Abstract
Sepsis is defined as the systemic inflammatory response of the human host that is triggered by an invading pathogen. Despite tremendous advances in both our knowledge of and treatment strategies for this syndrome, sepsis remains among the major causes of morbidity and mortality in children worldwide. Thus, we hypothesize that an improved mechanistic understanding obtained via basic and translational science will continue to identify novel therapeutic targets and approaches. As a result, given the central importance of the alterations in gene expression in regulating the human host’s physiologic response to a pathogen, we review those complex factors: genetics, transcriptional expression and epigenetics, which regulate unique gene expression patterns in pediatric sepsis and septic shock. We anticipate that emerging data from genetic, genomic and other translation studies in pediatric sepsis will advance our biologic understanding of this response and undoubtedly identify targets for newer therapies.
Platforms for studying gene expression response to sepsis
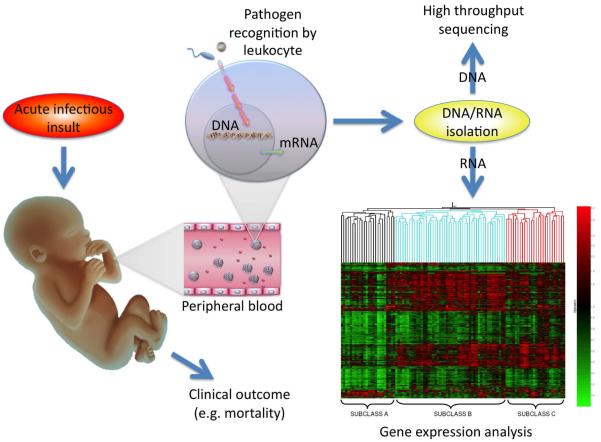
Sepsis reflects an extraordinarily complex response of the host to an invading pathogen. Historically, pre-clinical and clinical studies in sepsis have generally examined only one to a few genes and also typically focused analyses only on increased expression levels in affected hosts as compared to healthy cohorts. The development of high throughput sequencing capability has afforded the ability to determine genetic influences on disease phenotypes. Further, evolution of microarray technology provided an unprecedented opportunity to examine thousands of gene products simultaneously by measuring genome-wide mRNA expression—both increased and equally important, decreased levels--in clinical samples from affected individuals to be compared to expression levels in unaffected individuals. As a result, investigators have leveraged this approach to more comprehensively understand the pathobiology of pediatric septic shock as a means of both discovery and novel hypothesis generation. The overall approach to conducting array based studies is outlined in Figure 1 using RNA-based arrays as an example. While the technical details of this approach are beyond the scope of this article, in general, microarray technology utilizes a solid support (often glass or silicone) “chip” upon which large numbers (up to thousands) of nucleic acid sequences are immobilized at known locations 1. These sequences can represent any number of targets of inquiry, including specific gene sequences, so-called expressed sequence tags which represent genes of unknown function, or sequences representing gene polymorphisms (reviewed below). Robotic technology can apply the sequences to the solid support by a process termed “spotting,” or an array of shorter, oligonucleotide probes (usually 20 to 80 nucleotide base pairs long) can be synthesized directly on the solid support to create a “chip” containing a high density array of sequences that can be used for large-scale analyses of the targeted genome 2. Together these technologies are affording pediatric investigators the opportunity to examine genetic influences on outcomes from sepsis, as well as gene expression profiles that may provide novel mechanistic insight into functional pathways involved in the pathobiology of sepsis.
Figure 1.
Following exposure to an acute infectious insult, the pediatric patient mounts an immune response to defend against microbial invasion. Pathogen recognition receptors on local and peripheral leukocytes are activated by the presence of microbial products. Leukocyte activation results in transcription of genomic DNA to messenger RNA that leads to protein production (e.g. interleukins, acute phase reactants). Isolation of nucleic acids permits high throughput sequencing of DNA and expression profiling of mRNA via array-based analyses. The presence of altered DNA sequences (e.g. polymorphisms) and patterns of mRNA production can then be correlated with a clinical outcome such as mortality to gain insight into genetic influences on and involved pathways in this complex pathobiology.
Genetic Factors Influencing the Host Response
Susceptibility to sepsis and the clinical course of patients with sepsis are both highly heterogeneous, raising the strong possibility that the host response to infection is, at least in part, influenced by heritable factors (i.e. genetics) 3. A landmark study by Sorensen et al, published more than 20 years ago, provides strong evidence linking genetics and susceptibility to infection 4. A longitudinal cohort of more than 900 adopted children born between 1924 and 1926 and both their biologic and adoptive parents were followed through 1982. If a biologic parent died of infection before the age of 50 years, the relative risk of death from infectious causes in the child was 5.8 (95% C.I. 2.5 to 13.7), which was higher than for all other causes studied. In contrast, the death of an adoptive parent from infectious causes did not confer a greater relative risk of death in the adopted child leading these investigators to hypothesize that there exists a strong link between genetics and susceptibility to infection.
As a result, recent investigations have attempted to identify a link between genetics and predisposition to sepsis by largely focusing on genome-wide association studies (GWAS) and gene polymorphisms. A gene polymorphism is defined as the regular occurrence (>1%), in a population, of two or more alleles at a particular chromosome location. The most frequent type of polymorphism is called a single nucleotide polymorphism (SNP): a substitution, deletion, or insertion of a single nucleotide that occurs in approximately 1 per every 1,000 base pairs of human DNA. SNPs can result in an absolute deficiency in protein, an altered protein, a change in the level of normal protein expression, or no discernible change in protein function or expression. As these studies have proceeded, a number of SNPs in several genes responsible for regulating the inflammatory, coagulation, and other key immune responses to sepsis have been identified to be associated with variable outcomes in the setting of infections (Reviewed: 5-9). Here, we highlight some key examples of this genetic regulation of the host responses in sepsis.
The signaling mechanisms involved in pathogen recognition, the immune response, and inflammation have been reviewed extensively 10-15 and SNPs have been identified in many of the genes involved in these signaling mechanisms. For example, mutations in TLR-4, a key pathogen recognition receptor, have been shown to increase susceptibility to infections secondary to gram-negative organisms 16. While several SNPs in the TLR-4 receptor gene have been described, few have been found to be associated with an increased risk of septic shock or septic shock-related mortality in children. For example, an adenine for guanine substitution 896 base pairs downstream of the transcription start site for TLR-4 (+896) results in replacement of aspartic acid with glycine at amino acid 299 (Asp299Gly). The Asp299Gly polymorphism has been associated with reduced expression and function of the TLR-4 receptor in vitro 16, 17. Furthermore, adults who carry the Asp299Gly polymorphism appear to be at increased risk for septic shock and poor outcome in several cohort studies 9, 18, 19. While children who carry the Asp299Gly polymorphism appear to be at increased risk of urinary tract infection, this SNP does not appear to influence either the susceptibility or severity of meningococcal septic shock in children 20, 21. These results were further corroborated in a cohort study involving over 500 Gambian children 22.
SNPs related to other members of this LPS-receptor complex (e.g. CD14, MD-2, and MyD88) have been studied in adult populations, but no such studies have yet to be performed in children 19, 23-26. SNPs in other classes of Toll-like receptors have also been studied. For example, gene polymorphisms of TLR-2, the primary pattern recognition receptor for gram-positive bacteria, have been associated with increased risk of infection in both children and adults 27-29.
Several SNPs affecting cytokine expression have been described, though gene association studies in critically ill adults with septic shock have been conflicting 7, 8, 30. For example, two allelic variants of the TNF-α gene have been described: the wild-type allele TNF1 (guanine at −308A), and TNF2 (adenosine at −308A). The TNF2 allele has been associated with higher expression of TNF-α and increased susceptibility to septic shock and mortality in at least one study involving critically ill adults 31. Nadel and colleagues found an increased risk of death in critically ill children with meningococcal septic shock who carried this TNF2 allelic variant 32. Several additional SNPs in TNF-β, IL-1, IL-6, IL-8, and IL-10 have also been shown to influence susceptibility and severity of septic shock in children 33-39. Thus, there are genetically-based influences on the expression of many cytokines established to play key roles in the response to sepsis which may profoundly influence clinical outcomes in this setting.
Dysregulation of the coagulation cascade also plays an important role in the pathophysiology of septic shock and several studies have examined polymorphisms of key genes involved in coagulation. The 4G allele of a deletion/insertion (4G/5G) SNP in the promoter region of the plasminogen-activator inhibitor type-1 (PAI-1) gene has been associated with higher plasma concentrations of PAI-1. The 4G allele increases susceptibility to and severity of septic shock, as well as increasing the risk of mortality in children with meningococcal septic shock 40-43. In addition a SNP in the protein C promoter has been associated with susceptibility to meningococcemia and illness severity in children 44.
SNPs in genes involved in phagocytosis and the complement cascade have also been studied in the context of septic shock. SNPs that affect function have been described in virtually all family members of the Fcγ receptor, and several of these SNPs have been associated with susceptibility to meningococcal sepsis, severity of meningococcemia, and poor outcome from meningococcal septic shock 45-51. Finally, an SNP of the bactericidal permeability increasing protein (BPI) gene has also been associated with increased the mortality from septic shock in children 52.
Together, these specific examples demonstrate how variations in an individual’s genetic sequences can profoundly influence the expression of the encoded gene. This genetic influence on expression may have causal effects on the outcomes of individuals infected by a given pathogen. As more studies aim to prove such links between SNPs, gene expression and susceptibility and/or outcome of pediatric septic shock, the studies need to be carefully considered and evaluated. With respect to evaluating the validity of these studies, the ideal genome-wide association study requires several important qualities that have been emphasized in the genetics literature 53, 54. Important factors to evaluate include: matching by ethnic-geographic origin and other potentially confounding variables, validation by replicate studies, analysis limited to the primary hypothesis and avoidance of post-hoc analyses and multiple comparisons, reporting of associations as odds ratios with confidence limits all complemented by a commitment to publish both positive as well as sound, negative genetic association studies in order to avoid publication bias. By following these and other established principles, teams of investigators executing replicate studies should succeed in identifying important genetic influences on gene expression that ultimately alters the outcome—favorably as well an unfavorably—in the setting of sepsis.
Genome-wide Expression Profiling in Children with Septic Shock
The first publication involving genome-wide expression patterns in pediatric septic shock consisted of 42 children with septic shock that were compared to 15 normal controls 55. RNA samples were derived from whole blood samples obtained within 24 hours of admission to the pediatric intensive care unit with septic shock. As would be predicted, this comparison between two biological extremes (i.e. normal vs. septic shock), demonstrated a large number of genes (>2,000) that were differentially expressed / repressed in patients with septic shock, relative to normal controls. Also as would be expected, the upregulated genes in the patients with septic shock corresponded significantly with functional annotations related to immunity and inflammation.
The most novel finding of this initial genome-wide expression profiling study surrounded the genes that were significantly repressed in the patients with septic shock, relative to controls (>1,000 genes). Surprisingly, a large number of these repressed genes corresponded to functional annotations related to zinc biology. Thus, this initial approximation of the genomic response of pediatric septic shock was characterized by repression of a large number of genes that either directly participate in zinc homeostasis, or directly depend on zinc homeostasis for normal functioning. Consistent with this observation, the patients with septic shock that did not survive had significantly lower serum zinc levels compared to the patients who survived. These data have generated the hypothesis that altered zinc homeostasis may play a role in the pathobiology of septic shock, and is consistent with the known links between zinc and immune function 56. Recently, Knoell and colleagues independently corroborated this hypothesis by demonstrating that zinc depletion led to increased death in a murine model of sepsis and that zinc supplementation partially reversed this phenotype 57.
The second microarray-based study in pediatric septic shock focused on longitudinal expression profiles 58 from RNA samples on 30 children with septic shock obtained within 24 hours of admission to the PICU (“day 1”) and 48 hours later (“day 3”) compared to those from 15 normal controls. The upregulated genes in the patients with septic shock again corresponded to multiple signaling pathways and gene networks associated with inflammation and immunity. One of the most notable signaling pathways represented by the upregulated gene lists was the canonical anti-inflammatory cytokine, IL-10 with a large number of genes being persistently upregulated in children with septic shock on both days 1 and 3.
The downregulated gene expression patterns in this longitudinal study again demonstrated large scale repression of genes corresponding to zinc-related biology. This pattern of repression persisted, to a similar degree, from day 1 to day 3. Another notable gene repression pattern involved T-cell receptor signaling and the antigen presentation pathway. Again, this pattern of gene repression was evident on day 1, and persisted well into day 3. In summary, this study demonstrated persistent repression of genes corresponding to the adaptive immune system. This observation suggests that the pathobiology of pediatric septic shock may represent a failure of the adaptive immune system and is well in line with recent experimental and clinical data 59-67.
The translational approaches reviewed above continue to provide unique opportunities for advancing the diagnostics, prognostics and therapies in pediatric and neonatal septic shock. To that end, the third microarray-based study in pediatric septic shock focused on validation of previous observations by way of formal class prediction modeling and by the application of alternative filtering and statistical approaches 68. This study made use of the original data as the “training data set” and a new cohort of patients with septic shock enrolled through on-going accrual as the “test” or “validation” data set. Using two distinct class prediction algorithms, the gene list derived from the training data set was able to identify the separate cohort of patients in the validation data set with 100% accuracy. In addition, the application of alternative gene filtering and statistical approaches to the validation cohort of patients yielded similar observations to that derived from the original data. Namely, the validation data cohort of children with septic shock was also characterized by large scale repression of genes corresponding to zinc-related biology and adaptive immunity. These data strengthen the validity of previous observations regarding the genome-level response of pediatric septic shock.
Genome-wide expression studies in adults with sepsis and septic shock have also demonstrated wide spread and early repression of adaptive immunity-related genes, but the observations regarding repression of zinc biology-related gene programs have not been reported in these adult-centered studies 69-72. While it is tempting to conclude that repression of zinc biology-related gene programs is a unique feature of pediatric septic shock, this assertion needs to be directly tested by conducting direct comparisons of adult and pediatric expression data based on similar microarray platforms, RNA sources, and bioinformatic approaches.
The most recent microarray-based studies in pediatric septic shock have begun to refine our understanding of two distinct questions: 1) are the reported gene expression profiles discussed above specific to patients with septic shock, or are they more generic manifestations of critical illness?; and 2) can gene expression profiling be leveraged to identify subsets or sub-classifications of children with septic shock?
The question of specificity was addressed by comparing the gene expression profiles between children with the systemic inflammatory response syndrome (SIRS) versus sepsis versus septic shock 73. These comparisons identified patterns of conserved gene expression across the pediatric SIRS, sepsis, and septic shock spectrum, particularly with regard to upregulation of genes corresponding to innate immunity. There were also several notable gene expression patterns that were unique and persistent in the patients with septic shock, relative to the patients with SIRS or sepsis. Specifically, patients with septic shock had the most prominent and persistent upregulation of genes within the IL-10 signaling pathway. In addition, repression of zinc biology- and adaptive immunity-related genes was most prominent and persistent in the patients with septic shock. These data indicate that the previous observations surrounding zinc biology and adaptive immunity are relatively specific to pediatric septic shock, rather than being generic manifestations of critical illness.
The more challenging question of sub-classification and prognostication was addressed by using microarray data from 98 children with septic shock 74. Through a discovery-oriented filtering approach and unsupervised hierarchical gene clustering, 3 putative subclasses of children with septic shock were identified based solely on their gene expression profiles. One subgroup was characterized by significantly greater repression of genes corresponding to zinc biology and adaptive immunity. Notably, patients in this subgroup were significantly younger, more severely ill, and had a significantly higher mortality rate compared to the other 2 subgroups. These data suggested that clinically relevant, molecular sub-classification may be possible through gene expression profiling.
Contribution of Epigenetics to the Regulation of the Host Response in Sepsis
As reviewed in the prior section, the successful application of a genomic approach in pediatric sepsis has provided novel insight as to the gene expression patterns that characterize the complex biological human response to a pathogen. In the process of interrogating the entire genome through microarray and complex bioinformatic approaches, we have shown pediatric septic shock is characterized by early repression of a large number of genes—notably those related to T-cell receptor signaling and antigen presentation 55, 58, and significant upregulation of other sets of genes—notably components of the IL-10 signaling pathway. As a result, it is now incumbent on investigators to further understand the molecular mechanisms that either drive or repress the expression of these genes. Clearly, receptor activation, signal transduction pathways, and transcriptional activators, all affected by genetic polymorphisms will influence this expression. Importantly, more recent investigations—notably in developmental and cancer biology--have elucidated an additional mechanism that is capable of regulating the activation or inactivation of gene expression, termed epigenetics. Scientists have grappled with a specific definition for “epigenetics” since it incorporates a number of mechanisms. Perhaps the most accepted current operational definition of epigenetics is “a stably inherited phenotype resulting from changes in a chromosome without alterations in the DNA sequence” that was proposed by Berger and colleagues 75. At the most basic level, epigenetics refers to mechanisms by which expression of a gene encoded by a specific DNA sequence can be altered by changes on the chromatin induced by a variety of signals such as environmental exposures including infections.
This “epigenetic regulation” of gene expression is dependent upon modifications in the manner by which specific genes are packaged in chromatin and how the chromatin may subsequently be remodeled by post-translational modifications of the amino acids that make up histone proteins 76. This post-translational modification of the chromatin structure can give rise to heritable transcription state known as an “epigenetic” alteration, which in some cases appears capable of conferring long-term stabilization of a particular gene expression pattern 77. These epigenetic regulatory mechanisms can be related to post-translational modifications of histones with subsequent effects on transcriptional silencing/repression or activation 78. Though not specifically covered in this review, another epigenetic mechanism influencing gene expression is direct methlyation of DNA which can alter the binding of transcriptional activating factors. Finally, many investigators include the expression of microRNA, small segments of RNA that can bind to DNA to alter transcriptional activation of genes as an additional epigenetic mechanism.
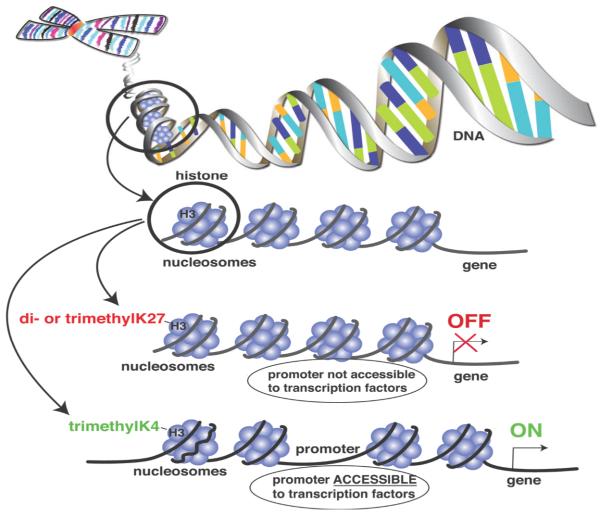
At the foundation of the maintenance of a particular gene expression “phenotype” that is potentially maintained over a long period of time is our understanding that DNA is incompletely stripped of its nucleosomes after replication. This mechanism allows post-translational modification of histones to serve as replicative templates to initiate identical modifications of new histones and results in the transfer of modified histones to daughter cells during replication to transfer the new “histone code”. This process is shown in Figure 2, in which nucleosomes are composed of DNA associated with 8 histones that can be targeted for a variety of post-translational modifications (PTMs). Post-translational modifications of histones include acetylation, methylation, and phosphorylation which are regulated by complex enzymatic machinery. Examples of this epigenetic regulation relevant to gene expression in sepsis include the finding that di- or trimethylation of lysine 27 (K27) of histone 3 (H3) keeps the chromatin in a conformation such that the promoter for a specific gene is not available to transcription factors and thus the gene is silenced (a.k.a. “gene-off”) 79. In contrast, methylation of lysine 4 on histone 3 is known to “open up” the conformation of the chromatin and allow transcription factors access to the promoter and initiation of gene expression (a.k.a. “gene-on”) 80. These epigenetic alterations are important as they can “stamp history” into the gene expression repertoire of cells in a heritable manner.
Figure 2.
Cartoon depiction of the relationship between DNA (chromatin) and interacting nucleosomes containing histone proteins. Histones can undergo post-translational modifications (e.g. methylation patterns) that alter the ability of transcriptional activation factors to access promoter regions. Depending on the pattern, these “methylation signatures” can result in either increased (e.g. me2H3K27) or repressed (e.g. me3H3K4) gene expression.
As reviewed above, IL-10 is a canonical signaling molecule for adaptive immunity that represses Th1 cytokine production, antigen presentation, and co-stimulatory molecules. Recent data have demonstrated that IL-10 is epigenetically regulated in the context of polarized Th1 versus Th2 helper cells 81. Specifically, association of the IL-10 promoter with “gene-on” histone modifications (H3K4me3 and AcH4) correlates with increased IL-10 production in Th2 cells, whereas association of the IL-10 promoter with the “gene-off” histone modification (histone deacetylase 1) correlates with decreased IL-10 production in Th1 cells.
IL-12 is also a canonical signaling molecule for the adaptive immune system that induces Th1 cytokine production and differentiation. Kunkel et al., recently demonstrated that dendritic cells of mice who survive an model of experimental sepsis possess a blunted adaptive immune response and acquire a phenotype consisting of decreased IL-12 production and increased IL-10 production 82. They further showed that the epigenetic signature on the promoters for the p35 and p40 subunits of IL-12 are consistent with an increased “gene-off” signal and a concomitant decreased “gene-on” signal 83, 84. Preliminary data as yet unpublished suggests that similar epigenetic regulation of IL-12 repression and IL-10 expression occurs in the circulating monocytes of patients with septic shock. Thus, it is very likely that substantial changes in epigenetic signatures occur across a broad spectrum of genes. These resulting signatures are likely to be pathogen-dependent, tissue specific and importantly, influenced by age and have long term effects on subsequent gene expression patterns. Therefore, it will be imperative that neonatal and pediatric clinician-scientists examine these changes in their cohort of patients with sepsis to fully understand how this regulation of gene expression influences both long and short-term clinical outcomes.
Utilizing Expression Data to Drive Stratification of Pediatric Septic Shock
While a certain gene expression pattern appears to predict a subclass of patients with worse outcomes from septic shock, it is also clear that substantial heterogeneity in gene expression occurs across a broad spectrum of clinical presentations fulfilling septic shock criteria. We hypothesize that the failure of the vast majority of interventional clinical trials in septic shock is not related to a fundamental flaw of the biological or physiological principle being tested, but rather, lies in the inability to effectively address the substantial heterogeneity that characterizes this syndrome. Septic shock is a heterogeneous syndrome with the potential to negatively and directly affect all organ systems. This heterogeneity consistently challenges investigators attempting to evaluate the efficacy of various experimental interventions. As astutely stated by Marshall, a key challenge in the field is to reduce and manage this heterogeneity by more effectively stratifying patients for the purposes of more rational and effective clinical research and clinical management 85. The concept of pre-intervention stratification in sepsis, and its positive impact on the efficacy of an experimental therapy, was very recently corroborated by Remick et al using a murine model of polymicrobial sepsis 86. In their study, mice subjected to cecal ligation and puncture were stratified to likelihood of dying based on serum IL-6 levels six hours after CLP and those with the highest predicted mortality were provided immune modulating therapy which had a significant impact on survival 86.
These pre-clinical studies support the concept of trying to stratify children with septic shock based upon early identification of septic shock sub-classes using genome-wide expression patterns. As described above, sub-classes of children with septic shock have been identified based exclusively on gene expression profiling conducted within 24 hours of admission and these sub-classes have highly relevant differences in illness severity and mortality 74. A similar strategy was recently demonstrated in adult patients suffering from trauma 87. As high throughput technologies evolve and validation studies are rigorously performed, the ability to conduct “real time” expression-based, sub-classification and stratification could very well become a clinical reality.
A further potential strategy for stratifying children with septic shock (and perhaps more technologically feasible in real-time at present) is to base such an approach on the ultimate proteins expressed as a result of the complex genetic and transcriptional machinery reviewed above. These proteins usually referred to as “biomarkers” can be readily measured in the blood compartment, thus providing a clinically feasible strategy for early stratification of patients. For example, interleukin-8 (IL-8) can be readily and rapidly measured in small volume blood samples. Recently, IL-8 was found to be a robust outcome biomarker in children with septic shock 88. Specifically, an IL-8 level, measured within 24 hours of admission to the PICU, was found to have a 95% negative predictive value for mortality in a derivation cohort of patients. Thus, this particular IL-8 level was able to predict survival in pediatric septic shock, with standard care, with 95% probability. The reliability of this assertion was supported by prospective, formal validation in an independent cohort which demonstrated an identically robust negative predictive value 88. A similar observation (98% negative predictive value for mortality) was found by measuring chemokine ligand 4 (CCL4) serum levels upon admission to the PICU with septic shock 89.
It has been proposed that these types of sepsis outcome biomarkers (i.e. biomarkers having high negative predictive values for mortality) could be used to stratify patients eligible for interventional septic shock trials 88, 89. Patients having a high likelihood of survival but otherwise meeting entry criteria for a given interventional trial could be excluded from the trial based on these biomarkers. Such a stratification strategy would serve to derive a study population with a more optimal risk-to-benefit ratio, thus improving the ability to demonstrate efficacy for a given test agent. This type of strategy would be particularly useful for a test agent carrying more than minimal risk.
While single biomarker-based patient stratification is clinically appealing, it may not be sufficiently robust to meet all clinical and research needs. Indeed, the aforementioned studies involving IL-8 and CCL4 had clinically unacceptable specificities, sensitivities, and positive predictive values, relative to the very high negative predictive values. In order to serve a wide range of clinical and research needs, the ideal biomarker-based stratification tool would simultaneously have high specificity, high sensitivity, high positive predictive value, and high negative predictive value.
Given the biological complexity of pediatric septic shock, a stratification strategy based on a panel of multiple biomarkers has more potential to meet the needs of an ideal biomarker-based stratification tool. We have recently launched a multi-institutional study to derive and validate a multi-biomarker-based stratification tool for pediatric septic shock. The foundation of this study is a panel of 15 biomarkers (Table 1) derived from a genome-wide expression database that identified differences in outcomes among nearly 100 children with septic shock (unpublished data). The database focused on the gene expression patterns representing the first 24 hours of admission to the PICU, which is an ideal time-frame for clinically useful stratification. An initial working list of candidate biomarkers was systematically and objectively derived using two complementary statistical tools to determine genes differentially regulated between survivors and non-survivors, and then class prediction modeling targeted at identification of “survivor” and “non-survivor” classes. The final panel of 15 biomarkers was refined from the initial working list based on biological plausibility in the context of sepsis, and the ability to readily measure the biomarkers (proteins) in blood samples. The resulting list will be used to derive the pediatric sepsis biomarker risk model (PERSEVERE: PEdiatRic SEpsis biomarkEr Risk modEl) that is intended to predict outcome and illness severity for patients with septic shock. This recently launched, multi-institutional study, PERSEVERE provides the unprecedented opportunity to test the hypothesis that biomarker determination, informed by prior gene expression studies, can provide a real-time decision and stratification tool that will impact the care of children with septic shock.
Table 1.
Proposed gene list of biomarkers for sepsis/septic shock derived from array-based determination of gene expression patterns
| Genbank # | Official gene symbol | Description / Gene Name |
|---|---|---|
| NM_002983 | CCL3 | chemokine (C-C motif) ligand 3, MIP1alpha |
| NM_005564 | LCN2 | lipocalin 2 (oncogene 24p3), NGAL |
| NM_002424 | MMP8 | matrix metallopeptidase 8 (neutrophil collagenase) |
| NM_020415 | RETN | resistin |
| AV726673 | THBS1 | thrombospondin 1 |
| J03189 | GZMB | granzyme B (cytotoxic T-lymphocyte-associated serine esterase 1) |
| NM_005346 | HSPA1B | heat shock 70kDa protein 1B |
| NM_000607 | ORM1 | orosomucoid 1 /// orosomucoid 2 |
| NM_002343 | LTF | lactotransferrin /// similar to lactotransferrin |
| NM_001972 | ELA2 | elastase 2, neutrophil |
| NM_000575 | IL1A | interleukin 1, alpha |
| AL133001 | SULF2 | sulfatase 2 |
| NM_006682 | FGL2 | fibrinogen-like 2 |
| NM_000584 | IL8 | interleukin 8 |
| NM_002984 | CCL4 | chemokine (C-C motif) ligand 4, MIP1beta |
Conclusion
Clinicians and investigators have enabled improved observational and interventional studies by defining consensus definitions that help identify the pediatric cohort affected by sepsis. Through these observations, it is clear that the neonate’s and child’s host response to an invading pathogen is an extraordinarily complex biologic process. This process will be influenced by both genetic as well as epigenetic mechanisms that ultimately impact on the gene expression patterns observed among affected hosts. It is hoped that understanding how these patterns vary by outcome inform clinical-investigators identify complementary stratification strategies (e.g. real-time biomarker determination) as well as novel pathways for therapeutic targeting. Only through successful collaboration among multiple centers will mechanistic, diagnostic and therapeutic studies be executed thereby affording our ability to substantially impact the outcomes of children affected by this complex clinical entity.
Acknowledgments
Grant support: Supported by K12HD047349 to TTC; R01GM064619_and 1RC1HL100474-01 to HRW; RO1HL097361, RO1GM066839 and UL1RR024986 to TPS.
Abbreviations
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association studies
- TLR
Toll like receptor
- SIRS
systemic inflammatory response syndrome
- PTM
post-translational modification
References
- 1.Wong HR, Shanley TP. Genetic Basis of Acute Lung Injury. Saunders; New York, NY: 2007. [Google Scholar]
- 2.Heller MJ. DNA microarray technology: devices, systems, and applications. Annu Rev Biomed Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 3.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2(12):967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318(12):727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 5.Arcaroli J, Fessler MB, Abraham E. Genetic polymorphisms and sepsis. Shock. 2005;24(4):300–312. doi: 10.1097/01.shk.0000180621.52058.e1. [DOI] [PubMed] [Google Scholar]
- 6.Waterer GW, Wunderink RG. Science review: Genetic variability in the systemic inflammatory response. Crit Care. 2003;7(4):308–314. doi: 10.1186/cc2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes CL, Russell JA, Walley KR. Genetic polymorphisms in sepsis and septic shock: role in prognosis and potential for therapy. Chest. 2003;124(3):1103–1115. doi: 10.1378/chest.124.3.1103. [DOI] [PubMed] [Google Scholar]
- 8.Stuber F, Klaschik S, Lehmann LE, Schewe JC, Weber S, Book M. Cytokine promoter polymorphisms in severe sepsis. Clin Infect Dis. 2005;41(Suppl 7):S416–420. doi: 10.1086/431991. [DOI] [PubMed] [Google Scholar]
- 9.Barber RC, Chang LY, Arnoldo BD, Purdue GF, Hunt JL, Horton JW, et al. Innate immunity SNPs are associated with risk for severe sepsis after burn injury. Clin Med Res. 2006;4(4):250–255. doi: 10.3121/cmr.4.4.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bone RC. Sepsis and coagulation. An important link. Chest. 1992;101(3):594–596. doi: 10.1378/chest.101.3.594. [DOI] [PubMed] [Google Scholar]
- 11.Deitch EA. Animal models of sepsis and shock: A review and lessons learned. Shock. 1998;9:1–11. doi: 10.1097/00024382-199801000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Flierl MA, Schreiber H, Huber-Lang MS. The role of complement, C5a and its receptors in sepsis and multiorgan dysfunction syndrome. J Invest Surg. 2006;19(4):255–265. doi: 10.1080/08941930600778263. [DOI] [PubMed] [Google Scholar]
- 13.Funk DJ, Parrillo JE, Kumar A. Sepsis and septic shock: a history. Crit Care Clin. 2009;25(1):83–101. viii. doi: 10.1016/j.ccc.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Oberholzer A, Oberholzer C, Moldawer LL. Sepsis syndromes: Understanding the role of innate and acquired immunity. Shock. 2001;16:83–96. doi: 10.1097/00024382-200116020-00001. [DOI] [PubMed] [Google Scholar]
- 15.Weighardt H, Holzmann B. Role of Toll-like receptor responses for sepsis pathogenesis. Immunobiology. 2007;212(9-10):715–722. doi: 10.1016/j.imbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25(2):187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz DA. The role of TLR4 in endotoxin responsiveness in humans. J Endotoxin Res. 2001;7(5):389–393. [PubMed] [Google Scholar]
- 18.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med. 2002;162(9):1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 19.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, et al. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186(10):1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 20.Karoly E, Fekete A, Banki NF, Szebeni B, Vannay A, Szabo AJ, et al. Heat shock protein 72 (HSPA1B) gene polymorphism and Toll-like receptor (TLR) 4 mutation are associated with increased risk of urinary tract infection in children. Pediatr Res. 2007;61(3):371–374. doi: 10.1203/pdr.0b013e318030d1f4. [DOI] [PubMed] [Google Scholar]
- 21.Read RC, Pullin J, Gregory S, Borrow R, Kaczmarski EB, di Giovine FS, et al. A functional polymorphism of toll-like receptor 4 is not associated with likelihood or severity of meningococcal disease. J Infect Dis. 2001;184(5):640–642. doi: 10.1086/322798. [DOI] [PubMed] [Google Scholar]
- 22.Allen A, Obaro S, Bojang K, Awomoyi AA, Greenwood BM, Whittle H, et al. Variation in Toll-like receptor 4 and susceptibility to group A meningococcal meningitis in Gambian children. Pediatr Infect Dis J. 2003;22(11):1018–1019. doi: 10.1097/01.inf.0000095431.15606.68. [DOI] [PubMed] [Google Scholar]
- 23.Hubacek JA, Stuber F, Frohlich D, Book M, Wetegrove S, Rothe G, et al. The common functional C(−159)T polymorphism within the promoter region of the lipopolysaccharide receptor CD14 is not associated with sepsis development or mortality. Genes Immun. 2000;1(6):405–407. doi: 10.1038/sj.gene.6363691. [DOI] [PubMed] [Google Scholar]
- 24.Gibot S, Cariou A, Drouet L, Rossignol M, Ripoll L. Association between a genomic polymorphism within the CD14 locus and septic shock susceptibility and mortality rate. Crit Care Med. 2002;30(5):969–973. doi: 10.1097/00003246-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Heesen M, Bloemeke B, Schade U, Obertacke U, Majetschak M. The −260 C-->T promoter polymorphism of the lipopolysaccharide receptor CD14 and severe sepsis in trauma patients. Intensive Care Med. 2002;28(8):1161–1163. doi: 10.1007/s00134-002-1389-0. [DOI] [PubMed] [Google Scholar]
- 26.Gu W, Shan YA, Zhou J, Jiang DP, Zhang L, Du DY, et al. Functional significance of gene polymorphisms in the promoter of myeloid differentiation-2. Ann Surg. 2007;246(1):151–158. doi: 10.1097/01.sla.0000262788.67171.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutukculer N, Yeniay BS, Aksu G, Berdeli A. Arg753Gln polymorphism of the human toll-like receptor-2 gene in children with recurrent febrile infections. Biochem Genet. 2007;45(7-8):507–514. doi: 10.1007/s10528-007-9091-0. [DOI] [PubMed] [Google Scholar]
- 28.Tabel Y, Berdeli A, Mir S. Association of TLR2 gene Arg753Gln polymorphism with urinary tract infection in children. Int J Immunogenet. 2007;34(6):399–405. doi: 10.1111/j.1744-313X.2007.00709.x. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68(11):6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Deventer SJ. Cytokine and cytokine receptor polymorphisms in infectious disease. Intensive Care Med. 2000;26(Suppl 1):S98–102. doi: 10.1007/s001340051125. [DOI] [PubMed] [Google Scholar]
- 31.Mira JP, Cariou A, Grall F, Delclaux C, Losser MR, Heshmati F, et al. Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. Jama. 1999;282(6):561–568. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 32.Nadel S, Newport MJ, Booy R, Levin M. Variation in the tumor necrosis factor-alpha gene promoter region may be associated with death from meningococcal disease. J Infect Dis. 1996;174(4):878–880. doi: 10.1093/infdis/174.4.878. [DOI] [PubMed] [Google Scholar]
- 33.McArthur JA, Zhang Q, Quasney MW. Association between the A/A genotype at the lymphotoxin-alpha+250 site and increased mortality in children with positive blood cultures. Pediatr Crit Care Med. 2002;3(4):341–344. doi: 10.1097/00130478-200210000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Read RC, Cannings C, Naylor SC, Timms JM, Maheswaran R, Borrow R, et al. Variation within genes encoding interleukin-1 and the interleukin-1 receptor antagonist influence the severity of meningococcal disease. Ann Intern Med. 2003;138(7):534–541. doi: 10.7326/0003-4819-138-7-200304010-00009. [DOI] [PubMed] [Google Scholar]
- 35.Endler G, Marculescu R, Starkl P, Binder A, Geishofer G, Muller M, et al. Polymorphisms in the interleukin-1 gene cluster in children and young adults with systemic meningococcemia. Clin Chem. 2006;52(3):511–514. doi: 10.1373/clinchem.2005.058537. [DOI] [PubMed] [Google Scholar]
- 36.Lehrnbecher T, Bernig T, Hanisch M, Koehl U, Behl M, Reinhardt D, et al. Common genetic variants in the interleukin-6 and chitotriosidase genes are associated with the risk for serious infection in children undergoing therapy for acute myeloid leukemia. Leukemia. 2005;19(10):1745–1750. doi: 10.1038/sj.leu.2403922. [DOI] [PubMed] [Google Scholar]
- 37.Michalek J, Svetlikova P, Fedora M, Klimovic M, Klapacova L, Bartosova D, et al. Interleukin-6 gene variants and the risk of sepsis development in children. Hum Immunol. 2007;68(9):756–760. doi: 10.1016/j.humimm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Balding J, Healy CM, Livingstone WJ, White B, Mynett-Johnson L, Cafferkey M, et al. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes Immun. 2003;4(8):533–540. doi: 10.1038/sj.gene.6364020. [DOI] [PubMed] [Google Scholar]
- 39.Artifoni L, Negrisolo S, Montini G, Zucchetta P, Molinari PP, Cassar W, et al. Interleukin-8 and CXCR1 receptor functional polymorphisms and susceptibility to acute pyelonephritis. J Urol. 2007;177(3):1102–1106. doi: 10.1016/j.juro.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 40.Westendorp RG, Hottenga JJ, Slagboom PE. Variation in plasminogen-activator-inhibitor-1 gene and risk of meningococcal septic shock. Lancet. 1999;354(9178):561–563. doi: 10.1016/S0140-6736(98)09376-3. [DOI] [PubMed] [Google Scholar]
- 41.Haralambous E, Hibberd ML, Hermans PW, Ninis N, Nadel S, Levin M. Role of functional plasminogen-activator-inhibitor-1 4G/5G promoter polymorphism in susceptibility, severity, and outcome of meningococcal disease in Caucasian children. Crit Care Med. 2003;31(12):2788–2793. doi: 10.1097/01.CCM.0000100122.57249.5D. [DOI] [PubMed] [Google Scholar]
- 42.Geishofer G, Binder A, Muller M, Zohrer B, Resch B, Muller W, et al. 4 G/5G promoter polymorphism in the plasminogen-activator-inhibitor-1 gene in children with systemic meningococcaemia. Eur J Pediatr. 2005;164(8):486–490. doi: 10.1007/s00431-005-1673-4. [DOI] [PubMed] [Google Scholar]
- 43.Hermans PW, Hibberd ML, Booy R, Daramola O, Hazelzet JA, de Groot R, et al. Meningococcal Research Group 4 G/5G promoter polymorphism in the plasminogen-activator-inhibitor-1 gene and outcome of meningococcal disease. Lancet. 1999;354(9178):556–560. doi: 10.1016/s0140-6736(99)02220-5. [DOI] [PubMed] [Google Scholar]
- 44.Binder A, Endler G, Rieger S, Geishofer G, Resch B, Mannhalter C, et al. Protein C promoter polymorphisms associate with sepsis in children with systemic meningococcemia. Hum Genet. 2007;122(2):183–190. doi: 10.1007/s00439-007-0392-5. [DOI] [PubMed] [Google Scholar]
- 45.van Sorge NM, van der Pol WL, van de Winkel JG. FcgammaR polymorphisms: Implications for function, disease susceptibility and immunotherapy. Tissue Antigens. 2003;61(3):189–202. doi: 10.1034/j.1399-0039.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 46.Bredius RG, Derkx BH, Fijen CA, de Wit TP, de Haas M, Weening RS, et al. Fc gamma receptor IIa (CD32) polymorphism in fulminant meningococcal septic shock in children. J Infect Dis. 1994;170(4):848–853. doi: 10.1093/infdis/170.4.848. [DOI] [PubMed] [Google Scholar]
- 47.Domingo P, Muniz-Diaz E, Baraldes MA, Arilla M, Barquet N, Pericas R, et al. Associations between Fc gamma receptor IIA polymorphisms and the risk and prognosis of meningococcal disease. Am J Med. 2002;112(1):19–25. doi: 10.1016/s0002-9343(01)01047-6. [DOI] [PubMed] [Google Scholar]
- 48.Platonov AE, Kuijper EJ, Vershinina IV, Shipulin GA, Westerdaal N, Fijen CA, et al. Meningococcal disease and polymorphism of FcgammaRIIa (CD32) in late complement component-deficient individuals. Clin Exp Immunol. 1998;111(1):97–101. doi: 10.1046/j.1365-2249.1998.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drogari-Apiranthitou M, Fijen CA, Thiel S, Platonov A, Jensen L, Dankert J, et al. The effect of mannan-binding lectin on opsonophagocytosis of Neisseria meningitidis. Immunopharmacology. 1997;38(1-2):93–99. doi: 10.1016/s0162-3109(97)00081-7. [DOI] [PubMed] [Google Scholar]
- 50.van der Pol WL, Huizinga TW, Vidarsson G, van der Linden MW, Jansen MD, Keijsers V, et al. Relevance of Fcgamma receptor and interleukin-10 polymorphisms for meningococcal disease. J Infect Dis. 2001;184(12):1548–1555. doi: 10.1086/324662. [DOI] [PubMed] [Google Scholar]
- 51.Fijen CA, Bredius RG, Kuijper EJ. Polymorphism of IgG Fc receptors in meningococcal disease. Ann Intern Med. 1993;119(7 Pt 1):636. doi: 10.7326/0003-4819-119-7_part_1-199310010-00026. [DOI] [PubMed] [Google Scholar]
- 52.Michalek J, Svetlikova P, Fedora M, Klimovic M, Klapacova L, Bartosova D, et al. Bactericidal permeability increasing protein gene variants in children with sepsis. Intensive Care Med. 2007;33(12):2158–2164. doi: 10.1007/s00134-007-0860-3. [DOI] [PubMed] [Google Scholar]
- 53.Stalets E, Wong HR. Critically associating. Crit Care Med. 2009;37(4):1492–1493. doi: 10.1097/CCM.0b013e31819d2be0. [DOI] [PubMed] [Google Scholar]
- 54.Cooper DN, Nussbaum RL, Krawczak M. Proposed guidelines for papers describing DNA polymorphism-disease associations. Hum Genet. 2002;110(3):207–208. doi: 10.1007/s00439-001-0672-4. [DOI] [PubMed] [Google Scholar]
- 55.Wong HR, Shanley TP, Sakthivel B, Cvijanovich N, Lin R, Allen GL, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30(2):146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rink L, Haase H. Zinc homeostasis and immunity. Trends Immunol. 2007;28(1):1–4. doi: 10.1016/j.it.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 57.Knoell DL, Julian MW, Bao S, Besecker B, Macre JE, Leikauf GD, et al. Zinc deficiency increases organ damage and mortality in a murine model of polymicrobial sepsis. Crit Care Med. 2009;37(4):1380–1388. doi: 10.1097/CCM.0b013e31819cefe4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shanley TP, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Doctor A, et al. Genome-Level Longitudinal Expression of Signaling Pathways and Gene Networks in Pediatric Septic Shock. Mol Med. 2007;13(9-10):495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayala A, Chaudry IH. Immune dysfunction in murine polymicrobial sepsis: mediators, macrophages, lymphocytes and apoptosis. Shock. 1996;6(Suppl 1):S27–38. [PubMed] [Google Scholar]
- 60.Felmet KA, Hall MW, Clark RS, Jaffe R, Carcillo JA. Prolonged lymphopenia, lymphoid depletion, and hypoprolactinemia in children with nosocomial sepsis and multiple organ failure. J Immunol. 2005;174(6):3765–3772. doi: 10.4049/jimmunol.174.6.3765. [DOI] [PubMed] [Google Scholar]
- 61.Hotchkiss RS, Swanson PE, Knudson CM, Chang KC, Cobb JP, Osborne DF, et al. Overexpression of Bcl-2 in transgenic mice decreases apoptosis and improves survival in sepsis. J Immunol. 1999;162(7):4148–4156. [PubMed] [Google Scholar]
- 62.Hotchkiss RS, Swanson PE, Freeman BD, Tinsley KW, Cobb JP, Matuschak GM, et al. Apoptotic cell death in patients with sepsis, shock, and multiple organ dysfunction. Crit Care Med. 1999;27(7):1230–1251. doi: 10.1097/00003246-199907000-00002. [DOI] [PubMed] [Google Scholar]
- 63.Hotchkiss RS, Tinsley KW, Karl IE. Role of apoptotic cell death in sepsis. Scand J Infect Dis. 2003;35(9):585–592. doi: 10.1080/00365540310015692. [DOI] [PubMed] [Google Scholar]
- 64.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 65.Hotchkiss RS, Coopersmith CM, Karl IE. Prevention of lymphocyte apoptosis--a potential treatment of sepsis? Clin Infect Dis. 2005;41(Suppl 7):S465–469. doi: 10.1086/431998. [DOI] [PubMed] [Google Scholar]
- 66.Hotchkiss RS, Osmon SB, Chang KC, Wagner TH, Coopersmith CM, Karl IE. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J Immunol. 2005;174(8):5110–5118. doi: 10.4049/jimmunol.174.8.5110. [DOI] [PubMed] [Google Scholar]
- 67.Hotchkiss RS, McConnell KW, Bullok K, Davis CG, Chang KC, Schwulst SJ, et al. TAT-BH4 and TAT-Bcl-xL peptides protect against sepsis-induced lymphocyte apoptosis in vivo. J Immunol. 2006;176(9):5471–5477. doi: 10.4049/jimmunol.176.9.5471. [DOI] [PubMed] [Google Scholar]
- 68.Cvijanovich N, Shanley TP, Lin R, Allen GL, Thomas NJ, Checchia P, et al. Validating the genomic signature of pediatric septic shock. Physiol Genomics. 2008;34(1):127–134. doi: 10.1152/physiolgenomics.00025.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pachot A, Cazalis MA, Venet F, Turrel F, Faudot C, Voirin N, et al. Decreased expression of the fractalkine receptor CX3CR1 on circulating monocytes as new feature of sepsis-induced immunosuppression. J Immunol. 2008;180(9):6421–6429. doi: 10.4049/jimmunol.180.9.6421. [DOI] [PubMed] [Google Scholar]
- 70.Pachot A, Lepape A, Vey S, Bienvenu J, Mougin B, Monneret G. Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett. 2006;106(1):63–71. doi: 10.1016/j.imlet.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176(7):676–684. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 72.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. Gene-expression profiling of peripheral blood mononuclear cells in sepsis. Crit Care Med. 2009;37(3):882–888. doi: 10.1097/CCM.0b013e31819b52fd. [DOI] [PubMed] [Google Scholar]
- 73.Wong HR, Cvijanovich N, Allen GL, Lin R, Anas N, Meyer K, et al. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37(5):1558–1566. doi: 10.1097/CCM.0b013e31819fcc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. 2009;7:34. doi: 10.1186/1741-7015-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 77.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 78.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 79.Cao S, Fernandez-Zapico ME, Jin D, Puri V, Cook TA, Lerman LO, et al. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J Biol Chem. 2005;280(3):1901–1910. doi: 10.1074/jbc.M407941200. [DOI] [PubMed] [Google Scholar]
- 80.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 81.Su RC, Becker AB, Kozyrskyj AL, Hayglass KT. Epigenetic regulation of established human type 1 versus type 2 cytokine responses. J Allergy Clin Immunol. 2008;121(1):57–63. e53. doi: 10.1016/j.jaci.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111(4):1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression following a severe innate immune response. Blood. 2007 doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marshall JC. Sepsis: rethinking the approach to clinical research. J Leukoc Biol. 2008;83(3):471–482. doi: 10.1189/jlb.0607380. [DOI] [PubMed] [Google Scholar]
- 86.Osuchowski MF, Connett J, Welch K, Granger J, Remick DG. Stratification is the key: inflammatory biomarkers accurately direct immunomodulatory therapy in experimental sepsis. Crit Care Med. 2009;37(5):1567–1573. doi: 10.1097/CCM.0b013e31819df06b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warren HS, Elson CM, Hayden DL, Schoenfeld DA, Cobb JP, Maier RV, et al. A genomic score prognostic of outcome in trauma patients. Mol Med. 2009;15(7-8):220–227. doi: 10.2119/molmed.2009.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wong HR, Cvijanovich N, Wheeler DS, Bigham MT, Monaco M, Odoms K, et al. Interleukin-8 as a stratification tool for interventional trials involving pediatric septic shock. Am J Respir Crit Care Med. 2008;178(3):276–282. doi: 10.1164/rccm.200801-131OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nowak JE, Wheeler DS, Harmon KK, Wong HR. Admission chemokine (C-C motif) ligand 4 levels predict survival in pediatric septic shock. Pediatr Crit Care Med. 2009 doi: 10.1097/PCC.0b013e3181b8076c. [DOI] [PMC free article] [PubMed] [Google Scholar]