Abstract
The yeast Sln1p sensor kinase is best known as an osmosensor involved in the regulation of the hyperosmolarity glycerol mitogen-activated protein kinase cascade. Down-regulation of Sln1 kinase activity occurs under hypertonic conditions and leads to phosphorylation of the Hog1p mitogen-activated protein kinase and increased osmotic stress-response gene expression. Conditions leading to kinase up-regulation include osmotic imbalance caused by glycerol retention in the glycerol channel mutant, fps1 (Tao, W., Deschenes, R. J., and Fassler, J. S. (1999) J. Biol. Chem. 274, 360–367). The hypothesis that Sln1p kinase activity is responsive to turgor was first suggested by the increased Sln1p kinase activity in mutants lacking Fps1p in which glycerol accumulation leads to water uptake. Also consistent with the turgor hypothesis is the observation that reduced turgor caused by treatment of cells with nystatin, a drug that increases membrane permeability and causes cell shrinkage, reduced Sln1p kinase activity (Tao, W., Deschenes, R. J., and Fassler, J. S. (1999) J. Biol. Chem. 274, 360–367; Reiser, V., Raitt, D. C., and Saito, H. (2003) J. Cell Biol. 161, 1035–1040). The turgor hypothesis is revisited here in the context of the identification and characterization of the cell wall gene, CCW12, as a determinant of Sln1p activity. Results of this analysis suggest that the activity of the plasma membrane localized Sln1p is affected by the presence or absence of specific outer cell wall proteins and that this effect is independent of turgor.
Little is known about how cellular osmosensors monitor changes in the osmolarity of the environment. Yeast sensors of hyper-osmotic stress are located in the plasma membrane and include the histidine kinase, Sln1p (3, 4), as well as the four transmembrane domain protein, Sho1p (5, 6). These proteins have distinct relationships to the osmotic response HOG1 MAP2 kinase pathway that they regulate. Sho1p is involved in recruiting the Pbs2p MAPK kinase to the cell surface (6), whereas Sln1p is the initiating member of a two-component type phospho-relay cascade that negatively regulates the HOG pathway (3). Msb2p, a putative third osmosensor, encodes a protein with a single transmembrane domain and a large putative extracellular domain that works in parallel with Sho1p (7).
The Sln1p sensor kinase has three activity states. Under normal growth conditions the activity of the Sln1p kinase is modest. Modest activity is required for viability, because loss of Sln1p kinase activity (as in the null mutant) leads to inappropriate and lethal activation of the HOG pathway (8). Hyper-osmotic conditions shift the kinase to a low activity state. As Sln1p and other members of the SLN1 phospho-relay accumulate in the dephosphorylated form, the HOG pathway becomes activated, ultimately increasing expression of the osmotic response genes that are required for survival during osmotic stress (3). In addition to inactivation by hyper-osmotic conditions, the Sln1p kinase can also be stimulated. An increase in kinase activity leads to shutdown of the HOG pathway and activation of the Skn7p transcription factor (9). Sln1p kinase activation is apparent by the increase in expression of SLN1-SKN7 response genes such as OCH1 (10) and NCA3.
The role of the SLN1-SKN7 pathway is not well understood. Although the sensitivity of skn7Δ mutants to oxidative stress reflecting a separate SLN1-independent role for this transcription factor in the oxidative stress-response has been reported (11), cells lacking SKN7 are neither sensitive to hyper- nor hypo-osmotic conditions. A role for the SLN1-SKN7 pathway in cell wall integrity was suggested by the original isolation of skn7 mutants on the basis of their resistance to killer factor (kre, for killer resistance) (12). A large body of data link Skn7p to the PKC cell wall integrity pathway (10, 13-15) and suggest that the SLN1-SKN7 and the PKC pathways may function in parallel to protect cells from lysis because of the build up of turgor pressure.
The role of turgor pressure in Sln1p kinase regulation was first suggested by the increased Sln1p kinase activity in mutants lacking the major glycerol efflux channel, Fps1p (1). The absence of the glycerol channel leads to osmotic imbalance because of the accumulation of intracellular glycerol and the subsequent uptake of water (1, 16-18). Also consistent with a mechanical signal involving the presence or absence of pressure against the wall is the observation that reduced turgor caused by treatment of cells with reagents such as nystatin, which increases membrane permeability and causes cell shrinkage, caused a reduction in Sln1p kinase activity and a consequent increase in Hog1p activity in the absence of an osmotic stimulus (2).
In a recent screen of the yeast deletion collection for mutants with increased SLN1-SKN7 pathway activity, we identified a specific component of the cell wall that affects the activity of the SLN1-SKN7 pathway. Although genes involved in cell wall integrity had previously been identified as targets of the pathway (10), the role of specific wall components in osmotic stress sensing and signaling has not received much attention. Here we find that the relative abundance in the wall of the abundant GPI-anchored wall mannoprotein, Ccw12p, affects SLN1-SKN7 pathway activity. A working model in which the Ccw12 protein plays a special role in maintaining the Sln1 kinase in the repressed state is proposed. Conditions leading to reductions or loss of the Ccw12 protein from the cell wall are predicted to cause activation of the SLN1-SKN7 pathway.
EXPERIMENTAL PROCEDURES
Strains
Strains were constructed for these experiments or are from the Fassler laboratory collection (Table 1). Disruption of CCW12, SED1, and YLR111W and PPZ1 and PPZ2 in the S288C background (JF1565) was accomplished by PCR-mediated one-step gene disruption with a kanamycin (CCW12 and YLR111W) or HIS3 (SED1) cassette flanked by homologous tails. CCW12-F–300 (5′-ATAGGATCCCCATTTCGCGGCCAC-3′) and CCW12-R+1000 (5′-CGGATATCAGACACCACACACATCAAGTTTG-3′) primers were used to amplify a CCW12-KanR PCR fragment; YLR111W-F (5′-AGCGGATCCAGTGTAAGAGGTGACAGAGTG-3′) and YLR111W-R (5′-GATGAGCTCAACTGTAAACAGGTACAATGC-3′) primers were used to amplify a YLR111W-KanR PCR fragment; PPZ1-F–242 (5′-AAGCACAATACAATAGCTTCCA-3′) and PPZ1-R+2291 (5′-GTATAGAGCGAATGAGCATTCA-3′) primers were used to amplify a PPZ1-KanR PCR fragment; and PPZ2-F–202 (5′-TGCGTAAGAAATACACATATAGT-3′) and PPZ2-R+2231 (5′-CTATAACGACAACTTTATCGGCGA-3′) primers were used to amplify a PPZ2-KanR PCR fragment. Templates were genomic DNA prepared from the appropriate deletions from the Research Genetics disruption collection in strain BY4742 (19). SED1-1F-pRS (ACTACAAAGACAAGCAAAATAAAATACGTTCGCTCTATTAAGCTGTGCGGTATTTCACACCG) and SED1+1017-pRS (GAAAGCATTAAGAAGGCGGATGTGTCAAACACCACCGTAGATTGTACTGAGAGTGCAC) primers were used with pRS313 (Stratagene) template to generate a SED1-HIS3 PCR fragment. Disruptions were confirmed by genomic PCR. The ppz1 and ppz2 disruption strains were crossed to generate the double mutant.
TABLE 1.
Strains used in this study
| Genotype | Derivation | |
|---|---|---|
| JF1565 | MATα canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | Ref. 1 |
| JF1732 | MATα fps1Δ::LEU2 canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | fps1Δ::LEU2 derivative of JF1565 (1) |
| JF1904 | MATα skn7Δ::TRP1 canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | skn7Δ::TRP1 derivative of JF1565 (10) |
| JF1919 | MATΔ ssk1::LEU2 canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | Ref. 45 |
| JF2007 | MATα sln1ΔLEU2 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 pRS426-PTP2 | Also known as RJY1428 (21) |
| JF2008 | MATα sln1ΔLEU2 sho1ΔLEU2 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 pRS426-PTP2 | sho1Δ::LEU2 derivative of JF2007; one-step replacement |
| JF2154 | MATα, ssk1Δ::LEU, ypd1Δ::KanMX, sln1Δ::TRP1, his3Δ200, leu2Δ1, ura3-52, trp1Δ63, lys2Δ202 | |
| JF2162 | MATa canR cyhR, skn7Δ::TRP1, ssk1Δ::LEU2, his3Δ200, leu2Δ1, ura3-52, trp1Δ63, lys2Δ202 | |
| JF2181 |
MATa ssk1Δ::LEU2 sln1Δ::TRP1 canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 |
Product of a cross between JF2154 and JF2161 |
| JF2278 | MATα ccw12Δ::KanMX canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | ccw12Δ::KanMX derivative of JF1565; one-step replacement |
| JF2279 | MATα, skn7Δ::TRP1, ccw12Δ::KanMX canR cyhR his3Δ200 leu2Δ1 ura3-52 lys2Δ202 | ccw12Δ::KanMX derivative of JF1904; one-step replacement |
| JF2300 | MATα ssk1Δ::LEU2 sln1Δ::TRP1 ccw12Δ::KanMX canR cyhR his3Δ200 ura3-52 lys2Δ202 | ccw12Δ:KanMX derivative of JF2181; one-step replacement |
| JF2301 | MATα ssk1Δ::LEU2 ccw12Δ::KanMX canR cyhR his3Δ200 ura3-52 lys2Δ202 trp1Δ63 | ccw12Δ:KAN derivative of JF1919; one-step replacement |
| JF2336 | MATα skn7D427N canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | Derivative of JF1904; one-step replacement using fragment from pSL237 |
| JF2337 | MATα sed1Δ::HIS3 canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | sed1Δ::HIS3 derivative of JF1565; one-step replacement |
| JF2338 | MATα sed1Δ::HIS3 ccw12Δ::KanMX canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ202 | ccw12Δ:KanMX derivative of JF2337; one-step replacement |
| JF2368 | MATa sln1ΔLEU2 his3Δ200 leu2Δ1 ura3-52 trp163 ccw12Δ::KanMX pRS426-PTP2 | ccw12Δ::KAN derivative of JF2007; one-step replacement |
| JF2437 | MATa ppz1Δ::KAN canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 | ppz1Δ::KAN derivative of JF1562; one-step replacement |
| JF2438 | MATα ppz2Δ::KAN canR cyhR his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2Δ201 | ppz2Δ::KAN derivative of JF1565; one-step replacement |
| JF2441 | MATα ppz1Δ::KanMX ppz2Δ::KanMX his3Δ200 leu2Δ1 ura3-52 trp1Δ63 | Product of cross between JF2437 and JF2438 |
Plasmids
Plasmids were constructed for these experiments or are from the Fassler laboratory collection (Table 2). pHJ1470 was constructed by introduction of a 2.73-kb PCR product containing the complete AHP1, CCW12, YLR111W, and partial YLR112W open reading frames into ClaI- and EcoRV-digested pRS313 (CEN HIS3) (20). pHJ1497 and pHJ1498 are subclones of pHJ1470 in pRS313. A 1.6-kb NcoI fragment encompassing the open reading frames of CCW12 and the adjacent hypothetical open reading frame YLR111W was cloned into Litmus28 (New England Biolabs) to generate plasmid pJF1558. The 1.6-kb fragment was isolated by XhoI and StuI digestion and introduced into XhoI-SmaI cut pRS313 vector to generate pSS1559. pSS1560 was generated by site-directed mutagenesis of the start codon of YLR111W in pSS1558 using primers YLR111W-12F (5′-GGTGGCCGCGATATCGGGAATTCCAAAAACAAACGG-3′) and YLR111W-12R (5′-CCGTTTGTTTTTGGAATTCCCGATATCGCGGCCACC-3′). The mutagenized 1.6-kb fragment was then cloned into pRS313 to generate pSS1560. pCLM1774 is a derivative of pCLM994 (21), in which amino acids 76–305 were deleted using a two-step PCR protocol. The product was cloned into pCLM994 using EcoRI and StuI sites. Primer sequences for the two-step PCR are available on request.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Ref. |
|---|---|---|
| pSL1156 | CEN URA3 plasmid with OCH1 promoter fragment −154 to + 26 (minimal promoter) cloned upstream of the lacZ gene | 10 |
| pZL1320 | CEN URA3 plasmid with OCH1 promoter fragments −336 to +20 cloned upstream of the lacZ gene | 10 |
| pHJ1470 | CCW12 locus including AHP1, CCW12, YLR111W, and HOG1 in pRS313 (CEN, HIS3) | |
| pHJ1497 | CCW12 in pRS313 (CEN, HIS3) | |
| pHJ1498 | YLR111W in pRS313 (CEN, HIS3) | |
| pSS1559 | CCW12-YLR111w in pRS313 (CEN, HIS3) | |
| pSS1560 | ATG mutant of YLR111w in pRS313 (CEN, HIS3) | |
| pSS1564 | 3× HA after codon 30 in CCW12 in pRS313 (CEN, HIS3) | |
| pSS1881 | SLN1-GFP in pRS313 (CEN, HIS3) | |
| pSS1904 | Sln1ΔECD(aa76–305)-GFP in pRS313 (CEN, HIS3) | |
| pHJ1604 | SED1 in pRS313 (CEN, HIS3) | |
| pHJ1606 | SED1 in pRS423 (2μ, HIS3) | |
| pSL232 | SKN7 in pRS315 (CEN, LEU2) | 46 |
| pSL237 | skn7D427N in pRS315 (CEN, LEU2) | 46 |
| pCLM994 | SLN1-6x myc in pRS313 (CEN, HIS3) | 21 |
| pCLM1774 | Sln1ΔECD (amino acids 76–305)-6x Myc in pRS313 (CEN, HIS3) | |
| pCLM1994 | sln1D1144N-6x Myc in pRS313 (CEN, HIS3) |
Deletion Screen
Each individual strain in the arrayed haploid deletion collection (Research Genetics) was transformed with the POCH1-lacZ reporter, pJF1320 (10), using an adaptation of the micro-scale protocol of the Frozen-EZ yeast transformation kit (Zymo Research Corp.). Briefly, cells of each deletion strain were made competent following growth in a microtiter dish by pelleting the cells, discarding the supernatant, washing the pellet in EZ1 solution, and resuspending the cells in EZ2 + EZ3 + DNA. Following incubation at 30 °C for 45–90 min, cells were spread over one-quarter of an SC-Ura plate. After working out the procedure manually, all but the spreading was accomplished robotically. Transformants were replica-plated to 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) plates, and those exhibiting an elevated blue color were further tested for the specificity of the response using pJF1156 (POCH1-lacZ, a lacZ reporter regulated by the basal OCH1 promoter) and activities determined in liquid assays.
High Copy Suppressor Screen
The ccw12 deletion strain was transformed with the YEp13 high copy library. Transformants were replica-plated for resistance to hygromycin B. Hygromycin B-resistant suppressors were also isolated by direct selection on hygromycin B plates. Candidate high copy suppressor plasmids were isolated from yeast, amplified in Escherichia coli, and reintroduced into the mutant. Plasmids that improved the hygromycin B phenotype of the ccw12 mutant upon retransformation were sequenced, and suppressor genes were identified by comparison of the sequence with the yeast genome data base.
Tests of SLN1-SKN7 Activation by Hypotonic Conditions and by Zymolyase Treatment
For hypotonic conditions, log phase cultures grown at 30 °C in YPD were diluted 1:1 with water. Aliquots removed at various times following dilution were subjected to filtration and the filters immediately frozen at −70 °C. Cells from frozen filters were resuspended in water and collected by low speed centrifugation for RNA or protein extraction. For zymolyase treatment, 1 unit/ml zymolyase was directly added to log phase cultures growing in YPD after adjusting the pH of the culture with Na2HPO4 to a final concentration of 20 mm (2). Zymolyase negative controls were also adjusted to 20 mm Na2HPO4. Reducing agents were not added. Following incubation at 30 °C for 60 min, cells were harvested by centrifugation and stored at −80 °C until needed. Where plasmid retention was an issue, cultures were grown in selective media overnight and then subcultured in YPD for approximately two generations prior to harvesting at log phase.
HOG Pathway Activation
Cultures were grown selectively overnight and subcultured in YPD. At a density of ~1 × 107 cells/ml, 4 m NaCl in YPD was added to a final concentration of 0.4 m. Ten-ml aliquots were taken at 0, 2, 4, and 10 min. Aliquots were filtered and the filters immediately placed on dry ice. Cells were washed in 10 mm Tris, pH 8, 1 mm EDTA, pelleted, and disrupted in 16% trichloroacetic acid plus glass beads. Following centrifugation of the lysate, the pellet was resuspended in loading buffer (150 mm Tris base, 10 mm EDTA, 5% β-mercaptoethanol, 15% glycerol), boiled 10 min, and subjected to electrophoresis on 10% SDS-polyacrylamide gels.
Intracellular Glycerol Measurements
Intracellular glycerol was assayed enzymatically with a commercial glycerol determination kit (Roche Applied Science). Glycerol levels were normalized to cell dry weight. Extracts were prepared from log phase cultures grown in selective media. Pelleted cells were washed once with chilled selective media and once with 2 ml of 0.5 m Tris-HCl, pH 7.4, and resuspended in 2 ml of 0.5 m Tris-HCl, pH 7.4. 500 μl of the cell suspension was heated at 100 °C for 10 min and pelleted at 5000 rpm for 5 min, and the super-natant was used to determine glycerol levels as per the manufacturer's instructions. Cell dry weight was determined by filtering 1.5 ml of the cell suspension through pre-weighed 0.45-μm cellulose nitrate filters (Whatman). After drying at 70 °C for 2 h, filters were weighed, and the difference in weight was used to calculate cell dry weight. Intracellular glycerol levels were expressed as the average of six different assays using at least three different colonies per strain.
Plate Assays
Log phase cultures (~107 cells/ml) were diluted in YPD, and 100, 10−1 , 10−2, and 10−3 dilutions were spotted on plates containing YPD or YPD plus hygromycin B (15–50 μg/ml), Calcofluor White (30 μg/ml), or Congo Red (30 μg/ml). Survival following hypo-osmotic stress was performed using log phase cells grown in YPD plus 1 m sorbitol. Cultures were diluted in the same medium and spotted on YPD with (iso-osmotic) or without sorbitol (hypo-osmoti)c and incubated for 48 h at 30 °C.
β-Galactosidase Assays
Cultures were grown selectively at 30 °C and harvested at 107 cells/ml. Yeast protein extracts were prepared by glass bead lysis and extracts cleared by centrifugation. Activities were calculated in Miller units (22), normalized to protein levels, and expressed as the average of four to six assays using at least three independent colonies or transformants.
Northern (RNA) Hybridization Analysis
Total RNA was prepared using the hot acidic phenol method (23) from cultures grown to ~107 cells/ml in suitable medium. 20 μg of total RNA was loaded per lane of a 1% formaldehyde, 1χ MOPS, 1% aga-rose gel and electrophoresis carried out at 75 V for 4 h. Hybridization was performed with PerfectHyb hybridization buffer (Sigma) as directed by the manufacturer. 32P-Labeled probes were prepared by using random primers with Prime-It random priming labeling kit (Stratagene) and the PCR fragments listed. Signals were detected and quantified by PhosphorImager analysis.
Western Analysis
Extracts were prepared from frozen cell pellets by glass bead breakage in protein extraction buffer (30 mM Tris, pH 8.5, 5 mM EDTA, 3 mM DTT, 5% glycerol, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor mixture (Sigma)). 20 μg of cleared (13,000 rpm, 5 min) extract was subjected to SDS-polyacrylamide (29:1 acrylamide:bisacrylamide) gel electrophoresis (10% running gel and 4% stacking gel). Separated proteins were electroblotted to nitrocellulose filter paper (0.45 μm; Schleicher & Schuell). The blot was stained with Ponceau S, washed, and scanned. Following destaining, the blot was blocked with 5% skim milk, incubated with the appropriate antibody in 5% bovine serum albumin, washed, incubated with a 1:1000 dilution of the appropriate peroxidase-conjugated IgG (Sigma), and exposed to film. Antibodies included phospho-p38 antibody (1:1000, Cell Signaling); yC-20 anti-Hog1 antibody (1:2000, Santa Cruz Biotechnology); 12CA5 anti-HA antibody (1:1000, Roche Applied Science); antiphospho-p44/p42 MAP kinase antibody (1:1000, Cell Signaling); and anti-Myc antibody (1:1000, Cell Signaling). Blots were developed using the SuperSignal bioluminescence kit (Pierce).
Dithiothreitol Extraction of Ccw12-HA from Intact Cells
The cells pellet from a 10-ml culture at ~107cells/ml was washed with 1:5 volume 0.25 mM Tris, pH 8.0, and resuspended in 1:10 volume of the same buffer. 1 m DTT was added to the cell suspension to a final concentration of 5mM and incubated at 4 °C with shaking for 2 h. Cells were collected by low speed centrifugation, and the supernatant was analyzed. Cell pellets from DTT-extracted cells were used in preparing the “post-DTT” cell extract.
Fluorescence Microscopy
Log phase cultures expressing GFP fusions were pelleted and resuspended in residual culture medium. Cells were observed with a Leica DM RBE microscope and a Leica ×100 PL Fluotar 1.3 NA objective lens. Fluorescence images were captured using a Photonic Science digital charge-coupled device camera system. Images were processed and edited using Adobe Photoshop.
RESULTS
CCW12 Involvement in SLN1-SKN7 Signaling
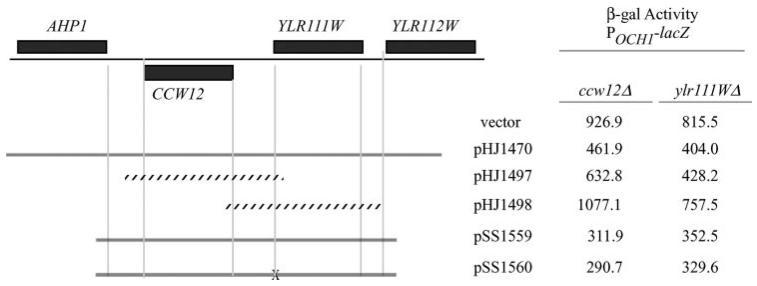
A screen of the yeast deletion collection (Research Genetics) based on increased expression of a lacZ reporter driven by the promoter of the SLN1-SKN7 target gene, OCH1 (POCH-lacZ) (10), led to the identification of the YLR111W open reading frame and the CCW12 gene (YLR110C) as potential regulators of SLN1-SKN7 pathway activity.3 The short distance (330 bp) between ATGs of the two divergently transcribed open reading frames suggested that one deletion might eliminate the regulatory region required for expression of the other. To determine the open reading frame responsible for normal SLN1-SKN7 pathway activity, subclones were tested for complementation (Fig. 1). A subclone containing CCW12 and associated upstream regulatory sequence partially complemented the elevated reporter gene expression seen in the ccw12 and ylr111w mutants (vector); however, the same was not true of YLR111W. Because full complementation of the phenotype required both the CCW12 and YLR111W open reading frames, the pertinence of the YLR111W open reading frame was more directly tested by introducing a mutation into its predicted start codon (Fig. 1). The ATG mutant (pSS1560) complemented the ccw12 deletion phenotype to the same extent as the equivalent construct lacking the mutation (pSS1559), confirming that activation of SLN1-SKN7 signaling in the YLR111W deletion mutant was indirect.
FIGURE 1. Involvement of the CCW12 locus in SLN1-SKN7 pathway activation.
Subclones containing portions of the left arm of chromosome XII were tested for complementation of the pathway activation phenotype of the ccw12 and ylr111w deletion mutants from the Research Genetics Collection. The effects of subclones on pathway activity was measured using the pathway-responsive POCH1-lacZ reporter, pJF1320 (10). β-Galactosidase (β-gal) activity values are the average of three or more transformants and are represented as Miller units normalized to protein. Standard deviations are less than 25%. pSS1560 is identical to pSS1559, but with a mutation of the putative translational start codon of YLR111W, which is marked by the X. Solid lines, clones that complement; stippled lines, clones that complement poorly or not at all.
The relevance of the ccw12 mutation to SLN1-SKN7 pathway activity was further investigated by RNA analysis of the SLN1-SKN7 target genes, OCH1 (10) and NCA3. NCA3 (nuclear control of ATPase) is a nuclear gene involved in the mitochondrial expression of specific subunits of the F0-F1-ATP synthase (24) but also localizes to the cell wall and playsa role in septation (25, 26). NCA3 was identified as a target of the SLN1-SKN7 pathway in microarray experiments4 and is more abundant and therefore easier to measure than OCH1. As expected for mutations increasing SLN1-SKN7 pathway activity, expression of the OCH1 and NCA3 genes was elevated in the ccw12 mutant (Fig. 2). The activating effect of ccw12Δ was dependent on the phosphorylatable aspartate (Asp-427) in the receiver domain of Skn7p. No activation because of the ccw12Δ mutation was observed in the skn7D427N (D/N) mutant in which aspartate 427 is replaced with the nonphosphorylatable asparagine (Fig. 2). The increase in expression of OCH1 in the ccw12Δ strain was also dependent on SLN1 (Table 3). Strains lacking the SSK1 gene were used in examining the role of SLN1, because the ssk1 deletion mutation suppresses the lethality of the sln1Δ mutation (3). The ccw12-mediated elevation in expression of the POCH-lacZ reporter was eliminated in the ccw12 sln1 ssk1 triple mutant (Table 3). Although changes in expression related to the SSK1 and SLN1 (and SKN7) alleles were evident in the reporter assays, these changes did not interfere with detection of a ccw12 phenotype. For example, although POCH-lacZ activity decreased from 360.8 to 136.6 because of the ssk1Δ mutation; the ccw12 mutation nevertheless caused a 3.5-fold increase to 474.9 in the ssk1Δ background (Table 3).
FIGURE 2. ccw12Δ-mediated activation of the SLN1-SKN7 target genes, NCA3 and OCH1, and its dependence on SKN7.
Northern (RNA) hybridization analysis of the NCA3 and OCH1 expression in CCW12 skn7Δ (JF1904, lane 1) and ccw12Δ skn7Δ (JF2279, lanes 2– 4) strains containing different alleles of SKN7 carried on CEN plasmids (pSL232 (SKN7+), pSL237 (skn7D4227N), or pRS315 (skn7Δ)). DED1 hybridization is shown as a normalization control.
TABLE 3.
SKN7 and SLN1 dependence of ccw12 activation of the OCH1 SLN1-SKN7 pathway target gene
| Genotype |
β-Galactosidase activity (S.D.),a (POCH1-lacZb) |
Induction (ccw12Δ/CCW12) |
|---|---|---|
| SKN7 CCW12c | 320.8 (25.8) | 1.0 |
| SKN7 ccw12Δ | 538.9 (72.4) | 1.7 |
| skn7D427N ccw12Δ | 123.5 (30.1) | 0.4 |
| skn7Δ ccw12Δ | 26.5 (10.2) | 0.08 |
| SLN1 SSK1 CCW12 d | 360.8 (71.2) | 1.0 |
| SLN1 SSK1 ccw12Δ | 743.7 (59.9) | 2.1 |
| SLN1 ssk1Δ CCW12 | 136.6 (30.1) | 1.0 |
| SLN1 ssk1Δ ccw12Δ | 474.9 (44.3) | 3.5 |
| sln1Δ ssk1Δ CCW12 | 40.3 (13.9) | 1.0 |
| sln1Δ ssk1Δ ccw12Δ | 19.1 (0.8) | 0.5 |
β-Galactosidase activities are given in Miller units normalized to protein concentration and represent the averages of three measurements. Standard deviations are given in parentheses.
Each strain is transformed with the CEN based POCH1-lacZ, pZL1320 (10).
CCW12 skn7Δ, JF1904; ccw12 skn7Δ (JF2279) strains carry the wild type (pSL232) or D427N (pSL237) alleles of SKN7 on CEN plasmids, and strains lacking SKN7 carry the empty pRS315 vector.
Strains include CCW12 SSK1 SLN1, JF1565; CCW12 ssk1Δ SLN, JF1919; ccw12Δ SSK1 SLN1, JF2278; ccw12Δ ssk1Δ SLN1, JF2301; CCW12 ssk1Δ sln1Δ, JF2181; ccw12Δ ssk1Δ sln1Δ, JF2300.
Changes in Turgor Are Not Sufficient for SLN1-SKN7 Pathway Activation
The possibility that up-regulation of the Sln1 kinase is attributable to increased intracellular pressure in the ccw12 mutant as it appears to be in the fps1 mutant (1) was evaluated by testing the response of the SLN1-SKN7 pathway to other treatments known to affect turgor. The PKC cell wall integrity MAPK pathway is activated in response to increased turgor caused by a wide variety of conditions (27, 28). Two of these, hypo-osmotic shock and the ppz1 ppz2 double mutant which accumulates intracellular K+, were used in evaluating the response of the SLN1-SKN7 pathway to turgor.
Cultures grown in rich media (YPD) were diluted 1:1 with water. This treatment has been shown to activate the PKC pathway very quickly (27). Phosphorylation of the Slt2p/Mpk1p MAP kinase in the PKC pathway within 2 min of exposing cultures to a dilute environment confirms that this treatment does cause an increase in turgor (Fig. 3C). Because the SLT2/MPK1 gene is also a target of the PKC pathway (29), its expression was examined as a base line for normal PKC pathway target gene induction kinetics (Fig. 3A, bottom panel). SLT2/MPK1 expression was elevated 2-fold in one experiment (Fig. 3A) and 3.5-fold in a replicate experiment at the 10-min time point.
FIGURE 3. PKC-activating treatments, including hypotonic stress and accumulation of intracellular K+, fail to cause rapid activation of the SLN1-SKN7 pathway.
A, Northern (RNA) hybridization analysis of SLT2, OCH1, and NCA3 expression at indicated times following 1:1 dilution of the media with water. Strains included the skn7Δ strain, JF1904 carrying SKN7+ (pSL232), and skn7D427N (pSL237) plasmids. Hybridization to the SLN1-SKN7-independent PGK1 gene was used as a normalization control. Normalized values are shown below each strip. B, Northern (RNA) hybridization analysis of NCA3 expression in wild type (WT) (JF1565), ccw12Δ (JF2278), and ppz1Δ ppz2Δ (JF2441) strains. Hybridization to the SLN1-SKN7-independent CDC33 gene was used as a normalization control. C, Western analysis of phospho-Slt2p in wild type (JF1565) at times following 1:1 dilution of the media with water. D, Western analysis of phospho-Slt2p in wild type (JF1565), ccw12Δ (JF2278), and ppz1Δ ppz2Δ (JF2441) strains. Ponceau S staining is included to show equivalent loading of protein extracts in each lane.
To determine the effect of an increase in turgor sufficient to activate the PKC pathway on SLN1-SKN7 target genes, NCA3 and OCH1 gene expression was monitored over time. NCA3 levels were increased ~2-fold at the 30-min time point. This increase was not observed in a strain carrying the skn7D427N allele indicating that the increase was SKN7 Asp-427-dependent. OCH1 levels were increased only very slightly to 1.2- or 1.3-fold and then restored to untreated levels by 30 min (Fig. 3A).
The PPZ1 and PPZ2 phosphatases are regulators of K+ and pH homeostasis (30), and the double mutant exhibits elevated intracellular K+ levels (30) leading to water uptake. An associated increase in cell size and PKC pathway activation (Fig. 3D) has been observed (29), consistent with an increase in turgor in the mutant. If SLN1-SKN7 pathway activation in ccw12 and fps1 mutants and zymolyase-treated cells is attributable to changes in membrane tension, the ppz1, ppz2 double mutant would be expected to activate the pathway. No increase in the expression of NCA3 (Fig. 3B) or OCH1 (data not shown) was observed in the ppz1 ppz2 double mutant, despite the expected increase in NCA3 (Fig. 3B) and OCH1 (not shown) expression in the ccw12 mutant. Taken together, the absence of an effect on SLN1-SKN7 target gene expression in the ppz1 ppz2 double mutant combined with the slow and modest increases observed following hypo-osmotic stress suggests that simple elevation in turgor is not sufficient for Sln1p kinase stimulation.
Ccw12p Does Not Regulate the Fps1p Glycerol Channel
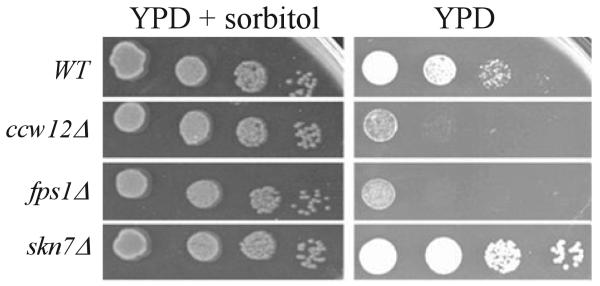
To test the possibility that Ccw12p might regulate activity of the Fps1p glycerol channel, loss of which activates the SLN1-SKN7 pathway (1), intracellular glycerol levels were measured in the ccw12 mutant. In the absence of the glycerol channel, accumulated glycerol cannot be released to the medium, and the resulting increase in turgor causes a 100-fold decrease in viability upon hypotonic shift (16, 18). The ccw12 mutant likewise exhibited loss of viability upon hypotonic shock. A shift from growth media containing 1 M sorbitol to media lacking sorbitol led to comparable losses in viability in both the ccw12 and the fps1 mutants (Fig. 4). Unlike the fps1Δ mutant, which exhibits a 4-fold increase in intracellular glycerol relative to wild type (16), the ccw12 mutant showed no elevation in glycerol levels despite activating the SLN1-SKN7 pathway to an extent equivalent to fps1Δ (Table 4). These results suggest that activation of the SLN1-SKN7 pathway in strains lacking the Ccw12 protein is not the result of changes in intracellular glycerol concentration, but it may instead be due to a weakened cell wall or to the absence of the Ccw12 protein itself.
FIGURE 4. The ccw12 mutant is sensitive to hypo-osmotic stress.
Wild type (WT) (JF1565), ccw12Δ (JF2278), skn7Δ (JF1904), and fps1Δ (JF1732) strains grown to log phase in YPD containing 1 M sorbitol were serially diluted in the same media and spotted on YPD plates with or without 1 M sorbitol. A representative experiment is shown. Plates were scanned after 48 h at 30 °C.
TABLE 4.
Effect of ccw12 on intracellular glycerol
| Genotypea | Glycerolb (S.D.) |
Relative glycerol |
β-Galactosidase activityc (S.D.) POCH1-lacZd |
Relative POCH1-lacZ |
|---|---|---|---|---|
| Wild type | 6.3 (1.6) | 1.0 | 113.1 (19.5) | 1.0 |
| ccw12Δ | 7.5 (1.4) | 1.2 | 312.6 (14.7) | 2.8 |
| fps1Δ | 23.9 (3.2) | 4.1 | 298.5 (45.8) | 2.6 |
Wild type, JF1565; ccw12Δ, JF2278 and fps1ΔA, JF1732 are shown.
Glycerol (mg/g) values are the average of six determinations per strain.
β-Galactosidase values are given in Miller units normalized to protein concentration and are the average of three measurements per strain.
Each strain is transformed with the CEN based POCH1-lacZ reporter, pZL1320 (10).
Activation of the SLN1-SKN7 Pathway Is Not a General Feature of Cell Wall Mutants
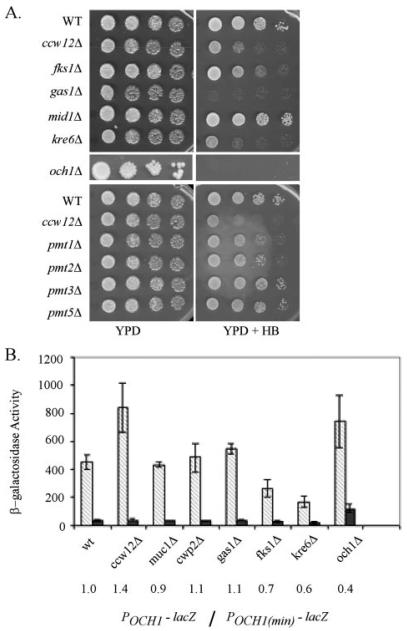
To determine whether the SLN1-SKN7 pathway responds to all major cell wall perturbations or exclusively to the absence of Ccw12p, the effect on the SLN1-SKN7 pathway activity of wall gene mutations, mutations in genes affecting the wall, or mutations in genes encoding proteins similar to Ccw12p was evaluated. Included in the survey were genes showing partial sequence similarity to CCW12 (FLO1, SED1, AGA1, FIG 2, PRY3, FLO10, YDR124C, SPI1, and WSC4), genes encoding proteins involved in mannosylation (MNN1, MNN2, MNN9, OCH1, PMI1, PMI2, PMI3, PMI5, and PMI6), and genes encoding proteins with other cell wall functions (FKS1, β-1,3-glucan synthase subunit; KNR4, regulation of cell wall synthesis and possible coordination with cell cycle; KRE6, β-1,6-glucan synthase; and GAS1, β-1,3-glucanosyltransferase). Several of the genes tested (SED1, AGA1, SPI1, MUC1, and CWP2) encode GPI-anchored mannoproteins like Ccw12p. Deletion mutants were tested for POCH-lacZ reporter gene activation. A minimal reporter (POCH(min)>-lacZ) was used as a control for nonspecific activation. SLN1-SKN7 pathway activation was evaluated by comparing changes in POCH-lacZ reporter gene expression to changes in POCH(min)-lacZ reporter expression in each mutant relative to the wild type strain. SLN1-SKN7 pathway activation was not elevated in any of the wall mutants listed with the exception of the ccw12 mutant. Among cell wall mutants, the ccw12 mutant is especially sensitive to the aminoglycoside antibiotic, hygromycin B (HB), suggesting that it has reduced levels of cell surface glycosylation that lead to an increase in permeability to the drug (31). Wall mutants exhibiting comparable HB sensitivity (and presumably equivalent types of wall perturbations) were therefore of special interest (Fig. 5A). Fig. 5B shows all mutants with an HBS phenotype as well as a sampling of others tested. An apparent increase in the och1 mutant was found to be nonspecific (see elevated POCH(min)-lacZ and a relative OCH1/minimal reporter ratio of 0.4). These data suggest that the SLN1-SKN7 pathway activation phenotype is not a generic consequence of reduced wall integrity but rather a specific consequence of the loss of the Ccw12 protein.
FIGURE 5. Hygromycin B-sensitive wall mutants do not activate the SLN1-SKN7 pathway.
Strains were Kan::MX marked deletions from the haploid Research Genetics Deletion Collection assembled based on their known functions in wall integrity and tested for hygromycin B sensitivity and activation of the SLN1-SKN7 pathway-dependent POCH1-lacZ reporter gene. A, hygromycin B sensitivity test. Strains were grown to log phase in YPD and 10-fold dilutions spotted on YPD or YPD containing 25 μg/ml hygromycin B plates, which were incubated at 30 °C for 48 h and then scanned. B, β-galactosidase was measured in a at least three transformants carrying the POCH1-lacZ reporter, pZL1320 (diagonal striped bars), or the POCH1(min)-lacZ reporter, pSL1156 (dark bars). Activity is represented in Miller units and normalized to protein concentration. The number below the plot is the ratio of average activity for the mutant (POCH1 reporter/POCH1(min) control reporter) over the average activity for a wild type (WT) strain measured in the same experiment.
Suppression of ccw12 Wall and Activation Phenotypes by SED1 Overexpression
Insight into the role of Ccw12p in SLN1 signal transduction was sought by looking for genes that are able to complement the HBS phenotype of the ccw12 mutant. The strongest high copy suppressors carried the SED1 gene. Subclones containing only the SED1 gene partially suppressed the HB sensitivity and reporter gene activation phenotypes of the ccw12 mutant (Fig. 6A). The lower SED1 dosage (centromere (CEN)) was somewhat more effective than high dosage (2 μm) suggesting that there is an optimal stoichiometry for SED1 in the wall. SED1 encodes a highly glycosylated, glucanase-extractable, GPI-anchored cell wall protein that is most abundant in the walls of stationary phase cells (32). To investigate the role of SED1 and CCW12 in wall integrity and SLN1-SKN7 pathway regulation, the effects of single and double deletions were compared. Resistance to hygromycin B and SLN1-SKN7 pathway activity measured by the POCH1-lacZ reporter was normal in the sed1Δ strain (Fig. 6B). In contrast, ccw12Δ phenotypes, including HBS and SLN1-SKN7 pathway activation, were exacerbated in the absence of SED1 (Fig. 6) consistent with previous reports that elevated SED1 expression in mutants lacking GPI-anchored wall proteins such as Ccw12p has a compensatory effect (33).
FIGURE 6. SED1 overexpression suppresses the drug sensitivity and SLN1-SKN7 pathway activation phenotypes of a ccw12 mutant.
A, growth of the ccw12Δ mutant strain, JF2278 carrying a CEN-based (pHJ1604) or 2-μm based (pHJ1606) SED1 expression plasmid, was assayed on YPD plates or YPD plates containing 25 μg/ml HB, 30 μg/ml Congo Red (CR), or 20 μg/ml CFW. The same strain carrying a complementing CCW12 plasmid (pSS1559) or empty vector (pRS313) is shown for comparison. SLN1-SKN7 pathway activation was measured using the POCH1-lacZ reporter, pZL1320 (10). β-Galactosidase (β-gal) activity values are the average of three of more transformants and are represented in Miller units normalized to protein. Standard deviations (SD) are shown in parentheses. B, SED1 contributes to cell wall integrity of log phase cells in the absence of Ccw12p. Wild type (WT) (JF1565), sed1Δ (JF2337), ccw12Δ (JF2278), and ccw12Δsed1Δ (JF2338) strains carrying the POCH1-lacZ reporter, pZL1320 (10), were grown selectively, diluted into YPD media, and spotted on YPD plates containing 0 (YPD) or 15 mg/ml HB. β-Galactosidase activity is the average of three of more transformants and is represented in Miller units and normalized to protein concentrations. Standard deviations are shown in parentheses.
Perturbation of the Wall with Low Level Zymolyase Treatment Causes SLN1-SKN7 Pathway Activation
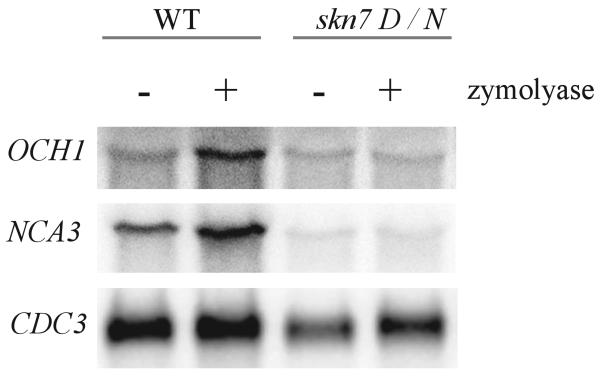
To investigate whether more general cell wall insults such as enzymatic weakening of the cell wall might affect the pathway, wild type cells were treated with low levels of zymolyase (β1–3-glucanase with trace amounts of alkaline protease and mannanase). Treatment was kept well below concentrations needed to digest the wall, and reducing agents that are normally added to stimulate zymolyase activity were omitted (34). There was no loss of light scattering at A600, consistent with minimal damage to the wall. These zymolyase concentrations nonetheless caused induction of SLN1-SKN7-dependent target genes, NCA3 and OCH1, to an extent comparable with that seen in the ccw12 mutant. Induction by zymolyase was dependent on SKN7 and on the phosphorylatable aspartate, Asp-427. skn7D427N mutants did not exhibit an increase in NCA3 expression after zymolyase treatment (Fig. 7).
FIGURE 7. Activation of the SLN1-SKN7 pathway by zymolyase treatment.
Northern (RNA) hybridization analysis of RNA was prepared from wild type (wt) (JF1565) and skn7D427N (JF2336) cultures after 60 min of treatment with 1 unit/ml zymolyase (+) or without treatment (−). OCH1 and NCA3 expression was normalized to expression of the SLN1-SKN7-independent CDC33 gene.
Zymolyase-treated Cells and fps1 Mutants Have Reduced Levels of Wall-associated Ccw12
Because zymolyase is an impure glucanase preparation with a contaminating protease component, it has the potential to digest outer wall proteins. The effect of zymolyase on SLN1-SKN7 activity might therefore be attributable to loss of the Ccw12 protein from the cell wall. To test this possibility, wall-associated Ccw12p levels in zymolyase-treated cells were compared with levels in untreated cells.
The GPI anchor of the Ccw12 (covalent cell wall) protein is believed to be responsible for its association with the wall (35). Consistent with this, the protein can be extracted using glucanase in the form of a very large (>300 kDa) polydisperse band, which includes remnants of the glucan-chitin meshwork from the wall (33). A 58-kDa cytoplasmic Ccw12p species is observed in cleared crude cell extracts. This species is larger than the 16-kDa encoded protein because of extensive O-mannosylation and the presence of a GPI anchor (33). Unexpectedly, we found that treatment of intact cells with the reducing agent, dithiothreitol, extracted a similar ~58-kDa Ccw12p species from wild type cell walls (Fig. 8A, lanes 1 and 2) suggesting that a fraction of wall-associated Ccw12p is not actually GPI anchored but may be associated with the wall via disulfide bonds.
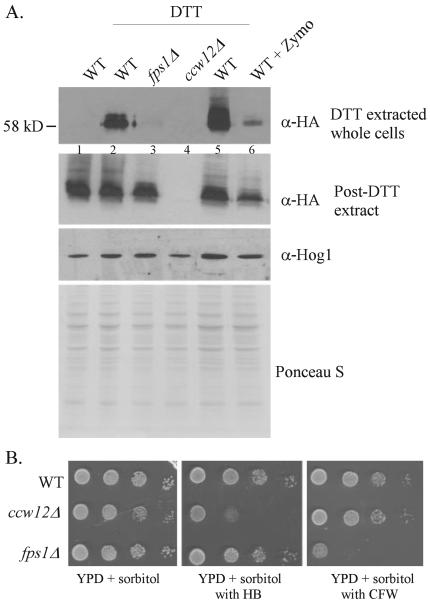
FIGURE 8. Zymolyase treatment and the fps1 mutation cause reductions in wall-associated Ccw12p.
Anti-HA Western analysis of Ccw12 protein levels in strains carrying the CCW12-HA expression plasmid pSS1564. A, levels of cell wall-associated Ccw12p in various strains were compared by Western analysis following recovery of surface proteins upon (5 mm) DTT treatment and in extracts prepared from the DTT-treated cell pellet. The α-Hog1 Western and Ponceau S staining show even loading of protein extracts in each lane. Equivalent volumes of the DTT supernatants were loaded in the top panel. Wild type (WT), ccw12Δ (JF2278) carrying pSS1564 (CCW12-HA); ccw12Δ, JF2278 carrying vector; fps1Δ, ccw12Δfps1Δ (JF2442) carrying pSS1564 (CCW12-HA). Cells were treated with 1 unit of zymolyase 100T (US Biological) prior to DTT extraction in lane 6. B, strains were grown to log phase, and 10-fold dilutions were spotted on YPD + 0.5 m sorbitol plates and the same type of plate but containing 25 μg/ml HB or 30 μg/ml CFW. Plates were incubated at 30 °C for 48 h prior to scanning.
A major reduction in DTT-extractable Ccw12 protein was observed in cells treated with the low levels of zymolyase sufficient to activate the SLN1-SKN7 pathway (Fig. 8A, lanes 5 and 6). That the CCW12 gene is expressed normally under these conditions is shown by comparing levels of the 58-kDa Ccw12p species in cleared crude cell extracts prepared from cells of each genotype following their treatment with DTT. Protein levels in each extract were normalized using anti-Hog1p reactivity and separately by Ponceau S staining (Fig. 8A, 3rd and 4th panels).
If reduced wall-associated Ccw12p were responsible for SLN1-SKN7 pathway activation, a similar reduction in wall-associated Ccw12p would be expected in the fps1 mutant. Our analysis of DTT-extractable Ccw12p shows that the fps1 mutant lacks detectable DTT-extractable Ccw12 protein (Fig. 8A, lane 3).
Although a glycerol channel defect is not expected to cause wall abnormalities, the fps1 mutant does exhibit sensitivity to the chitin-binding drug, CFW (Fig. 8B). Sensitivity to CFW is indicative of increased chitin levels, a phenotype common in mutants with reduced wall integrity. Although the CFWS phenotype in the fps1 mutant differs from the HBS phenotype of the ccw12 mutant, the reduction in Ccw12 protein in walls of the fps1 mutant is indicative of a correlation between activation of the pathway and reduction in Ccw12p levels in the wall.
The Sln1 Extracellular Domain Is Required for Pathway Activation by Wall Perturbants
If the state of the cell wall plays a role in Sln1p kinase regulation, detection of that signal is expected to involve the extracellular (periplasmic) domain (ECD) of Sln1p. An ECD deletion encompassing amino acids 76–305, roughly 80% of the domain, was constructed. Despite the large deletion, the ECD mutant was viable, showed partial localization to the cell periphery (Fig. 9A), and exhibited Hog1p phosphorylation kinetics similar to wild type upon treatment with salt (Fig. 9C). Because mutants lacking Sln1p kinase activity are inviable because of constitutive activation of the HOG1 pathway, the viability of the ECD mutant suggests that basal kinase levels are not substantially affected. Likewise, normal Hog1p phosphorylation kinetics in the ECD mutant suggests that Sln1p inactivation is normal. To investigate the effect of the ECD mutation on Sln1p activation, the effect of the activating ccw12 mutation and of zymolyase treatment on OCH1 and NCA3 levels were compared in SLN1 and sln1ΔECD strains. Both types of activation were reduced or eliminated in the ECD deletion mutant (Fig. 9D). Although Western analysis revealed a reduction in the expression of the ECD mutant relative to wild type (Fig. 9B), there was no detectable effect on basal NCA3 or OCH1 expression (Fig. 9D), suggesting that the reduced protein levels are nonetheless sufficient for normal levels of signaling. The requirement for the extracellular domain of Sln1p is consistent with models of activation involving physical contact between Sln1p and some component of the wall which remains to be defined.
FIGURE 9. The extracellular domain is required for Sln1 kinase activation by wall perturbations.
A, representative field showing localization of SLN1-GFP (pSS1881) or sln1ΔECD-GFP (pSS1904). BF, bright field. B, Western analysis showing reduced levels of sln1ΔECD compared with full-length SLN1. SLN1, JF2007 (sln1Δ) carrying full-length SLN1-myc (pCLM994); sln1ΔECD, JF2007 carrying sln1ΔECD-myc (pCLM1774). C, comparison of Hog1 phosphorylation kinetics in SLN1 (JF2008 (sln1Δsho1Δ) carrying full-length (pCLM994)) and sln1ΔECD (JF2008 (sln1Δsho1Δ) carrying sln1ΔECD (pCLM1774)) strains following addition of 0.4 m NaCl. D, Northern (RNA) hybridization analysis of RNA prepared from wild type cultures from the sln1Δ strain, JF2007, or sln1Δccw12Δ strain, JF2368, transformed with the SLN1 expression plasmids, pCLM994 (SLN1) or pCLM1774 (sln1ΔECD), and treated with 1 unit/ml zymolyase (+) where indicated. NCA3 expression was normalized to expression of the SLN1-SKN7-independent CDC33 gene. The extent of induction by zymolyase is shown relative to the untreated SLN1 or sln1ΔECD strains.
Physiological Role for the SLN1 Pathway in Cells Lacking a Full Complement of Wall Proteins
Involvement of the SLN1-SKN7 pathway in wall composition is suggested by the increased resistance to hygromycin B in sln1* mutations that activate the pathway and modest sensitivity to the drug in an skn7D427N mutant (10). This is consistent with SLN1-SKN7 pathway regulation of the OCH1 gene encoding an α1,6-mannosyltransferase involved in N-glycosylation of wall mannoproteins (10).
The modest hygromycin B sensitivity of the skn7D427N mutant suggests that the role of the SLN1-SKN7 pathway in the integrity of a relatively static wall is minor. To investigate whether the SLN1-SKN7 pathway may assume more importance during wall remodeling, the effect of the SLN1 phospho-relay mutation, sln1D1144N, was examined in the ccw12 mutant. The lethality of the sln1D1144N mutation was suppressed in these strains by the presence of a plasmid expressing the PTP2 Hog1p phosphatase. Like the skn7D427N mutation (10), the sln1D1144N mutation alone exhibited only modest sensitivity to hygromycin B (reflecting wall glycosylation) and Congo Red (reflecting chitin level abnormalities). In the absence of the Ccw12 protein, however, the sln1D1144N mutation caused striking wall phenotypes. The effect of the sln1 mutation was most apparent on media containing 1 m sorbitol as osmotic support (Fig. 10), which corrects hygromycin B and Congo Red phenotypes attributable to the absence of the Ccw12 protein. These observations point to a specific role for the SLN1-SKN7 pathway in a compensatory wall formulation that may be elicited by the absence of Ccw12 protein.
FIGURE 10. The importance of the SLN1-SKN7 pathway increases under wall stress.
Strains JF2007 (sln1Δ) and JF2368 (sln1Δccw12Δ) carrying the PTP2 expression plasmid to allow viability were transformed with plasmids pCLM994 (SLN1+) or pCLM1994 (sln1D1144N) and grown to log phase in YPD or YPD + 1 m sorbitol, and serial dilutions were spotted onto YPD, YPD + hygromycin B (25 μg/ml), and YPD + Congo Red (30 μg/ml) plates to which sorbitol (1 m) was added where indicated (bottom). Plates were photographed after 48 h of incubation at 30 °C.
DISCUSSION
The yeast wall determines the shape and the integrity of the organism during growth and cell division. The relatively simple composition of the yeast wall, 40% mannoproteins, 60% β-glucan (85% long chain, and 15% short chain), and 2% chitin (36), nonetheless provides the necessary stability against tension caused by high internal hydrostatic pressure. This property of the wall is presumed to be due to the formation of interlinkages between the individual wall components to create a highly branched meshwork. A structure consisting of covalently linked β-1,3- and β-1,6-glucan, chitin, and mannoproteins has been reported (37, 38), and a stable phosphodiester linkage has been proposed to connect the mannoproteins to the rest of the meshwork (39).
CCW12 encodes one of three covalently linked cell wall proteins present in the most highly phosphorylated cell wall fraction of yeast cells (35). The ccw12 mutant exhibits reduced agglutination and mating (35) possibly due to inaccessibility of agglutinins at the cell surface or to other changes in the structure of the cell wall. Increased sensitivity to Calcofluor White, Congo Red (35), hygromycin B, and low doses of zymolyase5 (40) suggest extensive alterations in cell wall composition and structure in the ccw12 mutant.
The absence of the abundant Ccw12 protein from the cell surface activates the SLN1-SKN7 pathway, causing 2–3-fold increase in transcription of the OCH1 and NCA3 target genes in a SLN1 and SKN7 Asp-427-dependent manner. The isolation of SED1 as a multicopy suppressor of the ccw12 wall and activation phenotypes was predicted from previous reports in which SED1 was found to be up-regulated in mutants with cell wall instability caused by the absence of GPI-anchored mannoproteins, including Ccw12p (33). Like Ccw12p, Sed1p is a predicted GPI-anchored cell wall mannoprotein. In addition, conserved sequences (36% identity over 74 residues of the 133 amino acids Ccw12 (41)) in the two proteins suggest possible functional redundancy. Unlike CCW12, SED1 is most highly expressed in stationary phase (32) and is regulated by the PKC pathway (42) in response to cell wall stress (43, 44). Our finding that SED1 overexpression partially suppresses both the wall phenotypes of the ccw12 mutant and the SLN1-SKN7 pathway activation phenotype indicates that these two phenotypes are inter-dependent and that SLN1-SKN7 pathway activation may therefore be attributable to the wall defects of the ccw12 mutant. The observation that the ccw12sed1 double deletion exhibits both enhanced sensitivity to cell wall perturbants as well as increased activation of SLN1-SKN7 target genes further supports the conclusion that pathway activation is a direct consequence of wall defects because of the absence of specific mannoproteins on the outer surface of the cell.
Although osmotic imbalance and increased turgor in the fps1 mutant was thought to be responsible for SLN1-SKN7 pathway activation (1), more extensive testing shows that this is not a sufficient condition for Sln1p kinase activation. For example, hypotonic stress conditions sufficient to cause immediate activation of the PKC MAPK pathway cause a slow and relatively weak response in SLN1-SKN7 target gene expression. Similarly, SLN1-SKN7 target genes are not activated in the ppz1ppz2 double mutant, in which elevated intracellular K+ levels cause increased phosphorylation of the Slt2p/Mpk1p MAPK. These observations suggest that changes in turgor are unlikely to be the proximate cause of activation.
The observation that zymolyase-treated cells and the fps1 mutant both have reduced levels of wall-associated Ccw12p leads to the alternative explanation that Ccw12p plays a specific role in regulating Sln1p kinase activity. Because soluble Ccw12p levels are unaffected in the fps1 mutant (Fig. 8A), the effect is not on expression or protein stability. The basis for the reduction in cell wall association of Ccw12p in the fps1 mutant will require further investigation.
The increase in Sln1p kinase activity in response to Ccw12p loss required the extracellular domain. Because absence of the extracellular domain did not affect viability (SLN1 is an essential gene), the kinetics of Hog1 phosphorylation in response to salt or basal activity of SLN1-SKN7-dependent genes (Fig. 9), we conclude that the ECD is required for activation of the kinase, but it is less critical for its inactivation, which is needed to trigger Hog1p phosphorylation.
The role of the cell wall in Sln1p kinase regulation has been hinted at previously. Cells lacking walls were found to activate the HOG1 pathway (2) and thus inactivate the Sln1p kinase. Because wall removal might mimic a reduction in turgor, these results were interpreted to mean that turgor is important for Sln1 kinase activation. In this study we find that the presence or absence of the Ccw12 protein in the wall, but not changes in turgor, nor other types of alterations in the wall, affects the activation of the Sln1 kinase. Our recent observation5 that small deletions of specific ECD sequences reduce Sln1p kinase activation by zymolyase treatment are consistent with models in which loss of Ccw12p permits an interaction between the Sln1p ECD and one or more molecules associated with the inner face of the wall that are required for activation. Because Ccw12p is itself localized to the outer face of the wall, the interaction with Sln1p is unlikely to be direct. Consistent with the conclusion that the Ccw12p and Sln1 proteins do not interact are results of random mutagenesis of the CCW12 coding region in which 100% (20 out of 20) SLN1-SKN7 activating mutations isolated were found to be changes in the cysteine 72 residue of the Ccw12 protein that caused a reduction in CCW12 expression.5 We therefore favor the interpretation that the loss of Ccw12p from the wall leads to the appearance of novel inner wall or periplasmic molecules that are poised to contact the Sln1 ECD thus causing its activation.
Our study of the Ccw12 wall protein also provides insight into the physiological role of the SLN1-SKN7 pathway. The mild hygromycin B phenotype seen in skn7D427N (10) and sln1D1144N (Fig. 10) mutants suggests that the SLN1-SKN7 pathway plays a minor role in cell wall integrity under normal growth conditions. However, the pathway appears to become more important when the wall is reformulated as it is expected to be in the absence of Ccw12p or in zymolyase-treated cells. In the ccw12Δ background, loss of SLN1-SKN7 pathway activity because of the sln1D1144N mutation caused striking wall phenotypes, especially on media containing 1 m sorbitol as osmotic support. Sorbitol corrected the hygromycin B and Congo Red phenotypes caused by the ccw12 mutation alone but was unable to correct the growth defect caused by the sln1 mutation. Hence, SLN1-SKN7 pathway activity appears to gain importance under conditions requiring wholesale wall remodeling. We therefore propose that Ccw12p levels may be a marker for conditions requiring wall remodeling activity.
Acknowledgments
We are grateful for the continuing technical support of Greg Gingerich and previous lab members Hyunjeong Jeong and Zhijian Li who provided excellent assistance with aspects of this work.
Footnotes
This work was supported by the National Institutes of Health Grant GM56719 and by a predoctoral fellowship from the Center for Biocatalysis and Bioprocessing (to S. S. N.).
The abbreviations used are: MAP, mitogen-activated protein; HOG, hyperosmolarity glycerol; MAPK, MAP kinase; HA, hemagglutinin; DTT, dithiothreitol; GFP, green fluorescent protein; GPI, glycosylphosphatidylinositol; CFW, Calcofluor White; ECD, extracellular domain; PKC, protein kinase C; HB, hygromycin B; CEN, centromere.
Z. Li and J. S. Fassler, unpublished data.
J. S. Fassler, O. Carmel-Harel, G. Storz, and A. Gasch, unpublished data.
S. S. Narang and J. S. Fassler, unpublished observations.
REFERENCES
- 1.Tao W, Deschenes RJ, Fassler JS. J. Biol. Chem. 1999;274:360–367. doi: 10.1074/jbc.274.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiser V, Raitt DC, Saito H. J. Cell Biol. 2003;161:1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda T, Wurgler-Murphy SM, Saito H. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 4.Ostrander DB, Gorman JA. J. Bacteriol. 1999;181:2527–2534. doi: 10.1128/jb.181.8.2527-2534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeda T, Takekawa M, Saito H. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 6.Reiser V, Salah SM, Ammerer G. Nat. Cell Biol. 2000;2:620–627. doi: 10.1038/35023568. [DOI] [PubMed] [Google Scholar]
- 7.O'Rourke SM, Herskowitz I. Mol. Cell. Biol. 2002;22:4739–4749. doi: 10.1128/MCB.22.13.4739-4749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ota IM, Varshavsky A. Proc. Natl. Acad. Sci. U. S. A. 1992;89:2355–2359. doi: 10.1073/pnas.89.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fassler JS, Gray WM, Malone CL, Tao W, Lin H, Deschenes RJ. J. Biol. Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Dean S, Li Z, Horecka J, Deschenes RJ, Fassler JS. Mol. Biol. Cell. 2002;13:412–424. doi: 10.1091/mbc.01-09-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krems B, Charizanis C, Entian KD. Curr. Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- 12.Brown JL, North S, Bussey H. J. Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberts AS, Bouquin N, Johnston LH, Treisman R. J. Biol. Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- 14.Brown JL, Bussey H, Stewart RC. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketela T, Green R, Bussey H. J. Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luyten K, Albertyn J, Skibbe WF, Prior BA, Ramos J, Thevelein JM, Hohmann S. EMBO J. 1995;14:1360–1371. doi: 10.1002/j.1460-2075.1995.tb07122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutherland FCW, Lages F, Lucas C, Luyten K, Albertyn J, Hohmann S, Prior BA, Kilian SG. J. Bacteriol. 1997;179:7790–7795. doi: 10.1128/jb.179.24.7790-7795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamas MJ, Luyten K, Sutherland FC, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Kilian SG, Ramos J, Gustafsson L, Thevelein JM, Hohmann S. Mol. Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- 19.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 20.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao W, Malone CL, Ault AD, Deschenes RJ, Fassler JS. Mol. Microbiol. 2002;43:459–473. doi: 10.1046/j.1365-2958.2002.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. p. 354. [Google Scholar]
- 23.Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc.; New York: 1989. [Google Scholar]
- 24.Pelissier P, Camougrand N, Velours G, Guerin M. Curr. Genet. 1995;27:409–416. doi: 10.1007/BF00311209. [DOI] [PubMed] [Google Scholar]
- 25.Mouassite M, Camougrand N, Schwob E, Demaison G, Laclau M, Guerin M. Yeast. 2000;16:905–919. doi: 10.1002/1097-0061(200007)16:10<905::AID-YEA584>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Velours G, Boucheron C, Manon S, Camougrand N. FEMS Microbiol. Lett. 2002;207:165–172. doi: 10.1111/j.1574-6968.2002.tb11046.x. [DOI] [PubMed] [Google Scholar]
- 27.Davenport KR, Sohaskey M, Kamada Y, Levin DE, Gustin MC. J. Biol. Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 28.Kamada Y, Jung US, Piotrowski J, Levin DE. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 29.Merchan S, Bernal D, Serrano R, Yenush L. Eukaryot. Cell. 2004;3:100–107. doi: 10.1128/EC.3.1.100-107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yenush L, Mulet JM, Arino J, Serrano R. EMBO J. 2002;21:920–929. doi: 10.1093/emboj/21.5.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean N. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. J. Bacteriol. 1998;180:3381–3387. doi: 10.1128/jb.180.13.3381-3387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagen I, Ecker M, Lagorce A, Francois JM, Sestak S, Rachel R, Grossmann G, Hauser NC, Hoheisel JD, Tanner W, Strahl S. Mol. Microbiol. 2004;52:1413–1425. doi: 10.1111/j.1365-2958.2004.04064.x. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura K, Yashushi Y. Agri. Biol. Chem. 1981;45:1761–1766. [Google Scholar]
- 35.Mrsa V, Ecker M, Strahl-Bolsinger S, Nimtz M, Lehle L, Tanner W. J. Bacteriol. 1999;181:3076–3086. doi: 10.1128/jb.181.10.3076-3086.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klis FM, Mol P, Hellingwerf K, Brul S. FEMS Microbiol. Rev. 2002;26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 37.Kollar R, Reinhold BB, Petrakova E, Yeh HJ, Ashwell G, Drgonova J, Kapteyn JC, Klis FM, Cabib E. J. Biol. Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 38.Lipke PN, Ovalle R. J. Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapteyn JC, Ram AF, Groos EM, Kollar R, Montijn RC, Van Den Ende H, Llobell A, Cabib E, Klis FM. J. Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragni E, Sipiczki M, Strahl S. Yeast. 2007;24:309–319. doi: 10.1002/yea.1465. [DOI] [PubMed] [Google Scholar]
- 41.Hardwick KG, Boothroyd JC, Rudner AD, Pelham HR. EMBO J. 1992;11:4187–4195. doi: 10.1002/j.1460-2075.1992.tb05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jung US, Levin DE. Mol. Microbiol. 1999;34:1049–1057. doi: 10.1046/j.1365-2958.1999.01667.x. [DOI] [PubMed] [Google Scholar]
- 43.Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM. J. Biol. Chem. 2003;278:34998–35015. doi: 10.1074/jbc.M306291200. [DOI] [PubMed] [Google Scholar]
- 44.Lagorce A, Hauser NC, Labourdette D, Rodriguez C, Martin-Yken H, Arroyo J, Hoheisel JD, Francois J. J. Biol. Chem. 2003;278:20345–20357. doi: 10.1074/jbc.M211604200. [DOI] [PubMed] [Google Scholar]
- 45.Lu JM, Deschenes RJ, Fassler JS. Eukaryot. Cell. 2003;2:1304–1314. doi: 10.1128/EC.2.6.1304-1314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]