Abstract
Enoyl-ACP reductases catalyze the final step in the elongation cycle of the bacterial fatty acid biosynthesis (FAS-II) pathway. Currently four distinct enoyl-ACP reductases have been identified, which are the products of the fabI, fabL, fabK and fabV genes. The FabV enoyl-ACP reductase is the most recent member of this enzyme class and was originally identified in Vibrio cholerae by Cronan and coworkers [Massengo-Tiasse and Cronan (2008) Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase, J. Biol. Chem. 283, 1308–1316]. In the present work a detailed kinetic analysis of the mechanism of the FabV enzyme from Burkholderia mallei (bmFabV) has been undertaken, which reveals that bmFabV catalyzes a sequential Bi Bi mechanism with NADH binding first and NAD+ dissociating last. The enzyme is a member of the short chain dehydrogenase/reductase superfamily in which the catalytic tyrosine (Y235) and lysine (K244) residues are organized in the consensus Tyr-(Xaa)8-Lys motif. The role of these active-site residues has been investigated using site-directed mutagenesis which has shown that both Y235 and K244 are involved in acid/base chemistry during substrate reduction. Sequence alignment and site-directed mutagenesis also identify a second lysine in the active site (K245) that has an important role in binding of the enoyl substrate. Due to interests in developing inhibitors of bmFabV, a detailed analysis of the inhibition of the enzyme by triclosan has been conducted showing that triclosan is a competitive inhibitor with respect to NADH and an uncompetitive inhibitor with respect to the substrate 2-dodecenoyl-CoA (Ki = 0.4 µM). Combined with fluorescence binding experiments, it is concluded that triclosan binds to the enzyme-NAD+ product complex which is in rapid and reversible equilibrium with other intermediates on the reaction pathway.
The bacterial fatty acid biosynthesis (FAS-II) pathway is a validated yet relatively unexploited target for antimicrobial development (1–2). Fatty acids are important components of the cell membrane and are essential for bacterial viability (3–4). While eukaryotes synthesize fatty acids using a multi-domain enzyme complex (FAS-I), the FAS-II pathway found in prokaryotes consists of individual enzymes that catalyze each step of fatty chain elongation (5–7). Consequently, the fundamental structural differences that exist between the FAS-I and FAS-II pathways supports the proposal that compounds can be developed that selectively target bacterial fatty acid biosynthesis.
The majority of the FAS-II enzymes are essential for bacterial viability (8) and detailed kinetic studies coupled with high resolution crystal structures have provided a solid foundation for the development of compounds that target this pathway (2, 9). In particular, the enoyl-ACP reductase, which catalyzes the last reaction in each elongation circle, has been the most heavily targeted component of the pathway based on the discovery that antibacterial compounds such as triclosan and isoniazid target this enzyme (10–13). The FabI enoyl-ACP reductase, exemplified by the enzyme from Escherichia coli, was initially considered to be the only reductase in bacteria (14), and extensive efforts have been made to develop inhibitors of this enzyme from organisms such as Mycobacterium tuberculosis and Staphylococcus aureus (15–16). However, interests in targeting FabI for the development of an agent with activity against both S. aureus and Streptococcus pneumonia were reduced by the discovery of FabK, an alternative flavin-dependent enoyl-ACP reductase from S. pneumonia which was insensitive to the lead FabI inhibitor triclosan (17). At around the same time Rock and coworkers also discovered a third enoyl-ACP reductase in Bacillus subtilis (FabL) which was homologous to FabI (18).
Recently, a fourth isoenzyme (FabV) was identified by Cronan’s lab from the Gram-negative bacterium Vibrio cholerae (vcFabV) (19). vcFabV, like FabI and FabL, is a member of the short-chain dehydrogenase/reductase (SDR) superfamily although it is significantly larger than other enzymes in this family. However, there is no convincing sequence homology between FabV and the FabI and FabL enzymes. Subsequent sequence similarity studies revealed that FabV is well conserved among a variety of organisms including several clinically important pathogens, such as Yersinia pestis, Pseudomonas aeruginosa and Burkholderia species (19).
To date, no detailed kinetic study has been conducted on any of the FabV enzymes. Here we report the cloning and expression of bmFabV from Burkholderia mallei, the category B pathogen that causes glanders (20), and demonstrate that this enzyme is a NADH dependent enoyl-ACP reductase. The impact of site-directed mutagenesis on catalytic activity is consistent with the knowledge that bmFabV is a member of the short chain dehydrogenase reductase superfamily. Finally, since Cronan and coworkers have reported that the antibacterial compound triclosan is only a weak inhibitor of vcFabV (19), we have studied in detail the mechanism of bmFabV inhibition by this diphenyl ether.
Materials and Methods
Materials
His-bind Ni2+-NTA resin was purchased from Invitrogen while a QuikChange site-directed mutagenesis kit was obtained from Stratagene. Biotinylated thrombin and streptavidin agarose were from Novagen. Triclosan was a gift from Ciba. trans-2-Dodecenoic acid was purchased from TCI. All other chemical reagents were obtained from Sigma-Aldrich.
Synthesis of trans-2-Dodecenoyl-CoA (DD-CoA) and Lauryl-CoA
DD-CoA and lauryl-CoA were prepared from trans-2-dodecenoic acid and lauric acid, respectively using the mixed anhydride method (21). Product formation was confirmed by ESI mass spectrometry.
Cloning, Expression and Purification of bmFabV
The entire putative fabV gene from Burkholderia mallei ATCC 23344 (NCBI Reference Sequence: YP_102617.1) was amplified using the primers listed in Table 1 and inserted into Novagen pET15b vector using the 5’ NdeI and 3’ BamHI restriction sites (underlined) so that a His-tag was encoded at the N-terminus of the coding sequence. After purification from XL1Blue cells (Stratagene) using a DNA purification and gel extraction kit (Qiagen Inc) the correct sequence of the insert was confirmed using ABI DNA sequencing.
Table 1.
Primers Used for Cloning and Mutagenesis
| Name | Sequence a |
|---|---|
| bmFabV forward | 5’GGAATTCCATATGATCATCAAACCGCGCGTACGC3’ |
| bmFabV reversed | 5’CGCGGATCCTCATTCGATCAGATTCGGAATCCG3’ |
| Y235A forward | 5’GTCACGCACGACATCGCCTGGAACGGCTCGAT3’ |
| Y235A reversed | 5’ATCGAGCCGTTCCAGGCGATGTCGTGCGTGAC3’ |
| Y235S forward | 5’ACGCACGACATCTCCTGGAACGGCT3’ |
| Y235S reversed | 5’AGCCGTTCCAGGAGATGTCGTGCGT3’ |
| K244A forward | 5’ATCGGCGAAGCGGCGAAAGATCTCGACC3’ |
| K244A reversed | 5’GGTCGAGATCTTTCGCCGCTTCGCCGAT3’ |
| K244R forward | 5’GATCGGCGAAGCGAGGAAAGATCTCGACC3’ |
| K244R reversed | 5’GGTCGAGATCTTTCCTCGCTTCGCCGATC3’ |
| K245M forward | 5’CGAAGCGAAGATGGATCTCGACCGC3’ |
| K245M reversed | 5’GCGGTCGAGATCCATCTTCGCTTCG3’ |
| K244A/K245A forward | 5’ATCGGCGAAGCGGCGGCAGATCTCGACCG3’ |
| K244A/K245A reversed | 5’CGGTCGAGATCTGCCGCCGCTTCGCCGAT3’ |
| ftuACP forward | 5’GGAATTCCATATGAGTACACATAACGAAGATTCTAAA3’ |
| ftuACP reversed | 5’CCGCTCGAGACCTACATCTTTAGATTCGATATA-3’ |
Restriction sites and mutated sites are shown in underline.
Protein expression was performed using E. coli BL21(DE3)pLysS cells. After transformation, a single colony was used to inoculate 10 ml of Luria Broth (LB) media containing 0.2 mg/ml ampicillin in a 50 ml falcon tube, which was then incubated overnight at 37°C in a floor shaker. The overnight culture was then used to inoculate 1 l of LB media containing ampicillin (0.2 mg/ml) which was incubated on an orbital shaker 37°C until the optical density at 600 nm (OD600) increased to around 1.0. Protein expression was induced by adding 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) and the culture was then shaken at 25°C for 16 h. Cells were harvested by centrifugation at 5,000 rpm for 25 min at 4 ºC. The cell paste was then resuspended in 30 ml of His-binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris HCl, pH 7.9) and lysed by sonication. Cell debris was removed by centrifugation at 33,000 rpm for 60 min at 4 ºC. The resulting supernatant was loaded onto a His-bind column (1.5 cm × 15 cm) containing 4 ml of His-bind resin (Novagen) that had been charged with 9 ml of charge buffer (Ni2+). The column was washed with 60 ml of His-binding buffer and 30 ml of wash buffer (60 mM imidazole, 0.5 M NaCl, 20 mM Tris HCl, pH 7.9). Subsequently, the protein was eluted using a gradient of 20 ml binding buffer and 40 ml elute buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris HCl, pH 7.9). Fractions containing bmFabV were collected and imidazole was removed using a Sephadex G-25 column (1.5 cm × 55 cm long) using PIPES buffer (30 mM PIPES, 150 mM NaCl, 1.0 mM EDTA pH 8.0) as the eluent. The purity of the protein was shown to be >95% by 12% SDS-PAGE, which gave an apparent molecular weight of ~45 kDa. The concentration of the protein was determined by measuring the A280 and using an extinction coefficient (ε280) of 42,650 M−1cm−1 calculated from the primary sequence. The enzyme was stored at −80°C after flash freezing with liquid N2.
To remove the N-terminal His-tag, 1 µl of biotinylated thrombin was added to 1 ml of bmFabV (20 µM) in PIPES buffer and the reaction mixture was incubated at room temperature for 16 h. Streptavidin agarose (20 µl) was then added and the solution was incubated for an additional hour, after which the agarose was removed by centrifugation at 13,000 rpm for 10 min. The supernatant was further purified by chromatography on a Sephadex G-25 column (0.5 cm × 15 cm long) using PIPES buffer as the eluent, and fractions containing bmFabV lacking the His-tag were pooled and analyzed by 12% SDS-PAGE.
Site-Directed Mutagenesis, Expression and Purification of bmFabV Mutants
The bmFabV mutants Y235A, Y235S, K244A, K244R, K245M and K244A/K245A were prepared using the QuikChange mutagenesis kit from Stratagene with the primers listed in Table 1. The sequence of each mutant plasmid was confirmed by ABI DNA sequencing and the expression and purification of all the mutants followed the same procedure as that described above for the wild-type enzyme.
Cloning, Expression and Purification of ftuACP
The open reading frame (NCBI Reference Sequence: YP_170325) which encodes the putative acyl carrier protein (ACP) in Francisella tularensis SCHU S4 was amplified using the primers listed in Table 1. The protein product of this ORF shows 68% identity and 79% similarity to the ACP from Burkholderia mallei ATCC 23344. The purified PCR product was inserted into the Novagen pET23b vector using the 5’ BamHI and 3’ XhoI restriction sites (underlined) so that with a His-tag was encoded at the C-terminus of the protein. The Sequence of the construct was confirmed by ABI DNA sequencing.
Expression and purification of ftuACP followed a similar protocol to that described above for bmFabV except that 0.1 M potassium phosphate buffer, pH of 8.0 was used for G-25 chromatography. The purified protein was analyzed by 15% SDS-PAGE and MALDI-TOF mass spectrometry. The concentration of the protein was determined by measuring the absorption at 280 nm and by using an extinction coefficient of 2,560 M−1cm−1 calculated from the primary sequence. The protein was stored at 4°C for at over 3 months without losing any activity.
Preparation of Crotonyl-ACP (Cr-ACP)
Purified ftuACP was concentrated to 900 µM in the reaction buffer (0.1 M potassium phosphate solution at pH of 8.0, 3 ml) and an equimolar amount of dithiothreitol was added to the solution. The reaction mixture was then stirred under nitrogen at 0°C for 2 h to ensure complete reduction of the ACP thiol group, after which a 1.5-fold molar excess of crotonic anhydride was added to the reaction mixture. After stirring for 15 min at 0°C, small molecules were removed from the Cr-ACP by chromatography on a Sephadex G-25 column (0.5 cm × 15 cm) using PIPES buffer as the eluent..
Steady-state Kinetic Analysis
Steady-state kinetic parameters were determined at 25°C in 30 mM PIPES buffer pH 7.9 containing 1.0 mM EDTA and 0.62 mM NaCl. The optimum value for ionic strength was determined by varying the concentration of NaCl from 0 to 700 mM while the pH optimum was obtained by varying the pH of the reaction mixture from 6.6 to 8.5. Initial velocities were determined using a Cary 300 Bio (Varian) spectrophotometer to monitor the oxidation of NADH to NAD+ at 340 nm (ε = 6300 M−1cm−1). Initial characterization of the enzyme mechanism was performed in reaction mixtures containing 5 nM bmFabV and measuring initial velocities at several fixed concentrations of NADH (33, 110 and 250 µM) and by varying the concentration of DD-CoA (1.5–35 µM), or at a fixed concentration of DD-CoA (6, 12 and 18 µM) and by varying the concentration of NADH (10–352 µM). Double reciprocal plots were then used to differentiate between Ping-Pong or ternary-complex mechanisms.
To further investigate the binding order of substrates, product inhibition studies were performed in which each substrate concentration was varied in the presence of several fixed concentrations of one of the products, NAD+ (0, 50 and 110 µM) or lauryl-CoA (0, 50 and 100 µM). The type of inhibition in each case was subsequently determined using a Lineweaver-Burk plot.
Finally the kinetic data in the absence of products were globally fit to the equation for the steady-state sequential Bi Bi mechanism (equation 1) to determine the Km values for DD-CoA and NADH.
| (1) |
In equation 1, v is the initial velocity, Vmax is the maximum velocity, [A] and [B] are the concentration of the two substrates, KA and KB are the Michaelis constants for A and B respectively, and KiA is the dissociation constant for A. Data analysis was performed using GraFit 4.0 (Erithacus)
Km values of wild-type bmFabV toward DD-CoA and NADH were also determined by varying the concentration of one substrate at a fixed, saturating, concentration (> 10 × Km) of the second substrate. Data sets were then fit to the Michaelis-Menten equation (equation 2) using GraFit (Erithacus).
| (2) |
Km values determined using the above method were found to be very close to those obtained from global fitting. Consequently, all subsequent measurements of Km utilized initial velocities determined at fixed saturating concentrations of one substrate while the concentration of the other was varied.
Circular Dichroism Spectroscopy
The far-UV CD spectra of the wild-type bmFabV protein and its mutants were recorded in 50 mM Tris buffer pH 7.9 at 25°C at a concentration of 10 µM using an AVIV 62 DS spectrometer equipped with a Peltier temperature control unit. Data analysis was performed using Microsoft Excel.
Fluorescence Titration of Triclosan Binding to wild-type bmFabV
Equilibrium fluorescence titrations were conducted using a Spex FL3–21 Fluorolog-3 spectrofluorimeter. One µl aliquots of triclosan (20.0 mM stock in DMSO) were added to a 1 ml solution of enzyme (2 µM) in the same buffer as that used in the steady-state kinetic experiments. The excitation wavelength was 295 nm (5 nm slit width), and the emission wavelength was fixed at 335 nm (1 nm slit width). Dilution of protein concentration was controlled to minimum (< 1%) and the change in fluorescence as a function of triclosan concentration was fit to a quadratic equation (equation 3).
| (3) |
In equation 3, Fe and F1 are the fluorescent intensity in the presence and absence of enzyme respectively, Fe,max and F1,max are the maximum fluorescence intensity in the presence and absence of enzyme, respectively, Kd is the dissociation constant, [E]0 is the total enzyme concentration and [L] is the amount of triclosan added to the reaction buffer. Data fitting was performed using Grafit.
Progress curve analysis
Progress curve analysis was used to determine if triclosan was a slow onset inhibitor of bmFabV. FabI from F. tularensis (ftuFabI) was used as a model system since it has been shown that triclosan is a slow onset inhibitor of ftuFabI. In the assay, a very low concentration of enzyme (2 nM) and high concentration of substrate (200 µM of DD-CoA and 250 µM of NADH) were used so that initial velocities were linear over a period of 1 h. Since triclosan and other diphenyl ether inhibitors bind to enoyl-ACP reductases in the presence of the oxidized cofactor, 100 µM of NAD+ was added to the reaction so that [NAD+] was effectively constant during progress curve data collection. The concentration of triclosan was 60 µM, and data were collected for 1 h to ensure that the system had reached the steady state. Grafit was used for the data fitting.
Inhibition of bmFabV by Triclosan
Steady state kinetics revealed that triclosan is an uncompetitive inhibitor of bmFabV with respect to DD-CoA and a competitive inhibitor with respect to NADH. Initial velocities were measured at a fixed concentration of NADH (250 µM) or DD-CoA (35 µM) and at various concentrations of the second substrate and inhibitor. The equilibrium constant for the uncompetitive inhibition of bmFabV by triclosan (Kii) was calculated using equation 4 for the data collected at a fixed concentration of NADH,
| (4) |
where [S] is the concentration of DD-CoA, Km is the Michaelis-Menten constant for DD-CoA, Vmax is the maximum velocity, [I] is the concentration of inhibitor added and Kii is the inhibition constant.
Similarly, the equilibrium constant for the competitive inhibition of bmFabV by triclosan (Kis) was calculated using equation 5 for the data collected at a fixed concentration of DD-CoA,
| (5) |
where [S] is the concentration of NADH, Km is the Michaelis-Menten constant for NADH, Vmax is the maximum velocity, [I] is the concentration of inhibitor added and Kis is the inhibition constant.
Results and Discussion
Bioinformatic Analysis
In order to identify putative enoyl-ACP reductase homologues in B. mallei, the sequence of FabI from E. coli (ecFabI), FabL from B. subtilis, FabK from S. pneumoniae and FabV from Vibrio cholerae (vcFabV) were used as templates for a BLAST analysis of the B. mallei genome (ATCC 23344). Two open reading frames were identified as ecFabI homologues, one in chromosome 1 (63% identify to ecFabI) and the second in chromosome 2 (42% identity to ecFabI), while a homologue of vcFabV (bmFabV) was also located on chromosome 1 (56% identity to vcFabV). Subsequently the vcFabV homolog was cloned, expressed and purified, giving a protein with a MW of ~45 kDa on SDS PAGE, consistent with the expected MW of 44,965 Da for His-tagged bmFabV. The enzyme was shown to catalyze the NADH-dependent reduction of DD-CoA, demonstrating that bmFabV has in vitro enzymatic activity characteristic of an enoyl-ACP reductase.
Kinetic Mechanism
The optimal ionic strength (0.62 mM) and pH (7.9) values for the enzyme catalyzed reaction were determined by measuring initial velocities at saturating levels (> 10 × Km) of both DD-CoA and NADH. The Michaelis-Menten constants for DD-CoA and NADH were then determined by varying the concentration of one substrate at a saturating concentration of the second substrate. The values of kcat, Km,NADH, and Km,DD-CoA obtained using this method were 930 ± 6 min−1, 28.7 ± 1.2 µM and 2.4 ± 0.1 µM, respectively, which are close to the values obtained from global fitting (see below). Due to reports that vcFabV had a much higher catalytic activity without the C-terminal His-tag (19), we also determined the kinetic parameters of bmFabV after thrombin was used to remove the N-terminal His-tag. Subsequently we found that bmFabV lacking the N-terminal His-tag had kcat and Km,DD-CoA values of 1728 ± 10 min−1 and 4.4 ± 0.3 µM (Table 2), respectively, indicating that the His-tag did not have a dramatic effect on activity. Consequently, all subsequent kinetic experiments were performed using enzymes bearing an N-terminal His-tag.
Table 2.
Kinetic Parameters for Wild-type and Mutant bmFabV Enzymes
| Enzyme | kcat (min−1) | Km (µM) | kcat/Km (µM−1min−1) | |
|---|---|---|---|---|
| DD-CoA | NADH | DD-CoA | ||
| wild-type without His-tag |
1728 ± 10 | 4.4 ± 0.3 | ND | 392 ± 27 |
| wild-type with His- tag |
1242 ± 30 | 2.5 ± 0.4 | 23 ± 3 | 497 ± 79 |
| Y235A | 5.0 ± 0.1 | 2.7 ± 0.2 | 23 ± 2 | 1.8 ± 0.1 |
| Y235S | 6.1 ± 0.3 | 2.2 ± 0.1 | 6 ± 1 | 2.8 ± 0.1 |
| K244A | 11 ± 1 | 7.2 ± 0.6 | 66 ± 2 | 1.5 ± 0.1 |
| K244R | 1.3 ± 0.1 | 7.6 ± 0.7 | 69 ± 1 | 0.17 ± 0.01 |
| K245M | 18 ± 1 | 28 ± 2 | 28 ± 2 | 0.64 ± 0.03 |
| K244A/K245A | ND | ND | 65 ± 4 | 0.023 ± 0.002 |
To demonstrate that the recombinant enzyme is also capable of catalyzing the reduction of ACP substrates, the ACP from F. tularensis SCHU S4 (ftuACP), which is 68% identical to the putative ACP from B. mallei, was expressed and purified. ESI mass spectrometry revealed that the purified ftuACP was entirely in the holo form, and a chemical coupling reaction with crotonic anhydride was subsequently used to synthesize Cr-ACP. The values of kcat, Km,NADH, and Km,cr-ACP obtained were 1299 ± 29 min−1, 31 ± 4 µM and 25.1 ± 2 µM, respectively.
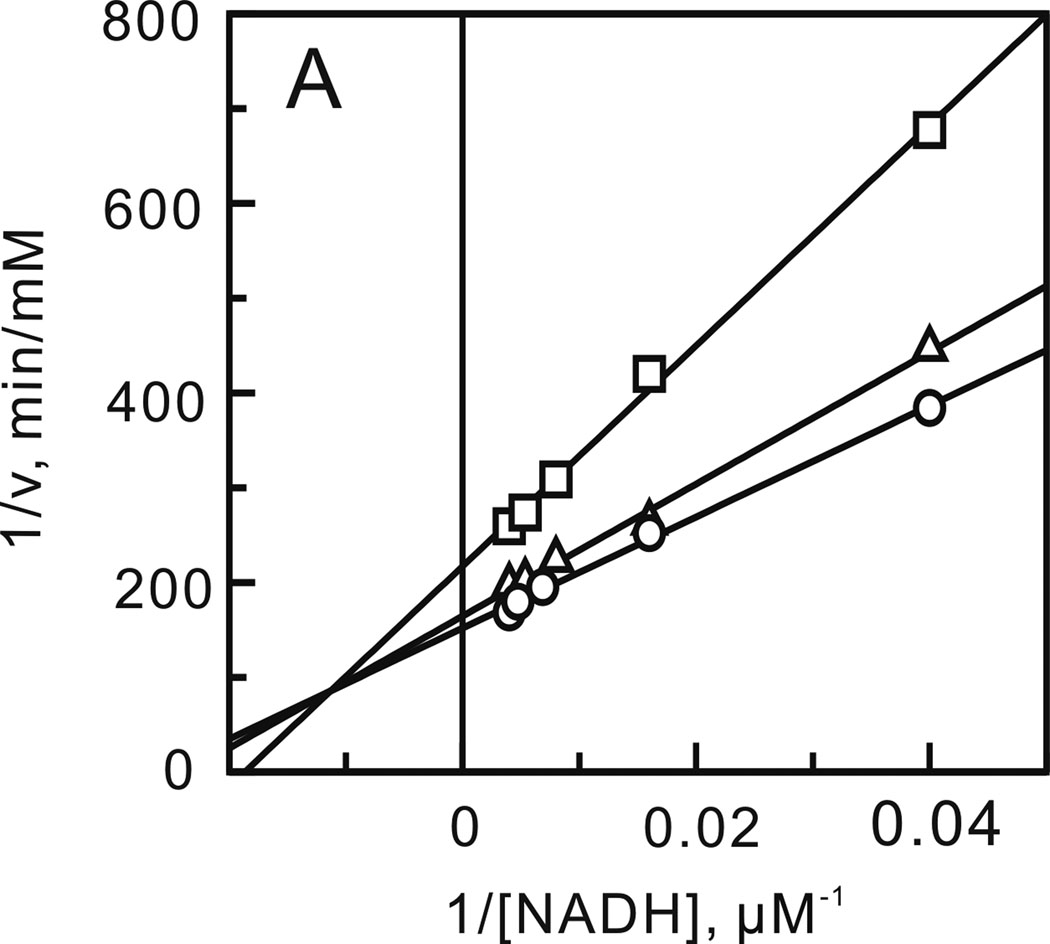
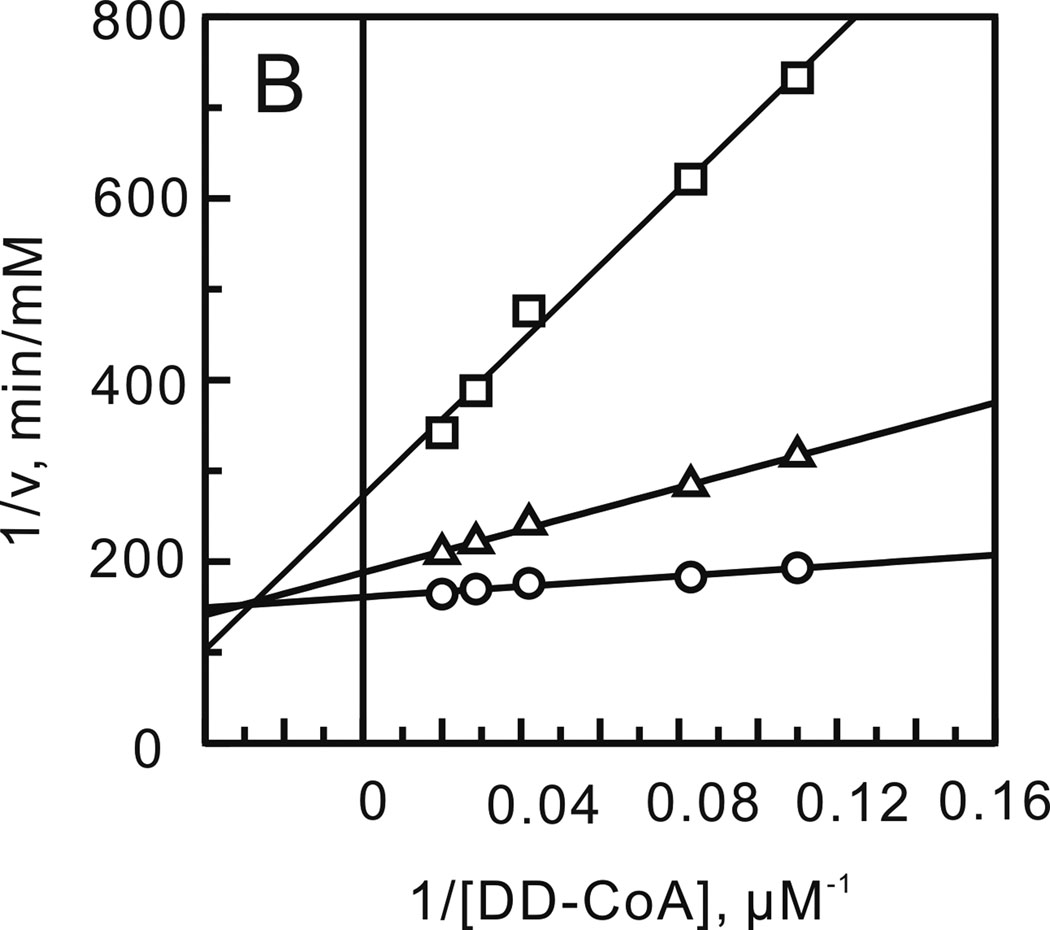
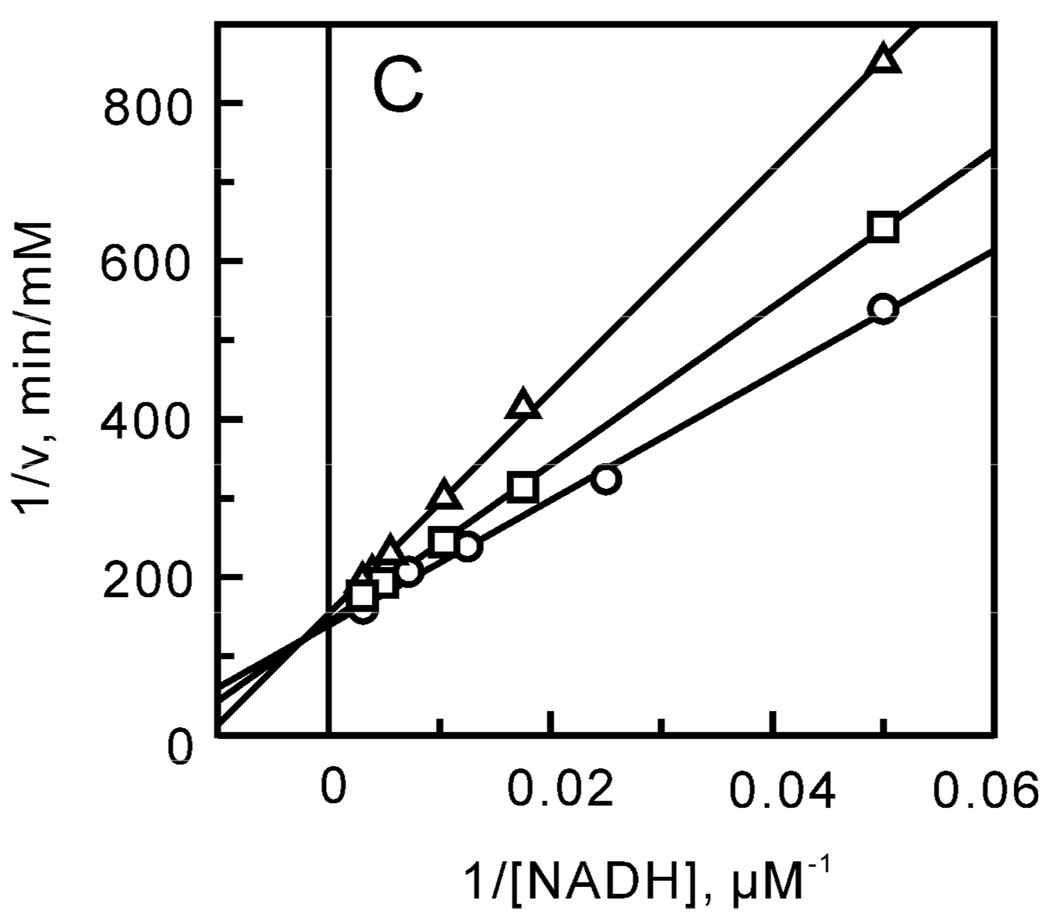
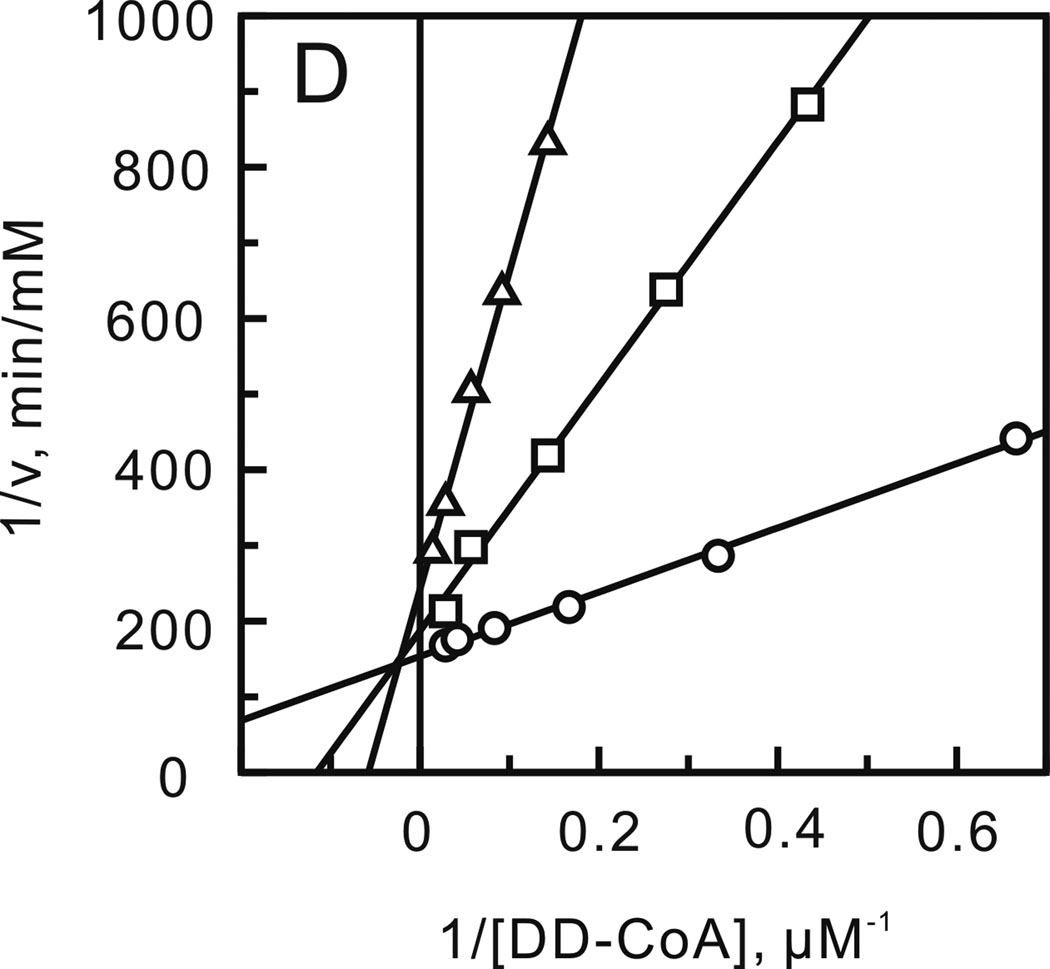
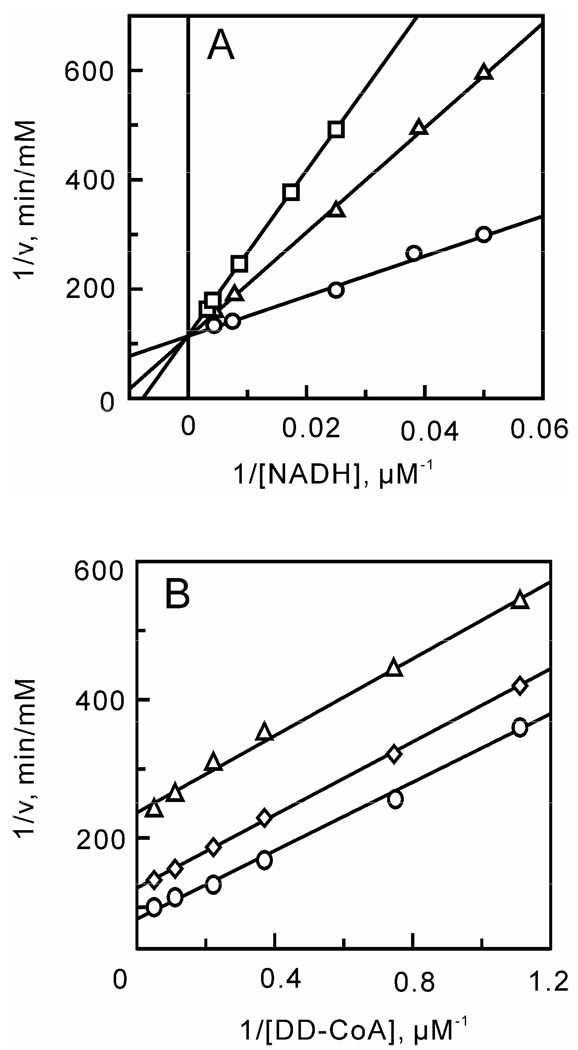
After optimizing the ionic strength and pH, a matrix of initial velocities was measured at different concentrations of DD-CoA and NADH, and the kinetic data were then analyzed using double-reciprocal plots. Similar to other enoyl-ACP reductases, these studies confirmed that bmFabV catalyzes substrate reduction through a ternary-complex mechanism, since all the lines in the double-reciprocal plot intersect with each other at a single point (Figure 1) (22–25). To further investigate the order of substrate binding, product inhibition studies using NAD+ and lauryl-CoA were performed. Analysis of the results from double-reciprocal plots indicated that NAD+ was a competitive inhibitor with respect to NADH, but was a non-competitive inhibitor with respect to DD-CoA. However, when NADH or DD-CoA was varied in the presence of lauryl-CoA, mixed patterns of inhibition were observed for both substrates (Figure 2). This combination of product inhibition patterns is indicative of a sequential Bi Bi mechanism with NADH binding to the enzyme first. The ternary complex steady-state kinetic parameters were subsequently obtained by globally fitting the initial velocity data to equation 1 for an ordered Bi Bi mechanism, giving Ki,NADH 13.0 ± 4.1 µM, Km,NADH 23.2 ± 3.4 µM, Km,DD-CoA 2.5 ± 0.4 µM and kcat 1242 ± 30 min−1 (Table 2). Due to the similarity in kinetic parameters obtained from global fitting and when only one of the substrates was varied at a saturating level of the second substrate, we subsequently analyzed the impact of site-directed mutagenesis by determining kinetic parameters at a single fixed concentration of one substrate while varying the second substrate.
FIGURE 1.
Two-substrate steady-state kinetics. Initial velocity patterns: (A) 1/v versus 1/[NADH] double-reciprocal plots in which the DD-CoA concentration was fixed at 6 (□), 12 (Δ) and 18 µM (○) and (B) 1/v versus 1/[DD-CoA] double-reciprocal plots in which the NADH concentration was fixed at 33 (□), 110 (Δ) and 250 µM (○).
FIGURE 2.
Product inhibition studies to determine the substrate binding order. Assays were performed by varying the concentration of one substrate at a fixed concentration of the second substrate and in the presence of one of the products of the reaction. (A) NADH varied in the presence of NAD+ (0 (□), 55 (○) and 110 µM (Δ)) with DD-CoA fixed at 35 µM. (B) DD-CoA varied in the presence of NAD+ (0 (○), 55 (Δ) and 110 µM (□)) with NADH fixed at 250 µM. (C) NADH varied in the presence of lauryl-CoA (0 (○), 50 (□) and 100 µM (Δ)) with DD-CoA fixed at 35 µM. (D) DD-CoA varied in the presence of lauryl-CoA (0 (○), 50 (□) and 100 µM (Δ)) with NADH fixed at 250 µM.
Taken together, the steady state kinetic data reveal that bmFabV functions as an enoyl-ACP reductase, possessing even higher catalytic activity than other enoyl-ACP reductases characterized previously (21, 23–24, 26). bmFabV catalyzes substrate reduction through a sequential Bi Bi mechanism with NADH binding first. This suggests that NADH forms part of the enoyl substrate binding site as seen in several other FabI enoyl-ACP reductases (22, 25). In addition, we found that the N-terminal His-tag in bmFabv did not have a dramatic effect on catalytic activity as reported for the C-terminal His-tag in vcFabV.
Active-site Residues and Catalytic Mechanism
The FabI and FabV enoyl-ACP reductases are members of the short-chain dehydrogenase/reductase (SDR) superfamily (27–30). In this family a catalytic tyrosine and lysine are found in a Tyr-(Xaa)n-Lys motif, where n is generally 3 for the dehydrogenases and 6 for enoyl-ACP reductases. The variation in n is thought to relate to the difference in chemistry between reduction of a carbonyl group, and a carbon-carbon double bond conjugated to a thioester. Analysis of the bmFabV sequence suggests the presence of an even more extended active-site motif, Tyr-(Xaa)8-Lys, involving Y235 and K244. In addition, unlike other SDR reductases, K244 is followed by a second lysine (K245), which is conserved in the FabV homologues from a variety of organisms including Burkholderia sp., Yersinia sp. and Clostridium acetobutylicum (Figure 3).To investigate the roles of these residues in catalysis, the following mutant enzymes were expressed and purified: Y235A, Y235S, K244A, K244R, K245M and K244A/K245A. CD spectra of the wild-type and mutant proteins were superimposable (data not shown), suggesting that the alteration in catalytic activity resulting from mutagenesis was not a result of major structural changes in the protein. Subsequently the kinetic parameters of the mutant enzymes were determined (Table 2).
FIGURE 3.
Sequence alignment of active site residues of the enoyl-ACP reductases from Escherichia coli, Bacillus subtilis, Burkholderia mallei, Yesinia pestis, Clostridium acetobutylicum, Pseudomonas aeruginosa, Variovorax paradoxus, Methylobacillus flagellatus, Erwinia tasmaniensis and Vibrio cholerae. Conserved residues are labeled with an asterix. The sequence alignment was performed using Clustal W (48), and the figure was made using Jalview (49).
Y235 in bmFabV is homologous to the conserved tyrosine that is found in all members of the SDR superfamily (21, 31). Replacement of this conserved tyrosine with phenylalanine in the dehydrogenases, such as 3β/17β-hydroxysteroid dehydrogenase, results in an enzyme with virtually no activity (31), consistent with the notion that this residue plays a critical role in substrate oxidation/reduction. In contrast, equivalent mutations in the FabI enzymes from E. coli (ecFabI; Y156F) and M. tuberculosis (InhA; Y158F) result in much more modest decreases in kcat values of 7- and 21-fold, without substantially affecting the Km value for the substrate (21, 32). These data have raised questions concerning the importance of the conserved tyrosine in the enoyl-ACP reductases (33), and indeed Anderson and coworkers have suggested that the enolate formed during reduction of the enoyl thioester does not need to be stabilized to a great extent by the enzyme (34). Consistent with this idea, the Y158S InhA enzyme has wild-type activity (21), although it now appears that a second tyrosine in ecFabI (Y146) may be involved in transition state stabilization (33). This latter residue, which constitutes the third residue in the SDR ‘triad’, is a phenylalanine in mycobacterial FabI enzymes and is a serine in the dehydrogenases (21, 31).
Returning to bmFabV, replacement of Y235 with alanine gave a kcat/Km value of 1.8 ± 0.1 µM−1min−1, which is 280-fold lower than the wild-type enzyme. This decrease in kcat/Km primarily results from a reduction in kcat, suggesting that Y235 plays a crucial role in substrate reduction. To further investigate the catalytic role of Y235, this residue was replaced by a serine. However, unlike InhA, this mutant could not restore the activity of bmFabV, and instead resulted in an enzyme with similar activity to the Y235A mutant. Although the pKa values of the Y235 hydroxyl group and of the S235 mutant in the enzyme are unknown, the pKa values of these side chains differ by ~ 4.5 log in solution, suggesting that the acidity of Y235 is important for the function of this residue in catalysis.
In the SDR enzymes, the primary function of the active site lysine is to bind the cofactor through hydrogen bonds formed with the nicotinamide ribose. In addition, this lysine is also thought to interact directly with the conserved tyrosine in the dehydrogenases, facilitating protonation of the substrate carbonyl by the tyrosine (31). In bmFabV, the K244A mutant had kcat, Km,DD-CoA and Km,NADH values of 11 ± 1 min−1, 7.2 ± 0.6 µM and 66 ± 2 µM. This mutation thus causes small (3-fold) increases in the Km values for both substrates, together with a 110-fold decrease in the kcat value for substrate reduction. The corresponding mutation in InhA (K165A) resulted in an enzyme that was unable to bind cofactor. However, in the case of InhA, replacement of K165 with arginine completely restored activity. In contrast the K244R bmFabV enzyme had a kcat value 950-fold smaller than that of wild-type enzyme and was thus even less active that the K244A mutant. The modest changes in Km values coupled with the 102–103-fold reduction in kcat values caused by mutation of K244 (Table 2) thus support differing roles for the conserved lysine in FabI and FabV. In FabV it appears that K244 plays a much more direct role in facilitating substrate reduction than does the corresponding residue in FabI where it functions primarily in cofactor binding. Indeed the function of both Y235 and K244 in bmFabV is more akin to the role played by the conserved tyrosine and lysine residues in the dehydrogenases than in the FabI enzymes.
In bmFabV, a second lysine (K245) is found adjacent to K244. This lysine-lysine sequence is not observed in other enoyl reductases, but is conserved in FabV from a variety of organisms (Figure 3). In order to probe the role of K245, the K245M and K244A/K245A mutants were generated. While the Km value for NADH was unchanged in the K245M mutant, the Km value for DD-CoA was increased 10-fold compared to the wild-type enzyme. This mutation also caused a 70-fold reduction in the kcat value for substrate reduction. Although the K244A mutation only resulted in a small 3-fold increase in Km,DDCOA, combination of K244A with K245A resulted in an enzyme for which the Km value for DD-CoA has increased to a value that cannot be measured. In contrast the Km values for NADH in the single and double mutants are very similar, suggesting that K245 plays an important role in binding the enoyl substrate.
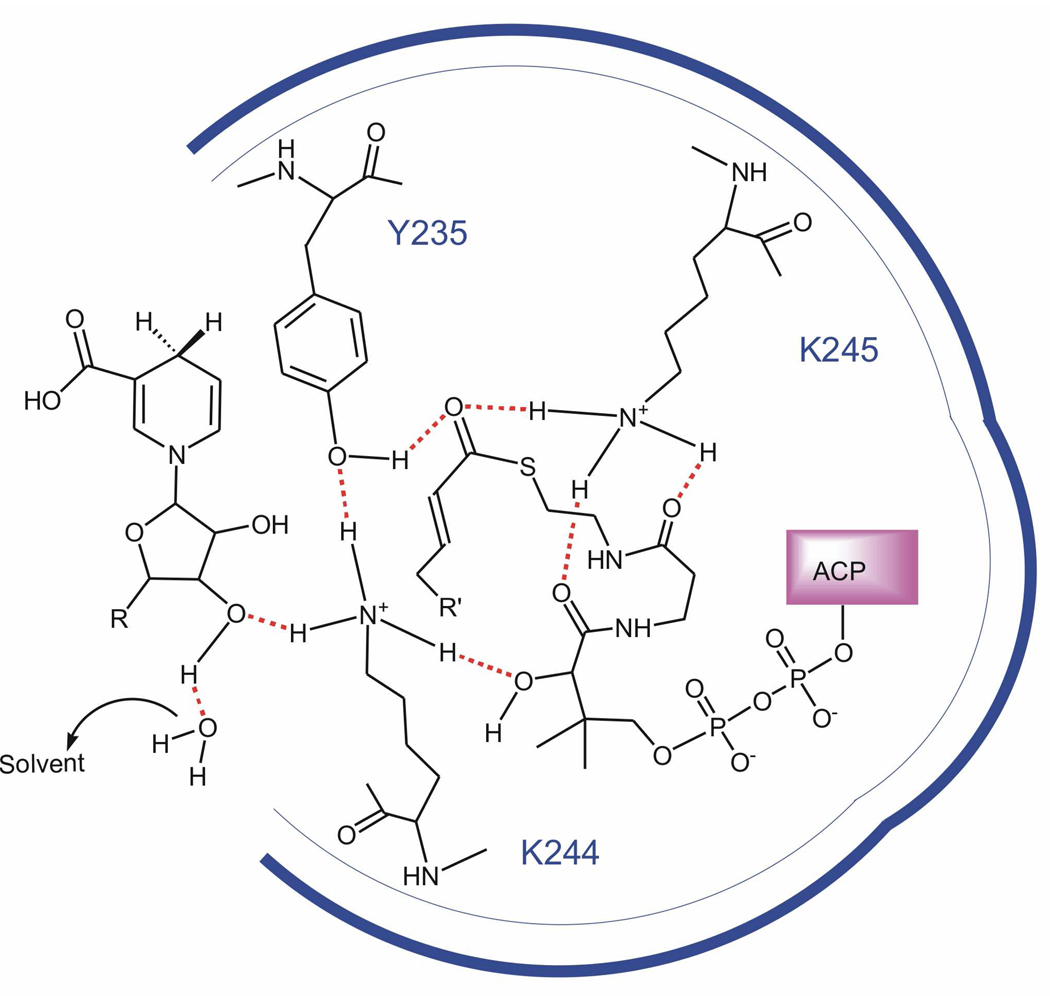
In summary, we have identified three important active site residues in bmFabV using site directed mutagenesis. The kinetic data support a model in which Y235, K244 and K245 form a hydrogen bond network that is important for substrate reduction. By analogy to the reaction mechanism in the SDR dehydrogenases, we suggest that Y235 is hydrogen bonded to K244/K245, an interaction that would lower the pKa of Y235 and facilitate the ability of this residue to either stabilize and/or protonate the enolate intermediate formed during substrate reduction. In addition, K244 is thought to interact with both cofactor and acyl substrate, while K245 interacts solely with the acyl substrate. A schematic diagram showing these proposed interactions is given in Figure 4. This figure has been adapted from the proposed catalytic mechanism of 3β/17β-hydroxysteroid dehydrogenase in which the conserved tyrosine and lysine residues function together to protonate/deprotonate the substrate (35). The interactions that we propose for FabV differ somewhat from those in other FabI enoyl reductases where the conserved lysine is thought to only be involved in cofactor binding.
FIGURE 4.
Proposed hydrogen bonding network between the three active site residues (Y235, K244 and K245), cofactor and the enoyl–ACP substrate. Y235 stabilizes the transition state of the enoyl-ACP substrate through a hydrogen bond with the carbonyl group while K244 interacts directly with Y235 as proposed in the SDR dehydrogenases. K244 is also shown interacting with the NADH ribose hydroxyl and the secondary hydroxyl group in the ACP pantetheine, in order to account for the role of this residue in binding cofactor and substrate. K245 is shown interacting with the thioester carbonyl group, to account for the effect of mutating this residue on kcat, and also with the ACP pantetheine.
Inhibition of bmFabV by Triclosan
The diphenyl ether triclosan is a potent inhibitor of the FabI enzyme from a variety of organisms. Studies with the FabI enzymes from E. coli and F. tularensis have revealed that triclosan is a slow-onset inhibitor of these organisms, and it is thought that the slow step in formation of the final E-I* complex involves ordering of a substrate recognition loop that closes over the active site after triclosan binding (15, 26, 36–39). The high affinity of triclosan for the ecFabI and ftuFabI, as well as related FabIs such as the enzyme from S. aureus, is generally a hallmark of the FabI enoyl-ACP reductases, with the outlier being the InhA enzyme from M. tuberculosis. Triclosan inhibits InhA with a Ki value of only 0.2 µM (40) compared to the Ki values of 7 pM, 50 pM and 5 nM for ecFabI, ftuFabI and saFabI, respectively (15, 24, 32, 41). In addition triclosan is not a slow-onset inhibitor of InhA in contrast to the other enzymes. However a structure-based approach has successfully resulted in diphenyl ether-based inhibitors of InhA with improved affinity (42) and that are slow-onset inhibitors of the enzyme (43). An important distinction between the FabI and FabK enoyl-ACP reductases is the insensitivity of the latter to triclosan (17), and Cronan and coworkers have shown that vcFabV is only a weakly inhibited by triclosan (50% inhibition at 12 µM) (19). Due to our success in developing high affinity inhibitors of InhA, we were thus very curious to study the inhibition of bmFabV by triclosan.
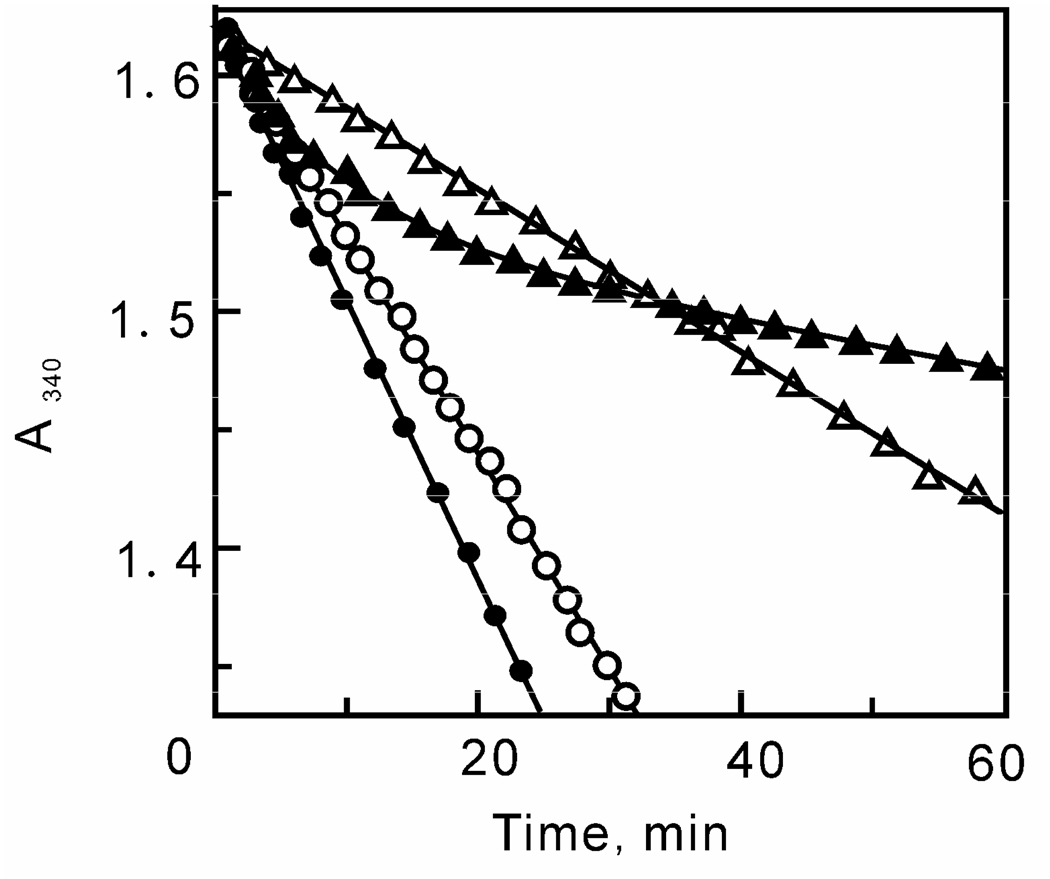
Previous research in our lab has shown that triclosan is a slow onset inhibitor of ftuFabI (41). In a standard progress curve analysis assay, a linear decrease in [NADH] is observed in the absence of triclosan while a curved line is seen in the presence of inhibitor (Figure 5). This is a characteristic of slow onset inhibition, since rapid reversible inhibitors are expected to also yield a linear decrease in substrate concentration under similar condition. In the case of ftuFabI, the initial velocity decreases exponentially with time until a final steady-state velocity is achieved (44). However, in the studies with bmFabV, the [NADH] decreased linearly with time, which strongly suggests that triclosan is a rapid, reversible inhibitor of bmFabV (Figure 5).
FIGURE 5.
Progress curve analysis of ftuFabI and bmFabV. Progress curves for ftuFabI (2 nM) in the presence of DD-CoA (200 µM), NADH (250 µM) and either 0 (●) or 60 nM (▲) triclosan. Progress curves for bmFabV (2 nM) in the presence of DD-CoA (200 µM), NADH (250 µM) and either 0 (○) or 2 µM (∆) triclosan.
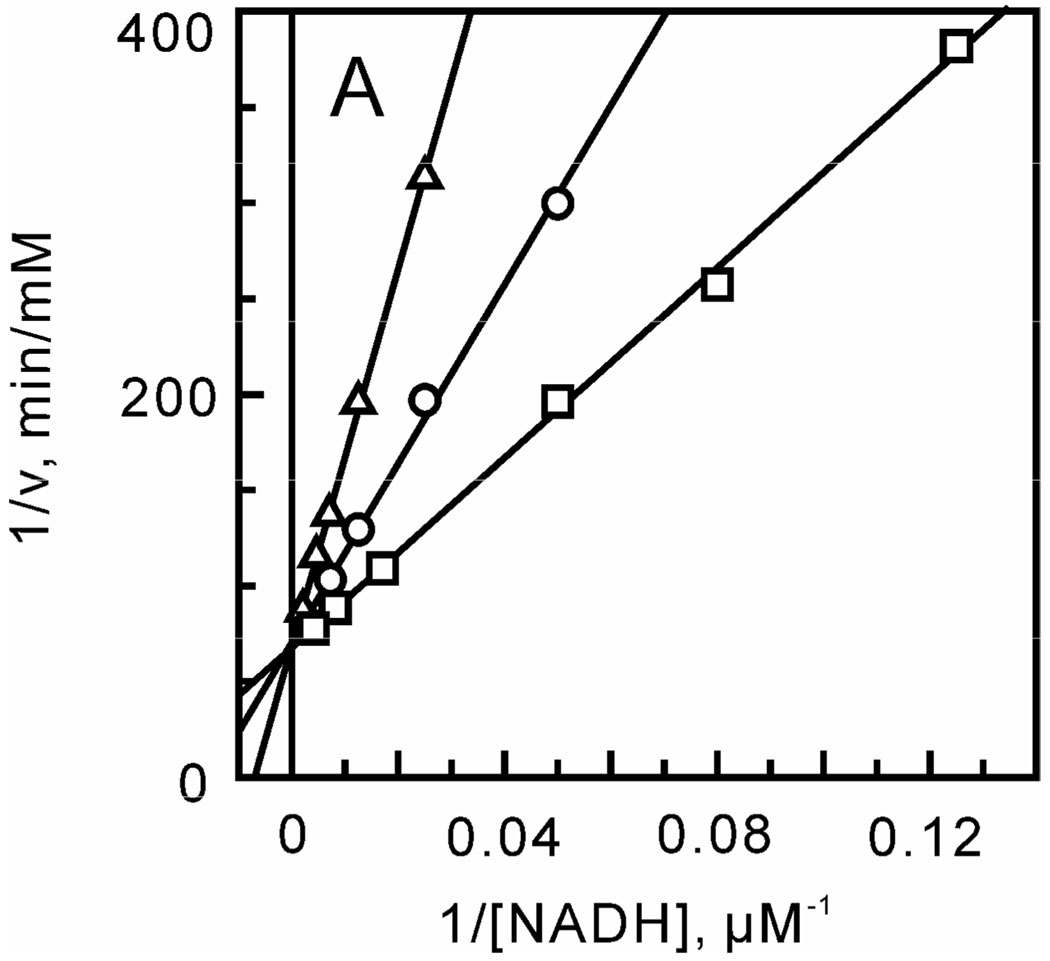
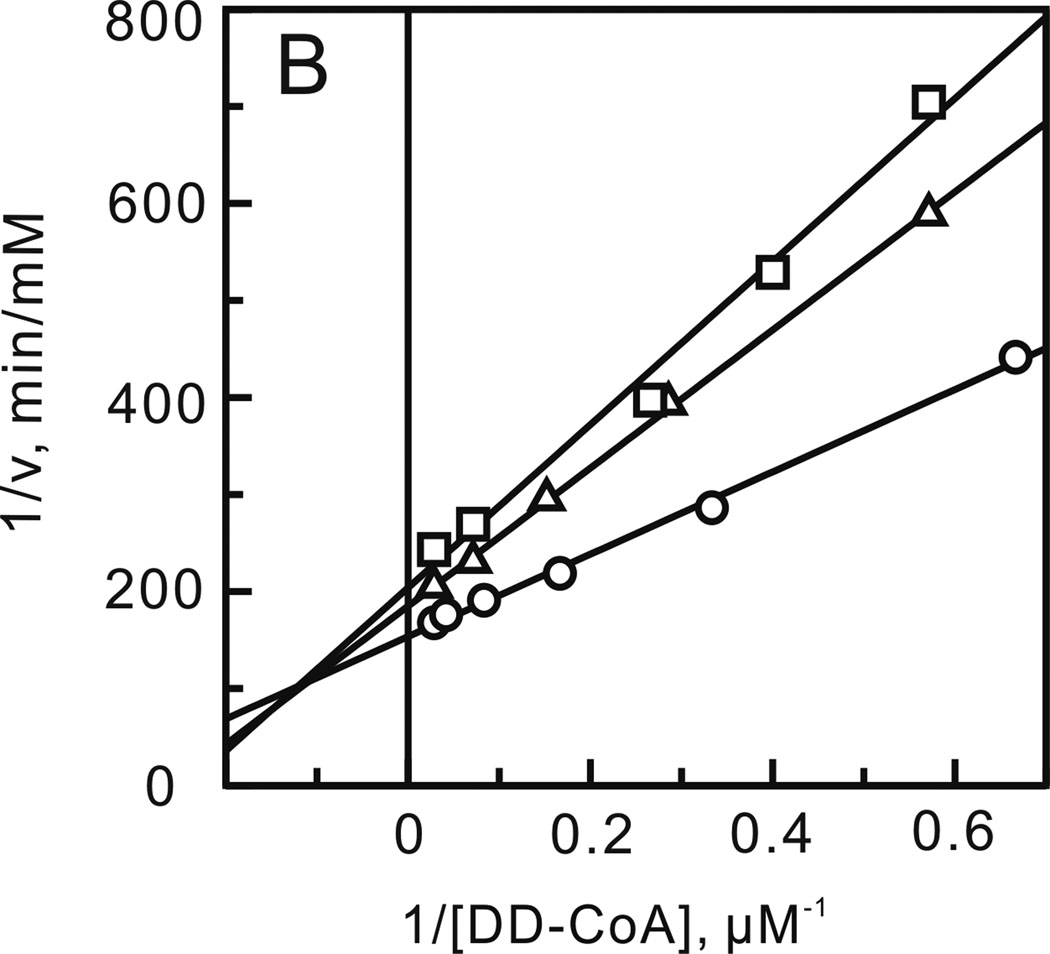
Initial velocities were subsequently obtained under a range of substrate and inhibitor concentrations, and data analysis revealed that triclosan is an uncompetitive inhibitor with respect to DD-CoA (Kii = 0.42 ± 0.02) and a competitive inhibitor with respect to NADH (Kis = 0.48 ± 0.04 µM) (Figure 6). Uncompetitive inhibition with respect to DD-CoA indicates that triclosan binds after DD-CoA. Since we have already shown that bmFabV catalyzes the reaction through a sequential mechanism with NADH binding first and NAD+ dissociating last, triclosan then can only bind to the substrate ternary complex (E-NADH-DD-CoA), the product ternary complex (E-NAD+-lauryl-CoA) or the binary product complex (E-NAD+). In addition, since triclosan is a competitive inhibitor with respect to NADH, triclosan must bind to either the NADH binding site in the free enzyme or to the E-NAD+ complex. Taken together, we can conclude that triclosan binds to the E-NAD+ product complex as observed for the FabI class of enoyl-ACP reductases (15). In support of this hypothesis, fluorescence titration experiments demonstrated that the Kd value for the interaction of triclosan with the free enzyme is larger than 100 µM, which indicates that triclosan binds only weakly to the free enzyme. Finally, the effect of NAD+ on triclosan binding was investigated by adding NAD+ to the reaction mixture. When the inhibition of bmFabV by triclosan was measured by varying the concentration of NADH in the presence of 100 µM NAD+, a 4-fold decrease in the Kis value (0.11 µM) was observed. This means that NAD+ can potentiate the binding of triclosan to the enzyme by forming an E-NAD+-triclosan ternary complex. However, the formation and dissociation of this ternary complex must be rapid and reversible, occurring without ordering of the substrate binding loop. The mechanism of inhibition of bmFabV by triclosan thus closely follows that characterized previously for InhA (40).
FIGURE 6.
Mechanism of bmFabV inhibition by triclosan. Assays were performed by varying the concentration of one substrate at a fixed concentration of the second substrate constant and at various concentrations of triclosan. (A) NADH varied in the presence of triclosan (0 (○), 0.8 (∆) and 1.6 µM (□)) with DD-CoA fixed at 35 µM. (B) DD-CoA varied in the presence of triclosan (0 (○), 0.8 (◊) and 1.6 µM (∆)) with NADH fixed at 250 µM.
The observation that triclosan is a rapid, reversible inhibitor of bmFabV with a Ki value of only 0.4 µM is consistent with the suggestion, based on studies with vcFabV, that the FabV enzyme class is relatively insensitive to this antibacterial diphenyl ether (19). This proposal has received additional support from the recent observation that deletion of FabV in P. aeruginosa (paFabV) increased the sensitivity of this organism to triclosan by more than 2000-fold, suggesting that resistance of P. aeruginosa to triclosan is due to the relative insensitivity of paFabV to triclosan rather than to efflux (45). This latter observation is of high interest since, like B. mallei, P. aeruginosa contains both FabI and FabV homologues. However, given our success at developing potent diphenyl ether inhibitors of InhA (42–43), a FabI enzyme that is also relatively insensitive to triclosan (Ki = 0.2 µM (40)), we are confident that the studies reported in the present work will provide a firm foundation for the development of antibacterials that target pathogens with FabV enoyl-ACP reductases such as B. mallei, Y. pestis, and P. aeruginosa, as well as the closely related category B pathogen B. pseudomallei, which contains a FabV homologue that is 99% identical to bmFabV (46–47).
Conclusion
In summary, studies on the mechanism of the reaction catalyzed by bmFabV supports a sequential Bi Bi mechanism in which NADH binds first to the free enzyme. Site directed mutagenesis suggests that the two conserved active-site residues in the Tyr-(Xaa)8-Lys motif play a more significant role in the chemical steps of substrate reduction than in the FabI class of enoyl-ACP reductases, and that their function is more in line with that of the homologous residues in the SDR dehydrogenase family members. In addition, a second lysine in the active site has been identified and shown to play a key role in acyl substrate binding. Finally, detailed inhibition studies have revealed that triclosan binds to the bmFabV-NAD+ product complex through a rapid, reversible mechanism of inhibition that closely resembles the inhibition of InhA by triclosan, with a Ki value of 0.4 µM.
Footnotes
This work was supported by NIH grant AI44639.
REFERENCES
- 1.Campbell JW, Cronan JE., Jr Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- 2.Heath RJ, White SW, Rock CO. Lipid biosynthesis as a target for antibacterial agents. Prog Lipid Res. 2001;40:467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 3.Egan AF, Russell RR. Conditional mutations affecting the cell envelope of Escherichia coli K-12. Genet Res. 1973;21:139–152. doi: 10.1017/s001667230001332x. [DOI] [PubMed] [Google Scholar]
- 4.Turnowsky F, Fuchs K, Jeschek C, Hogenauer G. envM genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1989;171:6555–6565. doi: 10.1128/jb.171.12.6555-6565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res. 2003;42:289–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- 6.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim Biophys Acta. 1996;1302:1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 8.Heath RJ, Rock CO. Fatty acid biosynthesis as a target for novel antibacterials. Curr Opin Investig Drugs. 2004;5:146–153. [PMC free article] [PubMed] [Google Scholar]
- 9.White SW, Zheng J, Zhang YM, Rock The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 10.McMurry LM, Oethinger M, Levy SB. Triclosan targets lipid synthesis. Nature. 1998;394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 11.Heath RJ, Yu YT, Shapiro MA, Olson E, Rock CO. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem. 1998;273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 12.Musser JM, Kapur V, Williams DL, Kreiswirth BN, van Soolingen D, van Embden JD. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 13.Tonge PJ, Kisker C, Slayden RA. Development of modern InhA inhibitors to combat drug resistant strains of Mycobacterium tuberculosis. Curr Top Med Chem. 2007;7:489–498. doi: 10.2174/156802607780059781. [DOI] [PubMed] [Google Scholar]
- 14.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J Biol Chem. 1994;269:5493–5496. [PubMed] [Google Scholar]
- 15.Lu H, Tonge PJ. Inhibitors of FabI, an enzyme drug target in the bacterial fatty acid biosynthesis pathway. Acc Chem Res. 2008;41:11–20. doi: 10.1021/ar700156e. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YM, White SW, Rock CO. Inhibiting bacterial fatty acid synthesis. J Biol Chem. 2006;281:17541–17544. doi: 10.1074/jbc.R600004200. [DOI] [PubMed] [Google Scholar]
- 17.Heath RJ, Rock CO. A triclosan-resistant bacterial enzyme. Nature. 2000;406:145–146. doi: 10.1038/35018162. [DOI] [PubMed] [Google Scholar]
- 18.Heath RJ, Su N, Murphy CK, Rock CO. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J Biol Chem. 2000;275:40128–40133. doi: 10.1074/jbc.M005611200. [DOI] [PubMed] [Google Scholar]
- 19.Massengo-Tiasse RP, Cronan JE. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem. 2008;283:1308–1316. doi: 10.1074/jbc.M708171200. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan A, Kraus CN, DeShazer D, Becker PM, Dick JD, Spacek L, Bartlett JG, Byrne WR, Thomas DL. Glanders in a military research microbiologist. N. Engl. J. Med. 2001;345:256–258. doi: 10.1056/NEJM200107263450404. [DOI] [PubMed] [Google Scholar]
- 21.Parikh S, Moynihan DP, Xiao G, Tonge PJ. Roles of tyrosine 158 and lysine 165 in the catalytic mechanism of InhA, the enoyl-ACP reductase from Mycobacterium tuberculosis. Biochemistry. 1999;38:13623–13634. doi: 10.1021/bi990529c. [DOI] [PubMed] [Google Scholar]
- 22.Marcinkeviciene J, Jiang W, Kopcho LM, Locke G, Luo Y, Copeland RA. Enoyl-ACP reductase (FabI) of Haemophilus influenzae: steady-state kinetic mechanism and inhibition by triclosan and hexachlorophene. Arch Biochem Biophys. 2001;390:101–108. doi: 10.1006/abbi.2001.2349. [DOI] [PubMed] [Google Scholar]
- 23.Kapoor M, Dar MJ, Surolia A, Surolia N. Kinetic determinants of the interaction of enoyl-ACP reductase from Plasmodium falciparum with its substrates and inhibitors. Biochem Biophys Res Commun. 2001;289:832–837. doi: 10.1006/bbrc.2001.6061. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Sullivan TJ, Sekiguchi J, Kirikae T, Ojima I, Stratton CF, Mao W, Rock FL, Alley MR, Johnson F, Walker SG, Tonge PJ. Mechanism and inhibition of saFabI, the enoyl reductase from Staphylococcus aureus. Biochemistry. 2008;47:4228–4236. doi: 10.1021/bi800023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fawcett T, Copse CL, Simon JW, Slabas AR. Kinetic mechanism of NADH-enoyl-ACP reductase from Brassica napus. FEBS Lett. 2000;484:65–68. doi: 10.1016/s0014-5793(00)02128-1. [DOI] [PubMed] [Google Scholar]
- 26.Ward WH, Holdgate GA, Rowsell S, McLean EG, Pauptit RA, Clayton E, Nichols WW, Colls JG, Minshull CA, Jude DA, Mistry A, Timms D, Camble R, Hales NJ, Britton CJ, Taylor IW. Kinetic and structural characteristics of the inhibition of enoyl (acyl carrier protein) reductase by triclosan. Biochemistry. 1999;38:12514–12525. doi: 10.1021/bi9907779. [DOI] [PubMed] [Google Scholar]
- 27.Roujeinikova A, Levy CW, Rowsell S, Sedelnikova S, Baker PJ, Minshull CA, Mistry A, Colls JG, Camble R, Stuitje AR, Slabas AR, Rafferty JB, Pauptit RA, Viner R, Rice DW. Crystallographic analysis of triclosan bound to enoyl reductase. J Mol Biol. 1999;294:527–535. doi: 10.1006/jmbi.1999.3240. [DOI] [PubMed] [Google Scholar]
- 28.Baldock C, Rafferty JB, Stuitje AR, Slabas AR, Rice DW. The X-ray structure of Escherichia coli enoyl reductase with bound NAD+ at 2.1 A resolution. J Mol Biol. 1998;284:1529–1546. doi: 10.1006/jmbi.1998.2271. [DOI] [PubMed] [Google Scholar]
- 29.Rozwarski DA, Vilcheze C, Sugantino M, Bittman R, Sacchettini JC. Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate. J Biol Chem. 1999;274:15582–15589. doi: 10.1074/jbc.274.22.15582. [DOI] [PubMed] [Google Scholar]
- 30.Rafferty JB, Simon JW, Baldock C, Artymiuk PJ, Baker PJ, Stuitje AR, Slabas AR, Rice DW. Common themes in redox chemistry emerge from the X-ray structure of oilseed rape (Brassica napus) enoyl acyl carrier protein reductase. Structure. 1995;3:927–938. doi: 10.1016/S0969-2126(01)00227-1. [DOI] [PubMed] [Google Scholar]
- 31.Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 32.Sivaraman S, Zwahlen J, Bell AF, Hedstrom L, Tonge PJ. Structure-Activity Studies of the Inhibition of FabI, the Enoyl Reductase from Escherichia coli, by Triclosan: Kinetic Analysis of Mutant FabIs. Biochemistry. 2003;42:4406–4413. doi: 10.1021/bi0300229. [DOI] [PubMed] [Google Scholar]
- 33.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. Structure of acyl carrier protein bound to Fabi, the FASII enoyl reductase from Escherichia coli. J. Biol. Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fillgrove KL, Anderson VE. The mechanism of dienoyl-CoA reduction by 2,4-dienoyl-CoA reductase is stepwise: Observation of a dienolate intermediate. Biochemistry. 2001;40:12412–12421. doi: 10.1021/bi0111606. [DOI] [PubMed] [Google Scholar]
- 35.Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jornvall H, Oppermann U. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 36.Perozzo R, Kuo M, Sidhu AS, Valiyaveettil JT, Bittman R, Jacobs WR, Jr, Fidock DA, Sacchettini JC. Structural elucidation of the specificity of the antibacterial agent triclosan for malarial enoyl acyl carrier protein reductase. J Biol Chem. 2002;277:13106–13114. doi: 10.1074/jbc.M112000200. [DOI] [PubMed] [Google Scholar]
- 37.Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol. 1999;290:859–865. doi: 10.1006/jmbi.1999.2907. [DOI] [PubMed] [Google Scholar]
- 38.Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K. Structural basis for the variation in triclosan affinity to enoyl reductases. J Mol Biol. 2004;343:147–155. doi: 10.1016/j.jmb.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 39.Sivaraman S, Sullivan TJ, Johnson F, Novichenok P, Cui G, Simmerling C, Tonge PJ. Inhibition of the bacterial enoyl reductase FabI by triclosan: a structure-reactivity analysis of FabI inhibition by triclosan analogues. J Med Chem. 2004;47:509–518. doi: 10.1021/jm030182i. [DOI] [PubMed] [Google Scholar]
- 40.Parikh SL, Xiao G, Tonge PJ. Inhibition of InhA, the enoyl-reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry. 2000;39:7645–7650. doi: 10.1021/bi0008940. [DOI] [PubMed] [Google Scholar]
- 41.Lu H, England H, am Ende CW, Truglio JJ, Luckner S, Reddy BG, Marlenee N, Knudson SE, Knudson DL, Bowen RA, Kisker C, Slayden RA, Tonge PJ. Slow-Onset Inhibition of the FabI Enoyl Reductase from Francisella Tularensis: Residence Time and In Vivo Activity. ACS Chem. Biol. 2009;4:221–231. doi: 10.1021/cb800306y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan TJ, Truglio JJ, Boyne ME, Novichenok P, Zhang X, Stratton CF, Li HJ, Kaur T, Amin A, Johnson F, Slayden RA, Kisker C, Tonge PJ. High affinity InhA inhibitors with activity against drug-resistant strains of Mycobacterium tuberculosis. ACS Chem Biol. 2006;1:43–53. doi: 10.1021/cb0500042. [DOI] [PubMed] [Google Scholar]
- 43.Tonge PJ. Unpublished work. [Google Scholar]
- 44.Rawat R, Whitty A, Tonge PJ. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc Natl Acad Sci U S A. 2003;100:13881–13886. doi: 10.1073/pnas.2235848100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu L, Lin J, Ma J, Cronan JE, Wang H. The Triclosan Resistance of Pseudomonas aeruginosa PA01 is Due to FabV, a Triclosan-Resistant Enoyl-Acyl Carrier Protein Reductase. Antimicrob. Agents Chemother. 2009 doi: 10.1128/AAC.01152-09. Epub Nov 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nierman WC, DeShazer D, Kim HS, Tettelin H, Nelson KE, Feldblyum T, Ulrich RL, Ronning CM, Brinkac LM, Daugherty SC, Davidsen TD, Deboy RT, Dimitrov G, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Khouri H, Kolonay JF, Madupu R, Mohammoud Y, Nelson WC, Radune D, Romero CM, Sarria S, Selengut J, Shamblin C, Sullivan SA, White O, Yu Y, Zafar N, Zhou L, Fraser CM. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14246–14251. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng AC, Currie BJ. Melioidosis: Epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]