Abstract
The enzyme phosphopantothenoylcysteine synthetase (PPCS) catalyzes the nucleotide-dependent formation of phosphopantothenoylcysteine from (R)-phosphopantothenate and l-cysteine in the biosynthetic pathway leading to the formation of the essential biomolecule, coenzyme A. The Enterococcus faecalis gene coaB encodes a novel monofunctional PPCS which has been cloned into pET23a and expressed in E. coli BL21 AI. The heterologous expression system yielded 30 mg of purified PPCS per liter of cell culture. The purified enzyme chromatographed as a homodimer of 28 kDa subunits on Superdex HR 200 gel filtration resin. The monofunctional protein displayed a nucleotide specificity for cytidine-5’-triphosphate (CTP) analogous to that seen for bifunctional PPCS expressed by most prokaryotes. Kinetic characterization, utilizing initial velocity and product inhibition studies, found the mechanism of PPCS to be Bi Uni Uni Bi Ping Pong, with the nucleotide CTP binding first and CMP released last. Michaelis constants were 156, 17, and 86 µM for CTP, (R)-phosphopantothenate, and l-cysteine, respectively, and the kcat was 2.9 sec−1. [carboxyl-18O]Phosphopantothenate was prepared by hydrolysis of methyl pantothenate with Na18OH, followed by enzymatic phosphorylation with E. faecalis pantothenate kinase (PanK). The fate of the carboxylate oxygen of labeled phosphopantothenate, during the course of the PPCS-catalyzed reaction with CTP and l-cysteine, was monitored by 31P NMR spectroscopy. The results show that the carboxylate oxygen of the phosphopantothenate is recovered with the CMP formed during the reaction, indicative of the formation of a phosphopantothenoyl cytidylate catalytic intermediate, which is consistent with the kinetic mechanism.
Phosphopantetheine ((R)-3-hydroxy-4-(3-(2-mercaptoethylamino)-3-oxopropylamino)-2,2-dimethyl-4-oxobutyl dihydrogen phosphate) is an fundamental feature in many biological acyl transfer reactions (1–3). The molecule is found imbedded within coenzyme A (CoA), as well as on a post-translationally modified, conserved serine of acyl carrier protein (ACP) (1). Both CoA and ACP play essential roles in activating various acyl groups for enzyme catalyzed transfer in several reactions associated with intermediary metabolism and cell membrane assembly in living organisms (4). The critical nature of phosphopantetheine-containing molecules to the integrity and viability of cells makes the biosynthetic pathway leading to the production of these compounds an intriguing target for antimicrobial development (5, 6).
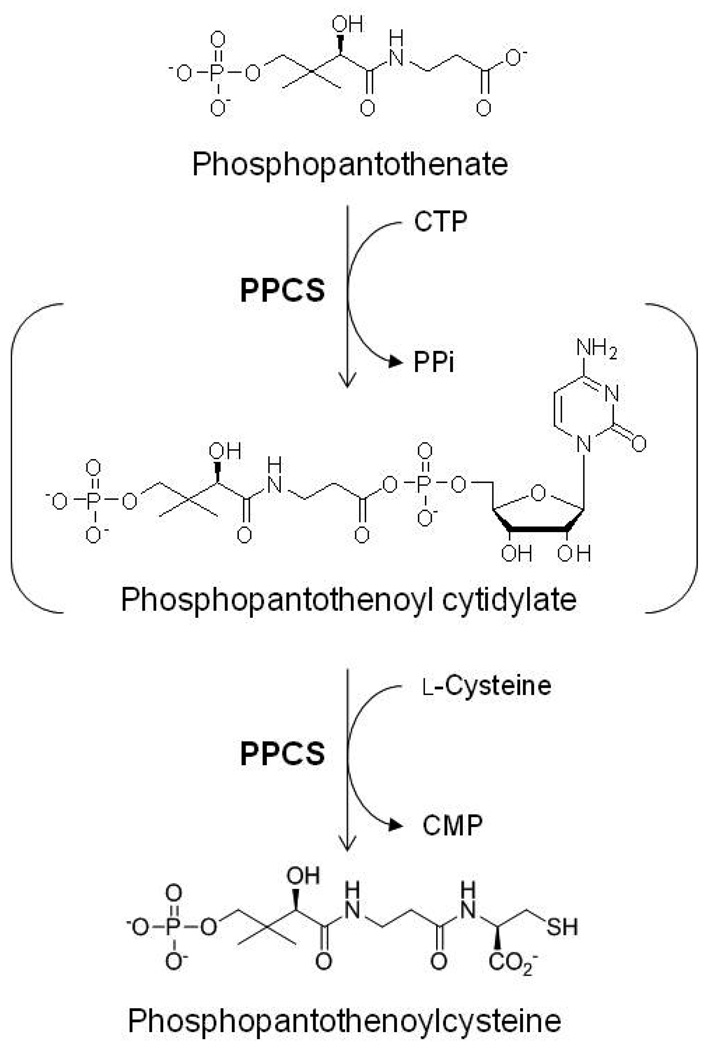
While it has been known for decades that CoA is biosynthesized from nucleotide triphosphate (NTP), d-pantothenate, and l-cysteine, the genes involved have only been identified in recent years (7–9). In eukaryotic systems that have been characterized, the phosphopantetheine portion of CoA is synthesized from pantothenate via three chemical reactions catalyzed by three polypeptides (Scheme 1) (10). First, pantothenate is phosphorylated by pantothenate kinase (PanK; EC 2.7.1.33; coaA), using ATP as the phosphate donor, to yield phosphopantothenate (PPA). This is followed by amide bond formation between the carboxyl group of PPA and the amino group of l-cysteine catalyzed by an ATP-dependent phosphopantothenoylcysteine synthetase (PPCS; EC 6.3.2.5; coaB). Subsequent decarboxylation of the carboxylic acid moiety on the ligated cysteine of phosphopantothenoylcysteine by phosphopantothenoylcysteine decarboxylase (PPCDC; EC 4.1.1.36; coaC) gives 4’-phosphopantetheine. In bacterial systems elucidated so far, the second and third chemical steps – the ligation and decarboxylation of cysteine by PPCS and PPCDC – are catalyzed by a single bifunctional polypeptide (9, 11). Furthermore, CTP, rather than ATP, is required by the bifunctional PPCS (7, 9).
Scheme 1.
PPCS from few species have been characterized. The proposed mechanism of PPCS involves the formation of an acyl cytidylate intermediate, 4’-phosphopantothenoyl cytidylate, between the carboxylate moiety of d-pantothenate and the α-phosphate of CTP, with the concomitant release of pyrophosphate. Formation of the product occurs by transfer of the nucleotide-activated acyl group to the amino group of l-cysteine, yielding phosphopantothenoylcysteine and CMP (Scheme 2). This proposed mechanism is supported by structural studies on the E. coli N210D PPCS domain, where the acyl cytidylate intermediate was observed in the active site when the crystal of the mutated PPCS was soaked with CTP and PPA (12). However, there has been no complete study on the kinetic mechanism of PPCS. In this paper, we overexpress, purify, and characterize PPCS from Enterococcus faecalis, which unlike most other bacterial PPCS, is encoded as a monofunctional protein and has similarly distant homology from both the monofunctional human PPCS protein and bifunctional bacterial PPCS domains (5, 10). In this study we use initial velocity and product competition experiments to elucidate the kinetic mechanism of PPCS from E. faecalis, and report for the first time the chemical mechanism and nucleotide specificity of a monofunctional bacterial PPCS.
Scheme 2.
MATERIALS AND METHODS
Materials
The chemicals used were of reagent grade or of the highest purity commercially available and were not further purified. Source 15Q, Source 15S, and Superdex 200 were from GE Healthcare. Calcium d-pantothenate, HEPES, Tris base, ATP, CTP, Pyrophosphate Reagent, ampicillin, sodium chloride, sodium hydroxide, methanol, formic acid, sodium pyrophosphate, dithiothreitol, l-cysteine, and 97% [18O] water were from Sigma-Aldrich. Lauria agar (Lennox), and Lauria broth (Lennox) were from Difco. Restriction and DNA modifying enzymes were from New England Biolabs. The Escherichia coli BL21 AI was obtained from Invitrogen and E. coli XL1 Blue was obtained from Stratagene. Polymerase Chain Reaction was performed using an Eppendorf Gradient Mastercycler and PfuTurbo Hotstart DNA polymerase (Stratagene). Oligonucleotides were synthesized by Invitrogen. Qiagen minispin kits were used for DNA purification and PCR reaction cleanup. Electrospray mass spectral analysis was performed at the University of Michigan Mass Spectrometry Facility, Department of Chemistry. DNA sequencing was performed by the University of Michigan, Biomedical Research Resources Core Facility.
Cloning, Overexpression, and Purification of E. faecalis PPCS
The coaB gene was amplified from E. faecalis genomic DNA (strain ATCC 700802), via PCR, using the forward primer GCGCCATATGGATGTTTTAGTTACTGCTGGCGG and the reverse primer GCGCCTCGAGTCATTGTTGTTCTCTCCATTTCTTTTC to introduce an NdeI site (shown underlined) upstream of the gene and an XhoI site downstream of the gene. The resulting PCR product, encoding the entire coaB gene, was digested with NdeI and XhoI, and ligated into pET23a(+) (Novagen) which had been restricted with NdeI and XhoI. The desired plasmid was designated pUMGD1 and the insert was confirmed by DNA sequencing. E. coli strain BL21 AI harboring the plasmid pUMGD1 was incubated in four 1 L flasks containing 250 ml each of LB-ampicillin media (5 g of NaCl, 5 g of yeast extract, 10 g tryptone, and 100 mg of ampicillin per L) at 37°C with vigorous shaking (250 rpm) until the optical density at 600 nm reached 0.6–0.8. The culture was cooled to 17°C and induced with a final concentration of 0.065% w/v l-arabinose. Incubation (17°C, 250 rpm) was continued overnight. Cells from the 1 L of culture were harvested by centrifugation at 10,000 × g, washed with 20 mM HEPES pH 8.0, and suspended in 80 ml of 20 mM HEPES pH 8.0. The cells were disrupted by French Press, and the resulting suspension was centrifuged at 20,000 × g for 30 minutes to separate the crude cytosol (supernatant) from cellular debris (pellet). The cytosol was chromatographed on a Source 15Q column (8 mL of resin per 250 mL of cell culture) equilibrated with 20 mM HEPES pH 8.0, washed with three column volumes of equilibration buffer, and eluted with a 10 column volume linear gradient of 0 – 0.5 M NaCl in 20 mM HEPES pH 8.0. Fractions were analyzed by SDS-PAGE and fractions containing PPCS were pooled, and passed through a 320 mL Superdex 200 prep grade column equilibrated in 20 mM HEPES pH 8.0 containing 150 mM NaCl. The purified fractions were dialyzed against 20 mM HEPES pH 8.0, and stored at −80°C.
Cloning, Overexpression, and Purification of the pyrophosphate dependent fructose-6-phosphate kinase (PPi-FPK)
The E. coli codon optimized PPi-PFK gene (DNA 2.0) of Propionibacterium freudenreichii, containing a NdeI site upstream and a XhoI site downstream of the gene, was digested with NdeI and XhoI and ligated into pET23a(+) via the same sites (13). The resulting vector was designated pUMJY901. Growth, expression, and purification protocols used were the same as for PPCS (above).
High Resolution Mass Spectrometry
The purified E. faecalis PPCS was dialyzed against H2O, and brought to 50/50 methanol/water containing 1% formic acid. The sample (final protein concentration of 0.2 mg/mL) was submitted for positive-ion ESI-MS.
Synthesis of 4’-phosphopantothenate
4’-Phosphopantothenate was synthesized via previously published procedures (9).
Synthesis of [carboxyl-18O]Phosphopantothenate
Methyl d-pantothenate (18.6 mg, 80 µmole) was treated with 0.4 M Na18OH in 200 µL final volume (8 µL of 10 M NaOH and 192 µL of 97% H2 18O for a final concentration of 93.2% 18O) for 2 hours at room temperature. The reaction was then neutralized by applying to an AG MP-50 resin spin column (H+ form; 1 mL) and centrifuging at 1000 × g for 2 minutes to yield [carboxyl-18O] d-pantothenate. The labeled pantothenate was then converted, biochemically, into [carboxyl-18O] d-phosphopantothenate by incubating at 30°C overnight in the presence of 1.1 equivalent ATP and 10 µM E. faecalis pantothenate kinase in 50 mM HEPES, pH 8.0 (5 mL final volume). The reaction was separated on a Source 15Q column (8 mL) via a linear gradient elution of 0–0.4 M NaCl in water. Fractions containing labeled phosphopantothenate, detected enzymaticly using the PPCS assay (described below), were combined and lyophilized. The lyophilized fractions were subjected to gel filtration on a BioGel P-2 column (240 mL) in water to remove NaCl. The relevant fractions were lyophilized and characterized. 1H NMR (500 MHz, D2O): δ0.82 (s, 3H), 0.99 (s, 3H), 2.41 (t, 2H), 3.39 (dd, 1H), 3.42 (dt, 2H), 3.75 (dd, 1H), 4.1 (s, 1H). 13C NMR (125 MHz, D2O): δ17.96 (s), 21.42 (s), 36.12 (s), 36.72 (s), 38.35 (d), 70.69 (d), 74.77 (s), 174.76 (s), 180.268 (s, caboxylate 18O), 180.295 (s, caboxylate 16O), ratio of carboxylate species 92 to 10 (18O to 16O).
18O Transfer Reaction
Reaction mixture of 10 mM CTP, 10 mM MgCl2, 12 mM [carboxyl-18O] d-phosphopantothenate, 10 mM l-cysteine, 10 mM DTT, and 2.4 µM E. faecalis PPCS in 500 µL of 100 mM Tris HCl pH 7.6, was incubated for 1 hour at 37°C. The reaction was then filtered through a Microcon YM-10 membrane (Amicon) at 13000 × g and 15°C for 30 minutes to remove the enzyme. To the filtrate was added 120 µL 2H2O, and the solution analyzed via 31P NMR. Analogous control experiments with unlabeled d-phosphopantothenate or without enzyme were also performed.
NMR spectroscopy
Proton-decoupled 31P NMR spectra were recorded on a Bruker DRX500 spectrometer (11.75 T) at a probe temperature of 298 K, tuned to 202.4 MHz, using 5-mm high resolution NMR tubes. Spectra were obtained with a spectral width of 80000 Hz, 1.0 s relaxation delay, and 32,768 complex points in the time domain using simultaneous detection of real and imaginary components. The time domain data were apodized with an exponential (0.5 Hz) prior to zero-filling followed by Fourier transformation. Chemical shifts are reported relative to an external sample of 10 mM inorganic phosphate (0.0 ppm) in 100 mM Tris-HCl (pH 7.6) and 10%2H2O.
Proton-decoupled 13C NMR spectra were recorded on a Bruker DRX500 spectrometer (11.75 T) at a probe temperature of 298 K, tuned to 125.7 MHz, using 5-mm high resolution NMR tubes. Spectra were obtained with a spectral width of 25000 Hz, 1.0 s relaxation delay, and 32,768 complex points in the time domain using simultaneous detection of real and imaginary components. The time domain data were apodized with an exponential (0.5 Hz) prior to zero-filling followed by Fourier transformation. Chemical shifts are reported relative to tetramethylsilane (0.0 ppm).
Enzyme Assay
The PPCS reaction was observed in the forward reaction via an enzyme linked assay, where the production of pyrophosphate from PPCS activity was coupled to the oxidation of NADH, which could be monitored as a disappearance of absorption at 340 nm. All of the components of the pyrophosphate detection system are available commercially as the Pyrophosphate Reagent (PR) from Sigma-Aldrich. It was necessary that additional units of PPi-PFK be added to the commercial preparation in order to use it as a continuous assay. With this modification PPCS activity could be observed in real time under our experimental conditions. The assays were performed on a SpectraMax M5 (Molecular Devices) microplate reader using 96-well half-area plates (Costar UV), where each assay has 100 µL final volume. Each vial of the PR was resuspended in 4.5 mL of 100 mM HEPES pH 7.6. The assay mix consisted of 30 µL PR, 1U of additional PPi-PFK, 10 mM DTT, and varying concentrations of the substrates (CTP, l-cysteine, and PPA) in a volume of 70 µL buffered in 50 mM Tris-HCl and pH 7.6. All assay mixes contained MgCl2 in 1:1 molar stoichiometry with nucleotide triphosphate. The assay mix and the enzyme solution (275 nM PPCS in 50 mM Tris-HCl pH 7.6), were incubated at 37°C for 15 min before the assay. Reactions were initiated by adding 30 µL of enzyme solution to the assay mix. The oxidation of NADH, monitored by a decreasing UV absorbance at 340 nm (ε = 6.22 mM−1cm−1), is measured over the course of the assay and the activity of the enzyme is calculated by adjusting for pathlength (0.5 cm) and taking into account that each mole of pyrophosphate produced leads to the oxidation of 2 moles of NADH. Assays were run in duplicates, with the average velocities reported.
Initial Velocity Studies
Pairwise analysis, where initial velocities were measured with varying concentrations of one substrate at different fixed concentrations of a second substrate while holding the third substrate at saturation, were conducted. For instance, initial velocities as a function [CTP] were measured at several fixed levels of [PPA] (0.6, 0.3, 0.15, 0.075, and 0.0375 mM) with constant level of [cysteine] (1 mM). Pairwise analyses were performed for all three substrates.
Product Inhibition Studies
Initial velocities were measured against different concentrations of one substrate with fixed concentrations of the other two substrates and different fixed concentrations of the product inhibitor. For example, initial velocities versus [CTP] were measured at 0.15 mM PPA, 1 mM cysteine, and several fixed levels of [CMP] (0, 250, 500, and 1000 µM).
Data Analysis
Initial velocity was first graphically analyzed as Lineweaver-Burk double reciprocal plots. Velocity versus the varying [substrate] data was fit to equation 1 to determine the Kmapp and kcatapp, which was in turn used to derive the fitting lines in the Lineweaver-Burke plots. The different patterns in the Lineweaver-Burke plots arising from the pairwise or inhibition analysis was used to determine which terreactant kinetic mechanism describes PPCS. The initial velocity data from the three sets of pairwise analysis were fit to the initial velocity equation (equation 2) describing the appropriate mechanism (Bi Uni Uni Bi Ping Pong) to determine the relevant kinetic parameters. Likewise, product inhibition data was fitted to the appropriate product inhibition equations (equations 3–5) to determine the relevant kinetic parameters.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
In theses equations v is initial velocity and Vmax is maximum velocity. KmA, KmB, and KmC are the Michaelis constants for substrates A, B, and C, respectively. KiA is the dissociation constant of substrate A and Kir is the inhibition constant of the product R.
RESULTS
Protein Expression and Purification of recombinant E. faecalis PPCS
PPCS is eluted from the Source 15Q anion exchange column at 200 mM NaCl. After anion exchange and gel filtration chromatography the synthetase was >98% pure as determined by SDS-PAGE. Approximately 30 mg of soluble, purified PPCS was obtained per liter of cell culture, with a specific activity of 5.2 U/mg. The native oligomerization state of PPCS was determined via gel filtration. The elution volume of the synthetase on a Superdex 200 HR column was compared to the elution volumes of known protein standard (Bio-Rad gel filtration standard) in order to determine its apparent molecular weight. PPCS eluted as a ~ 60 kDa molecule, suggestive of a homodimer (theoretical mass 57 kDa).
The purified protein was further analyzed by positive-ion ESI-MS, and found to have a molecular mass of 28415.0 Da. This mass corresponds to the calculated mass of the recombinant protein (28525.2 Da) associating with one molecule of sodium (23 Da), minus the amino terminal methionine (133.2 Da). The mass shows that E. faecalis PPCS has no attached prosthetic group, in agreement with previous studies performed on the E. coli synthetase.(9)
ATP/CTP Specificity of E. faecalis PPCS
Since the amino acid homology of E. faecalis PPCS is equally divergent from both human and E. coli PPCS, it was unclear whether the E. faecalis enzyme would be ATP or CTP selective. To determine its nucleotide specificity, the E. faecalis enzyme was assayed in the presence of either 10 mM ATP or CTP, 0.6 mM PPA, and 1 mM cysteine, representing saturating conditions for all substrates (particularly CTP, which is present at a concentration approximately 65 times its determined Km). Under these conditions, the ATP containing reaction showed only 8% of the activity of the CTP containing reaction, suggesting that the E. faecalis enzyme is highly selective for CTP like other bacterial PPCS.
Oxygen Transfer during PPCS Catalysis
Figure 1a shows the 13C NMR spectrum of [carboxyl-18O] PPA synthesized by hydrolysis of methyl phosphopantothenate with Na18OH (93% 18O). The inset shows two NMR resonances separated by 0.027 ppm, which correspond to [carboxyl-16O] PPA and [carboxyl-18O] PPA, and integrate to a ratio of 1 to 9.2, respectively. Thus, the labeled PPA contains 90% 18O distributed between the two carboxylate oxygens, consistent with the percentage of 18O (93%) contained in the saponification reaction. This compound was used as a substrate for the E. faecalis PPCS reaction to confirm the formation of a phosphopantothenoyl cytidylate intermediate. Figure 1b depicts the 31P NMR spectra of CMP formed during the PPCS catalyzed reaction. Two NMR resonances separated by 0.025 ppm were seen corresponding to CMP labeled with no and one 18O atom. The upfield shifted 18O labeled CMP resonance represents approximately 45% of the total CMP resonances, which is what would be expected from the transfer of one of the two equivalent carboxylate oxygens of PPA. The 31P NMR of the control reaction (Figure 1c), in which unlabeled PPA was used as a substrate for PPCS, shows a single resonance corresponding to unlabeled CMP. From these results it was determined that the E. faecalis PPCS catalyzed reaction proceeds through the formation of a phosphopantothenoyl cytidylate intermediate.
Figure 1. 13C and 31P NMR spectra.
(a) 13C NMR (proton-decoupled) spectrum of [carboxyl-18O]phosphopantothenate. (b) 31P NMR (proton-decoupled) spectrum of CMP produce during PPCS catalysis using [carboxyl-18O]phosphopantothenate. (c) 31P NMR (proton-decoupled) spectrum of CMP produce during PPCS catalysis using unlabeled phosphopantothenate. Experimental details are listed under Materials and Methods.
Enzyme Mechanism
In the double reciprocal plot of initial velocity against [CTP] at various [PPA] with saturating [cysteine], the fitted lines intersect to the left of the y-axis. Replots of intercept vs. 1/[PPA] and slope vs. 1/[PPA] both have a good linear fit (Figure 2). Thus, CTP and PPA are sequential with respect to each other, so in the overall mechanism both CTP and PPA must bind to the enzyme before a chemical reaction can occur. In the double reciprocal plot of initial velocity against [cysteine] at various [PPA], the fitted lines are parallel, with replot of intersect vs. 1/[PPA] having a good linear fit, consistent with a mechanism where PPA and cysteine are Ping-Pong with respect to each other (Figure 3). Thus, in the overall mechanism, there is a product release step between the binding of PPA and the binding of cysteine. Likewise, in the double reciprocal plot of initial velocity against [cysteine] at various [CTP], the fitted lines are parallel again, with a linear fit in the replot of intersect vs. 1/[CTP], consistent with a mechanism where PPA and CTP are Ping-Pong with respect to each other (Figure 4). Again, in the overall mechanism, there is a product release step between the binding of CTP and the binding of cysteine. Given that CTP and PPA must both bind to the enzyme before the first half reaction occurs, the only terreactant mechanism consistent with the data is the Bi Uni Uni Bi Ping Pong mechanism.
Figure 2. Pairwise analysis of CTP and PPA.
Initial velocity analysis of PPCS with varying [CTP] and [PPA] at 1 mM cysteine. (a) Double-reciprocal plot of velocity data. (b) Secondary plot of intercepts versus reciprocal [PPA]. (c) Secondary plot of slope versus reciprocal [PPA].
Figure 3. Pairwise analysis of cysteine and PPA.
Initial velocity analysis of PPCS with varying [cysteine] and [PPA] at 0.6 mM CTP. (a) Double-reciprocal plot of velocity data. (b) Secondary plot of intercepts versus reciprocal [PPA].
Figure 4. Pairwise analysis of cysteine and CTP.
Initial velocity analysis of PPCS with varying [cysteine] and [CTP] at 0.6 mM PPA. (a) Double-reciprocal plot of velocity data. (b) Secondary plot of intercepts versus reciprocal [CTP].
The product inhibition pattern of CMP with respect to the three substrates is examined to determine the sequence of CTP and PPA binding. In the double reciprocal plot of initial velocity against [CTP] at various [CMP], the fitted lines intersect on the y-axis, consistent with a mechanism where CMP is a competitive inhibitor with respect to CTP (Figure 5). In the double reciprocal plot of initial velocity against [PPA] at various [CMP], the fitted lines intersect in the second quadrant, consistent with CMP as a mixed inhibitor with respect to PPA (Figure 6). Finally, in the double reciprocal plot of initial velocity against [cysteine] at various [CMP], the fitted lines are parallel, consistent with CMP as an uncompetitive inhibitor with respect to cysteine (Figure 7). Together, the inhibition patterns are consistent with a Bi Uni Uni Bi Ping Pong mechanism where CTP is the first substrate to bind to the enzyme and CMP is the last product to be released (Scheme 3).
Figure 5. Product inhibition analysis of CMP versus CTP.
Initial velocity analysis of PPCS versus varying [CTP] at different fixed [CMP] with 0.15 mM PPA and 1 mM cysteine. Double-reciprocal plot of velocity data.
Figure 6. Product inhibition analysis of CMP versus PPA.
Initial velocity analysis of PPCS versus varying [PPA] at different fixed [CMP] with 0.3 mM CTP and 1 mM cysteine. Double-reciprocal plot of velocity data.
Figure 7. Product inhibition analysis of CMP versus cysteine.
Initial velocity analysis of PPCS versus varying [cysteine] at different fixed [CMP] with 0.3 mM CTP and 0.3 mM PPA. Double-reciprocal plot of velocity data.
Scheme 3.
The steady state kinetic parameters determined from fitting to the initial velocity equation of a Bi Uni Uni Bi Ping Pong mechanism, equation 2, are summarized in Table 1. The inhibition pattern of CMP with respect to the three substrates, as well as the calculated KiCMP (Kir in velocity equations) is summarized in Table 2.
Table 1.
Kinetic Parameters of PPCS at pH 7.6 and 37°C
| KiCTP | 652 ± 345 µM |
| KmCTP | 156 ± 17 µM |
| KmPPA | 17 ± 6 µM |
| Kmcys | 86 ± 7 µM |
| kcat | 2.9 ± 0.1 sec−1 |
Table 2.
Product Inhibition Patterns for PPCS
| inhibitor | variable substrate | fixed substrates | pattern | calc KiCMP |
|---|---|---|---|---|
| CMP | CTP | PPA, cysteine | comp | 766 ± 90 µM |
| CMP | PPA | CTP, cysteine | mixed | 522 ± 23 µM |
| CMP | cysteine | CTP, PPA | uncomp | 732 ± 97 µM |
DISCUSSION
Phosphopantothenoylcysteine synthetase catalyzes the second step of CoA biosynthesis, the amide bond formation between 4’-phosphopantothenate and l-cysteine (7, 9). Because of its essential nature in the growth and survival of all organisms, selective inhibitors of bacterial PPCS activity would be useful as broad spectrum antibiotics (5, 6). PPCS from E. faecalis represents a heretofore uncharacterized, new class of bacterial PPCS, found in several clinically and bioterrorism relevant pathogens, such as Streptococcus pneumoniae and Bacillus Anthracis (5, 10). This class of enzyme is uniquely monofunctional, in that it is not natively expressed as a protein fusion with phosphopantothenoylcysteine decarboxylase as seen in a majority of bacteria. Therefore, in order to gain a more complete description of bacterial PPCS to be used in inhibitor design, the E. faecalis enzyme has been cloned, expressed, purified, and subjected to kinetic and isotopic labeling studies.
The E. faecalis enzyme shows very distant sequence similarity to the PPCS domain of the bifunctional PPCDC/PPCS proteins, and in fact produce consistently higher similarity scores with the human monofunctional PPCS (5, 10). Nevertheless, our nucleotide utilization studies with the E. faecalis PPCS show that the enzyme is CTP-specific at physiological relevant concentrations. Thus, all bacterial PPCS enzymes characterized so far, whether expressed as a bifunctional polypeptide with PPCDC or natively expressed as a monofunctional protein, are CTP-specific. This is in stark contrast to the monofunctional human PPCS, which is reported to be ATP selective (5–7). The difference in nucleotide specificity between the bacterial and mammalian PPCS may have important implications in protein evolution and CoA regulation, as well as in developing selective bacterial PPCS inhibitors based upon differences in the nucleotide binding sites (10, 14).
To establish the formation of a phosphopantothenoyl cytidylate intermediate formed during the E. faecalis PPCS catalyzed reaction, heteronuclear NMR was preformed. 18O has a significant one-bond nuclear shielding effect upon carbon and phosphorus atoms to which it is directly bonded, resulting in an observable upfield shift in the NMR resonance of such nuclei (15–19). Isotopic-shifted heteronuclear NMR has been used in the present paper to study oxygen transfer during PPCS catalysis. When [carboxyl-18O]-labeled phosphopantothenate was incubated with CTP in the presence of PPCS, approximately half of the 18O ended up in the phosphate group of the CMP formed. This is consistent with our kinetic mechanism wherein a phosphopantothenoyl cytidylate intermediate is formed during the first half reaction with the release of pyrophosphate and subsequent transfer of the phosphopantothenoyl group to l-cysteine to yield phosphopantothenoylcysteine and CMP. Previous structural studies, using a mutant form of the E. coli PPCS domain which in non-crystallographic studies was unable to form phosphopantothenoylcysteine, also have shown a phosphopantothenoyl cytidylate intermediate formed within the active site upon crystallization in the presence of CTP and PPA (12).
Our steady-state initial velocity and product inhibition data for E. faecalis PPCS fit very well with a Bi Uni Uni Bi Ping Pong kinetic mechanism. These kinetic findings very nicely align with previous structural data from a series of crystal structures of an E. coli PPCS mutant (12). In the structural studies it was found that the dynamics of two protein loops, residues 284–299 which covers the binding site of the opposite dimer subunit, and residues 354–363 which forms a pocket around the terminal phosphate on PPA, control chemical reaction as well as substrate binding and product release. In the apo form, residues 284–299 and residues 354–363 are disordered, and the cleft where PPA binds is not well formed and almost entirely solvent exposed while the CTP binding pocket is mostly formed. In the CTP bound structure, residues 284–289, 298, and 299 become ordered and interact with the binding site formed from the other dimer (12). As a result, the PPA binding cleft is more defined. Also, the Kd determined from isothermal titration calorimetry (ITC; see Supporting Information) matches well with the dissociation constant of CTP calculated from our kinetic studies. Thus, consistent with our kinetic results, the ITC and structural data suggest that CTP binds first and is prerequisite to PPA binding.
The Ping-Pong portion of the kinetic mechanism also correlates well with the literature. In the CTP bound E. coli PPCS structure, the newly ordered Lys289 hydrogen bonds with the oxygen linking the α- and β-phosphates, serving to activate the pyrophosphate as a leaving group as well as to help orient where PPA binds to the enzyme for the subsequent attack on the α-phosphate of CTP to form the 4’-phosphopantothenoyl cytidylate intermediate. When both PPA and CTP are soaked into the crystal, the intermediate 4’-phosphopantothenoyl cytidylate is observed in the binding site, with pyrophosphate released (12). Our kinetic data show that PPA and cysteine have a Ping-Pong relationship, indicating that there is a product release step between the binding of PPA and the binding of cysteine. This is consistent with the release of PPi before the substrate cysteine binds and reacts.
In the acyl cytidylate intermediate bound structure, residues 354–363 become ordered and form a pocket around the terminal phosphate, making the PPA portion of the molecule no longer solvent accessible. Likewise, the CMP bound crystal structure obtained after all three substrates were soaked has residues 354–363 ordered, similar to the intermediate bound structure (12). Since CMP is still bound to the active site, while the other product phosphopantothenoylcysteine is not observed, phosphopantothenoylcysteine must be released first and CMP released last, same as the order determined via kinetics.
Overall, the enzyme mechanism of the monofunctional E. faecalis PPCS determined by our kinetic and isotopic studies is consistent with previous structural studies with the PPCS domain of the E. coli bifunctional protein. We have determined that CTP and PPA bind in an ordered fashion to the enzyme, and react to form the intermediate 4’-phosphopantothenoyl cytidylate with concomitant formation of pyrophosphate. Pyrophosphate is released before l-cysteine binds and reacts with the carbonyl moiety of the mixed anhydride to form the products phosphopantothenoylcysteine and CMP in the second half reaction. Further, product binding and substrate release is ordered, with CTP binding first and CMP released last.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Michael McLeish and Prof. Bruce Palfey for their helpful discussions.
Abbreviations
- ACP
acyl carrier protein
- ATP
adenosine-5’-triphosphate
- CoA
coenzyme A
- CTP
cytidine-5’-triphosphate
- DTT
dithiothreitol
- ESI-MS
electrospray ionization-mass spectroscopy
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- NADH
nicotinamide adenine dinucleotide
- PanK
pantothenate kinase
- PPA
phosphopantothenate
- PPCDC
phosphopantothenoylcysteine decarboxylase
- PPCS
phosphopantothenoylcysteine synthetase
- PPi
pyrophosphate
- PPi-FPK
pyrophosphate-dependent fructose-6-phosphate kinase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
This research was supported by the University of Michigan, College of Pharmacy.
Supported in part by U.S. Department of Homeland Security Fellowship administered by the Oak Ridge Institute for Science and Education.
Supported in part by National Institutes of Health Chemistry & Biology Interface Training Grant.
SUPPORTING INFORMATION AVAILABLE
Isothermal titration calorimetry data, SDS-PAGE of overexpressed proteins, and nucleotide specificity assay of monofunctional PPCS. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCE
- 1.Majerus PW, Alberts AW, Vagelos PR. Acyl Carrier Protein. Iv. the Identification of 4'-Phosphopantetheine as the Prosthetic Group of the Acyl Carrier Protein. Proc Natl Acad Sci U S A. 1965;53:410–417. doi: 10.1073/pnas.53.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MS, Bulawa CE, Raetz CR. The biosynthesis of gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J Biol Chem. 1985;260:15536–15541. [PubMed] [Google Scholar]
- 3.Brozek KA, Raetz CR. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J Biol Chem. 1990;265:15410–15417. [PubMed] [Google Scholar]
- 4.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes SY, Scholle MD, D'Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang YM, White SW, Rock CO. Inhibiting bacterial fatty acid synthesis. J Biol Chem. 2006;281:17541–17544. doi: 10.1074/jbc.R600004200. [DOI] [PubMed] [Google Scholar]
- 7.Brown GM. The metabolism of pantothenic acid. J Biol Chem. 1959;234:370–378. [PubMed] [Google Scholar]
- 8.Kupke T, Uebele M, Schmid D, Jung G, Blaesse M, Steinbacher S. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J Biol Chem. 2000;275:31838–31846. doi: 10.1074/jbc.M004273200. [DOI] [PubMed] [Google Scholar]
- 9.Strauss E, Kinsland C, Ge Y, McLafferty FW, Begley TP. Phosphopantothenoylcysteine synthetase from Escherichia coli. Identification and characterization of the last unidentified coenzyme A biosynthetic enzyme in bacteria. J Biol Chem. 2001;276:13513–13516. doi: 10.1074/jbc.C100033200. [DOI] [PubMed] [Google Scholar]
- 10.Genschel U. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol Biol Evol. 2004;21:1242–1251. doi: 10.1093/molbev/msh119. [DOI] [PubMed] [Google Scholar]
- 11.Kupke T, Hernandez-Acosta P, Steinbacher S, Culianez-Macia FA. Arabidopsis thaliana flavoprotein AtHAL3a catalyzes the decarboxylation of 4'-Phosphopantothenoylcysteine to 4'-phosphopantetheine, a key step in coenzyme A biosynthesis. J Biol Chem. 2001;276:19190–19196. doi: 10.1074/jbc.M100776200. [DOI] [PubMed] [Google Scholar]
- 12.Stanitzek S, Augustin MA, Huber R, Kupke T, Steinbacher S. Structural basis of CTP-dependent peptide bond formation in coenzyme A biosynthesis catalyzed by Escherichia coli PPC synthetase. Structure. 2004;12:1977–1988. doi: 10.1016/j.str.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Ladror US, Gollapudi L, Tripathi RL, Latshaw SP, Kemp RG. Cloning, sequencing, and expression of pyrophosphate-dependent phosphofructokinase from Propionibacterium freudenreichii. J Biol Chem. 1991;266:16550–16555. [PubMed] [Google Scholar]
- 14.Ostrander DB, O'Brien DJ, Gorman JA, Carman GM. Effect of CTP synthetase regulation by CTP on phospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 15.Hansen DE, Knowles JR. The stereochemical course of the reaction catalyzed by creatine kinase. J Biol Chem. 1981;256:5967–5969. [PubMed] [Google Scholar]
- 16.Hansen DE, Knowles JR. The stereochemical course at phosphorus of the reaction catalyzed by phosphoenolpyruvate carboxylase. J Biol Chem. 1982;257:14795–14798. [PubMed] [Google Scholar]
- 17.Kohlbrenner WE, Nuss MM, Fesik SW. 31P and 13C NMR studies of oxygen transfer during catalysis by 3-deoxy-D-manno-octulosonate cytidylyltransferase from Escherichia coli. J Biol Chem. 1987;262:4534–4537. [PubMed] [Google Scholar]
- 18.Dotson GD, Dua RK, Clemens JC, Wooten EW, Woodard RW. Overproduction and one-step purification of Escherichia coli 3-deoxy-D-manno-octulosonic acid 8-phosphate synthase and oxygen transfer studies during catalysis using isotopic-shifted heteronuclear NMR. J Biol Chem. 1995;270:13698–13705. doi: 10.1074/jbc.270.23.13698. [DOI] [PubMed] [Google Scholar]
- 19.Zheng R, Blanchard JS. Steady-state and pre-steady-state kinetic analysis of Mycobacterium tuberculosis pantothenate synthetase. Biochemistry. 2001;40:12904–12912. doi: 10.1021/bi011522+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.