Abstract
Purpose
In cases of multiple non-small cell lung cancer (NSCLC), clinicians must decide whether patients have independent tumors or metastases and tailor treatment accordingly. Decisions are currently made using the Martini and Melamed criteria, which are mostly based on tumor location and histological type. New genomic tools could improve the ability to assess tumor clonality.
Experimental Design
We obtained fresh-frozen tumors specimens from patients who underwent surgery on at least two occasions for presumptively independent NSCLC. We performed array comparative genomic hybridization (aCGH), mutational profiling of select genes, and detailed clinico-pathological review.
Results
We analyzed a total of 42 tumors from 20 patients (6 patients with synchronous tumors; 14 patients with metachronous tumors; 24 potential tumor pair comparisons); 22 tumor pairs were evaluable by aCGH. Surprisingly, classification based upon genomic profiling contradicted the clinico-pathologic diagnosis in 4 (18%) of the comparisons, identifying independent primaries in one case diagnosed as metastasis and metastases in 3 cases diagnosed as independent primaries. Matching somatic point mutations were observed in these latter 3 cases. Another 4 tumor pairings were assigned an “equivocal” result based on aCGH; however, matching somatic point mutations were also found in these tumor pairs. None of the tumor pairs deemed independent primaries by aCGH harbored matching mutations.
Conclusion
Genomic analysis can help distinguish clonal tumors from independent primaries. The development of rapid, inexpensive and reliable molecular tools may allow for refinement of clinico-pathologic criteria currently used in this setting.
Keywords: Non-small cell lung cancer, synchronous, metachronous, multiple primary lung cancers, array comparative genomic hybridization, mutational profiling
Introduction
Lung cancer is the leading cause of cancer-related death worldwide [1]. For patients with localized non-small cell lung cancers (NSCLCs), surgery remains the best curative option [2]. However, when two lung lesions are found, clinicians must decide whether patients have independent tumors or metastases and tailor treatment accordingly. With the rise in use of computed tomography scans of the chest for screening or surveillance, the identification of multiple primary lung cancers is becoming an increasingly common clinical problem [3].
In the 1970s, Martini and Melamed developed a set of clinical and pathological guidelines for the diagnosis of multiple primary lung cancer (Supplemental Table 1). The criteria rely upon tumor location and histology [4]. These do not incorporate any molecular biological tools [5]. Some studies have compared mutational and/or molecular profiles of NSCLC at candidate markers for this purpose [6–8], but only a few have actually correlated the molecular findings in the context of the Martini and Melamed criteria [9–11]. Importantly, these studies have analyzed only a limited number of genetic markers, theoretically not enough to distinguish independent clones from metastatic foci.
In this study, we performed a comprehensive molecular and clinico-pathological analysis of 20 patients originally presumed to have multiple primary lung cancers. Tumors were analyzed by array comparative genomic hybridization (aCGH), a method that allows identification of copy number changes across the genome [12]. By identifying precise regions of allelic gains and losses, aCGH has the potential to establish whether tumors are independent (i.e. lack matching gains/losses) or clonal (i.e. contain matching gains/losses) with very high confidence [13]. Tumors were also profiled for the presence of multiple somatic mutations commonly found in lung cancer.
Patients and Methods
Tissue procurement
Tumor specimens from patients with lung cancer who underwent surgical resection at Memorial Sloan-Kettering Cancer Center (MSKCC) from January 1999 to December 2007 were obtained with patients’ consent under institutional review board (IRB)-approved protocols (#92-055 and #06-107). During the study period, a total of 3846 NSCLC tumors were resected; frozen specimens from 1508 (39%) tumors were collected either in the operating room or in the Department of Pathology and stored at −80°C in institutional tumor banks. We included only those who received an operation for more than one NSCLC and who had available individual tumor specimens with >70% tumor content from at least two tumors banked separately. At the time of diagnosis, these patients were considered to have multiple primaries on a clinico-pathologic basis.
Clinical and pathological data review
To differentiate multiple primary NSCLCs from metastatic NSCLC, we used the clinico-pathological criteria established by Martini and Melamed (Supplemental Table 1) [4]. All available pathological materials were reviewed by an expert pathologist (W.D.T.) for tumor classification [14]. Disease status was staged according to the American Joint Committee on Cancer staging system for NSCLC [15].
Verification of the identity of paired samples
DNA was extracted from tissue specimens using a kit (DNeasy, Qiagen, Valencia, CA) or standard phenol extraction. In all cases, the banked tumors from individual patients were verified to belong to the same patient by genotyping of specimens for 23 highly informative single-nucleotide polymorphisms (SNPs) using mass spectrometry-based genotyping [16].
Genomic profiling
DNA was hybridized to Agilent 244K comparative genomic hybridization (CGH) arrays (Agilent Technologies, Santa Clara, CA) (See Supplemental methods). The resulting arrays consisted of markers that estimate copy number at each of 244K locations across the genome. The patterns of gains and losses for each pair of tumors were compared using statistical methodology developed specifically for this purpose.* The method calculates a measure that represents the relative chances that at least one of the observed concordant mutations is of clonal origin. Closely matching allelic changes contribute to a high value of this measure. These measures are then calibrated for the patients using a reference histogram based on the pairwise comparisons of all pairs of tumors that come from different patients in the study (which are definitively independent). The individual patients’ tumors are then classified as clonal metastases if the measure exceeds the upper limit of this reference histogram, and as independent if the measure falls below the 95th percentile of the reference distribution, with the intermediate results being considered “equivocal”.
Mutational profiling
Mutational profiling was performed using mass spectrometry-based genotyping (Sequenom, San Diego, CA) [17]. All tumor samples were analyzed for a total of 101 distinct activating mutations in 9 genes encoding components of the EGFR signaling pathway: EGFR, KRAS, HRAS, NRAS, BRAF, PIK3CA, AKT1, ERBB2, and MEK1 (Supplemental Table 2). Because the corresponding mutant protein products display gain of function and are often mutually exclusive [18, 19], these genetic alterations are considered to be ‘driver’ mutations required for both tumor initiation and maintenance. In addition, EGFR exon 19 and all exons encoding the tumor suppressor, P53, were analyzed utilizing exon-specific PCR amplification followed by dideoxynucleotide-based sequencing (Supplemental Methods).
Results
Patient characteristics
Between 1999 and 2007, 559 (14.5%) of 3846 patients underwent operations for multiple NSCLC; 175 had synchronous tumors, and 384 had metachronous tumors (Supplemental Figure 1). In total, 42 tumor samples were available from 20 patients, including 18 patients with 2 different tumors, and 2 individuals with 3 different tumors. In total, there were 24 possible tumor pair comparisons (8 in synchronous tumors and 16 in metachronous tumors).
Clinical characteristics of the 20 patients are listed in Table 1. Patients with synchronous tumors had bilateral disease; all but one had no local or regional lymph node invasion. Metachronous tumors occurred in the ipsilateral lung in 5 cases (same lobe: 1 case; different lobe: 4 cases), and in the contralateral lung in 9 cases; all but one tumor showed no lymph node involvement. The histologic type was similar in matching tumors for 19 pairs, and different for 5 pairs (Table 1).
Table 1.
Patient characteristics
| A. Synchronous multiple NSCLC | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Time interval (months) |
Size (cm) |
Tumor A | Stage | Size (cm) |
Tumor B | Stage | Size (cm) |
Tumor C | Stage | Martini Melamed criteria |
|||
| Location | Histology | Location | Histology | Location | Histology | |||||||||
| 1 | 0 | 1.0 | LLL | ADC | pT1N0 | 7.0 | RLL | ADC | pT2N0 | n/a | n/a | n/a | n/a | multiple primary |
| 2 | 0 | 1.5 | LLL | ADC | pT1N0 | 1.5 | RUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 3 | 0 | 1.8 | RUL | ADC | pT1N0 | 1.8 | LUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 4 | 0 | 1.7 | RLL | ADC | pT1N0 | 1.4 | LUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 5 | 0 | 2.0 | LLL | ADC | pT1N0 | 2.0 | LUL | LCNEC | pT1N0 | 2.5 | RUL | ADC | pT3N1 | multiple primary |
| 6 | 0 | 4.5 | RUL | ADC | pT2N0 | 4.5 | LLL | ADC | pT2N0 | n/a | n/a | n/a | n/a | metastases |
| B. Metachronous multiple NSCLC | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Time interval (months) |
Size (cm) |
Tumor A | Stage | Size (cm) |
Tumor B | Stage | Size (cm) |
Tumor C | Stage | Martini Melamed criteria |
|||
| Location | Histology | Location | Histology | Location | Histology | |||||||||
| 7 | 14.1 | 8.5 | LLL | ADC | pT2N0 | 3.0 | RML | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 8 | 55.1 | 3.5 | LUL | ADC | pT2N0 | 1.2 | RUL | LCNEC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 9 | 41.2 | 5.0 | RLL | ADC | pT2N0 | 3.8 | RUL | ADC | pT2N0 | n/a | n/a | n/a | n/a | multiple primary |
| 10 | 25.1 | 5.0 | LUL | SCC | pT3N0 | 2.0 | RLL | SCC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 11 | 42.7 | 3.1 | LUL | ADC | pT2N0 | 0.8 | RLL | ADC | pT2N0 | n/a | n/a | n/a | n/a | multiple primary |
| 12 | 23.0 | 1.6 | RUL | ADC | pT2N0 | 1.6 | LUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 13 | 18.9 | 7.5 | RLL | ADC | pT2N0 | 3.2 | RUL | ADC | pT2N0 | n/a | n/a | n/a | n/a | multiple primary |
| 14 | 18.6 | 2.2 | RUL | ADC | pT1N0 | 1.7 | LUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 15 | 50.2 | 1.5 | LUL | SCC | pT1N0 | 2.5 | RUL | SCC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 16 | 53.6 | 5.5 | LLL | SCC | pT2N0 | 1.8 | LUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 17 | 33.3 | 3.0 | LUL | SCC | pT2N0 | 1.0 | RUL | ADC | pT1N0 | n/a | n/a | n/a | n/a | multiple primary |
| 18 | 72.2 | 2.5 | LUL | ADC | pT1N0 | 2.9 | RLL | ADC | pT2N2 | n/a | n/a | n/a | n/a | multiple primary |
| 19 | 13.5 | 1.7 | LLL | SCC | pT1N0 | 2.1 | LLL | SCC | pT1N0 | n/a | n/a | n/a | n/a | metastases |
| 20 | 6.6*/11.5#/ | 4.0 | LLL | ADC | pT2N0 | 2.3 | LUL | ADC | pT1N0 | 3.2 | LUL | ADC | pT2N0 | multiple primary*,+/ |
| 18.1+ | metastases# | |||||||||||||
No patient presented with systemic metastases. Stage was assigned as per reference 15.
Legend: RUL: Right Upper Lobe; RLL: Right Lower Lobe; RML: Right Middle Lobe; LUL: Left Lower Lobe; LLL: Left Lower Lobe; ADC: adenocarcinoma; LCNEC: Large-Cell Neuro-Endocrine Carcinoma; n/a: not applicable.
No patient presented with systemic metastases. Stage was assigned as per reference 15.
Legend: RUL: Right Upper Lobe; RLL: Right Lower Lobe; RML: Right Middle Lobe; LUL: Left Lower Lobe; LLL: Left Lower Lobe; ADC: adenocarcinoma; SCC: Squamous Cell Carcinoma; LCNEC: Large-Cell Neuro-Endocrine Carcinoma; n/a: not applicable;
Tumor A vs. Tumor B
Tumor B vs. Tumor C
Tumor A vs. C.
Martini and Melamed characterization
Using the criteria established by Martini and Melamed (Supplemental Table 1), matching tumors were characterized as multiple primary NSCLCs in 7 of 8 synchronous tumor pairs and in 14 of 16 metachronous tumor pairs (Table 1). The remaining cases were classified as intrapulmonary metastases.
Genomic profiling
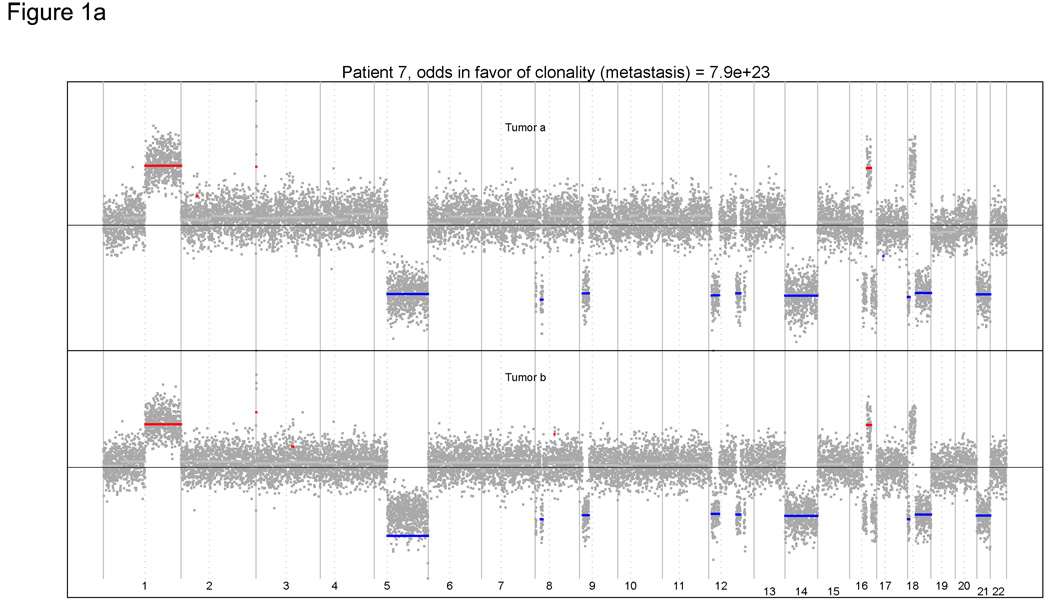
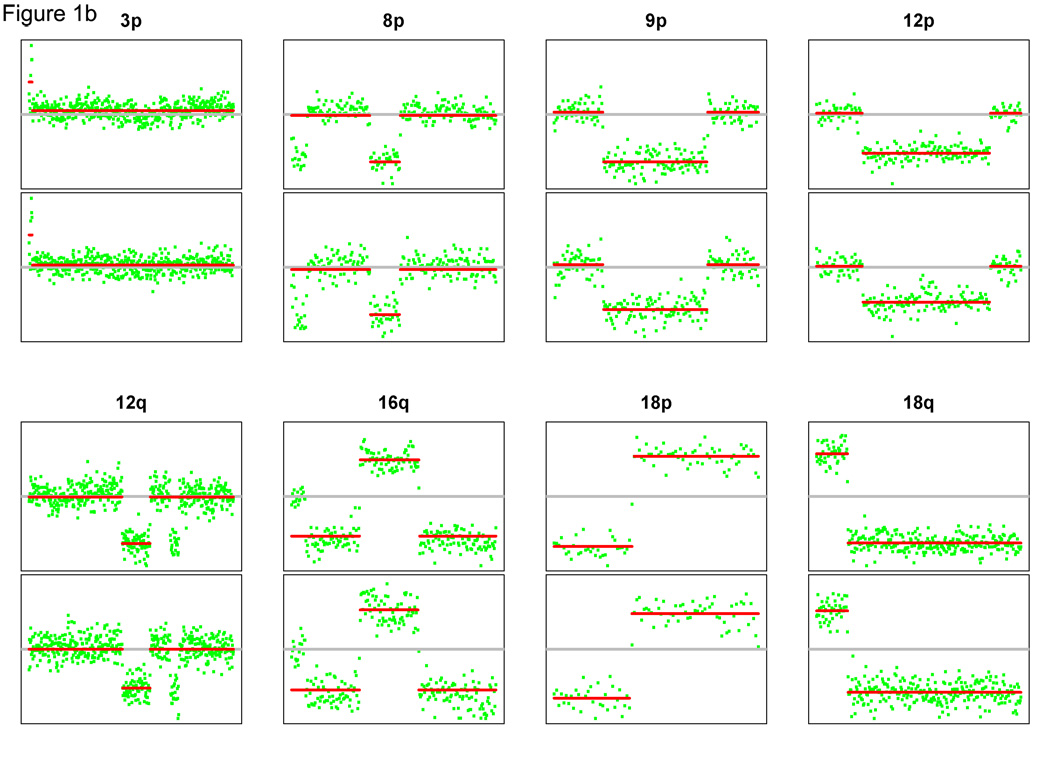
We performed aCGH on all 42 tumor samples, but had to eliminate two cases from evaluation because of poor array quality. Thus, 22 tumor pairs were evaluable by aCGH. Case 7 illustrates the power of this approach. The tumors were considered to be independent multiple primaries by clinical criteria (Table 1), but the genomic profiles were remarkably similar (Figure 1A). Both tumors displayed a matching amplification of the whole arm on 1q and matching whole arm losses on 5q, 14q and 21q. More strikingly, both tumors harbored within-arm gains and losses with plausibly identical start and stop locations. For example, closely matching allelic changes were identified on 8 separate chromosome arms (Figure 1B). Conversely, case 4 is an example where the patterns of gains and losses were seemingly random (Figure 2). These results suggest that the tumors arose independently and were thus multiple primaries.
Figure 1.
(A) Genomic profiles of multiple tumors from case 7. Both tumors displayed matching amplification and losses. (B): Closely matching allelic changes for 8 chromosome arms. The top and bottom panels represent the aCGH array segmentation for tumor A and B, respectively. Paired tumors exhibit similar genomic alterations, suggesting the diagnosis of metastases.
Figure 2.
Genomic profiles of multiple tumors from case 4. Profiles are different, suggesting the diagnosis of multiple primaries.
Overall, among the 22 paired comparisons, genomic profiling led to a diagnosis of metastases and multiple primaries in 4 and 14 cases, respectively. The conclusion was equivocal in the remaining 4 comparisons. Plots of allelic changes for all cases are displayed in Supplementary Figure 2, and the reference histograms are displayed in Supplemental Figure 3.
Mutational profiling
Results of mutational profiling are listed in Table 2. Matching point mutations were observed in 2 synchronous tumor pairs and in 6 metachronous pairs (including a matching KRAS mutation for all 3 tumors of case 20). Discordant mutations were observed in 9 paired tumors. Interestingly, the matched mutations occurred in the 4 paired tumors that were classified as metastases by genomic profiling (cases 6, 7, 9 and 13) and also in the 4 “equivocal” cases (comparison of tumors A and C for case 5 and all 3 pairings of case 20). Conversely, there were no matching mutations in any of the pairings diagnosed by genomic profiling as multiple primaries. Collectively, these data suggest that tumor mutation status is highly concordant with results from aCGH studies. They also suggest that our pre-determined methodological strategy for determining an “equivocal” region of diagnostic uncertainty may be inappropriately conservative in failing to classify cases as clonally related.
Table 2.
Mutational profiling of synchronous and metachronous multiple non-small cell lung cancer
| A. Synchronous multiple NSCLC | ||||||
|---|---|---|---|---|---|---|
| Case | Tumor | EGFR | KRAS | BRAF | PIK3CA | TP53 |
| 1 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 2 | A | wt | wt | wt | wt | wt |
| B | L858R | wt | wt | wt | wt | |
| 3 | A | wt | wt | wt | wt | wt |
| B | wt | G12D | wt | wt | wt | |
| 4 | A | wt | wt | wt | wt | C277F |
| B | L858R | wt | wt | wt | R273H | |
| 5# | A | wt | G12C | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| C | wt | G12C | wt | wt | wt | |
| 6# | A | wt | wt | V600E | wt | wt |
| B | wt | wt | V600E | wt | wt | |
| B. Metachronous multiple NSCLC | ||||||
|---|---|---|---|---|---|---|
| Case | Tumor | EGFR | KRAS | BRAF | PIK3CA | TP53 |
| 7# | A | wt | G12V | wt | wt | wt |
| B | wt | G12V | wt | wt | wt | |
| 8 | A | wt | G12F | wt | wt | Q38* |
| B | wt | wt | wt | wt | wt | |
| 9# | A | wt | G12D | wt | wt | wt |
| B | wt | G12D | wt | wt | wt | |
| 10 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 11 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 12 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 13# | A | wt | G12D | wt | wt | wt |
| B | wt | G12D | wt | wt | wt | |
| 14 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | H1047R | P71P | |
| 15 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 16 | A | wt | wt | wt | wt | wt |
| B | wt | G12D | wt | wt | wt | |
| 17 | A | wt | wt | wt | wt | R267P |
| B | wt | wt | wt | wt | wt | |
| 18 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 19 | A | wt | wt | wt | wt | wt |
| B | wt | wt | wt | wt | wt | |
| 20# | A | wt | G12D | wt | wt | wt |
| B | wt | G12D | wt | wt | wt | |
| C | wt | G12D | wt | wt | wt | |
HRAS, NRAS, AKT1, ERBB2, and MEK1 were wild-type (wt) in all tumors.
tumors with similar mutations.
HRAS, NRAS, AKT1, ERBB2, and MEK1 were wild-type (wt) in all tumors.
tumors with similar mutations;
truncation.
Comparison of genomic and mutational profiling with clinico-pathological criteria
The results from all of the classification schemes are summarized in Table 3. The genomic classification contradicted the clinical diagnosis in 4 (18%) of 22 comparisons, identifying clonal patterns (metastases) in 3 (16%) cases diagnosed as independent primaries (cases 7, 9 and 13), and no evidence of clonality in 1 case diagnosed clinically as a metastasis (case 19). If the equivocal cases are considered to be a tentative diagnosis of metastases, then the genomic approach contradicted the clinical diagnosis in 7 of 22 comparisons (32%).
Table 3.
Integration of genomic and mutational profiling with clinico-pathological characterization (using the Martini-Melamed criteria).
| A. Synchronous multiple NSCLC | |||
|---|---|---|---|
| Case | Genomic profiling | Clinical evaluation | Mutational profiling |
| 1 | multiple primary | multiple primary | n/a |
| 2 | multiple primary | multiple primary | different |
| 3 | multiple primary | multiple primary | different |
| 4 | multiple primary | multiple primary | different |
| 5 (A vs. B) | multiple primary | multiple primary | different |
| 5 (A vs. C) | equivocal | multiple primary | same |
| 5 (B vs. C) | multiple primary | multiple primary | different |
| 6 | metastases | metastases | same |
| B. Metachronous multiple NSCLC | |||
|---|---|---|---|
| Case | Genomic profiling | Clinical evaluation | Mutational profiling |
| 7* | metastases | multiple primary | same |
| 8 | multiple primary | multiple primary | different |
| 9* | metastases | multiple primary | same |
| 10 | multiple primary | multiple primary | n/a |
| 11 | --** | multiple primary | n/a |
| 12 | multiple primary | multiple primary | n/a |
| 13* | metastases | multiple primary | same |
| 14 | multiple primary | multiple primary | different |
| 15 | multiple primary | multiple primary | n/a |
| 16 | multiple primary | multiple primary | different |
| 17 | multiple primary | multiple primary | different |
| 18 | --** | multiple primary | n/a |
| 19* | multiple primary | metastases | n/a |
| 20 (A vs. B) | equivocal | multiple primary | same |
| 20 (B vs. C) | equivocal | metastases | same |
| 20 (A vs. C) | equivocal | multiple primary | same |
n/a: not applicable.
Indicates discrepant results between genomic profiling and clinical evaluation; n/a: not applicable.
We could not reach a genomic diagnosis in these two cases due to an excessively high level of noise in one of the paired arrays.
In metachronous multiple primary cases that were misclassified, the size of the first resected tumor was generally larger than the corresponding tumor in cases that were correctly classified (means 7.0 cm versus 3.2 cm, p = 0.04, Wilcoxon-Mann-Whitney Test, two-sided). Similarly, the misclassified cases had somewhat shorter intervals from first to second primary diagnosis, though this difference was not significant (means 25 mos. versus 37 mos. p = 0.21, Wilcoxon-Mann-Whitney Test).
Discussion
Emerging data indicate that molecular analysis of patterns of somatic changes within tumor DNA has the potential to improve diagnostic classifications of second tumors as either metastases or independent second primaries. Thus far, studies have focused primarily on cancer sites where second primaries are common, such as those involving the bladder, head and neck, and breast. The typical strategy has been to genotype markers at candidate loci where somatic mutations are known to occur commonly in the tumor under investigation. More recently, the use of aCGH has been applied to this problem, in recognition of the attractiveness of using a comprehensive genome-wide strategy [20, 21], especially in breast tumors [9, 13, 22–26]. Studies of lung cancer using this technology have been rare and not directly targeted at the classification of multiple lung tumors [27, 28]. Our goal in this study was to apply aCGH and mutational profiling to the problem of multiple primary NSCLCs to determine their usefulness in assessing clonality of tumors.
Our results provide strong evidence that genomic profiling can help distinguish clonal tumors from independent primaries. However, the application of aCGH in normal clinical practice would be challenging. First, the need for fresh frozen tissue would make the technique more immediately applicable for evaluating synchronous tumors. For metachronous tumors, frozen tissues would need to be stored and accessible in an easy manner. Patients who have tumors resected at different institutions would require additional coordination for genomic profiling. Second, standardized software for routine automated statistical comparisons of the array data would need to be widely available, although software for the application of the methods in this article is available from the authors. Third, aCGH is expensive and can be a time-consuming and labor-intensive procedure. Fourth, aCGH requires relatively large amounts of DNA and array quality can be variable, depending upon differences in tumor content, tissue handling, specimen storage, and specimen processing. For example, despite using fresh-frozen tumor samples in an experimental (non-clinical) setting, we found arrays from 2 of the 42 tumors to be of insufficient quality to reach a diagnosis with confidence. The requirement for abundant DNA also may preclude such analysis on DNA derived from needle biopsies prior to surgery, where such results may actually be more useful in deciding whether patients should undergo surgery in the first place.
A more practical solution may be to develop strategies based upon mutational profiling that can be performed using DNA extracted from formalin fixed, paraffin embedded tissue rather than frozen samples. Although mutational profiling is less comprehensive than aCGH, it may yet have sufficient sensitivity for improved diagnoses, as evidenced by the strong concordance of the genomic and mutational profiling results in this study. The strength of the evidence from individual mutational matches depends on the rarity of the mutation. For example, three of our matching cases involved matching KRAS G12V mutations. This specific KRAS alteration occurs at a frequency of 4.4% in lung adenocarcinomas [29]. Thus, the probability of a match in independent tumors, given that the first tumor has the mutation, is 0.044. Viewed in this context, the simultaneous independent occurrence of this mutation in all 3 tumors for case 20 is an unlikely event with a probability of 0.002 (0.0442), providing further evidence that these three tumors are of clonal origin. The corresponding population frequencies of the other matches in our study have been estimated to be 5.2% for KRAS G12D and 1.2% for BRAF V600E [30].
Conversely, mutational profiling is limited in practice by the infrequency of the occurrences of specific mutations. Indeed, for the panel of markers in our study, none of the mutations were observed for 20 of the 42 tumors, and none were observed in either tumor for 7 of the 20 patients. Mutational profiling would be more powerful if assays could detect all known loci that are commonly mutated at the somatic level in NSCLC. A panel of about 20 commonly occurring markers would have good statistical power for detecting clonal tumors, provided that the preponderance of the somatic events observed occur in the originating clonal cell [31]. Additionally, mutational profiling would allow the use of partially-fragmented DNA, such as can be obtained from paraffin-embedded tissue, or of whole-genome-amplified DNA, when starting tumor DNA quantities are limited (i.e. derived from bronchoscopic or needle biopsies). Importantly, one of us (ML) has demonstrated that similar results are obtained from mutation testing of EGFR and KRAS using matching formalin fixed and frozen specimens (M Ladanyi, unpublished data).
Our results suggest surprisingly that diagnostic misclassifications may occur frequently when using the Martini-Melamed guidelines with histologic classification according to traditional WHO criteria. Out of 22 evaluable comparisons of tumor pairs, the genomic classification contradicted the clinical diagnoses in 4 (18%), identifying clonal patterns of metastasis (as in the case highlighted in Figure 1) in 3 cases diagnosed as independent primaries. Based upon our statistical algorithm, it is highly unlikely that such identical or near-identical allelic changes would have occurred in the tumors independently by chance. In another 3 cases deemed multiple primaries by clinical criteria but diagnosed as equivocal by aCGH, the identification of matching somatic point mutations by mutational profiling suggested that the tumors were of clonal origin. Thus, potentially up to 7 of the 22 cases (32%) were misclassified by the traditional clinical/histologic criteria alone.
We recognize that we cannot state with absolute certainty that the diagnoses based on genomic profiling are “correct” and that the clinical diagnoses were “wrong”. However, the kinds of precise matching of regions of allelic losses and gains that we observed seem compelling as a definitive basis for diagnosis from a logical standpoint. Prospective studies with much larger sample sizes will be necessary to validate the usefulness of genomic analyses in relation to patient survival. We further note that this study was based upon a highly selected, small sample size. To be included for study, the metachronous patients all had to survive long enough to develop multiple primaries and undergo resection.
Distinguishing multiple primary tumors from metastases may actually not change the initial management of these patients, as surgery has been shown to increase survival in these instances [4, 32]. However, in the 3 cases initially considered as independent primaries, reclassification as metastases using genomic profiling could have led to consideration of adjuvant chemotherapy and/or to a closer surveillance monitoring plan. Conversely, patients found to have multiple primaries rather than metastases could be spared from adjuvant chemotherapy.
In summary, our results suggest that genome-wide examination of copy number changes using aCGH and mutational profiling of multiple loci can help distinguish multiple independent primaries from metastases. Molecular diagnostics such as these should be honed for practical application in the clinic.
STATEMENT OF TRANSLATIONAL RELEVANCE
The incidence of multiple primary lung cancers is rising. When patients are found to have multiple primary lung lesions, clinicians must decide whether they have independent tumors or metastases and tailor treatment accordingly. Decisions are currently made using clinical and pathological criteria. Here, we performed a comprehensive molecular and clinico-pathological analysis of 20 patients originally presumed to have multiple primary lung cancers. Tumors were analyzed by array comparative genomic hybridization (aCGH) and profiled for the presence of multiple somatic mutations. Surprisingly, classification based upon genomic/mutational profiling contradicted the clinico-pathologic diagnosis in 4 (18%) of the comparisons. Although the study involved limited numbers of samples, the results suggest that clinico-pathologic criteria used to distinguish multiple primaries from metastases could be further refined by the routine use of molecular profiling.
Supplementary Material
Acknowledgments
We thank Mark Kris for critical reading of the manuscript; MSKCC Tumor Procurement Service, Benjamin Golas and Bhuvanesh Singh for their assistance in the collection of samples; Agnes Viale from MSKCC Genomics Core Laboratory; Adriana Heguy from the Beene Translational Oncology Core Facility.
Financial support: This study was supported by the National Cancer Institute (CA124504; CBB), the HOPP Lung Cancer Research Fund (WP), the Rosalind Warren Memorial Fund (WP), and the MSKCC Geoffrey Beene Cancer Research Center (WP). Nicolas Girard is a recipient of travel grants from the Philippe Foundation, the College des Professeurs de Pneumologie (CEP)/AstraZeneca, and from the National Federation of French Comprehensive Cancer Centers/Fondation de France.
Footnotes
References
- 1.American Cancer Society: Cancer Facts and Figures 2007. Atlanta, GA: American Cancer Society; 2007
- 2.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 3.Vazquez M, Carter D, Brambilla E, et al. Solitary and multiple resected adenocarcinomas after CT screening for lung cancer: Histopathologic features and their prognostic implications. Lung Cancer. 2009 doi: 10.1016/j.lungcan.2008.08.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg. 1975;70:606–612. [PubMed] [Google Scholar]
- 5.Braakhuis BJ, Tabor MP, Leemans CR, van der Waal I, Snow GB, Brakenhoff RH. Second primary tumors and field cancerization in oral and oropharyngeal cancer: molecular techniques provide new insights and definitions. Head Neck. 2002;24:198–206. doi: 10.1002/hed.10042. [DOI] [PubMed] [Google Scholar]
- 6.Shin SW, Breathnach OS, Linnoila RI, et al. Genetic changes in contralateral bronchioloalveolar carcinomas of the lung. Oncology. 2001;60:81–87. doi: 10.1159/000055301. [DOI] [PubMed] [Google Scholar]
- 7.Dacic S, Ionescu DN, Finkelstein S, Yousem SA. Patterns of allelic loss of synchronous adenocarcinomas of the lung. Am J Surg Pathol. 2005;29:897–902. doi: 10.1097/01.pas.0000164367.96379.66. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Wang M, MacLennan GT, et al. Evidence for common clonal origin of multifocal lung cancers. J Natl Cancer Inst. 2009;101:560–570. doi: 10.1093/jnci/djp054. [DOI] [PubMed] [Google Scholar]
- 9.Huang J, Behrens C, Wistuba I, Gazdar AF, Jagirdar J. Molecular analysis of synchronous and metachronous tumors of the lung: impact on management and prognosis. Ann Diagn Pathol. 2001;5:321–329. doi: 10.1053/adpa.2001.29338. [DOI] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Yatabe Y, Koshikawa T, et al. Mutations of the P53 tumor suppressor gene as clonal marker for multiple primary lung cancers. J Thorac Cardiovasc Surg. 1997;114:354–360. doi: 10.1016/S0022-5223(97)70180-6. [DOI] [PubMed] [Google Scholar]
- 11.van Rens MT, Eijken EJ, Elbers JR, Lammers JW, Tilanus MG, Slootweg PJ. p53 mutation analysis for definite diagnosis of multiple primary lung carcinoma. Cancer. 2002;94:188–196. doi: 10.1002/cncr.10001. [DOI] [PubMed] [Google Scholar]
- 12.Cho EK, Tchinda J, Freeman JL, Chung YJ, Cai WW, Lee C. Array-based comparative genomic hybridization and copy number variation in cancer research. Cytogenet Genome Res. 2006;115:262–272. doi: 10.1159/000095923. [DOI] [PubMed] [Google Scholar]
- 13.Bollet MA, Servant N, Neuvial P, et al. High-resolution mapping of DNA breakpoints to define true recurrences among ipsilateral breast cancers. J Natl Cancer Inst. 2008;100:48–58. doi: 10.1093/jnci/djm266. [DOI] [PubMed] [Google Scholar]
- 14.Travis WB, Brambilla A, Muller-Hermelinck HK, et al., editors. WHO histological classification of tumours of the lung. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2004. p. 10. [Google Scholar]
- 15.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 16.Lee JC, Vivanco I, Beroukhim R, et al. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol. 2004;26:147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marks JL, McLellan MD, Zakowski MF, et al. Mutational analysis of EGFR and related signaling pathway genes in lung Adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4. PLoS ONE. 2007;2:e426. doi: 10.1371/journal.pone.0000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss MM, Kuipers EJ, Meuwissen SG, van Diest PJ, Meijer GA. Comparative genomic hybridisation as a supportive tool in diagnostic pathology. J Clin Pathol. 2003;56:522–527. doi: 10.1136/jcp.56.7.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wa CV, DeVries S, Chen YY, Waldman FM, Hwang ES. Clinical application of array-based comparative genomic hybridization to define the relationship between multiple synchronous tumors. Mod Pathol. 2005;18:591–597. doi: 10.1038/modpathol.3800332. [DOI] [PubMed] [Google Scholar]
- 22.Park SC, Hwang UK, Ahn SH, Gong GY, Yoon HS. Genetic changes in bilateral breast cancer by comparative genomic hybridisation. Clin Exp Med. 2007;7:1–5. doi: 10.1007/s10238-007-0123-1. [DOI] [PubMed] [Google Scholar]
- 23.Waldman FM, DeVries S, Chew KL, Moore DH, Kerlikowske K, Ljung BM. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J Natl Cancer Inst. 2000;92:313–320. doi: 10.1093/jnci/92.4.313. [DOI] [PubMed] [Google Scholar]
- 24.Agelopoulos K, Tidow N, Korsching E, et al. Molecular cytogenetic investigations of synchronous bilateral breast cancer. J Clin Pathol. 2003;56:660–665. doi: 10.1136/jcp.56.9.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang ES, Nyante SJ, Yi Chen Y, et al. Clonality of lobular carcinoma in situ and synchronous invasive lobular carcinoma. Cancer. 2004;100:2562–2572. doi: 10.1002/cncr.20273. [DOI] [PubMed] [Google Scholar]
- 26.Torres L, Ribeiro FR, Pandis N, Andersen JA, Heim S, Teixeira MR. Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Res Treat. 2007;102:143–155. doi: 10.1007/s10549-006-9317-6. [DOI] [PubMed] [Google Scholar]
- 27.Gallegos Ruiz MI, van Cruijsen H, Smit EF, et al. Genetic heterogeneity in patients with multiple neoplastic lung lesions: a report of three cases. J Thorac Oncol. 2007;2:12–21. [PubMed] [Google Scholar]
- 28.Knöel T, Schlüns K, Dietel M, Petersen I. Chromosomal alterations in lung metastases of colorectal carcinomas: associations with tissue specific tumor dissemination. Clin Exp Metastasis. 2005;22:533–538. doi: 10.1007/s10585-005-5239-7. [DOI] [PubMed] [Google Scholar]
- 29.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratilas CA, Hanrahan AJ, Halilovic E, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Begg CB, Eng KH, Hummer AJ. Statistical tests for clonality. Biometrics. 2007;63:522–530. doi: 10.1111/j.1541-0420.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliaro A, Filosso PL, Cavallo A, et al. The significance of intrapulmonary metastasis in non-small cell lung cancer: upstaging or downstaging? A re-appraisal for the next TNM staging system. Eur J Cardiothorac Surg. 2008;34:438–443. doi: 10.1016/j.ejcts.2008.03.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.