Abstract
IL-10 plays an essential part in controlling inflammation and instructing adaptive immune responses. Consequently, dysregulation of IL-10 is linked with susceptibility to numerous infectious and autoimmune diseases in mouse models and in humans. It has become increasingly clear that appropriate temporal/spatial expression of IL-10 may be the key to how IL-10 contributes to the delicate balance between inflammation and immunoregulation. The mechanisms that govern the cell type- and receptor-specific induction of IL-10, however, remain unclear. This is due largely to the wide distribution of cellular sources that express IL-10 under diverse stimulation conditions and in a variety of tissue compartments. Further complicating the issue is the fact that human IL-10 expression patterns appear to be under genetic influence resulting in differential expression and disease susceptibility. In this review, we discuss the cellular sources of IL-10, their link to disease phenotypes and the molecular mechanisms implicated in IL-10 regulation.
Keywords: IL-10, Inflammation, Human, Immunoregulation, Gene expression, Polymorphism, Transgenic, SNP
Introduction
In the absence of appropriate feedback control, inflammatory responses can lead to vast immunopathology and death. Due to its primary ability to restrain inflammation, IL-10 has been a topic of long-standing interest. The majority of the work describing the biological roles of IL-10 in vivo has been done in the mouse by taking advantage of robust models of infectious and autoimmune diseases. Numerous clinical observations, however, have validated mouse studies by linking IL-10 levels with disease outcomes. Likewise, disease association studies identifying correlations between single nucleotide polymorphisms (SNPs) around the IL10 gene and disease susceptibility (as well as IL-10 levels) have bolstered the belief that appropriate regulation of IL-10 expression is fundamental to governing host inflammatory responses.
IL-10 was originally described by its ability to inhibit T-helper (Th) 1 activation and Th1 cytokine production. Thus, IL-10 was previously named cytokine synthesis inhibitory factor (CSIF) [1] and was found to be expressed by a variety of cell types including macrophages, dendritic cell subsets, B cells, several T-cell subpopulations including Th2 and T-regulatory cells (Tregs) and NK cells [2]. It is now recognized that the biological effects of IL-10 are directed at antigen-presenting cells (APCs) such as macrophages and DCs. Thus, the actions of IL-10 on T-cell development and differentiation are largely indirect by inhibiting macrophage/dendritic cell activation and maturation [3]. IL-10 antagonizes the expression of MHC class II and the co-stimulatory molecules B7.1/B7.2 (CD80/CD86) as well as the pro-inflammatory cytokines IL-1β, IL-6, IL-8, TNF-α, and notably IL-12 [4, 5]. Inhibition of antigen-presenting cell function may be a predominant mechanism by which IL-10 regulates antigen-specific Th-cell populations and adaptive immune responses in general with the goal of limiting immunopathology [6]. However, the biological role of IL-10 is not limited to inactivation of APCs as IL-10 is also known to enhance B cell, granulocyte, mast cell, and keratinocyte growth/differentiation, as well as NK-cell and CD8+ cytotoxic T-cell activation [2, 5, 7–9]. In addition, IL-10 has been shown to enhance NK-cell proliferation and/or production of IFN-γ [10–12].
IL-10 has been implicated as a key regulator of host inflammatory responses during infection with a variety of parasitic, bacterial, viral, and fungal pathogens [reviewed in 13]. Although there is a clear association between IL-10 and disease susceptibility, less is known regarding the cellular sources of IL-10 which mediate disease phenotypes. This is complicated by the fact that IL-10 is regulated by various receptor systems and is expressed by a wide array of cell types. In the case of infectious disease in particular, anti-inflammatory properties of IL-10 creates somewhat of a paradox. On the one hand, the initiation of inflammatory responses is required for effective responses against harmful pathogens, but if left unchecked, can result in inflammatory disorders, autoimmunity and even some cancers. On the other hand, in IL-10 expression can facilitate pathogen survival and the establishment of persistent infection such as during Leishmania and LCMV infection [14, 15]. Therefore, resolving the cellular sources and temporal/spatial expression profiles of IL-10 in vivo remains a priority.
Cell-specific expression
T cells
Based on a large body of evidence, T cells are thought to be the main source of IL-10 in vivo and by utilizing various mouse models, the critical role of T cell-derived IL-10 has been clearly demonstrated both in maintaining immune homeostasis and in enabling microbial persistence [reviewed in 16]. IL-10-expressing CD4+ T cells come in a variety of flavors, but each is suspected of delivering IL-10 at sites of inflammation. An important question pertains to the developmental origins of IL-10 expressing T cells, and the advent of several IL-10 transgenic reporter mice in recent years has proven useful in tracking IL-10-producing cells in vivo and provided more insight into how and perhaps where these regulatory cells develop the capacity to express IL-10 [17]. Nonetheless, it is now accepted that IL-10 is expressed by subsets of all CD4+ T helper populations including Th1, Th2, and Th17 [18]. In addition, T regulatory (Treg) subsets are also a key source of IL-10 in vivo and play a central role in mediating inflammatory control functions [19].
The importance of IL-10 in such responses is particularly evident in the gut, where despite the large burden of commensal bacteria, there is a delicate balance of pro-inflammatory and anti-inflammatory cytokines which act in concert to maintain a steady state. In Il10−/− mice, however, the inflammatory balance is disrupted, and these mice exhibit exaggerated inflammatory responses in the intestinal mucosa and develop severe inflammatory bowel disease (IBD) [20, 21]. When Il10−/− animals are cleared of the gut flora, mice are free of IBD, indicating the indispensible role of IL-10 in brokering tolerance in the gut microenvironment [reviewed in 22]. This evidence is supported by studies that suggest that defective IL-10 expression is central to the pathogenesis of IBD in humans, while other reports have linked SNPs around the IL10 gene with IBD [23–25].
While the origins of IBD are unclear, T cell-derived IL-10 has been linked to the control of inflammation at mucosal surfaces and IL-10-producing regulatory T cells have been shown to protect against enterocolitis in mice [26]. Conditional deletion of the Il10 gene in CD4 T cells demonstrated a role for T cell-derived IL-10 in regulating inflammatory responses in the gut as these mice not only developed colitis, but also displayed augmented contact hypersensitivity reactions [27]. In addition, conditional deletion of Il10 in Foxp3+ Tregs results in a failure to regulate inflammation in the gut, skin, and lung [28] which ultimately makes a solid argument for the significance of CD4 T cell-derived IL-10 in limiting inflammatory responses particularly at “environmental surfaces”.
Other model systems have been instrumental in defining the importance of T cell-derived IL-10 in regulating effector T-cell responses. For example, in the absence of IL-10, infection with the intracellular parasite Toxoplasma gondii leads to severe immunopathology and eventual death due to a failure to down-regulate inflammatory and effector responses [reviewed in 29]. Again, T cells have been implicated as a critical source of IL-10, as T cell-specific deletion of IL-10 also results in immunopathology following infection with T. gondii [27]. Recent work indicates that T-bet+Foxp3− Th1 cells are in fact the predominant source of IL-10 during T. gondii infection and play a critical role in controlling the collateral damage associated with the Th1 response to this intracellular pathogen [30]. The significance of this work is indicated by the conclusion that a single CD4 subset (Th1 cells) appears to mediate resistance to the pathogen and limit immuno-pathology by acquiring the ability to express IL-10.
Conversely, under circumstances when there is inappropriate induction of IL-10, the host may be resistant to immunopathology yet susceptible to infection due to dampened inflammatory responses. As an example, transgenic mice that constitutively over-express human or mouse IL-10 are highly susceptible to infection with intracellular pathogens [31, 32]. This may be particularly relevant for pathogens that look to establish persistent infections. This is well illustrated by the discovery that several viral genomes contain highly homologous, functional IL-10 homologues including cytomegaloviruses [33], a pox virus [34], and various herpesviruses [35–38]. The capture of cellular IL10 genes (or mRNAs) by these viruses suggests a survival advantage for viruses that encode their own source of IL-10 to presumably impede host immune responses [39]. Similarly, the preferential induction and/or expansion of IL-10-expressing host cells is another tactic to repress immune responses that some viruses (such as lymphocytic choriomeningitis virus—LCMV) utilize to gain a selective survival advantage in vivo [15].
Along these lines, Leishmania parasites establish persistent infections in susceptible hosts, and IL-10 is a critical player in leishmaniasis pathology as lack of IL-10 confers resistance to disease [14, 40–42]. Although human forms of leishmanial infection are associated with elevated IL-10 levels [43], the cellular source(s) have been difficult to determine and direct evidence for the role of IL-10 in leishmaniasis is based primarily on mouse models. Nonetheless, in patients suffering from visceral leishmaniasis, splenic aspirates depleted of CD25+ cells expressed the highest IL-10 mRNA levels and were the main lymphocyte subset in the spleen [44]. In another report, increases in IL-10+ CD4 and CD8 T cells in patients correlated with active disease [45]. Likewise, in mouse models, T cell-derived IL-10 from CD4+ subsets also plays a critical role in pathogen persistence in Leishmania major [46] and L. donovani (a causative agent of visceral leishmaniasis) infection models [47]. Although multiple cell types express IL-10 during leishmania infections, recent findings suggest that IL-10-producing Th1 cells are key players in the IL-10-mediated immune suppression as CD4+CD25−Foxp3− Th1 cells were found to be necessary for suppression of acquired immunity in leishmania major-infected Rag−/− recipient mice [46]. Leishmania pathogenesis is discussed in more detail by S. Stäger in this issue.
Various CD4+ and even some IL-10-expressing CD8+ [48, 49] T-cell subpopulations have been implicated in contributing to if not mediating disease pathology. Yet it is still not clear how these subsets acquire the capacity to express IL-10 and whether innate-derived IL-10 fosters the development of IL-10-secreting T cells in vivo. In that regard, IL-12 and the IL-12 homolog IL-27 have been clearly linked to the development of IL-10-expressing T cells in both humans and mice [50–54]. In mouse and human T cells, IL-12 has been implicated in the differentiation of T-cell subpopulations that express both IL-10 and IFN-γ [55, 56]. As mentioned, these IL10+IFN-γ+ T cells are well described in response to infection with certain intracellular pathogens and are thought to involve the Th1 transcription factor T-bet [30, 57, 58]. Recent efforts suggest that Stat4-dependent IL-12 signaling works in conjunction with high antigen dose to preferentially induce IL-10 in Th1 cells [59]. While IL-12 appears to be required for the development of both IL-10 and IFN-γ expression in Th1 cells, the IL-27 pathway seems to have more broad ability to induce IL-10 expression from several CD4+ subsets namely Th0, Th1, Th2, and Th17 cells which could be in part a function of differential receptor distribution. Thus, T cell-derived IL-10 has been demonstrated to play an integral role in restricting host responses, sometimes to the detriment to the host. Nevertheless, other cellular sources of IL-10 have established links to disease susceptibility.
Innate APCs
Antigen-presenting cells (APCs) are another potentially an important source of IL-10 given that expression of IL-10 by innate cells may represent an early autocrine feedback mechanism to constrain activation of APCs thereby influencing the development of adaptive responses. Within the innate APC compartment, various cell types can be divided into distinct subpopulations based on IL-10 expression patterns. Most notably, monocytes, alternatively activated macrophages (AAM), type II macrophages, and some dendritic cell (DC) subsets including myeloid (but not plasmacytoid) DCs provide sources of IL-10 from the myeloid lineage [60, reviewed in 61].
The immunoregulatory function of myeloid-derived IL-10 in vivo is well characterized during sepsis and the anti-inflammatory response to lipopolysaccharide (LPS) [62, 63]. In animal models of endotoxic shock, blockade of IL-10 (with blocking antibodies or in Il10-deficient mice) results in uncontrolled systemic inflammatory responses typified by pro-inflammatory cytokines which leads to death [62, 64–68]. Treatment with IL-10 or IL-10-expressing cells reduces inflammatory cytokine production and prevents lethality [69–74]. Similarly, in human sepsis IL-10 administration inhibits pro-inflammatory cytokines [75] and blocks the activation of monocytes [76]. Human IL-10 protein has conserved biologic properties in mice and thus can control sepsis either by administration of recombinant human IL-10 or by transgenic over-expression of human IL-10 in the myeloid compartment [77, 78]. A clear role for myeloid-derived IL-10 in mediating the anti-inflammatory response to endotoxin was demonstrated in mice deficient for IL-10 specifically in myeloid cells [63]. Similarly, the skin irritation response to tetradecanoyl-phorbol acetate (TPA) is enhanced in Il10−/− and myeloid-specific Il10−/− [79] mice but not mice deficient in T cell-derived IL-10 [27].
IL-10-producing APCs have distinct biological features and gene expression profiles which are known to have a profound influence on the differentiation, maintenance, and function of various T-cell subsets [80]. Some macrophage subsets develop a high IL-10/low IL-12 phenotype in response to FcγR engagement which can lead to the skewing of adaptive immune responses [74]. In the context of IL-10-dependent phenotypes generally attributed to T cell-derived IL-10, including susceptibility to leishmania and maintenance of gut homeostasis, the supply of innate IL-10 should be considered in defining the phenotype. In fact, Leishmania-specific antibodies have also been shown to induce IL-10 production by macrophages through ligation of FcγRs with IgG-opsonized parasites [40]. Indeed, antibody-induced macrophage-derived IL-10 has been shown to play a very important role in disease progression in a low dose model of L. major [81]. Similarly, in experimental autoimmune encephalomyelitis (EAE) model of multiple sclerosis, type II monocytes were shown to have increased IL-10 (and TGF-β) production leading to the differentiation of Th2 and Tregs cells that modulated T cell-mediated inflammation in the brain [82].
As discussed previously, T cell-derived IL-10 is a critical factor influencing inflammatory responses in the gut. Recent studies, however, point to tissue-localized CD11b+ myeloid cells, which express IL-10 (and other anti-inflammatory molecules) in the lamina propria (LP) and induce Foxp3+ Tregs [83]. Another group detailed the influence of IL-10-expressing myeloid cells in maintaining Foxp3 expression in regulatory T cells in the context of gut inflammation, suggesting that maintenance of gut homeostasis depends on the carefully orchestrated temporal/spatial regulation of IL-10 from its numerous cellular sources [84]. Since APCs are critical for sensing potentially harmful pathogens, initiating inflammatory responses and directing adaptive immunity, it will be important to determine how innate sources of IL-10 influence host-adaptive responses to inflammation.
NK cells
Natural killer (NK) cells are another potentially important, yet underappreciated, innate source of IL-10. NK and NKT cells are integral parts of the innate immune system and are critical to host defense against infectious pathogens such as herpes viruses [85, 86], and choriomeningitis virus [87]. NKT cells are also a source of IL-10 production that has been suggested to be required for antigen-specific Treg differentiation and the induction of tolerance in some mouse models [88]. Both NKT and NK cells provide early sources of IFN-γ in vivo. As mentioned previously, IL-12 has been implicated in the regulation of IL-10 expression in T cells and similarly is a known regulator of NK cell-derived IL-10 [89]. Our group and others have shown that IL-2, IL-12, and the combination are potent regulators of IFN-γ expression in NK cells [90]. Interestingly, IL-2 [91] and IL-12 also induce IL-10 expression in NK cells, and the combination of IL-2 and IL-12 is synergistic for mouse and human IL-10 [92, 93]. We found this effect is dependent on Stat4 and identified a conserved STAT motif within a conserved non-coding sequence (CNS) in the 4th intron of the mouse and human IL10 genes which binds IL-12-induced Stat4. Unlike IL-12, we found that IL-27 did not induce IL-10 expression in NK cells, suggesting that the role of IL-27 in regulating IL-10 may be restricted to T cells.
The functional significance of NK cell-specific IL-10 is not clear, but several reports have suggested NK cells may have regulatory activity in certain contexts including pregnancy, during tumor development, and chronic hepatitis C infection [94–96]. By adding experimental evidence, a series of recent publications provide further indication that NK cell-derived IL-10 may contribute to disease susceptibility. In the case of experimental visceral leishmaniasis, Maroof et al. found that NKp46+CD49b+ NK cells are recruited to sites of inflammation (spleen and liver) and contributed to the inhibition of host protective immunity [97]. In addition, NK sources of IL-10 were implicated to provide protection to immunopathology associated with mouse cytomegalovirus (MCMV) infection [98]. Finally, in a comprehensive report, Perona-Wright and colleagues assessed the role of NK-specific IL-10 during infection based on whether the pathogen established local or disseminated infections. Their results suggest that in response to rapidly disseminating pathogens (such as Yersinia pestis, and T. gondii), NK cells quickly begin expressing IL-10 in an IL-12-dependent fashion. Interestingly, during locally restricted infections (influenza and a Y. pestis strain which cannot establish systemic infection) NK cells did not express IL-10 and enlisted other sources of the cytokine (Perona-Wright et al. unpublished data) (personal communication M. Mohrs). The biological significance of NK-derived IL-10 is not completely established but not surprisingly appears to be part of an important feedback mechanism during infection to restrict inflammatory responses perhaps at specific tissues sites [99].
B cells
IL-10 is also expressed by B cells, and negative regulatory roles for B cells have been implicated in a number of autoimmune and infectious diseases [reviewed in 100]. A recently reported B-cell Il10-deficient reporter mouse indicated a role for B cell-derived IL-10 in limiting virus-specific CD8+ T-cell responses [101]. Just as T-cell subsets seem to preferentially express IL-10, B-cell subpopulations (B1 and B2) are also known to differentially produce IL-10 [102, 103]. In particular, CD5+ (Ly1+) B cells produce more IL-10 than CD5− B cells [102, 104, 105]. More specifically, the CD1dhiCD5+ B-cell subset (B10 cells) has been suggested to account for the majority of B cell-derived IL-10 [106]. Other reports, however, indicate that the CD5− subset can also express IL-10 in response to LPS treatment [107], and in an IL-10-GFP reporter mouse, GFP expression was noted in multiple CD19+ B-cell subsets following LPS challenge. In addition, other Toll ligands such as TLR9 have been shown to regulate B-cell production of IL-10 [108], and incubation of B cells with Staphylococcus aureus also induces IL-10 expression [109]. Furthermore, transformed B-cell lines are known to produce significant amounts of IL-10 [110].
The role of B-cell production of IL-10 has been suggested as an autocrine mechanism to promote B-cell survival and effector function [104, 111]. However, as mentioned, IL-10-producing B cells can play an important role in directly suppressing immune responses and inflammation [101]. Thus, B cells express IL-10 in response to various signaling pathways, yet very little is known about the mechanisms that control B-cell expression of IL-10.
Genetic control of IL-10
A potentially important consideration in the analysis of human IL-10 expression is the well-described inter-individual variation in IL-10 production which is associated with single nucleotide polymorphisms (SNPs) in the IL-10 promoter [112–115]. As mentioned, there are links between absolute levels of IL-10 and disease susceptibility [114]. In addition, numerous studies have demonstrated that IL-10 promoter SNPs are also correlated with differential risk for a variety of infectious and autoimmune diseases and some cancers presumably due at least in part to altered IL-10 expression [112–120]. In fact, IL-10 production levels can be stratified from low to high [121] which is based on SNP haplotype blocks upstream of IL10 [122–124]. This may make evolutionary sense, as evidence suggests that the IL10 locus is under selective pressure from various pathogens [125, 126]. The indication that IL-10 production is determined on a genetic basis is supported by the concordance of IL-10 levels in monozygotic twins, which suggests that up to 70% of IL-10 production is genetically determined [122]. Another group, however, suggests that 50% of the variability observed in IL-10 expression patterns between people may be explained by genetic factors [127]. Nonetheless, the analysis of IL-10 regulation in humans is obscured by genetic as well as other host factors [127].
The majority of genetic association studies have focused on a series of 3 SNPs in the 5′ proximal region adjacent to IL10 consisting of –1082G/A (rs1800896), –819C/T (rs1800871), and –592C/A (rs1800872) [128]. These SNPs are in tight linkage disequilibrium which results in 3 well-defined haplotypes: ATA, ACC, and GCC. For example, a recent report has demonstrated that susceptibility to infection with L. braziliensis is linked to a certain polymorphism in the IL10 promoter, the IL10-819C/C genotype, which is associated with higher levels of IL-10 [124]. Previously, we identified that these IL10 alleles were predictive of HIV progression rates [119]. In addition, we found that the SNPs in this haplotype block display unique DNA–protein-binding patterns, which suggests that these allelic variants may influence gene expression. It is important to appreciate, however, that individual IL10 promoter SNPs are part of larger haplotypes that may extend several hundred kilobases over the entire lL10 gene cluster [123, 125, 129]. In fact, a recent study was unable to find a link between IL10 promoter SNPs and IBD, but did find a strong association between a SNP 3′ to the IL10 gene and ulcerative colitis [23]. Thus, SNPs with putative function may actually be in linkage disequilibrium or a part of a cumulative effect of the extended haplotype. Nevertheless, this highlights the need to characterize the molecular and genomic requirements for appropriate cell type-specific IL-10 expression in both mouse and human systems.
The hIL10BAC model
The importance of appropriate cell type-specific IL-10 regulation has been clearly demonstrated in the mouse, yet the contribution of cell-specific IL-10 expression in human disease is lacking. For obvious technical, ethical, and practical considerations, much of the work to characterize the cellular sources of IL-10 in vivo has been done in mice. As mentioned, however, the study of human IL-10 regulation is potentially complicated by interindividual variation in gene expression. To address the issue of tissue-specific human IL-10 regulation and function while avoiding variation introduced by host genetic factors, we created transgenic mice with a human bacterial artificial chromosome (BAC) in which the IL10 gene is positioned centrally (hIL10BAC) [50]. Our goal was not only to monitor tissue-specific human IL-10 expression, but also to determine the functional impact of genomic regulation of human IL-10 in vivo. Since human IL-10 protein has conserved biologic properties in mice [31, 77], our model offers the opportunity to monitor gene regulation and in vivo function simultaneously. We tested the function of the human IL10 BAC construct in vivo by reconstituting Il10−/− mice with the hIL10BAC transgene through backcrossing (Il10−/−/hIL10BAC). We found that Il10−/−/hIL10BAC mice (which only express human IL-10) competently regulate IL-10-target genes and normalize sensitivity to LPS-induced lethality observed in Il10−/− mice, presumably by human IL-10 derived from myeloid sources. Indeed, human IL-10 is faithfully expressed in hIL10BAC bone marrow-derived macrophages (BMM) and DCs. To examine the likely contribution of T cell-derived human IL-10 on disease susceptibility, we utilized the IL-10-dependent Leishmania donovani model of chronic infection. WT mice predictably exhibited high parasite burdens in the liver and spleen; however, we were surprised to find that both Il10−/− and Il10−/−/hIL10BAC mice efficiently cleared the parasites. Similarly, CD4+ T cells cultured with the IL-10-promoting cytokine IL-27 strongly enhanced mouse but not transgenic human IL-10 production.
We concluded that the human IL10 BAC construct encodes for weak IL-10 expression in T cells. We cannot completely rule out the possibility that the BAC is missing a key regulatory element(s) which governs IL-10 expression in T cells. However, BACs (and other large genomic DNA constructs) are generally recognized to contain most, if not all, regulatory regions to support proper cell type-specific transgene expression profiles [130]. An additional question is whether the mouse transcriptional machinery can appropriately regulate a large human DNA cassette. Multiple studies indicate that cell-specific gene expression can be transferred across species to mice [131, 132]. In fact, Frazer and colleagues showed that the gene encoding human apolipoprotein(a) is appropriately regulated in yeast artificial chromosome (YAC) transgenic mice, despite the fact that this gene is not present in the mouse genome [133]. Another recent study demonstrated that the mouse transcriptional machinery supports faithful species-specific gene expression profiles across an entire human chromosome [134].
There is no question that human T cells have the capacity to express IL-10, sometimes at high levels [55, 58, 135]. In naïve human CD4+ T cells, we observed inter-individual variation in IL-10 production in response to IL-27 co-culture ranging from low (as observed in hIL10BAC mice) to high [50]. Therefore, the hIL10BAC is a model for low T cell-derived IL-10 production. Another group has also described IL-27 regulation of IL-10 in human CD4+ T cells [51]. Another report suggests that high levels of T cell-derived human IL-10 may arise from specific cellular subsets including a recently described rare population of CD4+CD45RA−CD25−IL-7R− which produce high levels of IL-10 and IFN-γ [136].
There are still unresolved questions raised from the hIL10BAC model including if other tissue-compartments (beyond the spleen) can harbor T cells enriched for human IL-10 production, such as the gut. This could suggest the molecular mechanisms which regulate cell type-specific IL-10 may depend on the tissue source of the cell population and emphasize importance of the microenvironment in the development of IL-10-expressing cellular subsets. In addition, it will be important to determine whether other stimulation conditions can normalize the weak T cell-specific human IL-10 expression observed in hIL10BAC mice. Finally, this model may serve as a template to characterize the influence of non-coding SNPs on human IL-10 gene expression profiles and the subsequent influence on disease susceptibility.
IL-10 gene regulation
There is still much to learn about the genomic and molecular requirements for appropriate IL-10 expression, but it is clear that IL-10 is regulated at both the transcriptional and post-transcriptional levels [137, 138]. The majority of the work on IL-10 regulation, however, has been concentrated on acute transcription and only more recently on epigenetic events associated with cell type-specific expression. The net result of which is altered accessibility of DNA to transcriptional regulators which lead to competency or repression of gene expression. These approaches have been employed to dissect both epigenetic and transcriptional control mechanisms that determine tissue-specific expression of IFN-γ in Th1 cells and IL-4, IL-13, and IL-5 expression in Th2 subsets [139–141]. These robust approaches are just beginning to be applied to the regulation of the IL-10.
Signaling pathways, receptors, and transcription factors
Regarding transcriptional control of IL-10, toll receptor signaling in the myeloid compartment has been studied somewhat more extensively. The literature has implicated the ubiquitously expressed transcription factors Sp1 and Sp3 as dominate players in controlling transcription of the II10 gene at least in macrophages [142, 143]. The proximal promoter region of IL-10 has several GC-rich sequences, which the Sp-family of transcription factors preferentially bind. Several studies have documented that LPS-induced Sp1 and Sp3 can bind to these sequence motifs in the proximal IL-10 promoter [142, 144, 145]. Site-directed mutagenesis and deletion analysis of short reporter constructs indicates the involvement of these Sp-family sites in regulating IL-10 promoter activity in LPS-stimulated cell lines. Interestingly, both the p38 and ERK MAPK pathways have been linked to recruitment of Sp1 to the IL-10 promoter [144, 146, 147].
It is now evident that multiple regulatory pathways are involved in IL-10 regulation. For instance, analysis of the proximal IL-10 promoter revealed several putative cAMP-response elements (CRE) which bind cAMP-responsive element-binding protein/activating transcription factor (CREB/ATF) [148, 149]. Signal transducer and activator of transcription 3 (Stat3) is activated by IL-10 receptor signaling and is required for the anti-inflammatory properties of IL-10. Interestingly, Stat3 has also been linked to regulation of the IL10 gene via several different signaling pathways including IFN-α [150], LPS [151], and glucocorticoids [152]. In addition, the regulation of IL-10 expression in CD4+ T cells by IL-27 is at least partly dependent on Stat3 and Stat1 [52]. This is interesting given that Stat3 is critical for IL-10 receptor signaling and could suggest a positive feedback mechanism.
Other STAT family members have been implicated in IL-10 regulation. IL-12 has been shown previously to regulate IL-10 expression in human T-cell clones [55], and recent reports identified a role for IL-12-induced Stat4 in the development and maintenance of IL-10-expressing Th1 cells [59, 153]. This line of evidence was also reported to include Stat4-dependent notch signaling [154]. Our group identified a Stat4-binding element in the 4th intron of the mouse and human IL10 gene [93]. The dependence of Stat4 on IL-12-induced expression of IL-10 in NK cells was determined in Stat4−/− mice which failed to up-regulate IL-10 in response to IL-12 stimulation. Stat4 is a substrate for the map kinase p38 [155], and we also found that by blocking the p38 pathway, IL-12 regulation of IL-10 was impaired. In addition, another group shortly after identified the same intronic site bound IL-2-induced Stat5 in human T cells [156]. Interestingly, the STAT-motif is completely conserved between mouse and human, yet IL-2 does not induce the recruitment of DNA-binding complexes to this region in the mouse Il10 gene [93].
A recent study characterized a role for Ikaros (and GATA3) in regulating IL-10 in mouse Th2 cells where several Ikaros-binding elements were identified, and one of these conserved sites interestingly was in the 4th intron [157]. Still other groups have found members of the AP-1 family bind the IL-10 promoter in Th2 cells [158], and in a B-cell line, an NF-Y/HMG-I(Y) binding element has been identified [159]. The protooncogene c-Maf has also been implicated in regulating LPS induction of IL-10 expression, specifically in macrophages [160]. In addition, the MAPK c-Jun N-terminal kinase 1 (JNK1) has been implicated as a negative regulator of IL-10 [161]. Finally, members of the NF-κB family including p50 [162] and p65/RelA [163] have also been associated with LPS induction of IL-10 expression in APCs. Thus, it is clear that multiple receptors, signaling pathways and transcriptional activators play a part in the regulation of IL-10 transcription. Other data is continuing to emerge, suggesting that various epigenetic events may help to shape cell type-specific regulation of IL-10 expression.
Chromatin structure
The status of post-translational histone modifications adjacent to a gene locus are correlated with the ability of a gene to be transcribed and serves as a marker of epigenetic changes and perhaps disease status [164, 165]. Recently, some groups have begun to consider chromatin structure in the II10 locus, as it relates to cell type-specific IL-10 expression (Fig. 1). For instance, studies from the Mosser group indicate stimulation-specific (FcγR) induction of chromatin remodeling in the IL-10 promoter in macrophages [146]. In addition, this group found FcγR-inducible, ERK-dependent histone phosphorylation and Sp1 recruitment to the IL-10 promoter in macrophages [166]. Another study indicates the presence of Th2-specific chromatin structure, inducible histone modifications, and DNA methylation patterns within the II10 gene that correlate with IL-10 expression and localize to several conserved non-coding (CNS) regions in the II10 locus [167]. Other reports have identified intronic regions and/or sites 3′ to II10 that are remodeled in T cells which express IL-10 [158, 163]. Interestingly, Saraiva and colleagues have identified a lineage-specific DNaseI hypersensitivity site (HS) in macrophages that maps to a region 5′ to the Il10 gene in the promoter. This group has also implicated the Th2 transcription factor GATA3 in regulating mouse Il10 locus in T cells, despite the fact that GATA3 binding did not transactivate the IL-10 promoter [168] but was found to bind the Il10 promoter in Th2 cells [153].
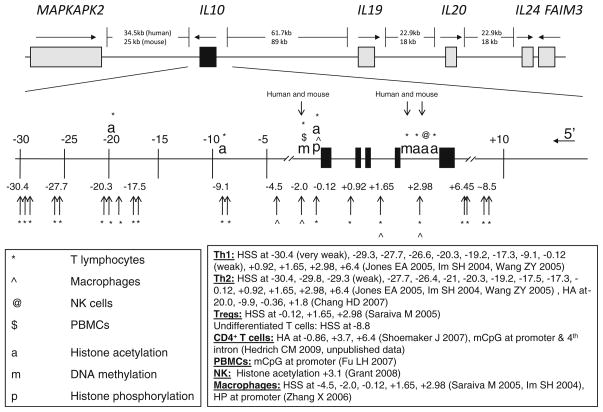
Fig. 1.
Known regulatory regions within and flanking the IL10 gene. In the top panel, the physical organization of the IL10 cluster is displayed, including intergenic regions and distances between all genes in the cluster. The lower panel displays regions of epigenetic remodelling that have been investigated by various groups and in different cellular subsets. The tables beneath the figure explain the used symbols and give an overview on the cellular subsets and specific epigenetic modifications in each investigated region
A more comprehensive analysis of the mouse II10 gene structure and expression focused on CD4+ Th1/Th2 cells in which the authors identified multiple DNase I hypersensitivity sites between the II10 and Il19 genes in the mouse and described three distal enhancer elements [169]. In addition, they demonstrated that some, but not all of the most highly conserved CNS regions between II10 and Il19 transcribe intergenic RNA in mouse T cells. This work provides an insight into Th1/Th2-specific IL-10 expression and chromatin structure between the murine II10 and ll19 genes.
Recently, Villagra showed that histone deacetylase 11 (HDAC11), by interacting with the distal segment of the promoter Il10, negatively regulated the expression of IL-10 in mouse and human APCs [170]. Along those lines, others have proposed that in models of inflammation-mediated tumorigenesis, inhibition of histone deacetylases resulted in a significant suppression of tumor growth in terms of size and number, along with reduced signs of inflammation [171]. It was suggested that as a result of deacetylase inhibition, increased acetylation of histone 3 and subsequent expression of IL-10 lead to reduced production of IFN-γ, enhanced apoptosis in lamina propria mononuclear cells and activation of NF-κB. It will be important to define the chromatin structure of the Il10 locus in the various IL-10-expressing cell types, as they acquire the ability to express IL-10 under different physiological conditions.
DNA methylation
Changes in cytidine phosphate guanosine (CpG) DNA methylation is a key mechanism controlling transcription while establishing stable heritable epigenetic marks [172]. At present, DNA methylation is the sole mechanism by which epigenetic information is faithfully propagated to the next generation [173]. Accumulating evidence indicates a role for DNA methylation in regulating cytokine expression largely based on studies in the Il4 and Ifng gene clusters [174]. In IL-4-expressing Th2 subsets, demethylation of regulatory regions of the Il4 gene was detected compared to Th1 and naïve CD4+ T cells [175, 176], whereas Ifng was selectively demethylated in IFN-γ-secreting Th1 cells [139, 177]. Less is known about the role of DNA methylation in cell type-specific regulation of IL-10 however.
Dong et al. [178] found no clear correlation between DNA methylation and IL-10 expression in selected conserved noncoding sequence (CNS) regions over the human IL10 cluster in Th1 and Th2 cells. Recently, another group demonstrated the involvement of CpG DNA methylation around a Stat5-binding element in the 4th intron of the human IL10 gene response to IL-2 stimulation in human T-cell lines [156]. In contrast, Szalmas showed that a high degree of DNA methylation in primary human “normal” and neoplastic cells, as well as in cell lines is associated with transcriptional silencing, whereas IL-10-expressing PBMCs showed low degrees of IL10 promoter methylation [179]. In agreement with these findings, another group [180] observed a negative correlation between IL10 promoter methylation and IL-10 expression in human PBMCs of RA patients and a higher degree of methylation of the IL10 promoter in RA patients compared to healthy controls.
In our hIL10BAC model, we observed disparities in CD4+ T cell-derived human and mouse IL-10 expression, and questioned if differential DNA methylation was involved. Across the mouse and human IL10 genes, there were no obvious patterns of CpG methylation in naïve and polyclonally activated CD4+ T cells. However, when compared to mouse Il10, the human gene showed higher levels of overall DNA methylation even following activation. Nonetheless, Th0 cells cultured in the presence of IL-27 displayed a site-specific reduction in DNA methylation of the mouse, but not human IL10 gene which correlated with high levels of mouse IL-10 expression. Demethylation occurred in the 4th intron previously reported to undergo activation- and differentiation-dependent epigenetic changes and transcription factor recruitment. Collectively, these results suggest that during T-cell activation, there are variable changes in DNA methylation patterns across the mouse and human IL10 genes. Our data, however, indicates a putative molecular mechanism by which IL-27 controls IL-10 induction at least in mouse CD4+ T cells (Hedrich, CM, unpublished data).
Intergenic transcription
Intergenic transcription is a potentially important, yet poorly understood mechanism to influence gene expression that is thought to be at the interface between chromatin remodeling and transcription of adjacent genes [181]. There is a strict correlation between chromatin structure (including hyperacetylation) and the presence of intergenic transcripts that is thought to be a mechanism to maintain an extended, “open” chromatin conformation [182]. This could permit interactions between distal regulatory elements and core promoters, potentially of co-regulated genes in a cluster [183, 184].
Tissue-specific intergenic transcription has been indicated to play a role in regulating gene expression in the Il4 gene cluster [185], and intergenic transcripts have also been detected between the mouse Il10 and Il19 genes [169]. In this study, the authors found that two enhancer regions contained in CNS sites around the mouse Il10 gene also express transcripts in Th2 cells and stimulated macrophages. Some other, but not all, CNS regions between Il10 and Il19 were also found to transcribe intergenic RNA.
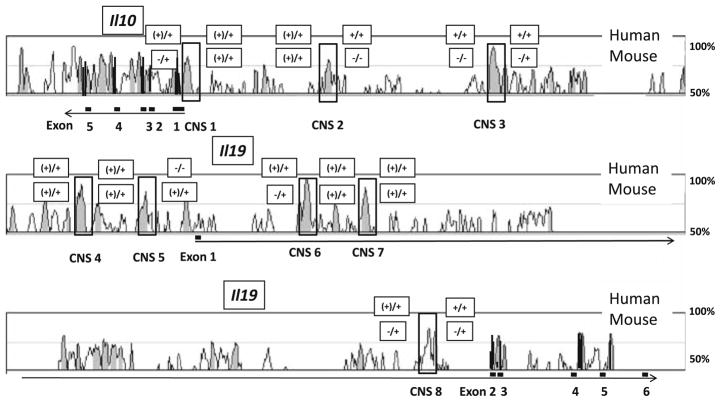
Since we have identified some differences between mouse and human IL-10 expression in hIL10BAC mice including a more prominent presence of constitutive human IL-10 mRNA in various tissues, we have begun to search for intergenic transcripts in the human IL10 locus in hIL10BAC mice [50]. We have found that in BMM from hIL10BAC mice, human and mouse intergenic transcripts are abundant both 5′ and 3′ of highly conserved CNS sites between IL10 and IL19 (Fig. 2). Interestingly, we find that some intergenic regions between mouse Il10 and Il19 have inducible transcripts, while human transcripts are present to a larger extent flanking all CNS sites tested with the exception of CNS 5. Some of the transcripts we detected map to CNS sites previously reported to transcribe intergenic RNA in mouse T cells [169]. It is not clear what these findings mean with respect to IL-10 regulation, but it is possible that alternative promoters may be involved in the complex cell type (and potentially species) -specific regulation of IL-10 expression beyond the TATA-containing proximal promoter currently identified.
Fig. 2.
Intergenic transcription around the mouse and human IL10 genes in BMM of hIL10BAC mice. Alignment of the human and mouse IL10 genes and surrounding sequences using human as the base genome for comparison (VISTA Genome Browser—http://pipeline.lbl.gov/cgi-bin/gateway2). The height of the peaks indicates the degree of sequence conservation. Noncoding sequences with a conservation degree of 70% or more over a length of 100 base pairs or more are shaded gray and indicated by black boxes and CNS numbers. Exons are shaded black and indicated by numbered black bars under the gene. The regions flanking the indicated CNS regions were analyzed for mouse and human transcripts and are indicated by boxes. DNA contamination was ruled out by including DNase treatment controls for each sample. The results are shown in boxes with thinner lines. Upper boxes indicate human transcripts, while lower boxes indicate mouse transcripts. −/+ represents no transcripts in non-stimulated (NS) cells but inducible transcripts after stimulation (LPS + IL-4). (+)/+ represents weak transcription in NS cells with inducible transcription after stimulation. −/− represents for no transcripts in NS cells or after stimulation. CNS3 here was reported as CNS-9 previously by Jones and Flavell and may act as enhancer element in Th1 and Th2 cells [169]. CNS4, which could be an origin of intergenic transcripts in human IL10 in macrophages, map to the previously reported CNS-20B that was shown to have basic promoter function in (transfected) D10 cells. CNS5 here maps to CNS-20A [169]
4th Intron
Although multiple regions around the Il10 gene have been implicated in the cell type-specific regulation of IL-10 expression, as mentioned, evidence is accumulating that the 4th intron in particular may harbor important regulatory sequences. This is based on results from several studies and the presence of two highly conserved CNS sites, multiple conserved transcription factor binding elements, and a prominent DNase I HSS site localize to this genomic region. As mentioned, we identified a conserved Stat4 DNA-binding element within the 4th intron of the Il10 gene that is located within a CNS (CNS + 3.10) of both the mouse and human IL10 genes [93]. In addition, we found that the CNS + 3.10 region is also a target for cytokine-induced chromatin remodeling based on the enhancement of histone H3 acetylation levels following cytokine stimulation. This is the same site that binds IL-2-induced Stat5 in human T cells [156], and another region of intron 4 binds Ikaros [157]. Meanwhile, Jones and Flavell reported the presence of a non-Th-lineage-specific DNaseI hypersensitivity site in the 4th intron at position +2.98 (HSS + 2.98) (Fig. 1) [169]. Although this is likely to be the same HS site reported by Im and colleagues (HSII) [167], there are inconsistencies as to whether this HS is Th-lineage specific. Furthermore, we found that IL-27 co-culture results in selective DNA demethylation of sites in the 4th intron of the mouse Il10 gene (Hedrich et al. manuscript submitted). It will, therefore, be interesting to determine whether the genomic regulatory elements within the 4th intron of the Il10 gene plays a prominent role in regulating signal- and/or cell type-specific IL-10 expression in mouse and human cells.
Overall, by using transgenic mice, targeted deletion strategies, fine mapping of transcription factor binding sites, and epigenetic signatures, the in vivo significance of various regulatory sites across the Il4 and Ifng clusters clearly demarcate permissive and repressive loci in Th1 and Th2 cells. This is a logical method to address the molecular mechanisms and genomic regions which control cell type-specific IL-10 expression. Given the diversity of IL-10 expression profiles within the CD4+ pool alone however, it may be more challenging to define the chromatin signatures unique to each subset.
Conclusions
The precise temporal/spatial control of IL-10 expression in vivo is a key to determining host responses to inflammation. In that regard, the cellular sources of IL-10 influence disease outcomes by carefully coordinating the localization and availability of IL-10 during immune responses. It will be important to resolve the molecular mechanisms including genomic boundaries, epigenetic modifications and transcriptional as well as posttranscriptional regulators which govern cell type-specific IL-10 expression. This insight will help to facilitate the development of treatment strategies aimed at regulating inflammation and perhaps persistent infections by manipulating IL-10 [186]. In addition, it will be critical to expand our understanding of how IL-10 is regulated in humans in relation to mice. To do this, a more complete understanding of how and to what extent SNPs influence IL-10 expression and disease susceptibility will be needed.
References
- 1.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–95. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore KW, De Waal MR, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 3.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–79. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 4.D’Andrea A, Aste-Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J Exp Med. 1993;178:1041–8. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–22. [PubMed] [Google Scholar]
- 6.Mocellin S, Marincola F, Rossi CR, Nitti D, Lise M. The multifaceted relationship between IL-10 and adaptive immunity: putting together the pieces of a puzzle. Cytokine Growth Factor Rev. 2004;15:61–76. doi: 10.1016/j.cytogfr.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Mocellin S, Panelli MC, Wang E, Nagorsen D, Marincola FM. The dual role of IL-10. Trends Immunol. 2003;24:36–43. doi: 10.1016/s1471-4906(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 8.Mocellin S, Panelli M, Wang E, Rossi CR, Pilati P, Nitti D, et al. IL-10 stimulatory effects on human NK cells explored by gene profile analysis. Genes Immun. 2004;5:621–30. doi: 10.1038/sj.gene.6364135. [DOI] [PubMed] [Google Scholar]
- 9.Boulland ML, Meignin V, Leroy-Viard K, Copie-Bergman C, Briere J, Touitou R, et al. Human interleukin-10 expression in T/natural killer-cell lymphomas: association with anaplastic large cell lymphomas and nasal natural killer-cell lymphomas. Am J Pathol. 1998;153:1229–37. doi: 10.1016/S0002-9440(10)65667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata Y, Foster LA, Kurimoto M, Okamura H, Nakamura RM, Kawajiri K, et al. Immunoregulatory roles of IL-10 in innate immunity: IL-10 inhibits macrophage production of IFN-gamma-inducing factors but enhances NK cell production of IFN-gamma. J Immunol. 1998;161:4283–8. [PubMed] [Google Scholar]
- 11.Cai G, Kastelein RA, Hunter CA. IL-10 enhances NK cell proliferation, cytotoxicity and production of IFN-gamma when combined with IL-18. Eur J Immunol. 1999;29:2658–65. doi: 10.1002/(SICI)1521-4141(199909)29:09<2658::AID-IMMU2658>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 12.Chakir H, Lemay AM, Webb JR. Cytokine expression by murine DX5+ cells in response to IL-12, IL-18, or the combination of IL-12 and IL-18. Cell Immunol. 2001;212:71–81. doi: 10.1006/cimm.2001.1844. [DOI] [PubMed] [Google Scholar]
- 13.Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–7. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 14.Nylen S, Sacks D. Interleukin-10 and the pathogenesis of human visceral leishmaniasis. Trends Immunol. 2007;28:378–84. doi: 10.1016/j.it.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 2007;15:143–6. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity. 2008;28:468–76. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol Rev. 2008;226:219–33. doi: 10.1111/j.1600-065X.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–8. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 19.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annu Rev Immunol. 2009;27:551–89. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 21.van Den BM, Bachmann MF, Kohler G, Barner M, Escher R, Zinkernagel R, et al. IL-4 and IL-10 antagonize IL-12-mediated protection against acute vaccinia virus infection with a limited role of IFN-gamma and nitric oxide synthetase 2. J Immunol. 2000;164:371–8. doi: 10.4049/jimmunol.164.1.371. [DOI] [PubMed] [Google Scholar]
- 22.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–38. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 23.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–23. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 24.Fowler EV, Eri R, Hume G, Johnstone S, Pandeya N, Lincoln D, et al. TNFalpha and IL10 SNPs act together to predict disease behaviour in Crohn’s disease. J Med Genet. 2005;42:523–8. doi: 10.1136/jmg.2004.027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leach MW, Davidson NJ, Fort MM, Powrie F, Rennick DM. The role of IL-10 in inflammatory bowel disease: “of mice and men”. Toxicol Pathol. 1999;27:123–33. doi: 10.1177/019262339902700124. [DOI] [PubMed] [Google Scholar]
- 26.Denning TL, Kim G, Kronenberg M. Cutting edge: CD4+CD25+ regulatory T cells impaired for intestinal homing can prevent colitis. J Immunol. 2005;174:7487–91. doi: 10.4049/jimmunol.174.12.7487. [DOI] [PubMed] [Google Scholar]
- 27.Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, Stenzel W, et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–97. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–58. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman LA, Hunter CA. The role of cytokines and their signaling pathways in the regulation of immunity to Toxoplasma gondii. Int Rev Immunol. 2002;21:373–403. doi: 10.1080/08830180213281. [DOI] [PubMed] [Google Scholar]
- 30.Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M, et al. Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intra-cellular protozoan infection. J Exp Med. 2007;204:273–83. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groux H, Cottrez F, Rouleau M, Mauze S, Antonenko S, Hurst S, et al. A transgenic model to analyze the immunoregulatory role of IL-10 secreted by antigen-presenting cells. J Immunol. 1999;162:1723–9. [PubMed] [Google Scholar]
- 32.Feng CG, Kullberg MC, Jankovic D, Cheever AW, Caspar P, Coffman RL, et al. Transgenic mice expressing human interleukin-10 in the antigen-presenting cell compartment show increased susceptibility to infection with Mycobacterium avium associated with decreased macrophage effector function and apoptosis. Infect Immun. 2002;70:6672–9. doi: 10.1128/IAI.70.12.6672-6679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci USA. 2000;97:1695–700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleming SB, McCaughan CA, Andrews AE, Nash AD, Mercer AA. A homolog of interleukin-10 is encoded by the poxvirus orf virus. J Virol. 1997;71:4857–61. doi: 10.1128/jvi.71.6.4857-4861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore KW, Vieira P, Fiorentino DF, Trounstine ML, Khan TA, Mosmann TR. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein–Barr virus gene BCRFI. Science. 1990;248:1230–4. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- 36.Rode HJ, Janssen W, Rosen-Wolff A, Bugert JJ, Thein P, Becker Y, et al. The genome of equine herpesvirus type 2 harbors an interleukin 10 (IL10)-like gene. Virus Genes. 1993;7:111–6. doi: 10.1007/BF01702353. [DOI] [PubMed] [Google Scholar]
- 37.Rivailler P, Jiang H, Cho YG, Quink C, Wang F. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein–Barr virus animal model. J Virol. 2002;76:421–6. doi: 10.1128/JVI.76.1.421-426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivailler P, Cho YG, Wang F. Complete genomic sequence of an Epstein–Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J Virol. 2002;76:12055–68. doi: 10.1128/JVI.76.23.12055-12068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001;9:86–92. doi: 10.1016/s0966-842x(00)01919-3. [DOI] [PubMed] [Google Scholar]
- 40.Miles SA, Conrad SM, Alves RG, Jeronimo SM, Mosser DM. A role for IgG immune complexes during infection with the intracellular pathogen Leishmania. J Exp Med. 2005;201:747–54. doi: 10.1084/jem.20041470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacks D, Noben-Trauth N. The immunology of susceptibility and resistance to Leishmania major in mice. Nat Rev Immunol. 2002;2:845–58. doi: 10.1038/nri933. [DOI] [PubMed] [Google Scholar]
- 42.Murphy ML, Wille U, Villegas EN, Hunter CA, Farrell JP. IL-10 mediates susceptibility to Leishmania donovani infection. Eur J Immunol. 2001;31:2848–56. doi: 10.1002/1521-4141(2001010)31:10<2848::aid-immu2848>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 43.Kurkjian KM, Mahmutovic AJ, Kellar KL, Haque R, Bern C, Secor WE. Multiplex analysis of circulating cytokines in the sera of patients with different clinical forms of visceral leishmaniasis. Cytometry A. 2006;69:353–8. doi: 10.1002/cyto.a.20256. [DOI] [PubMed] [Google Scholar]
- 44.Nylen S, Maurya R, Eidsmo L, Manandhar KD, Sundar S, Sacks D. Splenic accumulation of IL-10 mRNA in T cells distinct from CD4+CD25+(Foxp3) regulatory T cells in human visceral leishmaniasis. J Exp Med. 2007;204:805–17. doi: 10.1084/jem.20061141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganguly S, Das NK, Panja M, Pal S, Modak D, Rahaman M, et al. Increased levels of interleukin-10 and IgG3 are hallmarks of Indian post-kala-azar dermal leishmaniasis. J Infect Dis. 2008;197:1762–71. doi: 10.1086/588387. [DOI] [PubMed] [Google Scholar]
- 46.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–97. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stager S, Maroof A, Zubairi S, Sanos SL, Kopf M, Kaye PM. Distinct roles for IL-6 and IL-12p40 in mediating protection against Leishmania donovani and the expansion of IL-10+ CD4+ T cells. Eur J Immunol. 2006;36:1764–71. doi: 10.1002/eji.200635937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarnicki AG, Lysaght J, Todryk S, Mills KH. Suppression of antitumor immunity by IL-10 and TGF-beta-producing T cells infiltrating the growing tumor: influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J Immunol. 2006;177:896–904. doi: 10.4049/jimmunol.177.2.896. [DOI] [PubMed] [Google Scholar]
- 49.Noble A, Giorgini A, Leggat JA. Cytokine-induced IL-10-secreting CD8 T cells represent a phenotypically distinct suppressor T-cell lineage. Blood. 2006;107:4475–83. doi: 10.1182/blood-2005-10-3994. [DOI] [PubMed] [Google Scholar]
- 50.Ranatunga D, Hedrich CM, Wang F, McVicar DW, Nowak N, Joshi T, et al. A human IL10 BAC transgene reveals tissue-specific control of IL-10 expression and alters disease outcome. Proc Natl Acad Sci USA. 2009;106:17123–8. doi: 10.1073/pnas.0904955106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murugaiyan G, Mittal A, Lopez-Diego R, Maier LM, Anderson DE, Weiner HL. IL-27 is a key regulator of IL-10 and IL-17 production by human CD4+ T cells. J Immunol. 2009;183:2435–43. doi: 10.4049/jimmunol.0900568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJ, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8:1363–71. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 53.Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–9. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 54.Fitzgerald DC, Zhang GX, El-Behi M, Fonseca-Kelly Z, Li H, Yu S, et al. Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nat Immunol. 2007;8:1372–9. doi: 10.1038/ni1540. [DOI] [PubMed] [Google Scholar]
- 55.Gerosa F, Paganin C, Peritt D, Paiola F, Scupoli MT, Aste-Amezaga M, et al. Interleukin-12 primes human CD4 and CD8 T cell clones for high production of both interferon-gamma and interleukin-10. J Exp Med. 1996;183:2559–69. doi: 10.1084/jem.183.6.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Windhagen A, Anderson DE, Carrizosa A, Williams RE, Hafler DA. IL-12 induces human T cells secreting IL-10 with IFN-gamma. J Immunol. 1996;157:1127–31. [PubMed] [Google Scholar]
- 57.Pohl-Koppe A, Balashov KE, Steere AC, Logigian EL, Hafler DA. Identification of a T cell subset capable of both IFN-gamma and IL-10 secretion in patients with chronic Borrelia burgdorferi infection. J Immunol. 1998;160:1804–10. [PubMed] [Google Scholar]
- 58.Gerosa F, Nisii C, Righetti S, Micciolo R, Marchesini M, Cazzadori A, et al. CD4(+) T cell clones producing both interferon-gamma and interleukin-10 predominate in bronchoalveolar lavages of active pulmonary tuberculosis patients. Clin Immunol. 1999;92:224–34. doi: 10.1006/clim.1999.4752. [DOI] [PubMed] [Google Scholar]
- 59.Saraiva M, Christensen JR, Veldhoen M, Murphy TL, Murphy KM, O’Garra A. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–19. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boonstra A, Rajsbaum R, Holman M, Marques R, Asselin-Paturel C, Pereira JP, et al. Macrophages and myeloid dendritic cells, but not plasmacytoid dendritic cells, produce IL-10 in response to MyD88- and TRIF-dependent TLR signals, and TLR-independent signals. J Immunol. 2006;177:7551–8. doi: 10.4049/jimmunol.177.11.7551. [DOI] [PubMed] [Google Scholar]
- 61.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg DJ, Kuhn R, Rajewsky K, Muller W, Menon S, Davidson N, et al. Interleukin-10 is a central regulator of the response to LPS in murine models of endotoxic shock and the Shwartzman reaction but not endotoxin tolerance. J Clin Invest. 1995;96:2339–47. doi: 10.1172/JCI118290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siewe L, Bollati-Fogolin M, Wickenhauser C, Krieg T, Muller W, Roers A. Interleukin-10 derived from macrophages and/or neutrophils regulates the inflammatory response to LPS but not the response to CpG DNA. Eur J Immunol. 2006;36:3248–55. doi: 10.1002/eji.200636012. [DOI] [PubMed] [Google Scholar]
- 64.Scumpia PO, Moldawer LL. Biology of interleukin-10 and its regulatory roles in sepsis syndromes. Crit Care Med. 2005;33:S468–71. doi: 10.1097/01.ccm.0000186268.53799.67. [DOI] [PubMed] [Google Scholar]
- 65.Ishida H, Hastings R, Thompson-Snipes L, Howard M. Modified immunological status of anti-IL-10 treated mice. Cell Immunol. 1993;148:371–84. doi: 10.1006/cimm.1993.1119. [DOI] [PubMed] [Google Scholar]
- 66.Marchant A, Bruyns C, Vandenabeele P, Ducarme M, Gerard C, Delvaux A, et al. Interleukin-10 controls interferon-gamma and tumor necrosis factor production during experimental endotoxemia. Eur J Immunol. 1994;24:1167–71. doi: 10.1002/eji.1830240524. [DOI] [PubMed] [Google Scholar]
- 67.Standiford TJ, Strieter RM, Lukacs NW, Kunkel SL. Neutralization of IL-10 increases lethality in endotoxemia. Cooperative effects of macrophage inflammatory protein-2 and tumor necrosis factor. J Immunol. 1995;155:2222–9. [PubMed] [Google Scholar]
- 68.Song GY, Chung CS, Chaudry IH, Ayala A. What is the role of interleukin 10 in polymicrobial sepsis: anti-inflammatory agent or immunosuppressant? Surgery. 1999;126:378–83. [PubMed] [Google Scholar]
- 69.Gerard C, Bruyns C, Marchant A, Abramowicz D, Vandenabeele P, Delvaux A, et al. Interleukin 10 reduces the release of tumor necrosis factor and prevents lethality in experimental endotoxemia. J Exp Med. 1993;177:547–50. doi: 10.1084/jem.177.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howard M, Muchamuel T, Andrade S, Menon S. Interleukin 10 protects mice from lethal endotoxemia. J Exp Med. 1993;177:1205–8. doi: 10.1084/jem.177.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bean AG, Freiberg RA, Andrade S, Menon S, Zlotnik A. Interleukin 10 protects mice against staphylococcal enterotoxin B-induced lethal shock. Infect Immun. 1993;61:4937–9. doi: 10.1128/iai.61.11.4937-4939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rogy MA, Auffenberg T, Espat NJ, Philip R, Remick D, Wollenberg GK, et al. Human tumor necrosis factor receptor (p55) and interleukin 10 gene transfer in the mouse reduces mortality to lethal endotoxemia and also attenuates local inflammatory responses. J Exp Med. 1995;181:2289–93. doi: 10.1084/jem.181.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato T, Murata A, Ishida H, Toda H, Tanaka N, Hayashida H, et al. Interleukin 10 reduces mortality from severe peritonitis in mice. Antimicrob Agents Chemother. 1995;39:1336–40. doi: 10.1128/aac.39.6.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerber JS, Mosser DM. Reversing lipopolysaccharide toxicity by ligating the macrophage Fc gamma receptors. J Immunol. 2001;166:6861–8. doi: 10.4049/jimmunol.166.11.6861. [DOI] [PubMed] [Google Scholar]
- 75.Pajkrt D, Camoglio L, Tiel-van Buul MC, de BK, Cutler DL, Affrime MB, Rikken G, van der PT, ten Cate JW, van Deventer SJ. Attenuation of proinflammatory response by recombinant human IL-10 in human endotoxemia: effect of timing of recombinant human IL-10 administration. J Immunol. 1997;158:3971–7. [PubMed] [Google Scholar]
- 76.Brandtzaeg P, Osnes L, Ovstebo R, Joo GB, Westvik AB, Kierulf P. Net inflammatory capacity of human septic shock plasma evaluated by a monocyte-based target cell assay: identification of interleukin-10 as a major functional deactivator of human monocytes. J Exp Med. 1996;184:51–60. doi: 10.1084/jem.184.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Latifi SQ, O’Riordan MA, Levine AD. Interleukin-10 controls the onset of irreversible septic shock. Infect Immun. 2002;70:4441–6. doi: 10.1128/IAI.70.8.4441-4446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manley MO, O’Riordan MA, Levine AD, Latifi SQ. Interleukin 10 extends the effectiveness of standard therapy during late sepsis with serum interleukin 6 levels predicting outcome. Shock. 2005;23:521–6. [PubMed] [Google Scholar]
- 79.Berg DJ, Leach MW, Kuhn R, Rajewsky K, Muller W, Davidson NJ, et al. Interleukin 10 but not interleukin 4 is a natural suppressant of cutaneous inflammatory responses. J Exp Med. 1995;182:99–108. doi: 10.1084/jem.182.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–12. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 81.Padigel UM, Farrell JP. Control of infection with Leishmania major in susceptible BALB/c mice lacking the common gamma-chain for FcR is associated with reduced production of IL-10 and TGF-beta by parasitized cells. J Immunol. 2005;174:6340–5. doi: 10.4049/jimmunol.174.10.6340. [DOI] [PubMed] [Google Scholar]
- 82.Weber MS, Prod’homme T, Youssef S, Dunn SE, Rundle CD, Lee L, et al. Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat Med. 2007;13:935–43. doi: 10.1038/nm1620. [DOI] [PubMed] [Google Scholar]
- 83.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–94. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 84.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–84. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–5. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 86.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 87.Cousens LP, Orange JS, Biron CA. Endogenous IL-2 contributes to T cell expansion and IFN-gamma production during lymphocytic choriomeningitis virus infection. J Immunol. 1995;155:5690–9. [PubMed] [Google Scholar]
- 88.Sonoda KH, Faunce DE, Taniguchi M, Exley M, Balk S, Stein-Streilein J. NK T cell-derived IL-10 is essential for the differentiation of antigen-specific T regulatory cells in systemic tolerance. J Immunol. 2001;166:42–50. doi: 10.4049/jimmunol.166.1.42. [DOI] [PubMed] [Google Scholar]
- 89.Peritt D, Robertson S, Gri G, Showe L, ste-Amezaga M, Trinchieri G. Differentiation of human NK cells into NK1 and NK2 subsets. J Immunol. 1998;161:5821–4. [PubMed] [Google Scholar]
- 90.Bream JH, Curiel RE, Yu CR, Egwuagu CE, Grusby MJ, Aune TM, et al. IL-4 synergistically enhances both IL-2- and IL-12-induced IFN-gamma expression in murine NK cells. Blood. 2003;102:207–14. doi: 10.1182/blood-2002-08-2602. [DOI] [PubMed] [Google Scholar]
- 91.Bodas M, Jain N, Awasthi A, Martin S, Penke Loka RK, Dandekar D, et al. Inhibition of IL-2 induced IL-10 production as a principle of phase-specific immunotherapy. J Immunol. 2006;177:4636–43. doi: 10.4049/jimmunol.177.7.4636. [DOI] [PubMed] [Google Scholar]
- 92.Loza MJ, Perussia B. The IL-12 signature: NK cell terminal CD56+ high stage and effector functions. J Immunol. 2004;172:88–96. doi: 10.4049/jimmunol.172.1.88. [DOI] [PubMed] [Google Scholar]
- 93.Grant LR, Yao ZJ, Hedrich CM, Wang F, Moorthy A, Wilson K, Ranatunga D, Bream JH. Stat4-dependent, T-bet-independent regulation of IL-10 in NK cells. Genes Immun. 2008;9:316–27. doi: 10.1038/gene.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vigano P, Gaffuri B, Somigliana E, Infantino M, Vignali M, Di Blasio AM. Interleukin-10 is produced by human uterine natural killer cells but does not affect their production of interferon-gamma. Mol Hum Reprod. 2001;7:971–7. doi: 10.1093/molehr/7.10.971. [DOI] [PubMed] [Google Scholar]
- 95.Barber MA, Zhang T, Gagne BA, Sentman CL. NK cells negatively regulate antigen presentation and tumor-specific CTLs in a syngeneic lymphoma model. J Immunol. 2007;178:6140–7. doi: 10.4049/jimmunol.178.10.6140. [DOI] [PubMed] [Google Scholar]
- 96.De MA, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, et al. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol. 2007;37:445–55. doi: 10.1002/eji.200635989. [DOI] [PubMed] [Google Scholar]
- 97.Maroof A, Beattie L, Zubairi S, Svensson M, Stager S, Kaye PM. Posttranscriptional regulation of II10 gene expression allows natural killer cells to express immunoregulatory function. Immunity. 2008;29:295–305. doi: 10.1016/j.immuni.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206:2235–51. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trinchieri G. Regulatory role of T cells producing both interferon gamma and interleukin 10 in persistent infection. J Exp Med. 2001;194:F53–7. doi: 10.1084/jem.194.10.f53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bouaziz JD, Yanaba K, Tedder TF. Regulatory B cells as inhibitors of immune responses and inflammation. Immunol Rev. 2008;224:201–14. doi: 10.1111/j.1600-065X.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 101.Madan R, Demircik F, Surianarayanan S, Allen JL, Divanovic S, Trompette A, et al. Nonredundant roles for B cell-derived IL-10 in immune counter-regulation. J Immunol. 2009;183:2312–20. doi: 10.4049/jimmunol.0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O’Garra A, Chang R, Go N, Hastings R, Haughton G, Howard M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur J Immunol. 1992;22:711–7. doi: 10.1002/eji.1830220314. [DOI] [PubMed] [Google Scholar]
- 103.Duan B, Morel L. Role of B-1a cells in autoimmunity. Autoimmun Rev. 2006;5:403–8. doi: 10.1016/j.autrev.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 104.Gary-Gouy H, Harriague J, Bismuth G, Platzer C, Schmitt C, Dalloul AH. Human CD5 promotes B-cell survival through stimulation of autocrine IL-10 production. Blood. 2002;100:4537–43. doi: 10.1182/blood-2002-05-1525. [DOI] [PubMed] [Google Scholar]
- 105.Amel Kashipaz MR, Huggins ML, Lanyon P, Robins A, Powell RJ, Todd I. Assessment of Be1 and Be2 cells in systemic lupus erythematosus indicates elevated interleukin-10 producing CD5+ B cells. Lupus. 2003;12:356–63. doi: 10.1191/0961203303lu338oa. [DOI] [PubMed] [Google Scholar]
- 106.Yanaba K, Bouaziz JD, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–50. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 107.Gieni RS, Umetsu DT, DeKruyff RH. Ly1- (CD5−) B cells produce interleukin (IL)-10. Cell Immunol. 1997;175:164–70. doi: 10.1006/cimm.1996.1060. [DOI] [PubMed] [Google Scholar]
- 108.Lenert P, Brummel R, Field EH, Ashman RF. TLR-9 activation of marginal zone B cells in lupus mice regulates immunity through increased IL-10 production. J Clin Immunol. 2005;25:29–40. doi: 10.1007/s10875-005-0355-6. [DOI] [PubMed] [Google Scholar]
- 109.Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168:5397–402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- 110.Benjamin D, Knobloch TJ, Dayton MA. Human B-cell interleukin-10: B-cell lines derived from patients with acquired immunodeficiency syndrome and Burkitt’s lymphoma constitutively secrete large quantities of interleukin-10. Blood. 1992;80:1289–98. [PubMed] [Google Scholar]
- 111.Ishida H, Hastings R, Kearney J, Howard M. Continuous anti-interleukin 10 antibody administration depletes mice of Ly-1 B cells but not conventional B cells. J Exp Med. 1992;175:1213–20. doi: 10.1084/jem.175.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eskdale J, Wordsworth P, Bowman S, Field M, Gallagher G. Association between polymorphisms at the human IL-10 locus and systemic lupus erythematosus. Tissue Antigens. 1997;49:635–9. doi: 10.1111/j.1399-0039.1997.tb02812.x. [DOI] [PubMed] [Google Scholar]
- 113.Lazarus R, Vercelli D, Palmer LJ, Klimecki WJ, Silverman EK, Richter B, et al. Single nucleotide polymorphisms in innate immunity genes: abundant variation and potential role in complex human disease. Immunol Rev. 2002;190:9–25. doi: 10.1034/j.1600-065x.2002.19002.x. [DOI] [PubMed] [Google Scholar]
- 114.Vicari AP, Trinchieri G. Interleukin-10 in viral diseases and cancer: exiting the labyrinth? Immunol Rev. 2004;202:223–36. doi: 10.1111/j.0105-2896.2004.00216.x. [DOI] [PubMed] [Google Scholar]
- 115.Harley JB, Gallagher G. Lupus and interleukin 10. J Rheumatol. 1997;24:2273–5. [PubMed] [Google Scholar]
- 116.Eskdale J, McNicholl J, Wordsworth P, Jonas B, Huizinga T, Field M, et al. Interleukin-10 microsatellite polymorphisms and IL-10 locus alleles in rheumatoid arthritis susceptibility. Lancet. 1998;352:1282–3. doi: 10.1016/S0140-6736(05)70489-X. [DOI] [PubMed] [Google Scholar]
- 117.Kube D, Rieth H, Eskdale J, Kremsner PG, Gallagher G. Structural characterisation of the distal 5′ flanking region of the human interleukin-10 gene. Genes Immun. 2001;2:181–90. doi: 10.1038/sj.gene.6363750. [DOI] [PubMed] [Google Scholar]
- 118.Anaya JM, Correa PA, Herrera M, Eskdale J, Gallagher G. Interleukin 10 (IL-10) influences auto-immune response in primary Sjogren’s syndrome and is linked to IL-10 gene polymorphism. J Rheumatol. 2002;29:1874–6. [PubMed] [Google Scholar]
- 119.Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci USA. 2000;97:14467–72. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang MH, Helzlsouer KJ, Smith MW, Hoffman-Bolton JA, Clipp SL, Grinberg V, et al. Association of IL10 and other immune response- and obesity-related genes with prostate cancer in CLUE II. Prostate. 2009;69:874–85. doi: 10.1002/pros.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Siebert JC, Inokuma M, Waid DM, Pennock ND, Vaitaitis GM, Disis ML, et al. An analytical workflow for investigating cytokine profiles. Cytometry A. 2008;73:289–98. doi: 10.1002/cyto.a.20509. [DOI] [PubMed] [Google Scholar]
- 122.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- 123.Mormann M, Rieth H, Hua TD, Assohou C, Roupelieva M, Hu SL, et al. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5:246–55. doi: 10.1038/sj.gene.6364073. [DOI] [PubMed] [Google Scholar]
- 124.Salhi A, Rodrigues V, Jr, Santoro F, Dessein H, Romano A, Castellano LR, et al. Immunological and genetic evidence for a crucial role of IL-10 in cutaneous lesions in humans infected with Leishmania braziliensis. J Immunol. 2008;180:6139–48. doi: 10.4049/jimmunol.180.9.6139. [DOI] [PubMed] [Google Scholar]
- 125.Wilson JN, Rockett K, Keating B, Jallow M, Pinder M, Sisay-Joof F, et al. A hallmark of balancing selection is present at the promoter region of interleukin 10. Genes Immun. 2006;7:680–3. doi: 10.1038/sj.gene.6364336. [DOI] [PubMed] [Google Scholar]
- 126.Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Riva S, Clerici M, et al. Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J Exp Med. 2009;206:1395–408. doi: 10.1084/jem.20082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Hohler T. Differential regulation of interleukin-10 production by genetic and environmental factors—a twin study. Genes Immun. 2002;3:407–13. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- 128.Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 129.Oleksyk TK, Shrestha S, Truelove AL, Goedert JJ, Donfield SM, Phair J, et al. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10:309–22. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sparwasser T, Eberl G. BAC to immunology—bacterial artificial chromosome-mediated transgenesis for targeting of immune cells. Immunology. 2007;121:308–13. doi: 10.1111/j.1365-2567.2007.02605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bonifer C, Vidal M, Grosveld F, Sippel AE. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990;9:2843–8. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Welstead GG, Iorio C, Draker R, Bayani J, Squire J, Vongpunsawad S, et al. Measles virus replication in lymphatic cells and organs of CD150 (SLAM) transgenic mice. Proc Natl Acad Sci USA. 2005;102:16415–20. doi: 10.1073/pnas.0505945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Frazer KA, Narla G, Zhang JL, Rubin EM. The apolipoprotein(a) gene is regulated by sex hormones and acute-phase inducers in YAC transgenic mice. Nat Genet. 1995;9:424–31. doi: 10.1038/ng0495-424. [DOI] [PubMed] [Google Scholar]
- 134.Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, et al. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–8. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 136.Haringer B, Lozza L, Steckel B, Geginat J. Identification and characterization of IL-10/IFN-gamma-producing effector-like T cells with regulatory function in human blood. J Exp Med. 2009;206:1009–17. doi: 10.1084/jem.20082238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–18. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Powell MJ, Thompson SA, Tone Y, Waldmann H, Tone M. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J Immunol. 2000;165:292–6. doi: 10.4049/jimmunol.165.1.292. [DOI] [PubMed] [Google Scholar]
- 139.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci USA. 2004;101:12622–7. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci USA. 2004;101:2440–5. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Makar KW, Wilson CB. DNA methylation is a nonredundant repressor of the Th2 effector program. J Immunol. 2004;173:4402–6. doi: 10.4049/jimmunol.173.7.4402. [DOI] [PubMed] [Google Scholar]
- 142.Brightbill HD, Plevy SE, Modlin RL, Smale ST. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J Immunol. 2000;164:1940–51. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 143.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–91. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 144.Ma W, Lim W, Gee K, Aucoin S, Nandan D, Kozlowski M, et al. The p38 mitogen-activated kinase pathway regulates the human interleukin-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J Biol Chem. 2001;276:13664–74. doi: 10.1074/jbc.M011157200. [DOI] [PubMed] [Google Scholar]
- 145.Steinke JW, Barekzi E, Hagman J, Borish L. Functional analysis of -571 IL-10 promoter polymorphism reveals a repressor element controlled by sp1. J Immunol. 2004;173:3215–22. doi: 10.4049/jimmunol.173.5.3215. [DOI] [PubMed] [Google Scholar]
- 146.Lucas M, Zhang X, Prasanna V, Mosser DM. ERK activation following macrophage FcgammaR ligation leads to chromatin modifications at the IL-10 locus. J Immunol. 2005;175:469–77. doi: 10.4049/jimmunol.175.1.469. [DOI] [PubMed] [Google Scholar]
- 147.Vega MI, Huerta-Yepaz S, Garban H, Jazirehi A, Emmanouilides C, Bonavida B. Rituximab inhibits p38 MAPK activity in 2F7 B NHL and decreases IL-10 transcription: pivotal role of p38 MAPK in drug resistance. Oncogene. 2004;23:3530–40. doi: 10.1038/sj.onc.1207336. [DOI] [PubMed] [Google Scholar]
- 148.Platzer C, Fritsch E, Elsner T, Lehmann MH, Volk HD, Prosch S. Cyclic adenosine monophosphate-responsive elements are involved in the transcriptional activation of the human IL-10 gene in monocytic cells. Eur J Immunol. 1999;29:3098–104. doi: 10.1002/(SICI)1521-4141(199910)29:10<3098::AID-IMMU3098>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 149.Brenner S, Prosch S, Schenke-Layland K, Riese U, Gausmann U, Platzer C. cAMP-induced Interleukin-10 promoter activation depends on CCAAT/enhancer-binding protein expression and monocytic differentiation. J Biol Chem. 2003;278:5597–604. doi: 10.1074/jbc.M207448200. [DOI] [PubMed] [Google Scholar]
- 150.Ziegler-Heitbrock L, Lotzerich M, Schaefer A, Werner T, Frankenberger M, Benkhart E. IFN-alpha induces the human IL-10 gene by recruiting both IFN regulatory factor 1 and Stat3. J Immunol. 2003;171:285–90. doi: 10.4049/jimmunol.171.1.285. [DOI] [PubMed] [Google Scholar]
- 151.Schaefer A, Unterberger C, Frankenberger M, Lohrum M, Staples KJ, Werner T, et al. Mechanism of interferon-gamma mediated down-regulation of interleukin-10 gene expression. Mol Immunol. 2009;46:1351–9. doi: 10.1016/j.molimm.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 152.Unterberger C, Staples KJ, Smallie T, Williams L, Foxwell B, Schaefer A, et al. Role of STAT3 in glucocorticoid-induced expression of the human IL-10 gene. Mol Immunol. 2008;45:3230–7. doi: 10.1016/j.molimm.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 153.Chang HD, Helbig C, Tykocinski L, Kreher S, Koeck J, Niesner U, et al. Expression of IL-10 in Th memory lymphocytes is conditional on IL-12 or IL-4, unless the IL-10 gene is imprinted by GATA-3. Eur J Immunol. 2007;37:807–17. doi: 10.1002/eji.200636385. [DOI] [PubMed] [Google Scholar]
- 154.Rutz S, Janke M, Kassner N, Hohnstein T, Krueger M, Scheffold A. Notch regulates IL-10 production by T helper 1 cells. Proc Natl Acad Sci USA. 2008;105:3497–502. doi: 10.1073/pnas.0712102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Visconti R, Gadina M, Chiariello M, Chen EH, Stancato LF, Gutkind JS, et al. Importance of the MKK6/p38 pathway for interleukin-12-induced STAT4 serine phosphorylation and transcriptional activity. Blood. 2000;96:1844–52. [PubMed] [Google Scholar]
- 156.Tsuji-Takayama K, Suzuki M, Yamamoto M, Harashima A, Okochi A, Otani T, et al. The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. J Immunol. 2008;181:3897–905. doi: 10.4049/jimmunol.181.6.3897. [DOI] [PubMed] [Google Scholar]
- 157.Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4(+) T cells. J Immunol. 2009;183:5518–25. doi: 10.4049/jimmunol.0901284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang ZY, Sato H, Kusam S, Sehra S, Toney LM, Dent AL. Regulation of IL-10 gene expression in Th2 cells by Jun proteins. J Immunol. 2005;174:2098–105. doi: 10.4049/jimmunol.174.4.2098. [DOI] [PubMed] [Google Scholar]
- 159.Lin SC. Identification of an NF-Y/HMG-I(Y)-binding site in the human IL-10 promoter. Mol Immunol. 2006;43:1325–31. doi: 10.1016/j.molimm.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 160.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–92. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tran EH, Azuma YT, Chen M, Weston C, Davis RJ, Flavell RA. Inactivation of JNK1 enhances innate IL-10 production and dampens autoimmune inflammation in the brain. Proc Natl Acad Sci USA. 2006;103:13451–6. doi: 10.1073/pnas.0601155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, et al. BCL-3 and NF-kappaB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J Biol Chem. 2004;279:49995–50003. doi: 10.1074/jbc.M404246200. [DOI] [PubMed] [Google Scholar]
- 163.Saraiva M, Christensen JR, Tsytsykova AV, Goldfeld AE, Ley SC, Kioussis D, et al. Identification of a macrophage-specific chromatin signature in the IL-10 locus. J Immunol. 2005;175:1041–6. doi: 10.4049/jimmunol.175.2.1041. [DOI] [PubMed] [Google Scholar]
- 164.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 165.Bjornsson HT, Fallin MD, Feinberg AP. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–8. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 166.Zhang X, Edwards JP, Mosser DM. Dynamic and transient remodeling of the macrophage IL-10 promoter during transcription. J Immunol. 2006;177:1282–8. doi: 10.4049/jimmunol.177.2.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Im SH, Hueber A, Monticelli S, Kang KH, Rao A. Chromatin-level regulation of the IL10 gene in T cells. J Biol Chem. 2004;279:46818–25. doi: 10.1074/jbc.M401722200. [DOI] [PubMed] [Google Scholar]
- 168.Shoemaker J, Saraiva M, O’Garra A. GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J Immunol. 2006;176:3470–9. doi: 10.4049/jimmunol.176.6.3470. [DOI] [PubMed] [Google Scholar]
- 169.Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005;175:7437–46. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- 170.Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Glauben R, Sonnenberg E, Zeitz M, Siegmund B. HDAC inhibitors in models of inflammation-related tumorigenesis. Cancer Lett. 2009;280:154–9. doi: 10.1016/j.canlet.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 172.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–48. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 173.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 174.Young HA, Bream JH. IFN-gamma: recent advances in understanding regulation of expression, biological functions, and clinical applications. Curr Top Microbiol Immunol. 2007;316:97–117. doi: 10.1007/978-3-540-71329-6_6. [DOI] [PubMed] [Google Scholar]
- 175.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–60. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 176.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc Natl Acad Sci USA. 2007;104:17052–7. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–42. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dong J, Ivascu C, Chang HD, Wu P, Angeli R, Maggi L, et al. IL-10 is excluded from the functional cytokine memory of human CD4+ memory T lymphocytes. J Immunol. 2007;179:2389–96. doi: 10.4049/jimmunol.179.4.2389. [DOI] [PubMed] [Google Scholar]
- 179.Szalmas A, Banati F, Koroknai A, Laszlo B, Feher E, Salamon D, et al. Lineage-specific silencing of human IL-10 gene expression by promoter methylation in cervical cancer cells. Eur J Cancer. 2008;44:1030–8. doi: 10.1016/j.ejca.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 180.Fu LH, Cong B, Zhen YF, Li SJ, Ma CL, Ni ZY, et al. Methylation status of the IL-10 gene promoter in the peripheral blood mononuclear cells of rheumatoid arthritis patients. Yi Chuan. 2007;29:1357–61. doi: 10.1360/yc-007-1357. [DOI] [PubMed] [Google Scholar]
- 181.Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–5. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 182.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–86. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 183.Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–8. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 184.Sawado T, Halow J, Bender MA, Groudine M. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 2003;17:1009–18. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, et al. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci USA. 2004;101:2446–51. doi: 10.1073/pnas.0308327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol Rev. 2008;223:114–31. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]