Abstract
Telomeres are composed of specialized chromatin that includes DNA repair/recombination proteins, telomere DNA-binding proteins and a number of three dimensional nucleic acid structures including G-quartets and D-loops. A number of studies suggest that the BLM and WRN recQ-like helicases play important roles in recombination-mediated mechanisms of telomere elongation or Alternative Lengthening of Telomeres (ALT), processes that maintain/elongate telomeres in the absence of telomerase. BLM and WRN localize within ALT-associated nuclear bodies in telomerase-negative immortalized cell lines and interact with the telomere-specific proteins POT1, TRF1 and TRF2. Helicase activity is modulated by these interactions. BLM functions in DNA double-strand break repair processes such as non-homologous end joining, homologous recombination-mediated repair, resolution of stalled replication forks and synthesis-dependent strand annealing, although its precise functions at the telomeres are speculative. WRN also functions in DNA replication, recombination and repair, and in addition to its helicase domain, includes an exonuclease domain not found in other recQ-like helicases. The biochemical properties of BLM and WRN are, therefore, important in biological processes other than DNA replication, recombination and repair. In this review, we discuss some previous and recent findings of human rec-Q-like helicases and their role in telomere elongation during ALT processes.
Keywords: TELOMERES, ALT, BLM HELICASE, WRN HELICASE
Unique DNA structures at the end of human chromosomes are known as telomeres, and contain repetitive (TTAGG)n sequences. Telomere lengths decline with each cell division at a rate of ~50–150 bp. Shortening of telomeres past a critical threshold length triggers replicative senescence, an irreversible state of growth arrest that may prevent subsequent chromosome instability. Human tumor cells escape the limitation imposed by cellular senescence and continue to grow. These cells usually have an activated telomere maintenance mechanism that is acquired by activation of a ribonucleoprotein complex containing the telomere replicating enzyme telomerase [Shay and Bacchetti, 1997]. In other human tumors, immortalized human cell lines and telomerase-null mouse cell lines [Bryan et al., 1995, 1997; Hande et al., 1999; Niida et al., 2000], telomere lengths are maintained through many population doublings (PDs) using one or more non-telomerase mechanisms known as Alternative Lengthening of Telomeres or ALT [Bryan and Reddel, 1997].
Cells using ALT pathways are characterized by heterogeneous telomere lengths. Normally, the length of telomeres varies between 2 and 15 kb in germline cells [Allshire et al., 1989; de Lange et al., 1990], whereas in human ALT cell lines, telomere length varies between 2 and over 50 kb [Bryan et al., 1995; Park et al., 1998]. ALT mechanisms most likely involve recombination-mediated DNA replication. The first evidence of a recombination-driven ALT pathway came from studies of the unicellular eukaryotic organism Saccharomyces cerevisiae. Such studies suggest two models of telomere length maintenance [Shampay et al., 1984; Walmsley et al., 1984]. One pathway uses an enzyme to add telomeric repeats in vivo [Shampay et al., 1984]; the other uses a recombination-mediated method for polymerization of telomeric repeats [Walmsley et al., 1984]. Most yeast lacking telomerase have shorter telomeres and ultimately die [Lundblad and Blackburn, 1993], although a small subset survive. Survival of these cells is dependent on the function of the recombination protein Rad52p [Lundblad and Blackburn, 1993; Teng and Zakian, 1999]. In addition to Rad52-dependence, these cells are characterized by long and heterogeneous telomeres that suggest gene-conversion events as one mechanism to form extra long telomeres. Cells using ALT also contain unique nuclear structures known as ALT-associated PML bodies (APBs) [Yeager et al., 1999]. APBs contain telomere-specific proteins, recombination proteins Rad51p and Rad52p [Yeager et al., 1999] and telomeric DNA. The presence of recombination proteins in APBs further support a recombination-mediated mechanism for telomere lengthening in ALT cells.
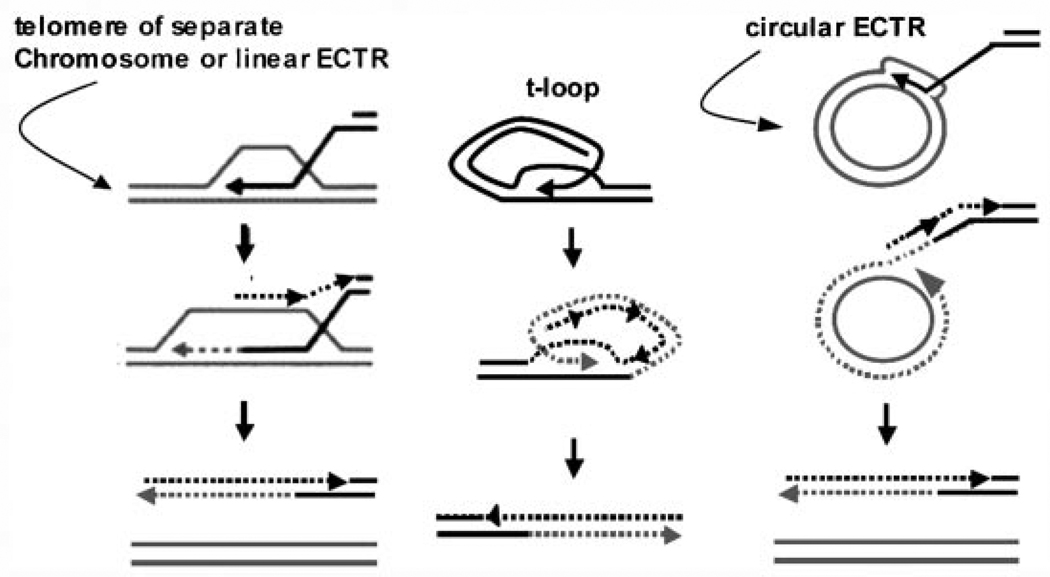
Telomeres in cells using ALT undergo continuous shortening and elongation, most likely occurring during recombination [Bryan et al., 1995]. Dunham et al. [2000] first demonstrated that recombination occurred between telomeric repeats in cells using ALT using a plasmid carrying a NEO-cassette integrated into telomeric DNA in a human cell line using ALT. Two cell clones derived from integration of this plasmid contained two and three chromosomes with tagged telomeres at the 23rd population doubling. The number of tagged telomeres in each clonal population increased to 10 after 40 population doublings. Plasmid integration into a subtelomeric region did not result in transfer or copying to any other chromosomes, suggesting that recombination events were telomere repeat-specific. Studies have also shown that homologous recombination is up-regulated at telomeres in cells using ALT, while the rate of recombination outside of the telomeres remained unchanged [Bechter et al., 2003; Bailey et al., 2004; Bechter et al., 2004; Londono-Vallejo et al., 2004]. The mechanism for the transfer of a telomere tag to multiple chromosomes may be accomplished by break-induced DNA replication (BIR) where telomeric sequences are copied between chromosomes, a process that may be facilitated by replication (Fig. 1A). However, telomere-telomere replication may not be the only mechanism for ALT. Replication may be initiated from the T-loop via intrachromosomal recombination events thereby allowing continuous replication by a rolling circle-type mechanism (Fig. 1B). ALT cells have high levels of linear and circular extrachromosomal telomeric repeat DNA [Tokutake et al., 1998]; such DNAs may represent by-products of telomeric recombination or may serve as additional templates for replication. Circular telomeric DNAs could facilitate telomere synthesis by rolling circle replication (Fig. 1C), as in K. lactis [Natarajan and McEachern, 2002].
Fig. 1.
Mechanisms of ALT. A: ALT may occur by BIR using a homologous chromosome-end or linear extra-chromosomal telomeric repeat (ECTR) DNA as a template. B: Alternatively, replication may be initiated directly from the T-loop or (C) using circular ECTR DNA as a template. Both B and C may facilitate continuous, rolling circle synthesis of new telomeric repeats.
PROTEINS INVOLVED IN ALT PATHWAYS
A number of proteins have been identified in APBs that may be involved in ALT mechanisms. PML nuclear bodies (PNBs), the nuclear matrix-associated subnuclear structures that exist in most cell types, are named for the promyelocytic leukemia or PML protein, a putative tumor suppressor. PNBs are highly dynamic structures, with protein components that vary with cell type, cell cycle, and in response to internal and external stimuli. APBs are composed of a subset of PNBs, present only in cells using ALT, and contain unique telomeric components including telomeric DNA (chromosomal and/or extrachromosomal), the telomere repeat binding proteins POT1, TRF1 and TRF2, and aggregates of proteins involved in homologous recombination (HR) repair, including RAD51, RAD52, RPA and the MRN complex [Yeager et al., 1999; Grobelny et al., 2000; Wu et al., 2000a]. Protein complexes that include the BLM helicase (the protein mutated in those with the inherited chromosome breakage syndrome Bloom’s syndrome or BS), topoisomerase IIIα and BLAP75 (BLM-associated polypeptide 75) [Mankouri and Hickson, 2007; Raynard et al., 2008] have also been implicated in ALT mechanisms. Topoisomerase IIIα localizes with TRF2 in ALT cells; siRNA (small interfering RNA)-mediated disruption of TOP3A topoisomerase IIIα reduces TRF2 levels, loss of G-strand overhangs and a reduction of ALT cell viability. These data suggest that topoisomerases may be necessary for telomere synthesis during ALT. That ALT mechanisms may involve recombination-mediated events suggests that DNA helicases may also play an important role during these processes.
Two members of the RecQ helicase family, BLM and WRN (the protein mutated in those with the inherited chromosome breakage syndrome Werner’s syndrome or WS), are localized at telomeres even though their exact function there remains unclear. WRN−/− cells are characterized by chromosome instability that leads to an increase in variegated translocation mosaicism, symptoms of premature aging and great predisposition to cancer, particularly to sarcomas. Loss of Wrn leads to instability in mouse cells, increased telomere sister chromatid exchange (T-SCE) rates and ready immortalization in culture [Laud et al., 2005]. Cells from WS individuals are not only characterized by similar types of genomic instability but also elevated rates of chromosomal translocations and deletions [Fukuchi et al., 1989] and accelerated loss of replicative capacity (and thus initiation of cellular senescence) [Faragher et al., 1993] that can be prevented by telomerase expression [Wyllie et al., 2000]. Increased cellular senescence and telomere shortening may be a direct cause of age-related pathologies in both WS individuals and the rest of the population, with the age of onset differing between these two groups. Cultured WS cells display extended S-phase [Poot et al., 1992], suggesting a function for WRN in S-phase. Although its expression peaks at G2/M-phases, the protein is expressed steadily throughout the cell cycle [Kitao et al., 1998]. Persons with BS (BLM−/−) also display cancer predisposition. Their cells are characterized by elevated levels of SCE [Chaganti et al., 1974], indicating that BLM is involved in regulating such exchanges although T-SCE is not elevated in BS cells [Londono-Vallejo et al., 2004]. Elevated T-SCE is prevalent in ALT-positive cells, raising questions of whether recQ-like helicases may be required during some mechanisms of ALT and what, if any, functional roles BLM and WRN have in these mechanisms.
Both WRN and BLM participate in ALT pathways in telomerase-negative cells, as both localize to APBs and associate with telomeric proteins TRF1 and TRF2 in ALT cells [Opresko et al., 2002; Stavropoulos et al., 2002; Lillard-Wetherell et al., 2004]. Furthermore, TRF2 stimulates the unwinding activity of the BLM and WRN helicases and allows prolonged unwinding of telomeric substrates [Opresko et al., 2002; Lillard-Wetherell et al., 2004]. Co-localization of BLM and TRF2 occurs within APBs and increases with bromodeoxyuridine (BrdU) incorporation during late S and G2-phases of the cell cycle when ALT is thought to occur [Lillard-Wetherell et al., 2004]. This overlap with BrdU incorporation suggests DNA synthesis of telomeric DNA within APBs. BLM also directly binds telomeric DNA within nuclear bodies, while deletion of the DNA interaction domain of BLM interrupts proper cell cycle progression [Schawalder et al., 2003]. WRN also specifically binds telomeric DNA sequences [Opresko et al., 2004] and co-localizes exclusively in ALT cells with alternative APB-associated proteins, including PML, RAD52 [Baynton et al., 2003] and NBS1 [Johnson et al., 2001]. POT1, which binds single stranded telomeric DNA and regulates telomere length, facilitates BLM and WRN unwinding activity of telomeric D-loop and forked duplexes structures, otherwise poor substrates. POT1 stimulates both BLM and WRN to unwind and release the single strand chromosome end from a telomeric D-loop structure [Opresko et al., 2005]. In addition, the MRN complex of MRE11, Rad50 and NBS1 localizes to telomeres, with Ku 70/80 and DNA-PKCS. WRN interacts with FEN1, a protein required for ALT telomere stability [Saharia and Stewart, 2009].
WRN and BLM are certainly involved in telomere maintenance. WS cells have normal telomere lengths, although passage in culture reveals elevated rates of telomere shortening and senescence induction at inconsistent telomere lengths [Schulz et al., 1996; Tahara et al., 1997]. Furthermore, cells lacking a functional WRN helicase increase sister telomere loss [Crabbe et al., 2004]. These cytogenetic abnormalities are all reversed by expression of telomerase [Wyllie et al., 2000; Crabbe et al., 2004], suggesting the necessity of WRN for proper telomeric maintenance in a telomerase-independent context. Experimental evidence suggests a direct interaction of BLM and WRN and similar mechanistic roles in DNA metabolism. Both proteins localize together in ALT cell lines [von Kobbe et al., 2002], although some co-localization occurs in telomerase-positive cell lines [Yankiwski et al., 2000]. Enzyme-linked immunosorbent assays (ELISA) have demonstrated that BLM and WRN directly interact. They are present in the same cellular fraction and co-immunoprecipitate from nuclear extracts. Binding assays mapped the N-terminal and RQC-containing regions of WRN as those required for BLM binding. No alteration of unwinding activity for BLM or WRN is detected when the proteins are added to in vitro reactions together, using both preferred and non-preferred DNA substrates. BLM, however, inhibits the in vitro exonuclease function of WRN [von Kobbe et al., 2002].
Unlike BLM, WRN does not appear necessary for ALT. Although numerous studies have localized WRN at the telomeres in ALT cells, their survival is possible without WRN. Immortalized WS cell lines exist without detectable telomerase activity, indicating the use of ALT pathways to maintain telomeres [Marciniak et al., 2005]. Laud et al. [2005] demonstrated that WRN deficiency in the context of telomere dysfunction also allows cells to engage ALT pathways. In addition, WS individuals have high incidence of sarcomas [Goto et al., 1996], a tumor type with an unusually high percentage of telomerase-negative tumors [Reddel, 2003; Montgomery et al., 2004].
BLM DIRECTLY AFFECTS TELOMERE LENGTH IN CELLS USING ALT
siRNA knockdown of BLM and telomere restriction fragment (TRF) length assays tested whether disruption of BLM expression alters telomere length in ALT cells [Bhattacharyya et al., 2009]. Telomere length attrition occurs rapidly in cells using ALT without BLM in comparison to scrambled controls. Cells immortalized with telomerase display no change in average telomere length whether BLM is present or not. Telomeres shorten in BLM-negative ALT cells at a rate of ~750 base pairs (bp)/PD, almost 10 times faster than what would be expected by the DNA end replication problem. This may be due to replication fork stalling and breaks within the telomeric DNA in absence of BLM. In general, telomere DNA is difficult to replicate and replication forks within telomeres often stall [Ohki and Ishikawa, 2004]. Thus, BLM may function to resolve stalled replication forks in addition to mediating ALT-specific recombination events. These data indicate a direct role for BLM in maintaining telomere length in cells using ALT.
BLM knockdown reduces the co-localization of BLM-associated proteins with TRF2 and APBs in ALT cells, although their overall level of expression remains unchanged. There are no significant changes in the co-localization pattern of these proteins with TRF2 and APBs in telomerase-positive cell lines [Bhattacharyya et al., 2009]. These data suggest that BLM might recruit its partners to telomeres during ALT.
BLM HELICASE AND ALT
Cytogenetic studies of BS cells demonstrate a high frequency of telomeric associations (TAs) between homologous chromosome arms [German, unpublished work; Lillard-Wetherell et al., 2004]. Other studies have identified telomere length hyper-variability in BS cells [Schawalder et al., 2003]. These data suggest that there are instabilities within telomeric repeats in cells without BLM and that BLM may be required for maintaining telomere structure and/or length. Immunofluorescence studies determined that BLM colocalizes with TRF1 and TRF2 in cells using ALT [Stavropoulos et al., 2002; Lillard-Wetherell et al., 2004], but not in telomerase-positive or primary cell lines. In vivo chromatin immunoprecipitation also demonstrated that BLM and TRF2 associate with telomeric DNA in ALT cells [Lillard-Wetherell et al., 2004]. Furthermore, TRF1 and TRF2 interact directly with BLM in vitro. As human telomeric DNA consists of repeated telomere sequences and 3′-overhang G-rich strand, the effects of TRF1 and TRF2 on BLM helicase activity were studied using DNA substrates that mimic telomeric DNA in vivo. BLM unwinding was stimulated by TRF2 but not TRF1. Relative amounts of TRF1 and TRF2 may also be important determinants for BLM stimulation, as equimolar amounts of TRF1 and TRF2 stimulated BLM helicase activity. This stimulation was lost when TRF1 was present in molar excess. These data suggest that TRF1 and TRF2 may function with BLM at different stages of telomere elongation or unwinding. TRF2 is believed to protect telomere ends within T-loops and facilitate formation of looped telomeric structures in vitro [Griffith et al., 1999]; TRF1 cannot promote loop formation but can mediate DNA bending and parallel pairing of telomere sequences [Bianchi et al., 1997; Griffith et al., 1998, 1999].
BLM binds and unwinds DNA “bubbles,” short regions of unpaired DNA flanked by fully duplexed DNA [van Brabant et al., 2000; Mohaghegh et al., 2001]. It has a strong affinity for binding and melting D-loops, structures that result from pairing of an invading DNA strand to a complementary strand within a bubble structure [van Brabant et al., 2000]. This activity is important for considering BLM function in all recombination-mediated processes, as there is similarity between such D-loops and the heteroduplex intermediates formed by RAD51 during recombination. BLM efficiently unwinds synthetic D-loop substrates in vitro; this unwinding is stimulated by TRF2 but not TRF1 [Lillard-Wetherell et al., 2004]. Additionally, BLM disrupts quadruplex or “G4” DNA, an unusually stable secondary structure that forms from non-canonical base pairing between guanine residues, either inter- or intra-strand [Sun et al., 1998]. Quadruplex-interacting ligands specifically inhibit unwinding of D-loops (bubble) and G4 duplex substrates by BLM [Li et al., 2001; Huber et al., 2002]. While quadruplexes form readily in vitro, data also indicate an enrichment of these structures at telomeres [Chang et al., 2004]. Hence, the unwinding of G-quadruplexes has unique implications for BLM function in maintaining the integrity of repetitive G-rich regions of the genome such as telomeres and ribosomal repeats [Schawalder et al., 2003]. These observations further support a function of BLM at the telomeres.
BLM is expressed in all tissues with active cell proliferation. It is barely detectable in cells during G1-phase of the cell cycle but dramatically increases during the S- and G2-phases [Dutertre et al., 2000]. As mentioned, BLM interacts with other proteins required for DNA replication/repair. It undergoes post-translational modifications that are necessary for its stability and activity. For example, BLM is hyper-phosphorylated during S-phase [Beamish et al., 2002]. It associates with and is phosphorylated by the ATM [Beamish et al., 2002] and ATR kinases [Franchitto and Pichierri, 2002; Davies et al., 2004] after specific types of DNA damage. The Fanconi anemia (FA) nuclear core complex is necessary for BLM phosphorylation and recruitment to nuclear foci in response to DNA crosslinking agents [Pichierri et al., 2004]. In vivo, BLM generally has a punctate nuclear distribution that partially overlaps with PML bodies. Upon treatment with DNA damaging agents or replication inhibitors, BLM associates at sites of DNA damage/repair with phosphorylated histone H2AX or RAD51 and at sites of stalled replication forks [Zhong et al., 1999; Yankiwski et al., 2000, 2001]. BLM is one of the first proteins to relocalize to sites of DNA damage/repair and bind directly to RAD51 [Wu et al., 2001]. BLM is also involved in the subsequent recruitment of p53 [Sengupta et al., 2003], BRCA1 [Davalos and Campisi, 2003] and MRN [Franchitto and Pichierri, 2002; Davalos and Campisi, 2003] to sites of stalled replication forks/DNA repair. Monoubiquitinated FANCD2 co-localizes and coimmunoprecipitates with BLM after replication arrest or treatment of cells with crosslinking agents [Meetei et al., 2003].
BLM is part of two protein super-complexes believed to function in DNA repair, called “BASC” and “BRAFT.” In addition to BLM and BRCA1, BASC (BRCA1-associated genome surveillance complex) includes the DNA mismatch repair proteins MSH2, MSH6 and MLH1, the DSB repair proteins ATM, RAD50, MRE11 and NBS1, and a single replication protein RFC [Wang et al., 2000]. As the name implies, BASC is a dynamic “genome-surveillance” complex that senses DNA damage and deploys appropriate proteins to mediate DNA repair. The BRAFT protein complex was identified by in vivo immunoprecipitation of BLM [Meetei et al., 2003]. BRAFT is named for its most 19 abundant protein components, including BLM, RPA, Fanconi anemia complementation (FANC) group proteins, and topoisomerase IIIα (TOPOIIIα). Like BASC, BRAFT most likely serves as scaffolding complex for DNA repair proteins. BLM and MLH1 are the sole components common to both BASC and BRAFT; therefore, each complex is likely to respond to unique types of DNA damage or cellular stress cues. Separate studies demonstrate that BLM interacts directly with BASC and BRAFT proteins, including MLH1, MSH6, ATM, RPA, FANCD2 and TOPOIIIα [Brosh et al., 2000; Wu et al., 2000b; Langland et al., 2001; Pedrazzi et al., 2001; Wu et al., 2001; Pichierri et al., 2004]. Hence, BLM is involved at several stages of DNA metabolism providing genomic stability and integrity. Although its function at the telomeres is not well understood, it is most likely acting in conjunction with one or more of the BASC or BRAFT proteins. Thus, our recent work tested the hypothesis that BLM may be a component of an ALT-specific complex whose formation and modification may occur dynamically during the strategic nucleic acid transactions that are required to protect the telomere: to align chromosome sequences at homologous telomeres, to permit strand invasion and elongation, and/or ultimately to disentangle telomeres.
BLM ASSOCIATES WITH ADDITIONAL PROTEINS IN ALT-SPECIFIC COMPLEXES
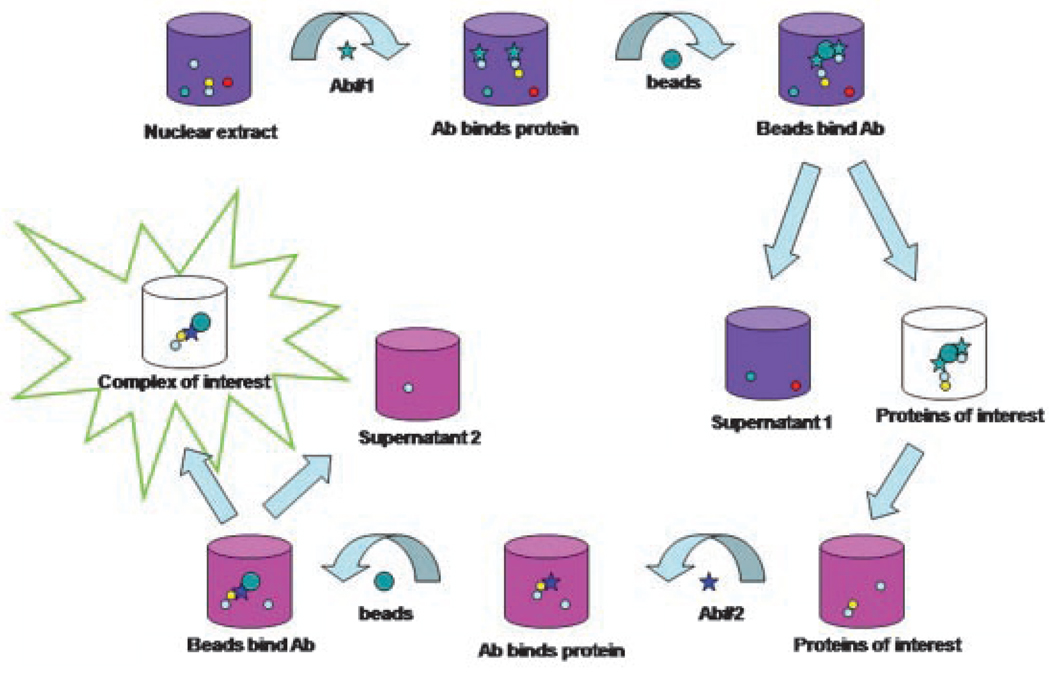
The first evidence for an ALT-specific complex containing BLM originated from the association of BLM with TRF2 in cells using ALT but not in telomerase-positive cells [Lillard-Wetherell et al., 2004]. Immunoprecipitation analysis was carried out in cells at the late S- and G2/M-phases of the cell cycle, when BLM expression is at its highest and telomere DNA synthesis is occurring. Double or sequential immunoprecipitation strategies isolated a more specific ALT complex of BLM and TRF2 [Bhattacharyya et al., 2009]. Antibodies specific for BLM and TRF2 were used to pull down BLMTRF2 complexes from ALT cells (Fig. 2), followed by the resolution of complexes by denaturing polyacrylamide gel electrophoresis to identify unique protein bands in cells using ALT. Mass spectrometric analysis determined protein identities: the telomerase-associated protein 1 (TEP-1), the spliceosome-associated protein 145 (SF3B2), HSP90 and topoisomerase IIα (TOPOIIα) in addition to BLM. These proteins, undetected in complexes from telomerase-positive cells, were validated by western analysis. Biochemical studies suggest that BLM associates with topoisomerase IIα in vivo via its N-terminus sequences [Russell et al., 2009]. This interaction enhances BLM helicase activity on a variety of recombination intermediates including bubble and X-junction substrates. Although, topoisomerase IIα is required for decatenation activity to resolve entangled sister chromatids during DNA replication [Deming et al., 2001], there are no changes in its decatenation activity following BLM interaction [Russell et al., 2009, in preparation]. As BLM associates with both TRF1 and TRF2 only in cells using ALT, and that a BLM-TRF2 complex was isolated from ALT cells, it was reasonable to assume a unique or common BLM-TRF1 complex might exist in ALT cells. Double immunoprecipitations using anti-BLM and anti-TRF1 antibodies identified a unique BLM-TRF1 complex in ALT cells. The previously identified HSP90 and TOPO IIα were detected along with TRF2 in the BLM-TRF1 complex, but not TEP1. The identification of two possible ALT-specific complexes, with and without TEP1, may be biologically relevant as these complexes could form dynamically. As TRF1 and TRF2 are telomere-specific proteins that associate with BLM in ALT cells, co-localization of the BLM-associated proteins with TRF1 and TRF2 would suggest a role in telomere maintenance. Immunofluorescence data suggest that all three proteins, TEP1, TOPOIIα and HSP90, co-localize with BLM, TRF1 and TRF2 in 80–90% of ALT cells analyzed, whereas co-localization was observed in less than 30% of telomerase-positive cells. These data implicate an ALT-specific role for the BLM-associated proteins.
Fig. 2.
Schematic showing the protocol for double immunoprecipitation. Nuclear extracts prepared from telomerase-positive and cells using ALT are incubated with one antibody overnight at 4°C, followed by addition of protein A/G beads. Antigen–antibody complexes are bound to the beads for 2 h at 4°C, washed and eluted with citrate buffer, pH 3. Immediately following elution, protein complexes are neutralized to pH 7 and a second antibody added. After overnight incubation, beads are re-added to capture enriched antigen–antibody complexes, which are then eluted using 2 × SDS-sample buffer.
A consistent feature of ALT cells is the presence of ALT-associated PML nuclear bodies or APBs [Yeager et al., 1999] in which telomeric DNA, telomeric proteins and homologous recombination proteins reside. Both BLM and TRF2 co-localize with PML in ALT cells, suggesting that BLM is a component of APBs [Lillard-Wetherell et al., 2004]. TOPOIIα, TEP1 and HSP90 associate with PML bodies similarly to BLM and TRF2; their localization at APBs increase as cells progress into the late S- and G2/M-phases of the cell cycle when ALT is thought to occur. Co-localization increases almost twofold from G1- to late S- and G2/M-phases. The cell cycle-specific incremental increases in co-localized foci suggests that not only do these proteins localize to APBs, but that they may be involved in the specific DNA-protein transactions required for telomere elongation during ALT. Telomerase-positive cells do not show similar protein localization, nor do they demonstrate any cell cycle-specific dynamic of co-localization. Proteins implicated in ALT, including BLM and TRF2, co-localize with sites of new DNA synthesis during late S- and G2/M-phases of the cell cycle [Lillard-Wetherell et al., 2004]. The newly identified proteins also co-localize with new DNA synthesis sites in ALT cells but not in telomerase-positive cells, suggesting a role in DNA synthesis during late S- and G2/M-phases when telomere DNA elongation occurs.
BLM-ASSOCIATED ALT-SPECIFIC PROTEINS AFFECT BLM FUNCTION IN VITRO
Not only do TEP1, TOPOIIα and HSP90 associate with BLM, TRF1 and TRF2 in vivo, they physically and functionally interact with BLM [Bhattacharyya et al., 2009]. Enzyme-linked immunosorbent assays (ELISA) confirmed direct physical interaction between these proteins and BLM in vitro. All three proteins interact with BLM specifically in a dose-dependent manner and independently of DNA. Helicase assays using telomeric and non-telomeric DNA substrates characterized the effects of BLM interactions with its newly identified partners [Bhattacharyya et al., 2009]. Non-telomeric substrates do not include telomere sequences and were used as a control to determine BLM sequence specificity towards telomeric substrates during unwinding. BLM unwinds both substrate types, but shows a preference for the telomeric-DNA substrate at higher concentrations higher than 15 nM. The unwinding kinetics of BLM at different concentrations were compared: high concentrations of BLM unwound the telomeric substrate more efficiently than the conventional 30′ overhang non-repetitive control substrate, with two- to sixfold enhancement in unwinding compared to the control.
DNA topoisomerases are nuclear enzymes necessary for topological interconversions of DNA during transcription, replication and chromosome segregation during both meiosis and mitosis [Wang, 1996; Champoux, 2001]; they may also influence telomere stability. In somatic and meiotic human cells, BLM associates and co-localizes with topoisomerase IIIα in promyelocytic leukemia (PML) protein nuclear bodies; this localization is disrupted in BS cells [Johnson et al., 2000]. The interaction of topoisomerase IIIα and BLM occurs in the amino-terminal region of BLM and is important for maintaining genomic stability in human cells [Wu et al., 2000b]. In vitro, equimolar TOPOIIα subtly alters the unwinding of telomeric substrates, especially during the initial stages of unwinding when TOPOIIα slows BLM unwinding kinetics. BLM unwinds non-telomeric substrates equally well with or without TOPOIIα, but less efficiently than the telomeric substrate. This suggests that BLM and TOPOIIα interactions may have different functions depending on the sequence and/or structure of DNA. Another recent study has suggested a role for topoisomerase IIIα in ALT [Temime-Smaali et al., 2008], as a direct interaction between TOPOIIIα and BLM was demonstrated in cells using ALT and in vitro. The inclusion of TOPOIIα as part of the BLM-associated ALT-specific complex supports the idea that topoisomerases are critically involved in pathways of telomere length maintenance in the absence of telomerase, most likely during the resolution of entangled telomere ends. Telomeres co-purify with nuclear matrix at places where TOPOIIα is present [de Lange, 1992], again suggesting that TOPOIIα associates with telomeres and provides a mechanism for untangling chromosome ends.
In contrast to TRF2 and TOPOIIα, HSP90 strongly inhibits BLM unwinding on both telomeric and non-telomeric substrates. HSP90 is a scaffolding protein primarily required for efficient telomerase assembly in vivo and in vitro [Holt et al., 1999]. As a component of the telomerase complex, HSP90 functions in the telomerase translocation step during telomere elongation, perhaps holding the complex together or assisting in a change of conformation to promote telomere lengthening [Holt et al., 1999]. HSP90 is also a component of the BLM-associated ALT-specific complex, and may be involved in ALT pathways. The precise role of HSP90 in the ALT pathway is unclear, although we speculate its requirement in maintaining the integrity of the BLM-TRF1 and BLM-TRF2 complexes. The strong inhibition of BLM helicase activity in the presence of HSP90 suggests that HSP90 may not be directly involved during the telomere unwinding stages of ALT pathway, but that it may act upstream of BLM-mediated telomere replication/recombination events by facilitating the formation of the BLM-associated ALT-specific complexes.
In vitro, TEP1 reduces BLM unwinding of telomeric substrates only during the early stages of unwinding [Bhattacharyya et al., 2009]. In the presence of TEP1, only 20% to 30% of the substrate is unwound by BLM compared to almost 70% with BLM alone. The reduced unwinding kinetics in the presence of TEP1 may be due to its binding to telomere sequence-containing DNA. The inhibition of BLM helicase activity by TEP1 is lost when the unwinding reactions proceed longer. Although TEP1 is associated with telomerase in human, mice and rat immortalized cell extracts [Liu et al., 2000], its precise role at the telomeres in telomerase-positive cells is not clearly understood. Murine studies indicate that Tep1 is not essential for mouse telomere length maintenance. TEP1 is a component of a large cytoplasmic RNP known as the vault particle [Kickhoefer et al., 1999]. At this time, TEP1 functions in the ALT pathways are unclear. However, the presence of TEP1 as a component of the BLM-TRF2 complex and its effects on in vitro BLM unwinding suggest a novel role in telomere maintenance by ALT pathways.
ADDITIONAL PROTEINS IN THE ALT-SPECIFIC COMPLEX
The shelterin proteins POT1, RAP1 and TIN2 are present in the BLM-associated ALT-specific complex with TRF2 and TRF1; they do not co-immunoprecipitate or co-localize with BLM in telomerase-positive cells [Bhattacharyya et al., 2009]. DNA mismatch repair proteins MSH2, MLH1 and PMS2, the MRN complex and RAD51 are not detected in the BLM-associated ALT-specific complexes. These data suggest that the BLM complexes identified in ALT cells are different from BASC and/or BRAFT. POLδ is also present [Bhattacharyya and Groden, unpublished data]. These BLM-associated proteins, including TRF1, TRF2 and TOPOIIα, may determine the structure of the uncapped telomere ends in ALT cells by facilitating the formation of T-loops and D-loops, and subsequently promoting telomere elongation.
CONCLUSIONS
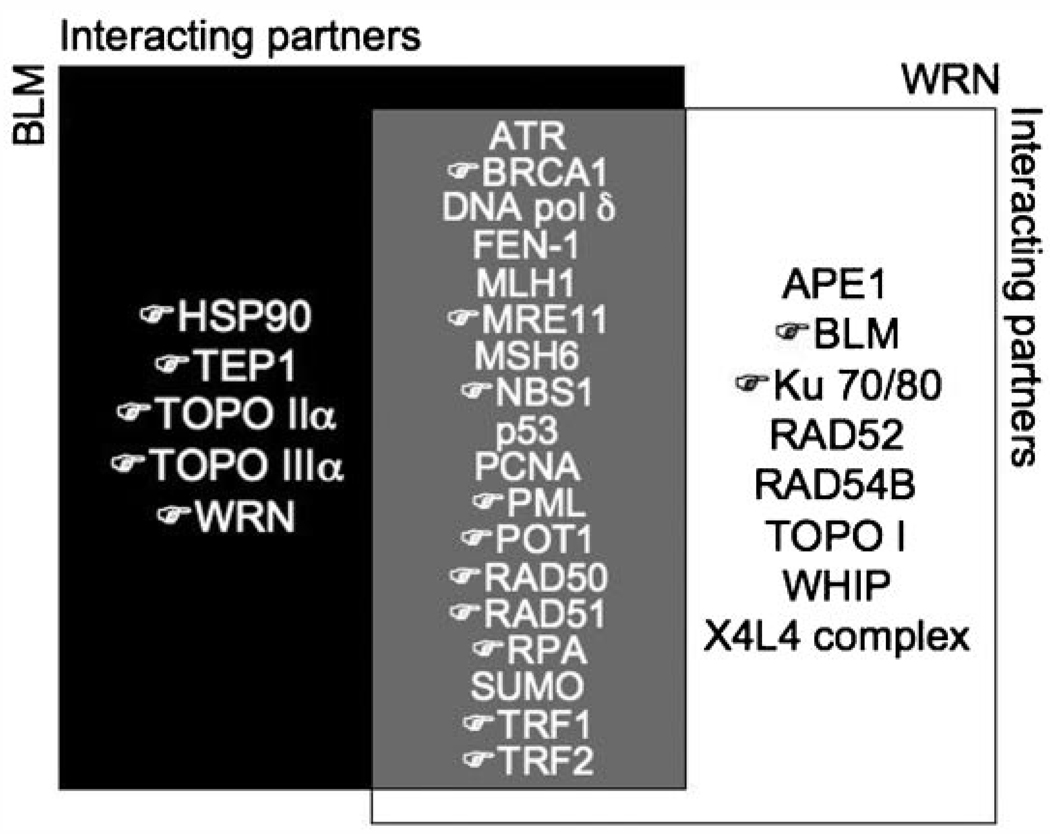
Future studies will identify additional complex members, and determine their effects on BLM or WRN function pertaining to telomere maintenance during ALT. Certainly, the identification of ALT-specific BLM-interacting proteins and subsequent structure–function analyses will continue to define the specifics of ALT mechanisms. Recent studies [Fan et al., 2009; Saharia and Stewart, 2009] have demonstrated that other previously unrecognized proteins, such as FANCD2 and FEN1, are also important for ALT mechanisms. It remains to be seen whether these proteins are present in the BLM-associated ALT complexes, and if so how they may affect BLM or WRN function at telomeres (Fig. 3).
Fig. 3.
Proteins that interact with BLM and/or WRN. Black region indicates proteins that interact with BLM only; while white region depicts proteins that interact with WRN only. Gray region in the center shows overlapping interacting proteins between BLM and WRN. (☞ ) indicates proteins known to be associated with telomeric DNA.
ACKNOWLEDGMENTS
We thank Groden lab members for their scientific contributions to the BLM and ALT research projects. This work was supported by NIH award CA-117898 and The Bloom’s Syndrome Foundation.
REFERENCES
- Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Goodwin EH. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 2004;32:3743–3751. doi: 10.1093/nar/gkh691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baynton K, Otterlei M, Bjoras M, von Kobbe C, Bohr VA, Seeberg E. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–36486. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF. Functional link between BLM defective in Bloom’s syndrome and the ataxia-telangiectasia mutated protein, ATM. J Biol Chem. 2002;28:28. doi: 10.1074/jbc.M203801200. [DOI] [PubMed] [Google Scholar]
- Bechter OE, Zou Y, Shay JW, Wright WE. Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep. 2003;4:1138–1143. doi: 10.1038/sj.embor.7400027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechter OE, Shay JW, Wright WE. The frequency of homologous recombination in human ALT cells. Cell Cycle. 2004;3:547–549. [PubMed] [Google Scholar]
- Bhattacharyya S, Keirsey J, Russell B, Kavecansky J, Lillard-Wetherell K, Tahmaseb K, Turchi JJ, Groden J. Telomerase-associated protein 1, HSP90, and topoisomerase IIalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem. 2009;284:14966–14977. doi: 10.1074/jbc.M900195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitro immortalised human cells. Eur J Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Chaganti RSK, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: Structure, function, and mechanism. Ann Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Davalos AR, Campisi J. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol. 2003;162:1197–1209. doi: 10.1083/jcb.200304016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc Natl Acad Sci USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res. 2009;37:1740–1754. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faragher RG, Kill IR, Hunter JA, Pope FM, Tannock C, Shall S. The gene responsible for Werner syndrome may be a cell division “counting” gene. Proc Natl Acad Sci USA. 1993;90:12030–12034. doi: 10.1073/pnas.90.24.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P. Bloom’s syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol. 2002;157:19–30. doi: 10.1083/jcb.200110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K, Martin GM, Monnat RJ. Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- Griffith J, Bianchi A, de Lange T. TRF1 promotes parallel pairing of telomeric tracts in vitro. J Mol Biol. 1998;278:79–88. doi: 10.1006/jmbi.1998.1686. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci. 2000;113:4577–4585. doi: 10.1242/jcs.113.24.4577. [DOI] [PubMed] [Google Scholar]
- Hande MP, Samper E, Lansdorp P, Blasco MA. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and HSP90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: Substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of Bloom syndrome protein with topoisomerase III alpha in somatic and meiotic cells. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer VA, Stephen AG, Harrington L, Robinson MO, Rome LH. Vaults and telomerase share a common subunit, TEP1. J Biol Chem. 1999;274:32712–32717. doi: 10.1074/jbc.274.46.32712. [DOI] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: Biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Bebenek K, Kunkel TA, Groden J. The Bloom’s syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem. 2001;276:30031–30035. doi: 10.1074/jbc.M009664200. [DOI] [PubMed] [Google Scholar]
- Laud PR, Multani AS, Bailey SM, Wu L, Ma J, Kingsley C, Lebel M, Pathak S, DePinho RA, Chang S. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Harrison RJ, Reszka AP, Brosh RM, Jr, Bohr VA, Neidle S, Hickson ID. Inhibition of the Bloom’s and Werner’s syndrome helicases by G quadruplex-interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- Liu Y, Snow BE, Hande MP, Baerlocher G, Kickhoefer VA, Yeung D, Wakeham A, Itie A, Siderovski DP, Landorp PM, et al. Telomerase-associated protein TEP1 is not essential for telomerase activity or telomere length maintenance in vivo. Mol Cell Biol. 2000;20:8178–8184. doi: 10.1128/mcb.20.21.8178-8184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: A DNA structure-specific ‘dissolvasome’? Trends Biochem Sci. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Marciniak RA, Cavazos D, Montellano R, Chen Q, Guarente L, Johnson FB. A novel telomere structure in a human alternative lengthening of telomeres cell line. Cancer Res. 2005;65:2730–2737. doi: 10.1158/0008-5472.CAN-04-2888. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E, Argani P, Hicks JL, DeMarzo AM, Meeker AK. Telomere lengths of translocation-associated and nontranslocation-associated sarcomas differ dramatically. Am J Pathol. 2004;164:1523–1529. doi: 10.1016/S0002-9440(10)63710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida H, Shinkai Y, Hande MP, Matsumoto T, Takehara S, Tachibana M, Oshimura M, Lansdorp PM, Furuichi Y. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: Characterization of an amplified telomeric DNA. Mol Cell Biol. 2000;20:4115–4127. doi: 10.1128/mcb.20.11.4115-4127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Ishikawa F. Telomere bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- Park KH, Rha SY, Kim CH, Kim TS, Yoo NC, Kim JH, Roh JK, Noh SH, Min JS, Lee KS, Kim BS, Chung HC. Telomerase activity and telomere lengths in various cell lines: Changes of telomerase activity can be another method for chemosensitivity evaluation. Int J Oncol. 1998;13:489–495. doi: 10.3892/ijo.13.3.489. [DOI] [PubMed] [Google Scholar]
- Pedrazzi G, Perrera C, Blaser H, Kuster P, Marra G, Davies SL, Ryu GH, Freire R, Hickson ID, Jiricny J, Stagljar I. Direct association of Bloom’s syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001;29:4378–4386. doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent Holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- Russell B, Keirsey J, Kavecansky J, Sandy A, Grierson P, Acharya S, Groden J. CHK1 phosphorylation of the BLM helicase regulates its interaction with topoisomerase IIα to prevent chromosome breakage. 2009 (in press) [Google Scholar]
- Saharia A, Stewart SA. FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene. 2009;28:1162–1167. doi: 10.1038/onc.2008.458. [DOI] [PubMed] [Google Scholar]
- Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the Bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakain VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–754. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- Tahara H, Tokutake Y, Maeda S, Kataoka H, Watanabe T, Satoh M, Matsumoto T, Sugawara M, Ide T, Goto M, Furuichi Y, Sugimoto M. Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner’s syndrome patients transformed by Epstein-Barr virus. Oncogene. 1997;15:1911–1920. doi: 10.1038/sj.onc.1201377. [DOI] [PubMed] [Google Scholar]
- Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis M, Londono-Vallejo A, Gilson E, Amor-Gueret M, Riou JF. Topoisomerase IIIα is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J. 2008;27:1523–1524. doi: 10.1038/emboj.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutake Y, Matsumoto T, Watanabe T, Maeda S, Tahara H, Sakamoto S, Niida H, Sugimoto M, Ide T, Furuichi Y. Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem Biophys Res Commun. 1998;247:765–772. doi: 10.1006/bbrc.1998.8876. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, physical and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- Walmsley RW, Chan CS, Tye BK, Petes TD. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated protein involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lee WH, Chen PL. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem. 2000a;275:30618–30622. doi: 10.1074/jbc.C000390200. [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouie H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000b;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci USA. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankiwski V, Noonan JP, Neff NF. The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biol. 2001;2:11. doi: 10.1186/1471-2121-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- Zhong S, Hu P, Ye TZ, Stan R, Ellis NA, Pandolfi PP. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]