Abstract
Activation of T cells is an essential step in the immunological response to infection. While activation of naïve T cells results in proliferation and slow differentiation into cytokine-producing effector cells, antigen engagement with memory cells leads to cytokine production immediately. Even though the cell surface signaling events are similar in both the cases, the outcome is different, suggesting that distinct regulatory mechanisms may exist downstream of the activation signals. Recent advances in the understanding of global epigenetic patterns in T cells have resulted in the appreciation of the role of epigenetic mechanisms in processes such as activation and differentiation. In this review we discuss recent data suggesting that naïve T cell activation, differentiation and lineage commitment results in epigenetic changes and a fine balance between different histone modifications is required. On the other hand, memory T cells are poised and do not require epigenetic changes for short-term activation.
Introduction
Upon positive and negative selection in the thymus, naive T cells enter circulation in the periphery. Those of them that encounter antigen displayed on the surface of antigen presenting cells (APCs) undergo massive clonal expansion and differentiate into effector cells that secrete inflammatory cytokines. Several cell types, including macrophages, dendritic cells and B cells, can play a role of APCs. The full activation of T cells, which leads to proliferation and cytokine expression, requires two separate signals. One is provided via interaction of T cell receptor (TCR) with major histocompatibility complex (MHC)-antigen complex on the surface of APC. The second signal or co-stimulation can be provided via interaction of CD28 receptor of the T cells with B7 ligands also on the surface of APCs. TCR and CD28 signaling leads to activation of protein kinase cascade, production of secondary messengers and activation of NFAT, NFkB and AP1 transcription factors (reviewed in [1,2]).
Depending on the type of infection and cytokine milieu, effector T cells can express specific characteristic sets of cytokines, which help to activate other components of the immune system and clear an infection (reviewed in [3]). In the presence of IL-l2 signaling, T cells polarize into T helper 1 cells (Th1), which express interferon-γ (IFN-γ). IL-4 signaling leads to the generation of T helper 2 cells (Th2) cells that express IL-4, −5 and −13. Transforming growth factor-β (TGF-β) is involved in the differentiation of T cells into both T helper 17 cells (Th17) that express IL-17 and induced regulatory T cells (iTreg) that are characterized by the expression of the transcription factor FOXP3 [4,5]. Th1 cells play a role in clearing intracellular pathogens. Th2 and Th17 cells mediate the immune responses against extracellular parasites. Regulatory T cells (Treg) cells are involved in the regulation of immune response [4–6]. Once the infection has been cleared, the majority of the effector cells are eliminated by apoptosis during the contraction phase of immune response, but some differentiate into memory cells, which can mount faster response upon re-encountering the antigen[7,8]. Interestingly, the T cells maintain their ability to alter their lineage specific expression patterns even after differentiation into specific lineages upon receiving the polarization signals [4,5,9].
Activation of T cells may occur in two different contexts, namely activation of naïve cells that have never encountered antigen and activation of antigen-experienced memory cells. In the case of naïve cells, activation leads to proliferation, often production of IL-2 and later polarization into effector cells that can produce characteristic cytokines such as IFN-γ and IL-4. Activation of memory cells leads to cytokine production much faster. The same signaling is believed to occur upon activation of both naïve and memory cells, although memory cells are thought to be less dependent upon co-stimulation. However, as discussed above, the outcome of this signaling is different between these cell types.
Both the T cell differentiation and cytokine production require tightly controlled gene expression, mainly regulated at the step of transcription. It is well established that the chromatin context has major impacts on transcriptional regulation. Recent studies have shown epigenetic regulation of chromatin structure to be an important contributing factor in maintaining cellular functions and homeostatsis. In this review we will discuss recent data suggesting that while alterations to histone modifications play an integral role in T cell differentiation from naïve to memory cells, the short-term activation and effector functions of memory cells do not require global epigenetic changes. The epigenetic conformation of T cells at different differentiation status is thus finely tuned to respond to external environmental cues.
Mapping the T cell epigenomes
In the eukaryotic nucleus DNA is compacted by wrapping 146 bp of DNA around the four core histone proteins H3, H4, H2A and H2B, which forms the basic structural unit of chromatin, the nucleosome. The N-terminal tails of histone proteins, which protrude from the nucleosomes can be covalently modified in a variety of ways such as acetylation, methylation and phosphorylation [10]. These modifications can have a direct effect on the gene expression by condensing or relaxing the chromatin leading to repression or activation, respectively. The effect can also be indirect where the histone modifications act as signals that are recognized by transcription factors or co-factors that subsequently regulate gene expression [10–12].
For the study of chromatin state on a genome-scale, recent years have seen development of novel techniques, most of which are based on chromatin immunoprecipitation (ChIP). Genome-wide analysis of epigenetic modifications began with the advent of ChIP-chip procedure where chromatin immunoprecipitated DNA fragments are detected by hybridization with DNA microarrays. This procedure has been used extensively to detect the localization of several proteins as well [13–15]. This was followed by sequencing based protocols such as Genome Wide Mapping Technique (GMAT) [16,17] and Serial Analysis of Chromatin Occupancy (SACO) [18]. Though they came in different flavors the principle behind all these procedures is the detection of chromatin immunopreciptated DNA by serial analysis of gene expression (SAGE) –like protocol. The newest tool in line is ChIP-Seq where ChIP DNA is directly sequenced using the next-generation massively parallel sequencing [19–25]. Application of ChIP-Seq has led to a superior understanding of several epigenetically regulated biological processes.
One of the biological systems that has been most studied using ChIP-Seq is T cells [19,20,26–29]. Among the various modifications of histones in T cells, methylation and acetylation have been extensively studied. While all acetylation marks are enriched in active chromatin regions, histone methylation is more complex since the different degrees of methylation has remarkably significant variations both in the genomic localization and in their effects on the gene expression. For example, histone H3 lysine 4 tri-methylation (H3K4me3) is highly enriched in an about 1 kb region surrounding the transcription start sites of active genes, H3K4me2 and H3K4me1 marks extend further into the gene bodies of transcribed genes. Similarly, H3K36me3 and H3K79me3 mark the transcribed regions of genes. However, they appear to localize to different regions with the former peaking close to the 3′ end of the genes and the latter peaking at the 5′ end of the transcribed genes. The role that these modifications play in gene regulation is still unclear. The methylation marks that are linked with gene silencing are H3K27me2/3, H3K9me2/3 and H4K20me3 [19,30]. The T cell epigenome has also been investigated using chromatin accessibility assays [26]. The data indicated that DNase I hypersensitive sites extensively correlated with the presence of “active” histone modifications including H3K4me3 and H3K4me1, which tend to mark regulatory DNA elements for transcription [16,26,31,32].
A study on CD4+ T cells, which characterized the genome-wide distribution of 19 histone lysine and arginine methylations, 18 histone lysine acetylations and H2AZ histone variant identified gene-specific combinatorial patterns of histone modifications in the human genome, which may be associated with distinct potential of transcription. Another interesting observation is that a group of 17 histone modifications co-exist at the promoters of 3,286 genes. These genes had higher expression levels compared to genes with no modifications [30]. The significance of the 17-histone modifications clustering in 25% of the genes can be appreciated from the fact that a mere 0–392 genes could be identified when just one of the 17 modifications is absent. A notable feature of these genes is the complete lack of repressive H3K27me3. The clustering of histone modifications suggests cooperation among various chromatin marks. This may also suggest redundancy of epigenetic regulation, which improves the robustness of the system through the reinforcement of chromatin modifications with similar functions. This further reinforces the notion that gene expression through epigenetic mechanisms is a network of independent but interconnected events [30,33]. However, the existence of any particular histone modification or a combination of these did not necessarily correlate with the level of gene expression, indicating that active histone modifications maintain the chromatin in a transcriptionally permissible state, which is acted upon by transcription factors to bring about the actual changes to gene expression.
T cell differentiation
Depending on the nature of the encountered antigen, CD4+ T cells undergo differentiation into various lineages. During the differentiation process, changes in chromatin structure occur through epigenetic mechanisms such as histone modifications, DNA methylation and generation of DNase I hypersensitive sites [6,34–37]. Differentiation of CD4+ T cells into their subtypes appears to be controlled by various layers of epigenetic modifications. The signature cytokine genes known to define lineage specificity, IFN-γ in Th1 cells, IL-4 in Th2 cells and IL-17 in Th17 cells, were marked with the active histone modification H3K4me3 in the appropriate lineages [34,38–40]. However, the repressive modification H3K27me3 clearly displays an additional level of regulation, as it does not always mark the genes that are not expressed in a particular lineage. While H3K27me3 marks IFN-γ in Th2 cells, it is completely absent in naturally-occurring regulatory T cells (nTreg) [41]. Two possible scenarios can explain this phenomenon: (1) repression of lineage inappropriate genes through other repressive epigenetic modifications such as H3K9me3 or DNA methylation. (2) maintenance of the potential for the expression of previously silent genes and lineage switch or epigenetic plasticity. On the other hand, H3K27me3 extensively marks IL17 in Th1, Th2 and Treg cells suggesting strong repression in these cells [41].
An even more striking phenomenon and one that argues for the second scenario, is the epigenetic modifications of transcription regulators of lineage specificity including Foxp3, RORγt, T-bet and GATA-3. The Foxp3 gene, which is important for Treg function, is marked by H3K4me3 in Treg cells. However, it is not marked by H3K27me3 in Th17 cells where it is silent [41], suggesting that the gene may be expressed under appropriate conditions in Th17 cells. Consistent with this, co-expression of Foxp3 and RORγt with the production of IL-17 has been demonstrated in a subset of T cells [42]. Moreover, nTreg cells could be induced to express IFN-γ and T-bet by IL-12 induction; and IL-6 mediated suppression of Foxp3 converts the Treg cells into IL-17 producing cells [41,43,44].
It has been puzzling how Th1 cells produce the Th2-specific cytokine IL-4 and Th2 cells produce the Th1-specific cytokine IFN-γ under appropriate stimulatory conditions [9]. The global epigenetic analysis of differentiated T helper cells has provided insights into this phenomenon [41]. Presence of bivalent modifications at genes encoding key transcriptional regulators is consistent with the plasticity of the differentiated T helper cells in expressing non-specific cytokines: the Tbx21 gene, which encodes Th1-specific T-bet, and the Th2-specific Gata3 gene are marked by both the activating H3K4me3 and the repressive H3K27me3 modifications in the lineages that do not express these factors [41]. The ability of Th17 cells, where Tbx21 is associated with bivalent modifications, to express IFN-γ again argues for the importance of bivalent domains in determining the expression pattern switch in the cells [5,36,41]. Through these studies it is clear that though the lineage commitment leads to expression of the lineage-specific cytokines, the silencing of the cytokines of alternative lineages is not always permanent. A potential to express the cytokines of opposing lineage is often maintained and can be realized in the presence of appropriate stimulus [36,45]. The, epigenetic bivalency at the genes encoding key regulators appears to maintain this plasticity in the cells.
Bivalent domains were previously shown to be associated with developmentally crucial genes in mouse embryonic stem cells where they keep genes repressed but prepared for future expression [46,47]. During differentiation the bivalent domains resolve to monovalency. As studies in CD4+ and CD8+ T cells have shown, bivalency can exist in non-ES cells as well [19,31,48]. The erythrocyte precursor CD133+ cells also possess bivalent histone modifications at several lineage specific genes. During its differentiation into CD36+ cells, genes specific for other lineages are shut down while the transcriptional activation of genes specific for CD36 lineage occurs [48]. The existence of the bivalent modification suggests that the genomic region bearing bivalent modifications is subject to a dynamic competition of both the activating and repressing processes [28]. We proposed previously that cell surface signaling could result in a shift of balance of H3K4me3 and H3K27me3 modifications and therefore lead to a change in gene expression [31]. Indeed, our recent study in CD8+ T cells showed that several inducible genes possess bivalent modifications in the resting state when they are silent [27]. Activation of the cells resulted in increased expression of several of these genes with a concomitant increase in the H3K4me3 levels, suggesting an opening of the chromatin structure. Thus bivalent histone modifications could form the basis for plasticity in T cell subsets. These studies show that bivalent domains function (1) in ES cells to silence genes involved in development [46]; (2) in differentiated cells that have the potential to undergo self renewal and further differentiation [48]; and in (3) terminally differentiated cells that undergo changes to the chromatin structure while responding to external stimuli [31]. Therefore, bivalent domains can operate in cells at all developmental stages where a dynamic change in the gene expression levels is required.
T cell activation
What happens to chromatin during activation of CD4+ T cells? Several groups have investigated changes in chromatin modifications at key cytokine genes. Agarwal and Rao [49] studied differentiation of mouse naïve T cells along Th1 or Th2 pathway and reported that activation of characteristic cytokine (Ifng or Il4) gene expression coincided with chromatin remodeling and demethylation at their promoters. Furthermore, differential histone acetylation at these promoters upon activation and differentiation in Th1 or Th2 direction was observed [50–52]. Interestingly, chromatin remodeling but not an increase in histone acetylation was observed at the IL-2 promoter upon its induction as a result of T cell activation [53]. This variation could be explained by the differences in the length of activation, initial cell state (memory vs. naïve) and acetylation levels of these gene promoters in resting cells (see below).
Our studies on global changes to the chromatin environment in the resting and short-term activated CD4+ T cells by ChIP-Seq showed that majority of the genes induced upon activation already had permissive chromatin environment even while being silent in the resting cells [54]. For example, E2F1, Il-2, IFN-γ and IL-4 genes already had their promoters marked with H3K4me3, H2A.Z and other modifications even though these genes were silent in resting cells [54]. Similar observations were made by others [55] [56]. In addition, RNA Polymerase II (Pol II) was present at the promoters of inducible genes while they were silent in the resting cells. Though these genes were not expressed, the presence of activating histone modifications and presence of Pol II suggested that these genes were poised for expression. Overall, about 20–30% of all silent genes were poised, but only a small fraction of them was induced during T cell activation.
Presence of Pol II at a large number of silent genes was previously reported by us and by others [19,57,58]. It is believed that so called promoter-proximal stalling of Pol II is due to failure of Pol II to transition from transcription initiation to processive elongation [59]. It was hypothesized that this transition is controlled by phosphorylation of Pol II C-terminal domain by positive transcription elongation factor P-TEFb [60], which is a complex of cyclin dependent kinase CDK9 and one of its cyclins: T1, T2 or K [61]. Interestingly, it was shown that levels and activity of both CDK9 and cyclin T1 strongly increase upon T cell activation [62,63], which likely explains how poised genes become expressed. How P-TEFb is targeted to poised genes during T cell activation is currently unknown.
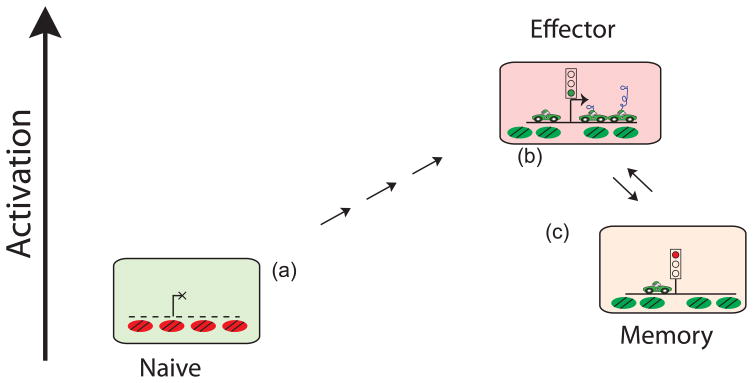
It remains unclear how poised genes become poised. One possibility is that poising is actually a memory of past transcription. In a genome-wide study in mouse resting naïve cells no H3K4me3 was observed at the Il2, Ifng and Il4 promoters [41]. Experiments performed on a single gene level also showed no histone acetylation in naïve cells and appearance of the modification upon T cell activation and differentiation [51,52]. Contrary to these results we observed that positive modifications were already present at cytokine genes. This difference is likely due to our experiments being performed on total CD4+ cells, which include both naïve cells, which never expressed cytokines and memory cells, where cytokine genes have been transcribed in the past. By inference we conclude that the poising signals observed in our study come from memory, rather than from naïve cells. Indeed, Messi et al.[64] observed differences in chromatin state at Ifng and Il4 promoters between naïve and memory T cells. These observations led us to propose a model in which transcriptional memory, particularly epigenetic marks on chromatin, forms the basis of immunological memory (Fig. 1). Upon activation, naïve cells go through several checkpoints, which are not completely understood yet, modify their chromatin and become cytokine-expressing effector cells. After infection clearance, effector cells die or become resting memory cells [7]. Cytokine gene expression ceases in memory cells, but positive chromatin modifications stay at key regulatory regions including promoters and enhancers, leaving them in a poised state. This means that when the antigen is encountered again these cells do not have to modify chromatin and can start to produce cytokines much faster. Indeed regulation of a cytokine gene expression by Pol II stalling has been recently reported during activation of macrophages[65,66].
Figure 1.
Model: Transcriptional Memory is a Basis of Immunological Memory. (a) In naïve T cells genes required for effector function are repressed by histone or DNA methylation (red nucleosomes). Upon activation, naïve T cells massively remodel their chromatin, recruit RNA Polymerase II and start to express cytokines (b). After elimination of infection surviving effector T cells become memory T cells (c). These cells stop expression of effector genes, but positive chromatin modifications (green nucleosomes) and sometimes RNA Polymerase II remain and keep the genes in a poised state. This allows cells to quickly express effector genes upon repeat infection.
Conclusion
Recent global studies have lead to an understanding of the epigenetic mechanisms in critical T cell functions such as differentiation and activation. Characterization of the genome-wide distribution of a number of histone modifications has provided new insights into the flexibilities of gene expression programs of various subsets of T cells, which were earlier considered terminally differentiated. More complete analysis of the T cell epigenome is required to understand how the cell fate decisions are made in response to various stimuli and to elucidate the mechanisms of specificity and plasticity of polarized cells. While the dynamics of epigenetic modifications in the process of differentiation is apparent, its role in activation of T cells remains relatively unclear. Global distribution of chromatin modifications and Pol II in purified naïve and memory populations will yield more information on whether epigenetic gene poising indeed forms the basis of immunological memory.
Acknowledgments
We thank Yrina Rochman for critical reading of this manuscript. This work was supported by the Intramural Research Program of the National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Crabtree GR. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 2.Koretsky GA. T Lymphocyte Signalling Mechanisms and Activation. In: Paul WE, editor. Fundamental Immunology. 6. 2006. pp. 346–375. [Google Scholar]
- 3.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YK, Mukasa R, Hatton RD, Weaver CT. Developmental plasticity of Th17 and Treg cells. Curr Opin Immunol. 2009;21:274–280. doi: 10.1016/j.coi.2009.05.021. [DOI] [PubMed] [Google Scholar]
- ••5.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reported the plasticity of Th17 cells during development, where an absence of TGF-β enhanced IFN-γ production in a STAT4 and T-bet dependent manner.
- 6.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 2008;452:356–360. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]
- 8.Intlekofer AM, Wherry EJ, Reiner SL. Not-so-great expectations: re-assessing the essence of T-cell memory. Immunol Rev. 2006;211:203–213. doi: 10.1111/j.0105-2896.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- •9.Krawczyk CM, Shen H, Pearce EJ. Functional plasticity in memory T helper cell responses. J Immunol. 2007;178:4080–4088. doi: 10.4049/jimmunol.178.7.4080. [DOI] [PubMed] [Google Scholar]; This study showed that both effector and central memory TH1 and Th2 cells retain the ability to produce cytokines of opposing lineages.
- 10.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S. Histone modifications: combinatorial complexity or cumulative simplicity? Proc Natl Acad Sci U S A. 2005;102:5308–5309. doi: 10.1073/pnas.0501853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 14.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 15.Hanlon SE, Lieb JD. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr Opin Genet Dev. 2004;14:697–705. doi: 10.1016/j.gde.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 18.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- ••19.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]; This is the first comprehensive genome-wide mapping of 20 histone methylation marks, histone variant H2A.Z, RNA polymerase II and the insulator binding protein CTCF in human T cells using ChIP-Seq.
- 20.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 22.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- ••23.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study mapped the genome-wide distribution of several histone trimethylation marks in mouse pluripotent and lineage-committed cells.
- 24.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J Cell Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araki Y, Wang Z, Zang C, Wood WH, 3rd, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that CTCF delineated active and repressive chromatin domains.
- 30.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provided the first genome-wide map of H3K9/K14ac, H3K4me3 and H3K27me3 modifications in human T cells. The study showed that the gene expression is dependent on the absolute and relative levels of H3K4me3 and H3K27me3 modifications.
- 32.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 33.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 34.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 35.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, Alt FW, et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 38.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- ••41.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study mapped the histone modifications H3K4me3 and H3K27me3 in naïve, Th1, Th2, Th17, iTreg and nTreg cells and showed that the key regulators of T cell differentiation lacked lineage specific epigenetic modifications. The study identified bivalently modified key regulators as one of the factors involved in generation of T cell plasticity.
- 42.Zhou L, Lopes JE, Chong MM, Ivanov, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••43.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated the antagonism between Treg and Th17 developmental programs which form the basis of the plasticity during differentiation. The study showed that Foxp3 inhibited Th17 differentiation while IL-6 induced reprogramming of Foxp3 cells.
- ••44.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25- Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]; This study showed that regulatory T cells can differentiate into Th17 cells.
- 45.Dorhoi A, Kaufmann SH. Fine-tuning of T cell responses during infection. Curr Opin Immunol. 2009;21:367–377. doi: 10.1016/j.coi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- •46.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]; This study provided the first evidence of the existence of H3K4me3 and H3K27me3 co-modified regions, termed bivalent domains.
- 47.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- ••48.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study eight histone modification marks along with the histone variant, H2A.Z and RNA Pol II were mapped genome-wide in human hematopoietic stem cells and the cells differentiated to erythrocyte precursors. The study revealed bivalency of several lineage specific genes in the hematopoietic stem cells.
- 49.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 50.Santangelo S, Cousins DJ, Winkelmann NE, Staynov DZ. DNA methylation changes at human Th2 cytokine genes coincide with DNase I hypersensitive site formation during CD4(+) T cell differentiation. J Immunol. 2002;169:1893–1903. doi: 10.4049/jimmunol.169.4.1893. [DOI] [PubMed] [Google Scholar]
- 51.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 52.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol Cell Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that active chromatin modifications were already in place for inducible genes even when the genes were silent in resting T cells.
- 55.Lim PS, Hardy K, Bunting KL, Ma L, Peng K, Chen X, Shannon MF. Defining the chromatin signature of inducible genes in T cells. Genome Biol. 2009;10:R107. doi: 10.1186/gb-2009-10-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith AE, Chronis C, Christodoulakis M, Orr SJ, Lea NC, Twine NA, Bhinge A, Mufti GJ, Thomas NS. Epigenetics of human T cells during the G0-->G1 transition. Genome Res. 2009;19:1325–1337. doi: 10.1101/gr.085530.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 61.Marshall RM, Grana X. Mechanisms controlling CDK9 activity. Front Biosci. 2006;11:2598–2613. doi: 10.2741/1994. [DOI] [PubMed] [Google Scholar]
- 62.Rice AP, Herrmann CH. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr HIV Res. 2003;1:395–404. doi: 10.2174/1570162033485159. [DOI] [PubMed] [Google Scholar]
- 63.Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 65.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]