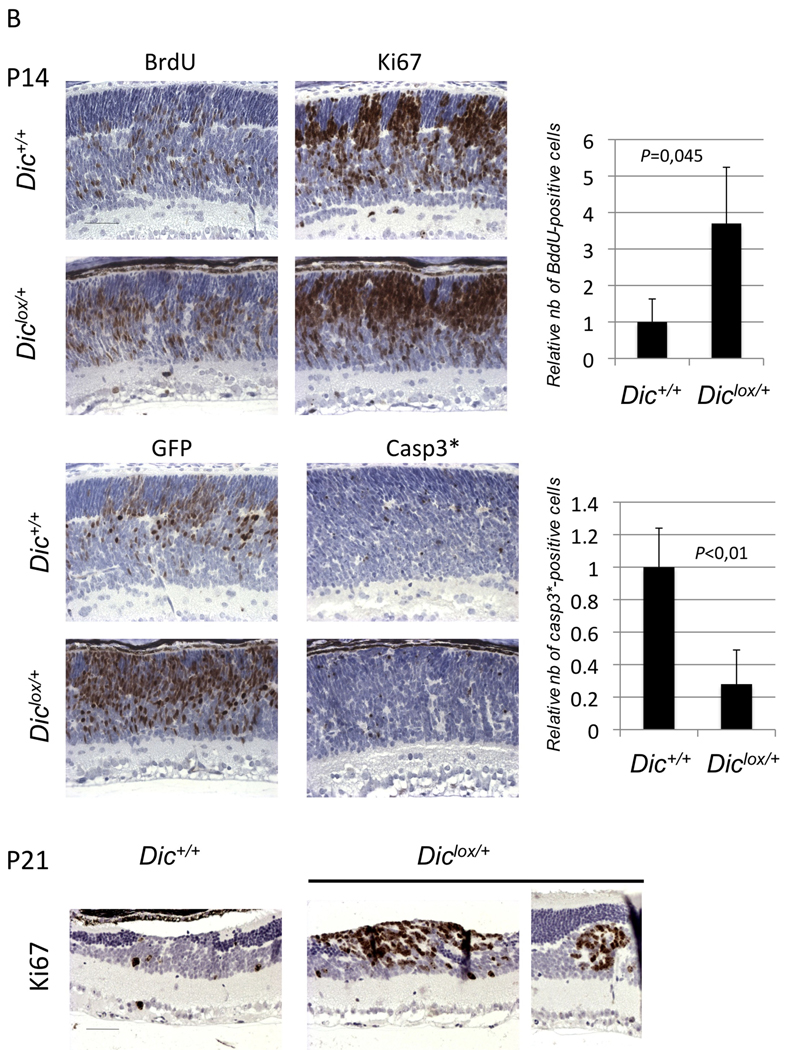
(A) Syntaxin, Chx10 and GFP immunostaining of Chx10Cre;Rblox/lox;p107−/−;Diclox/+ (Diclox/+) and Chx10Cre;Rblox/lox;p107−/−;Dic+/+ (Dic+/+) retinae at P45. (B) Proliferation and apoptosis in Chx10Cre;Rblox/lox;p107−/−;Diclox/+ (Diclox/+) and Chx10Cre;Rblox/lox;p107−/−;Dic+/+ (Dic+/+) at P14 and P21. BrdU incorporation, Ki67 and active Caspase3 immunostaining. Scale bars = 40µm. Quantification of BrdU and Caspase3 staining was performed on horizontal serial sections at the optic nerve level. The numbers (nb) of BrdU positive cells were normalized to the nb in Chx10Cre;Rblox/lox;p107−/−;Dic+/+(Dic+/+), which was set 1 (top right panel). The numbers of immunoreactive cells to active caspase3 staining were normalized to the numbers of GFP-positive cells in serial consecutive sections. The data are presented relative to the observed ratio GFP/Caspase3 in Chx10Cre;Rblox/lox;p107−/−;Dic+/+ (Dic+/+), which was set 1 (low right panel). Error bars represent standard deviation. P-values (Student’s t-test; N=3).
As inappropriate expansion of the immature retinal cells could be a consequence of increased cell proliferation and/or survival we measured cell proliferation and apoptosis using immunohistochemical assays. As previously reported, we found extensive BrdU incorporation in Chx10Cre; Rblox/lox; p107−/− retinae at P14, a time when retinogenesis is normally complete. This phenotype was significantly exacerbated in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ retinae (Figure 2B). Importantly, BrdU-positive cells were localized to the regions containing the GFP immunopositive cells and to where lamination was disrupted. These regions were also strongly positive for the proliferation marker Ki67 (Figure 2B, P14 and P21). BrdU- and Ki67-positive cells were found in all early dysplastic lesions and late (i.e. P200) retinoblastoma tumors (data not shown). Inappropriate cell proliferation leads to increased apoptosis in Chx10Cre; Rblox/lox; p107−/− retinae [17]. Accordingly, active-capase-3-positive cells could be detected in the Chx10Cre; Rblox/lox; p107−/− retinae (Figure 2D). There was no significant decrease per se in the number of cleaved-capase-3-positive cells in the Chx10Cre; Rblox/lox; p107−/−; Diclox/+ retinae (Figure 2D and 2E). However, as the number of GFP-positive cells is increased in these retinae compared to the Chx10Cre; Rblox/lox; p107−/− retinae, the data indicate that proportionally the number of dying Cre-positive cells is reduced in the Dicer heterozygous retinae.
Collectively, these data indicate that heterozygosity for a Dicer1 mutation promotes the switch from benign retinoma lesions to aggressive and invasive retinoblastoma. Monoallelic loss of Dic is sufficient to promote the expansion of retinal progenitor cells (with a bias towards the amacrine cell fate), which ultimately leads to the formation of early neoplastic lesions that progress into aggressive and metastatic tumors. The ability of these progenitor cells to form these aggressive tumors appears to be a consequence of both increased cell proliferation potential and resistance to apoptosis.
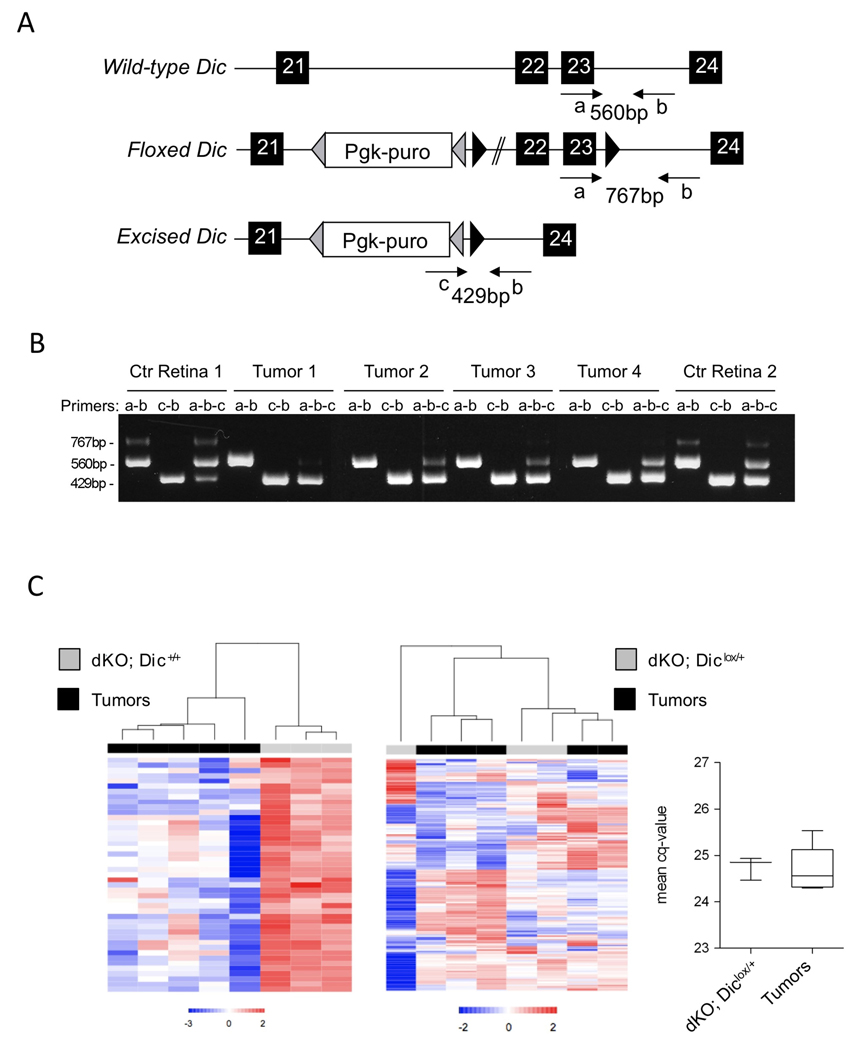
These data strongly support the view that Dicer1 functions in vivo as a haploinsufficient tumor suppressor gene. Considering that the dysplastic lesions in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ retinae are observed as early as P35, it is very unlikely that active selection against complete loss of the remaining wild-type Dic allele is required for tumorigenesis. However, this possibility had to be formally excluded experimentally. As expected, we obtain evidence of Cre-mediated recombination of the conditional Dic allele in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ normal retinae (Ctr Retinae, P20) and isolated tumors (Figure 3A). Importantly, the wild-type allele was retained in all tumors carefully dissected and genotyped (Figure 3A). Western blotting was further used to confirm that Dicer1 expression is retained in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ tumors (data not shown). To ensure that the remaining Dic allele is not functionally inactivated through mutations, we assessed the expression levels of all mature miRNAs in five different Chx10Cre; Rblox/lox; p107−/−; Diclox/+ tumors by RT-qPCR analysis. Consistent with a decrease of Dicer1 function we observed a global decrease in steady-state miRNA levels in all five tumors analyzed compared to the levels in Chx10Cre; Rblox/lox; p107−/−; Dic+/+ (Figure 3C, left panel). Importantly, all mature miRNAs that are expressed at high levels in Chx10Cre; Rblox/lox; p107−/−; Diclox/+ P20 retinae are expressed at comparable levels in all 5 tumors (Figure 3C, right panels). These analyses indicate that microRNA-processing is only partly impaired, but not completely disrupted, in these tumors. Finally, we directly assessed the consequences of complete Dicer1 inactivation in the retinoblastoma mouse model. While we confirmed that complete ablation of Dicer is well tolerated and only leads to progressive retinal degeneration [16] our data indicate that it dramatically affects retinal formation on the retinoblastoma-sensitized background. The retina of Chx10Cre; Rblox/lox; p107−/−; Diclox/lox is completely disorganized as early as P10 and completely degenerates soon after (Lambertz et al., manuscript in preparation). Consequently these mice are entirely protected from tumor formation (Figure 1A) but are at the same time completely blind. Together the data argue that while partial loss of Dicer function favors tumor formation, complete loss of Dicer is deleterious to retinoblastoma development further arguing in favor of a haploinsufficient tumor suppressor function of Dicer1.
Figure 3. Chx10Cre;Rblox/lox;p107−/−;Diclox/+ retinoblastoma tumors retain a wild-type and functional Dic allele.
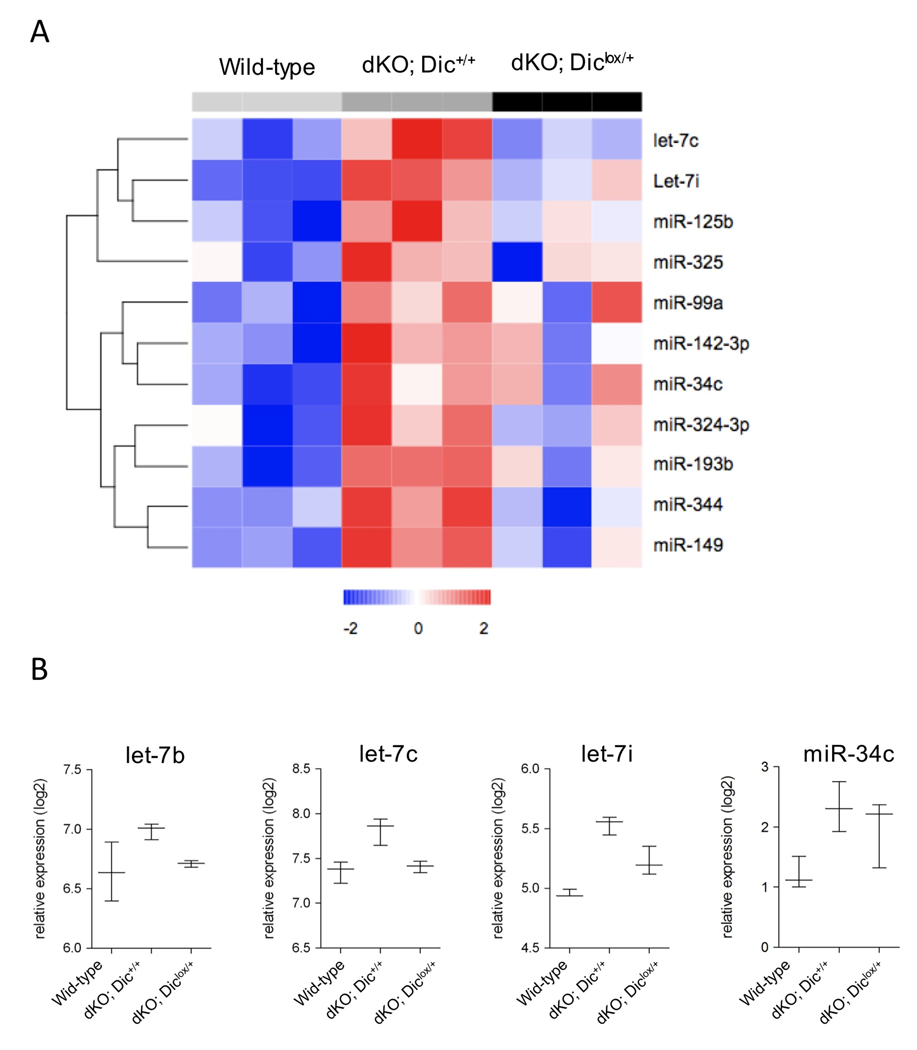
(A) Schematic representation of the Dic wild-type, floxed and Cre-excised alleles. (B) DNA was prepared from two Chx10Cre;Rblox/lox;p107−/−;Diclox/+ P20 retinae (Ctr Retina) and 10 isolated tumors (results from 4 are shown) and examined by PCR using the primers depicted in the top panel. (B) RT-qPCR analyses of mature miRNAs expression. Left panel: hierarchical clustering of miRNAs significantly down-regulated in five Chx10Cre;Rblox/lox;p107−/−;Diclox/+ tumours (black, dKO; Diclox/+) compared to Chx10Cre;Rblox/lox;p107−/−;Dic+/+ P20 retinae (grey, dKO; Dic+/+). Middle panel: heat map of all miRNAs expressed at higher levels than the mean in Chx10Cre;Rblox/lox;p107−/−;Diclox/+ P20 retinae shows comparable expression levels in both Chx10Cre;Rblox/lox;p107−/−;Diclox/+ P20 retinae and tumors. Right panel shows the mean cq-values for all miRNAs analyzed (522 different murine miRNAs) in both Chx10Cre;Rblox/lox;p107−/−;Diclox/+ P20 retinae and tumors.
These data also imply that a set of miRNAs, the function of which requires Dicer1 function, might act to prevent full-blown retinoblastoma formation on the Chx10Cre;Rblox/lox;p107−/− background. To identify such candidate tumor suppressor miRNAs we determined the expression profile of the entire miRNome in P20 retinae from wild-type (Cre-negative), Chx10Cre; Rblox/lox; p107−/−; Dic+/+ and Chx10Cre;Rblox/lox;p107−/−;Diclox/+ using LNA-based microarray and RT-qPCR approaches. Both types of analyses identified a common set of 11 miRNAs that are consistently up-regulated between wild-type and Chx10Cre; Rblox/lox; p107−/−; Dic+/+ (Figure 4A). Most interestingly, among them are 2 members of the let-7 family (let-7c and let7-i), the up-regulation of which and of another let-7 member, let-7b, was confirmed by independent Q-RT-PCR analysis (Figure 4B). Given that this up-regulation was significantly attenuated in the Dic heterozygous retinae and the recognized role of let-7 family members in tumor suppression [28], this observation raises the possibility that let-7 have a causal role in retinoblastoma formation as critical regulators of the switch from retinomas to retinoblastoma. Our list of 11 differentially expressed miRNAs also included miR-34c (Figure 4A and 4B). There was also a clear upregulation of miR-34b-3p in both microarray and RT-q-PCR analyses and a moderate, but reproducible, up-regulation of miR-34a was evident from the micro-array data (data not shown). Interestingly, the miR-34 family members have been identified as p53 targets and key mediators of its tumor suppressor function [29].
Figure 4. Search for putative tumor suppressor miRNAs in retinoblastoma.
(A) Heatmap of selected differentially expressed miRNAs in Chx10Cre-negative mice (light grey, wild-type), Chx10Cre;Rblox/lox;p107−/−;Dic+/+ (dKO; Dic+/+) and Chx10Cre;Rblox/lox;p107−/−;Diclox/+ P20 retinae (black, dKO; Diclox/+). (B) Expression analysis by RT-qPCR of let-7 family members (let-7b, let-7c and let-7i) and miR-34c.
Discussion
In order to address the importance of Dicer1 gene dosage in cancer development we used a preclinical mouse model of retinoblastoma [17]. Our data provide clear genetic evidence that Dicer1 function as a haploinsufficient tumor suppressor in vivo. In keeping with this observation, information in the public domain, e.g., Cancer Genome Project at the Sanger Institute [30] indicates that hemizygous deletions of DICER1 occur in 27% (207/761) of tumors derived from tissues of diverse origins such as central nervous system, lung, pancreas, soft tissues, breast, bone, haematopoietic or lymphoid. Importantly, consistent with our findings, homozygous deletions have never been observed in any of these 761 tumors. Very recently, heterozygous point mutations in DICER1 were reported in patients with pleuropulmonary blastoma [31]. However, DICER1 expression was retained in the mesenchymal tumor cells from these patients, again arguing against strong selective pressure for complete loss of Dicer function in human tumors. Thus, while the present study focuses on role of Dicer in a mouse model of retinoblastoma, there is evidence supporting a broad role for DICER1 as a haploinsufficient tumor suppressor in human cancer.
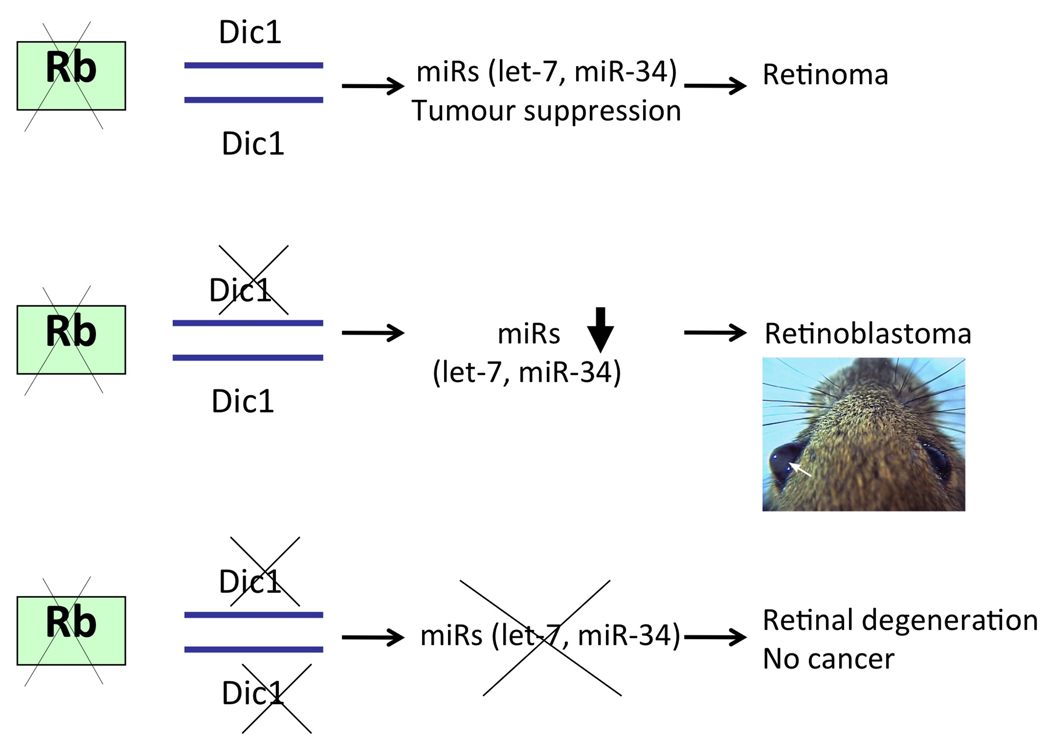
Kumar et al. attempted to investigate an etiological role for Dicer1 in a mouse model of cancer, in which Dicer1 was inactivated in a K-Ras-induced mouse model of lung cancer using intranasal infection with adenovirus expressing Cre [2]. Although it highlighted an important role for Dicer1 in tumor suppression, this elegant study did not resolve the important issue of gene dosage. The authors showed that tumorigenesis was enhanced on both Diclox/+ and Dicloxlox genetic backgrounds but they did not analyze the extent of Cre-mediated Dicer1 inactivation in these tumors. It is therefore still formally possible that lung tumors developed in the Dicloxlox mice only as a result of incomplete inactivation of Dicer function. Data from our retinoblastoma model clearly emphasize that only partial, rather than complete, inactivation of this process enhances tumorigenesis in vivo (Figure 5).
Figure 5. Monoallelic loss of Dicer enhances retinoblastoma formation.
Our data support a model in which monoallelic loss of Dicer1 cooperates with Rb inactivation in the formation of aggressive and invasive retinoblastoma. In contrast, biallelic loss of Dicer1 leads to retinal degeneration on a retinoblastoma-sensitized background and inhibition of tumor formation.
It is generally accepted that complete loss of tumor suppressor function through mutations and loss of heterozygosity is a pre-requisite for tumor development. To date, only a very small number of haploinsufficient tumor suppressor genes have been identified [32], mostly through the use of genetically-engineered mouse models. Our mouse genetic data identify DICER1 as another member of this group. As the list of haploinsufficient tumor suppressors grows hemizygous deletions at loci such as DICER1 should be considered as potential key pro-tumorigenic lesions.
Our data further establish a causative link between global down-regulation of miRNA-processing and cancer development. It is possible that the reduction of expression of only a subset of tumor suppressor miRNAs is the event that promotes tumorigenesis (Figure 5). In the context of retinoblastoma, our profiling data identified the let-7 family members as potential candidate tumor suppressors. Another sets of miRNAs that can account for the observed tumor suppressive activity of Dicer1 in retinoblastoma are the miR-34 family members. Given that these genes have been identified as p53 targets and are critical mediators of its activity [29] and the importance of the p53 tumor suppressor function in retinoblastoma [17, 21] it will be interesting to further assess genetically their functional relevance in the retinoblastoma mouse model. Of note, it has been shown that miR-34 family members are differentially expressed in human retinoblastoma cell lines and tumors compared to normal retina samples [33]. Exogenous miR-34a inhibited cell growth and/or increased apoptosis in retinoblastoma cells lines (Y73, Weri-Rb1) [33]. Together, these data identify the miR-34 family members as potential therapeutic targets for retinoblastoma. Interestingly, among the common putative mRNA targets of both let-7 and miR-34 family members is the N-myc oncogene. N-myc is indeed a validated miR-34a target (http://mirecords.umn.edu/miRecords/) and is a predicted target of let-7i (http://www.targetscan.org/). This gene is amplified in another mouse model of retinoblastoma [34] and in approximately 10% of human retinoblastomas [35]. These observations raise the possibility that members of both let-7 and miR34 families cooperate to restrain N-Myc oncogenic function in retinoblastoma.
Finally and importantly, our data argue against the development of DICER1-inactivating molecules as anti-cancer drugs. Indeed, even if complete Dicer inactivation seems to be deleterious for cancer development partial inactivation, at least in the context of retinoblastoma, promotes rather than inhibits tumor formation.
Materials and Methods
Mice
All animal experiments were performed in accordance with the guidelines of the University of Gent Animal Care and Use ethical Committee. BrdU (100 µg/g of body weight) was injected intraperitoneally 1hr prior to sacrifice.
Immunohistochemistry
Eyes were fixed overnight in 4% paraformaldehyde/PBS, and paraffin embedded. 5um sections were immunostained with the following antibodies: GFP (Santa Cruz Biotechnology, 1/100); BrdU (BD Pharmingen, 1/10); Ki-67 (DAKO, 1/30); cleaved caspase-3 (Cell Signaling, 1/100); syntaxin (Sigma, 1/20000); calretinin (Millipore, 1/700); Chx10 (Exalpha Biologicals, 1/100).
Recombination analysis
DNA was isolated from dissected retinea and isolated tumors using DNeasy Blood&Tissue Kit (Qiagen). Dicer1 recombination was analyzed by PCR using the following primers: a 5’-ATTGTTACCAGCGCTTAGAATTCC; c 5’-TCGGAAT AGGAACTTCGTTTAAAC and the reverse b primer 5’-GGGAGGTGTACGTCTA CAATT. PCR conditions were as follow: 1x precycle at 94°C for 3min and 30cycles of 94°C, 30sec; 60°C, 30sec; 72°C, 45sec.
microRNA Expression analyses
Total RNA was prepared from dissected retinea or isolated tumors using the miRNeasy kit (Qiagen) according to the manufacturer’s instructions. Profiling of these samples was performed by both micro-array (miRCURY LNA Array, Exiqon) and qPCR. The RT-qPCR expression profiling of 522 murine miRNAs and subsequent data normalization were performed as described previously[36, 37]. Differential miRNA expression was evaluated using Student’s t-test. Hierarchical clustering was performed with method Ward and distance Manhattan using R Bioconductor software.
Supplementary Material
Hematoxilin-eosin stain of P35 and P200 retinae show normal retinal lamination in Chx10Cre;Diclox/+ mice. ONL, outer nuclear layer; RPE, retinal pigment epithelium; INL, inner nuclear layer; GL, ganglion layer. Scale bars = 40µm.
Acknowledgments
We thank Natalie Farla and Rose Van Isacker for excellent technical assistance. We thank G. Hannon for providing the Dic floxed mince. J-C Marine received support from the EMBO Young Investigator program. This work was supported in part by Geconcerteerde Onderzoek Aangelegenheden (GOA, University Ghent, Belgium), and EU (FP7 program, ONCOMIRs, Contract 201102). This publication reflects only author’s views. The commission is not liable for any use that may be made of the information herein.
Footnotes
Conflicts of interest
The authors declare no conflict of interest
References
- 1.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 2.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 3.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem. 2009 doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 14.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Damiani D, Alexander JJ, O'Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Schweers B, Dyer MA. The first knockout mouse model of retinoblastoma. Cell Cycle. 2004;3:952–959. [PubMed] [Google Scholar]
- 18.Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- 20.DiCiommo D, Gallie BL, Bremner R. Retinoblastoma: the disease, gene and protein provide critical leads to understand cancer. Semin Cancer Biol. 2000;10:255–269. doi: 10.1006/scbi.2000.0326. [DOI] [PubMed] [Google Scholar]
- 21.Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 22.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 23.Dyer MA, Cepko CL. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J Neurosci. 2001;21:4259–4271. doi: 10.1523/JNEUROSCI.21-12-04259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuge K, Nakajima M, Uemura Y, Miki H, Uyama M, Tsubura A. Immunohistochemical features of the human retina and retinoblastoma. Virchows Arch. 1995;426:571–575. doi: 10.1007/BF00192111. [DOI] [PubMed] [Google Scholar]
- 25.Alexiades MR, Cepko CL. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. doi: 10.1242/dev.124.6.1119. [DOI] [PubMed] [Google Scholar]
- 26.Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D, Kalnins VI, McInnes RR. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 27.Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 28.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 29.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC). Chapter 10. Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don't need to go all the way. Biochim Biophys Acta. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Dalgard CL, Gonzalez M, deNiro JE, O'Brien JM. Differential microRNA-34a expression and tumor suppressor function in retinoblastoma cells. Invest Ophthalmol Vis Sci. 2009;50:4542–4551. doi: 10.1167/iovs.09-3520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hematoxilin-eosin stain of P35 and P200 retinae show normal retinal lamination in Chx10Cre;Diclox/+ mice. ONL, outer nuclear layer; RPE, retinal pigment epithelium; INL, inner nuclear layer; GL, ganglion layer. Scale bars = 40µm.