Abstract
A number of recent genome-wide association (GWA) studies have identified unequivocal statistical associations between inherited genetic variations, mostly single nucleotide polymorphisms (SNPs), and common complex diseases such as diabetes, cardiovascular disease, and obesity. Genotyping individuals for these variations has the potential to help redefine how pharmacologic agents undergo clinical development. By identifying carriers of known genomic variants that contribute to susceptibility, a high risk population can be defined as well as individuals with potential for a better response to a drug. We evaluated the potential utility that selecting individuals for a trial on the basis of genotype identified in contemporary GWA studies would have had on recently described clinical trials. We pursued this by constraining both the risks of a disease outcome associated with particular genotypes and overall drug responses to those actually observed in genetic association and clinical trial studies, respectively. We pursued these evaluations in the context of clinical trials investigating drugs for macular degeneration, obesity, heart disease, type II diabetes, prostate cancer and Alzheimer’s disease. We show that the increase in incidence of outcomes in trials restricted to individuals with specific genotypic profiles can result in substantial reductions in requisite sample sizes for such trials. In addition, we also derive realistic bounds for samples sizes for clinical trials investigating pharmacogenetic effects that leverage genetic variations identified in recent association studies.
Keywords: Polymorphism, Translational medicine, Drug validation, DNA sequencing, Study Design
INTRODUCTION
Despite the growing number of insights into the genetic basis of many common complex diseases resulting from genome wide association (GWA) and related studies (Manolio et al. 2008), there have been relatively few recent advances into the treatment or prevention of these diseases (e.g., Butler 2008; Liao et al. 2009). There are many reasons for this, with the most pronounced being that simply not enough time has elapsed since the identification of many disease-associated genetic variations for researchers to have evaluated or constructed, e.g., new therapeutic targets or diagnostic tests based on those genetic variations. In addition, even if new therapeutics or diagnostics could be easily and efficiently constructed based on knowledge of disease-associated genetic variations, these therapeutics and diagnostics would still need to be tested for their safety and efficacy, and this often comes at great expense and over long periods of time. Thus, it is unclear what, if any, immediate clinical benefits genetic association study results might have.
One area where the exploitation of the results of genetic association studies may have an immediate impact involves the design of clinical trials testing side effect profiles or efficacy of legacy or recently derived therapeutic agents Essentially, trial participants could be enrolled in clinical trials based on a priori knowledge of their genotypic profiles. Such studies could be pursued in the context of both preventive trials in which a drug or intervention is being tested to see if it staves off or prevents a particular adverse condition and curative trials in which a drug or intervention is being tested to see if it can ameliorate, or at least allow individuals to function with, a particular disease. There are at least two motivations for restricting entry into a clinical trial on the basis of genotypic profile. First, one could potentially ‘enrich’ the pool of individuals enrolled in a trial to those more susceptible to an outcome of interest based on high risk genotypes, and hence more likely to benefit from the therapeutic agent or drug being tested. This is particularly relevant for preventive trials. It is widely known that many preventive clinical trials (unknowingly) enroll individuals who ultimately are not at risk for developing the condition or manifesting the outcome that the drug or intervention of interest was designed to prevent. This is very clear in the case of cholesterol lowering drugs tested for their ability to prevent cardiac events such as myocardial infarction, as only a small fraction of individuals enrolled in relevant trials actually exhibit such events. Low event rates limit inferences about differential rates of events between drug classes or active compound/placebo groups without huge sample sizes (Muhkerjee and Topol 2002). Second, one could sample individuals with different genotypic profiles in roughly equal proportions under the assumption that the different genotypes might influence drug responsiveness. In this way, the clinical trial could be optimally designed to detect a pharmacogenetic effect. Although pharmacogenetic studies are usually considered in the context of curative trials, they could also be pursued in the context of a preventive trial if there is reason to believe that individuals with a certain genotypic profile are more likely to avoid the condition of interest via the drug or intervention under scrutiny.
Note that studies limiting participation in the trial to individuals with particular genotypic profiles is much different in orientation to studies involving the genotyping of individual participants in a trial a posteriori and then examining associations between drug response and genotype retrospectively (SEARCH Collaborative 2009; Karapetis et al. 2008; Brandt et al. 2007; Liggett et al. 2008). Recently, Simon and colleagues considered the influence of restricting a trial to entrants with a certain genotypic profile on the trial sample size, ultimately describing the theoretical relationship between study efficiency and the genotype-specific drug response (Simon and Maitournam 2006; Maitournam and Simon 2005). Although the studies by Simon and Maitournam shed enormous light on the contexts within which restricted sampling is likely to result in true efficiencies, little research has explored the potential utility of recently identified genetic variations in the design of clinical trials, either in the context of enriching a trial for individuals likely to benefit from a therapeutic agent by restricting entry into the trial based on genotypic risk or by contrasting response to a therapeutic agent as a function of genotype.
In this paper we consider the potential impact some recently identified genetic variations could have in the assessment of the efficacy and pharmacogenetic effects of different therapeutic agents used in preventive trials. We pursue these studies by asking what effects, if any, the use of genotype restricted sampling could have had on previously conducted clinical trials. We consider actual genetic variations identified in recent genetic association studies for this purpose (Manolio et al. 2008). We compute relevant sample size and efficiency calculations by constraining the effects of the genotype and drug responses considered to those observed in actual genetic association studies as well as actual clinical trials. With these constraints, we ultimately calculate a realistic estimate of the upper bound (in terms of efficiency) on the effect that genotype-restricted sampling can have on the sample size requirements of clinical trials that leverage genetic variations that are characteristic of those identified in recent GWA studies. Our studies point to the potential – and a way of assessing that potential – that genotype-restricted sampling can have on realistic future clinical trials. We also consider limitations of our studies as well as areas for further research.
METHODS
Genotype-Restricted Clinical Trials and Sample Size Calculations
As noted, the goal of our studies is to assess the utility of recently identified disease-associated genetic variations in the design of preventive clinical trials whose entry is limited to individuals with a particular genotypic profile. We chose actual historical preventive clinical trials whose design and conduct could have benefited from such genotype-based entry restrictions either to make the study more efficient by increasing the pool of individuals likely to develop the outcome for which a drug/intervention was designed to prevent or to test for pharmacogenetic effects. We chose clinical trials contrasting the frequency of a simple binary outcome measure among treated and non-treated. By considering data from actual clinical trials, we can constrain the impact that genotype-based sampling might have had on them in realistic ways. We used a simple contingency table framework for evaluating the impact of genotype-based sampling, as outlined in Table 1a. Values for rates of the outcome among individuals on placebo, Rp, and active drug/intervention, Ri, were obtained from the trials themselves. Genotype frequencies, α, and reported odds ratios for a particular genotype, ORg, measuring the strength of the association of that genotype to a specific outcome matching those investigated in the chosen clinical trials were obtained from the literature as well.
Table 1a.
Tabular Format for Evaluating Clinical Trials.
| Intervention | Placebo | |
|---|---|---|
| Disease | Ri | Rp |
| No Disease | 1 − Ri | 1 −Rp |
We assumed that among the individuals participating in a trial, the rate of the outcome among individuals on placebo, Rp, could be conceived of as a weighted average of the rates of the outcome among individuals with a certain genotype, , and without a certain genotype, , with weights given by the frequency of the genotype in the population at large; i.e., since the rate of the outcome among individuals on placebo should reflect the rate of the outcome in the population at large. Barring any substantial biases in the way subjects were recruited into the trial, the outcome rate among those on placebo is:
| (1) |
If the rate of the outcome among individuals treated with the placebo who also possess a particular genotype is known, then one can recover the rate among individuals without the genotype from equation 1:
| (2) |
The odds ratios reported in the literature measuring the strength of the association between a particular genotype and an outcome of interest is:
| (3) |
By substituting from equation 2 into equation 3 and solving for we get:
| (4) |
which can be solved through the use of the reported outcome rates from published clinical trials and the reported genotype frequencies and odds ratios from published association studies.
Typically, one would not know the rates of the outcome among individuals with, , and without, , the genotype, since this would be a function of the differential effect (if any) of the intervention among individuals with the genotype. If we assume that there is no differential effect (i.e., no pharmacogenetic effect of, e.g., a curative drug or preventive intervention) then effectively, , although and since, if the drug/intervention is not toxic, Ri< Rp. If there is a pharmacogenetic effect, then possibly and . Equations 1–4 can be used to assess situations involving a pharmacogenetic effect if assumptions are made about , although given the constraint imposed by equation 1, one might want to consider the restriction that , since violation of this assumption would suggest that the intervention actually carries an adverse potential or can be even considered ‘toxic’ for individuals without the genotype (i.e., the intervention actually raises the rate of the outcome for the individuals treated with the intervention to a level higher than when treated with the placebo).
To determine the number of individuals needed to be screened (i.e., genotyped) in order to identify a requisite number of individuals with a particular genotype for a genotype-restricted trial, we used the negative binomial distribution with frequency parameter equal to the genotype frequency. To compute sample size and power we used standard formula for comparing two proportions (equation 6.1 of Schlessleman .1982; equations 4.14 and 4.15 of Fleiss, Levin, and Paik 2003)., as well as standard formula for interaction analyses for the contingency table setting in Table 1b (pages 193–198 in Schlessleman .1982).
Table 1b.
Tabular Format for Evaluating Genotype Sampling-Based Clinical Trials.
| Intervention | Placebo | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | No Genotype | Genotype | No Genotype | |||||
| Disease |
|
|
|
|
||||
| No Disease |
|
|
|
|
||||
It is important to consider the assumption that there are not substantial differences or biases in the way subjects are selected for clinical trials as opposed to case/control-based genetic association studies. In many preventive trials, individuals deemed at high risk for the adverse condition or outcome hypothesized to be prevented by a particular drug or intervention are sampled. The criteria for entry into the trial therefore leverages knowledge of known risk factors for the adverse condition in question. Individuals chosen for a trial on the basis of known risk factors are thus not likely to be like control individuals in case/control studies, and possibly even unlike individuals chosen as representing prevalent cases of the condition in such studies. However, remarkably, many, though not all, genetic variations that have been identified as associated with particular disease-related conditions, such as myocardial infarction (MI) and type II diabetes, via GWA studies are not themselves associated – or at most only weakly associated – with many of the traditional risk factors for those diseases (see, for example, Talmud et al. 2008; Aulechenko et al. 2009; Bouatia-Naji et al. 2009; Lyssenko et al. 2008). In fact, it is relatively easy to verify this by simply comparing the most strongly associated variations with a particular disease condition with the most strongly associated variations with risk factors for those disease conditions using available databases (http://geneticassociationdb.nih.gov/). Thus, sampling individuals according to genotype can be considered a way of further enriching the sample for subjects predisposed to the adverse condition in question beyond whatever risk factors might also be considered. This ‘enrichment’ effect and its influence on sample size is a focus of our studies. Knowing that some genetic variations we assessed may be weakly associated with other risk factors used to select individuals for some of the preventive trials we considered, we suggest that our results on the potential effect of genotype-based sampling on the sample sizes of those studies be considered as upper bounds on efficiency.
In addition, we are making the implicit assumption that the accrual of new cases in a preventive trial who develop a certain condition and who have a certain genotype, relative to those without a certain genotype, will occur at rates consistent with the odds ratios quantifying the association between the genotype and the condition obtained from case/control studies of that genotype, though we constrain these values by the actual overall (i.e., final, end-of-trial) frequency of the outcome observed in the trial. Thus, we are making the assumption that the accrual of incident cases of a condition during a preventive trial among individuals with certain genotypes is proportional to odds ratios quantifying the strength of the association between prevalent cases manifesting the condition and the different genotypes. This is a fundamental assumption, but we think it is actually reasonable for a study of the possible effects of genotype restricted sampling in realistic clinical trial settings.
Recent Clinical Trials
We considered 5 historical clinical trials investigating drugs/interventions to prevent 5 different diseases. The disease outcomes and drugs examined in these studies are listed in Table 2 and include a study of Atorvastatin to prevent myocardial infarction (MI) (LaRossa et al. 2005; note that in the context of trials investigating drugs used to treat MI we also considered the following additional trials: Yusuf et al. 2000; Antiplatelet Trialists’ Collaborative 1994; Fibrinolytic Therapy Trialists Collaborative Group 1994; Kjekshus et al. 1995; The Epic Investigators 1994; Yusuf et al. 2001; The Cholesterol Treatment Trialists (CCT) Collaborators 2005); Finasteride to prevent prostate cancer (Thompson et al. 2003); a behavioral intervention to prevent obesity (Gortmaker et al. 1999); Metformin to prevent type II diabetes (Salpeter et al. 2008); and antioxidants to prevent Alzheimer’s disease (Heart Protection Study Collaborative Group 2002).
Table 2.
Example Clinical Trials Investigating the Ability of Certain Compounds for Disease Prevention
| Outcome | Intervention | Ref | I Rate | C Rate | Red | N I | N C | Pow N | R Pow/Act |
|---|---|---|---|---|---|---|---|---|---|
| Myocardial Infarction (MI) | Atorvastatin | 1 | 0.049 | 0.062 | 20.968 | 4995 | 5006 | 3830 | 0.766 |
| Prostate Cancer | Finasteride | 2 | 0.184 | 0.244 | 24.590 | 4368 | 4692 | 577 | 0.127 |
| Obesity* | Behavioral | 3 | 0.055 | 0.080 | 31.250 | 237 | 249 | 1243 | 5.110 |
| Type II Diabetes (T2D) | Metformin | 4 | 0.210 | 0.290 | 27.568 | 1556 | 1563 | 361 | 0.231 |
| Alzheimers* | Antioxidants | 5 | 0.234 | 0.244 | 4.098 | 10241 | 10228 | 22462 | 2.195 |
Key: Rate = incidence rate of the outcome among individuals on the active compound or the control compound for the study referenced; N= sample size for the group receiving the drug (“I”) and not receiving the drug (“C”). “Red” is the percentage reduction in disease due to the intervention. “Pow N” gives the sample sizes necessary for both the Intervention and Control groups in order to detect the observed effect assuming a power level of 0.8 and a type I error rate of 0.05. “R Pow/Act” provides the ratio of the computed sample size necessary to detect the observed effect (“Pow N”) to the actual total sample sized used in the studies.
The asterisk denotes studies that did not observe a significant difference in outcome rates between the intervention and control groups. References used in the Table are: 1: LaRosa et al. (2005); 2: Thompson et al. (2003); 3: Gortmaker et al. (1999); 4: Salpeter et al. (2008); 5: Heart Protection Study Collaborative Group (2002).
Also listed in Table 2 are the rates of the disease outcomes among those treated with the drug/intervention (‘I Rate’) and those treated with placebo (‘C Rate’) in columns 4 and 5, respectively; the overall reduction in disease outcome rates due to the drug/intervention (‘Red’) in column 6; the total number of participants in the drug/intervention (‘N I’) and placebo (‘N C’) arms of the trial in columns 7 and 8, respectively. The column labeled ‘Pow N’ gives the sample sizes necessary for both the Intervention and Control groups in order to detect the observed effect assuming a power level of 0.8 and a type I error rate of 0.05; and the column labeled ‘R Pow/Act’ provides the ratio of the computed sample size necessary to detect the observed effect (“Pow N”) to the actual total sample sized used in the studies. Note that since the behavioral intervention trial for obesity and the antioxidant trial for Alzheimer’s disease did not reveal a difference in outcome rates between the drug/intervention and placebo groups, a greater sample size is needed to detect the observed difference based on the observed rates of the outcomes in the drug/intervention and placebo arms of the trial.
Disease-Associated Genetic Variations
We considered a number of recently identified genetic variations that are associated with common chronic diseases in our assessment of the utility of genotype-based sampling for the clinical trials listed in Table 2. We specifically chose individual single nucleotide polymorphisms (SNPs) identified from GWA or other genetic association studies. Although the SNPs were chosen based on the strength of their association, many other genetic variations in linkage disequilibrium with the chosen SNPs also exhibit association with the diseases of interest. The genetic variations chosen for study included SNP rs10757278 in the 9p21 region found to be associated with MI (Helgodottir et al. 2007); SNP rs16901979 in the 8q24 region found to be associated with prostate cancer (Zheng et al. 2008); SNP rs1421085 in the FTO gene region found to be associated with obesity (Dina et al. 2007); SNP rs10811661 in the CDKN2A gene region found to be associated with type II diabetes (Diabetes Genetics Initiative 2007); and SNP rs4420638 in the APOE gene region found to be associated with Alzheimer’s disease (Coon et al. 2007). As an extension of our analyses for single SNP effects, we also considered the impact of multilocus-based genotype restricted sampling using the 5 SNPs tested for multilocus effects in the study of prostate cancer by Zheng et al. (2008). These SNPs, their frequencies, and their odds ratios for association are listed in columns 3, 5, 6 and 7 of Table 3.
Table 3.
Projected Sample Sizes for Genotype-based Trials that Test Compounds Listed in Table 1.
| Outcome | Intervention | SNP | Ref | Locus | Freq | OR | Gen N | Screen | R Act | R Pow |
|---|---|---|---|---|---|---|---|---|---|---|
| MI | Atorvastatin | rs10757278 | 1 | 9p21 | 0.22r | 1.72 | 2693 | 24485 ± 294 | 0.539 | 0.703 |
| Prostate | Finasteride | rs16901979 | 2 | 8q24 | 0.07d | 1.53 | 471 | 13463 ± 422 | 0.104 | 0.817 |
| Obesity | Behavioral | rs1421085 | 3 | FTO | 0.41a | 1.56 | 1006 | 4910 ± 84 | 4.142 | 0.809 |
| T2D | Metformin | rs10811661 | 4 | CDKN2A | 0.83a | 1.20 | 355 | 857 ± 3 | 0.228 | 0.984 |
| Alzheimers | Antioxidants | rs4420638 | 5 | APOE4 | 0.17a | 4.01 | 14238 | 167508 ± 904 | 1.391 | 0.634 |
Key: SNP = Single Nucleotide Polymorphism or locus identified in the study listed under the Ref column associated with the Outcome; Freq is the frequency of the risk genotype(s) with r=recessive, d=dominant, and a=additive allelic effects; “OR” is the effect of the allele on the outcome; “Gen N” denotes the Intervention and Control group sample sizes necessary for detecting a one sided test effect of the intervention at a power level 0.8 and type I error rate of 0.05 assuming that the intervention works just as well among the targeted subjects as the original study group referenced in Table 1. “Screen” gives the total number of individuals (and standard error) that must be screened (i.e., genotyped) in order to identify the requisite number with the appropriate genotype based on the mean negative binomial distribution. “R Act” and “R Pow” denote the ratio of the actual trial sample size and the projected sample size for the actual effect observed to the genotype-based sampling sample size, respectively relative to the original study listed in Table 2. Mode indicates the assumed genotypic or allelic effect: Rec=recessive; Dom=dominant; Add= additive (allelic); References in the Table are 1: Helgodottir et al. (2007); 2. Zheng et al. (2008); 3: Dina et al. (2007); 4: Diabetes Genetics Initiative (2007); 5: Coon et al. (2007).
RESULTS
Genotype-Based Sampling for Enrichment of the Trial for Susceptible Individuals
We computed the sample sizes necessary for conducting the trials described in Table 2 assuming that the trials could have been restricted to participants carrying relevant susceptibility genotypes at the SNP loci listed in columns 3–7 of Table 3. The basic intuition behind these calculations is that individuals with the susceptibility genotypes are more likely to develop the disease condition of interest; hence the event rates among the participants receiving the placebo will be larger than if the participants without the susceptibility genotype had been sampled. This increase in overall event rates will result in a power increase for detecting differences in event rates between the drug/intervention and placebo arms – given that the drug/intervention works – and hence require a smaller sample size for the study. Column 8 (‘Gen N’) of Table 3 provides the number of cases and controls needed for a study restricted to entrants with the susceptibility genotypes (i.e., twice the number in column 8 provides the total number of subjects needed for the study) based on equations 1–4. This number is contrasted with the actual number of participants in the original trial (column 10; ‘R Act’) and the number of participants needed for a study with power 0.8 to detect the effect reported in the original trial, assuming that the event rates are consistent with the original trial (column 11; ‘R Pow’).
Also provided in Table 3 is the number of individuals that would have to be screened in order to identify the appropriate number of individuals with the susceptibility genotype for the genotype-restricted trial (column 9; ‘Screen’). We have also included standard errors associated with the number of people who must be screened. Implied and overt assumptions associated with these calculations are considered in the Discussion section. It can be seen from Table 3 that limiting the trial to participants with the susceptibility genotypes can lead to more efficient studies, even in the context of designing studies investigating drugs/interventions that did not show efficacy in the original study (i.e., the obesity and Alzheimer’s disease studies). Note that these efficiency gains occur despite the fact the susceptibility genotypes do not have pronounced effect sizes (i.e., odd ratios) on relevant disease outcomes. However, the efficiency gains associated with genotype-restricted entry criteria do require the identification of individuals with the susceptibility genotypes via screening for individuals with the relevant genotypes, and this would add cost to the study.
We considered the evaluation of the effect of genotype-restricted entry for additional clinical trials investigating cholesterol lowering drugs and the prevention of MI. Sampling based on SNP rs10757278 (Helgodottir et al. 2007) was assumed in all the calculations. The results are described in Table 4 and follow the same format as in Table 3. Table 4 suggests that restricting entry into the trials on the basis of the rs10757278 genotype would have indeed resulted in efficiencies relative to the original trial as well as an appropriately powered, but non-genotype-restricted, trial based on the observed efficacy rates.
Table 4.
Projected Sample Sizes for Genotype-based Trials that Test Cholesterol Lowering Treatments for Preventing Myocardial Infarction (MI).
| Trial | Drug | Ref | I Rate | C Rate | N Int | N Ctrl | Pow N | Gen N | Screen | R Pow |
|---|---|---|---|---|---|---|---|---|---|---|
| HOPE | Ramipril | 1 | 0.099 | 0.123 | 4645 | 4652 | 2115 | 1554 | 14134 ± 223 | 0.735 |
| APTC | Aspirin | 2 | 0.024 | 0.035 | 48736 | 48882 | 2922 | 2012 | 18298 ± 254 | 0.689 |
| FTT* | Thrombolytics | 3 | 0.096 | 0.115 | 29315 | 29285 | 3228 | 2362 | 21475 ± 275 | 0.732 |
| 4S* | Simvastatin | 4 | 0.189 | 0.291 | 2275 | 2134 | 216 | 176 | 1604 ± 75 | 0.815 |
| EPIC | Abciximab | 5 | 0.057 | 0.086 | 1403 | 696 | 974 | 693 | 6306 ± 149 | 0.711 |
| CURE | Clopidogrel | 6 | 0.052 | 0.067 | 6258 | 6303 | 3071 | 2166 | 19694 ± 264 | 0.705 |
| 14 Total Trials | Many Statins | 7 | 0.044 | 0.062 | 45054 | 45002 | 1913 | 1342 | 12201 ± 207 | 0.701 |
Key: For the genotype-based trials all the calculations assumed sampling on the basis of homozygosity of the SNP rs10757278 at the 9p21 locus with frequency of 0.22, as in the first column of Table 2; Column headings are consistent with those provided in Tables 2 and 3.
The asterisk denotes studies for which total morbidity/mortality, and not just MI, was taken as the endpoint of interest. The first six studies are discussed in the review by Mukherjee and Topol (2002) and include references 1: Yusef et al. (2000); 2. Antiplatelet Trialists’ Collaborative (1994); 3: Fibrinolytic Therapy Trialists Collaborative Group (1994); 4: Kjekshus et al. (1995); 5: The Epic Investigators (1994); 6: Yusuf et al. (2001). The study reviewing 14 total trials (reference 7) is from the Cholesterol Treatment Trialists (CCT) Collaborators (2005).
Multilocus Genotype-Based Sampling
We also considered the effect of sampling for trials based on multilocus susceptibility genotypic profiles. Zheng et al. (2008) studied the risk of prostate cancer as a function of 5 susceptibility SNPs. Zheng et al. (2008) showed that individuals who possessed a greater number of the susceptibility genotypes had higher risk of prostate cancer. However, the frequency of individuals with greater numbers of these susceptibility genotypes is relatively small. We evaluated the effects of genotype-based sampling on the trial investigating finasteride listed in Tables 2–3 (Thompson et al. 2003). Zheng et al. (2008) also considered the inclusion of family history in assessing prostate cancer risk over-and-above the SNP information. We therefore included family history in our calculations. Table 5 describes the results and is constructed in the same manner as Tables 3 and 4. It can be seen from Table 5 that studies limiting entry to individuals with a greater number of susceptibility genotypes will lead to more efficient trials than the original trial. However, the number of individuals needed to be screened to identify carriers of the relevant susceptibility genotypes may be quite large, as expected, given the fact that individuals carrying multiple susceptibility genotypes are likely to be rare. It is possible to assess the potential impact of multilocus-based genotype-restricted sampling on clinical trials even if only single locus results are available by making assumptions about the combined effects of the individual loci, such as additivity or multiplicativity (Lu et al. 2009).
Table 5.
Projected Sample Sizes for Multilocus Genotype-based Trials that Test Some Compounds Listed in Table 1.
| Outcome | Drug | # of Fctrs | FH | Freq | OR | Gen N | Screen | R Act | R Pow |
|---|---|---|---|---|---|---|---|---|---|
| Prostate Cancer | Finasteride | ≥1 | − | 0.899 | 1.87 | 533 | 1234 ± 11 | 0.123 | 0.962 |
| Prostate Cancer | Finasteride | ≥2 | − | 0.531 | 2.13 | 468 | 1766 ± 39 | 0.104 | 0.813 |
| Prostate Cancer | Finasteride | ≥3 | − | 0.171 | 2.50 | 398 | 4661 ± 150 | 0.088 | 0.691 |
| Prostate Cancer | Finasteride | ≥4 | − | 0.022 | 4.47 | 377 | 34314 ± 1235 | 0.083 | 0.654 |
| Prostate Cancer | Finasteride | ≥1 | + | 0.850 | 2.25 | 535 | 1260 ± 14 | 0.118 | 0.929 |
| Prostate Cancer | Finasteride | ≥2 | + | 0.514 | 2.66 | 436 | 1700 ± 40 | 0.096 | 0.757 |
| Prostate Cancer | Finasteride | ≥3 | + | 0.214 | 2.91 | 380 | 3552 ± 114 | 0.084 | 0.659 |
| Prostate Cancer | Finasteride | ≥4 | + | 0.038 | 4.52 | 369 | 19448 ± 701 | 0.082 | 0.640 |
| Prostate Cancer | Finasteride | ≥5 | + | 0.003 | 9.46 | 471 | 314099 ± 10216 | 0.104 | 0.816 |
Key: Column titles are the same as in Table 3. The “# of Fctrs” column refers to the number of genetic factors (SNPs) with frequency and a cumulative effect given in the “Freq” and “Odds Ratio” columns as reported in the relevant reference. “FH” denotes that whether or not a positive Family History is included as one of the risk factors along with the SNPs. Data were taken from Zheng et al. (2008).
Genotype-Based Sampling for Pharamacogenetic Studies
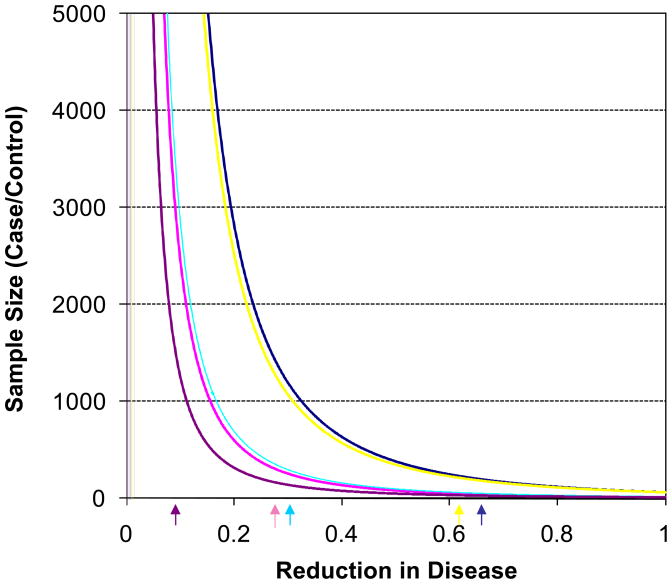
We considered the influence of genotype-based restricted sampling for pharmacogenetic studies investigating the drugs/interventions listed in Table 2. We first considered situations in which the reduction in outcomes for treated individuals carrying the susceptibility genotype was greater than the reduction associated with the treated individuals in the original trial. We constrained the total rate of events across treated and non-treated individuals in the trials to be consistent with reported total event rate in the trial. We used the calculations discussed in the Methods section and plotted the sample sizes necessary for the trials as a function of the fraction of the reduction in outcome among the treated individuals relative to the untreated groups (Figure 1). The sample size reflected on the y-axis in Figure 1 provides the number of cases and controls (i.e., twice the reported number gives the total sample size). We assumed a power level of 0.80 and type I error rate of 0.05 for the calculations. The arrows underneath the figure are the points beyond which the assumed rates for the outcome among treated individuals without the susceptibility genotype – based on the preservation of the observed rate of the outcome among all treated individuals in the trial (i.e., with and without the susceptibility genotype) to be consistent with the actual trial – would yield a rate among treated individuals without the genotype that would be greater than the rate for this group while on placebo, based on equation (1). Thus, the arrows under the figure essentially give points in the graph beyond which the calculated efficacy rates for the treated group with the genotype would suggest toxicity of the drug for those without the genotype given that the total number of event rates across both the treated and placebo groups is consistent with the original trial (see the Methods section).
Figure 1.
Sample size requirements to detect a difference in rates of disease outcomes between an intervention and control group using genotype-restricted samples based on the overall rates of the outcome for the trials listed in Table 2 and the genetic factors outlined in Table 3. The color coding is as follows: dark blue indicates the atorvastatin (MI) trial; pink indicates the finasterid (prostate) trial; yellow indicates the behavioral intervention (obesity) trial; light blue indicates the metformin (type II diabetes) trial; and purple indicates the antioxidant (Alzheimers) trial. The sample size provides the number of cases and controls (i.e., twice the reported number gives the total sample size). The figure relates the sample size needed to detect a rate difference with a power level of 0.80 and type I error rate of 0.05 as a function of the assumed decrease in the rate of the outcome among individuals with the genotype. Constraints on this rate, as well as the rate for those on placebo, were based on the observed rates and genotype effects in Tables 2 and 3 and the algebraic identities discussed in the text. The arrows underneath the figure are the rates of the outcome among treated individuals with the genotype that, based on the observed rate of the outcome among all treated individuals in the trial, would yield a rate among treated individuals without the genotype that would be greater than the rate for this group on placebo from equation (1).
To further investigate pharmacogenetic effects, we again constrained the total event rates considered in the calculations to the total event rates observed in the actual trials, as before (see the Methods section), we had to make a number of assumptions about the trial designs and genotype-specific event rates among individuals treated and not treated. As an initial analysis, we assumed that an equal number of individuals with and without the susceptibility genotype would be sampled for a pharmacogenetic trial and that interest was in testing a treatment × genotype interaction term in relevant contingency table analyses, as this analysis method has been considered in other theoretical studies of pharmacogenetic trial designs (Elston et al. 1999). In addition, we assumed that individuals harboring the susceptibility genotype would have an odds ratio of 0.8 to develop the condition of interest while on the drug whereas individuals without the susceptibility genotype would have an odds ratio to develop the outcome consistent with what was reported in the actual trial.
Table 6 describes some of the calculations for this pharmacogenetic trial setting given that actual trial data considered in Tables 2–4. The ‘NG Red’ and ‘G Red’ columns, respectively, provide the percent reduction in outcomes for treated individuals without (“NG”) and with (“G”) the genotype, respectively, given the assumed odds ratio of 0.8 for individuals with the susceptibility genotype while on the intervention and the overall disease incidences reported in Tables 3 and 4. The “PH Pow” column provides the post-hoc power to detect an interaction effect between the drug and genotype carrier status under the assumption that the odds ratio associated with the genotype in the intervention group is 0.8 (whereas, again, for control group the odds ratio is as reported in Tables 2 and 3 based on the actual sample size of the study and the frequency of genotypes in question). The “Pow N,” “Gen N,” and “R Pow” are the same as in Tables 2, 3, 4. Keep in mind that the “Gen N” column provides a sample size for a study determined assuming equal numbers of individuals both with and without the genotype in the intervention and control groups. Table 6 suggests that if pharmacogenetic effects of the type modeled here actually influence drugs tested historically, then designing a study with enrollment based on genotypic profile for recently discovered SNPs could lead to more efficient trials.
Table 6.
Projected Sample Sizes for Testing Pharmacogenetic Effects for the Drugs Listed in Tables 2, 3, and 4.
| Outcome | Drug | SNP | PH Pow | NG Red | G Red | Pow N | Gen N | Screen | R Pow |
|---|---|---|---|---|---|---|---|---|---|
| MI | Atorvastatin | rs10757278 | 0.995 | 5.590 | 53.903 | 1890 | 1228 | 11163 ± 198 | 0.650 |
| Prostate | Finasteride | rs16901979 | 0.978 | 21.771 | 52.154 | 2171 | 532 | 15199 ± 449 | 0.245 |
| Obesity | Behavorial | rs1421085 | 0.203 | 10.017 | 51.564 | 1465 | 1393 | 6795 ± 98 | 0.981 |
| T2D | Metformin | rs10811661 | 0.721 | 6.803 | 31.318 | 1886 | 1110 | 2674 ± 23 | 0.589 |
| Alzheimers | Antioxidants | rs4420638 | 1.000 | −24.281 | 58.779 | 130 | 71 | 835 ± 63 | 0.546 |
| MI | Ramipril | rs10757278 | 1.000 | 5.153 | 51.429 | 975 | 637 | 5790 ± 143 | 0.653 |
| MI | Aspirin | rs10757278 | 1.000 | 17.569 | 60.623 | 3783 | 2451 | 22281 ± 281 | 0.648 |
| MI | Thrombolytics | rs10757278 | 1.000 | 1.469 | 49.817 | 1003 | 655 | 5954 ± 145 | 0.653 |
| MI | Simvastatin | rs10757278 | 1.000 | 25.978 | 57.325 | 554 | 366 | 3327 ± 108 | 0.661 |
| MI | Abciximab | rs10757278 | 0.737 | 21.216 | 60.899 | 1635 | 1063 | 9663 ± 185 | 0.650 |
| MI | Clopidogrel | rs10757278 | 0.999 | 7.387 | 54.614 | 1785 | 1160 | 10545 ± 193 | 0.650 |
| MI | Many Statins | rs10757278 | 1.000 | 15.207 | 58.643 | 2096 | 1361 | 12371 ± 209 | 0.649 |
Key: SNP = Single Nucleotide Polymorphism or locus identified in the study listed under the Ref column associated with the Outcome (references are as in Tables 2, 3 and 2) or the number of SNPs defining increased risk for prostate cancer as defined in Table 4. “PH Pow” = Post Hoc power to detect an interaction effect between the drug and genotype carrier status under the assumption that the odds ratio associated with the genotype in the intervention group is 0.8 whereas for control group the odds ratio is as reported in Tables 2 and 3. “NG Red” and “G Red’ gives percent reduction in disease incidence for those without (“NG”) and with (“G”) the genotype given an odds ratio of 0.8 for the genotype in the intervention group and the overall disease incidences reported in Tables 3 and 4. “Pow N,” “Gen N,” and “R Pow” are as in Tables 2, 3, 4. The “Gen N” sample size was determined assuming an equal number individuals with and without the genotype and in the intervention and control groups.
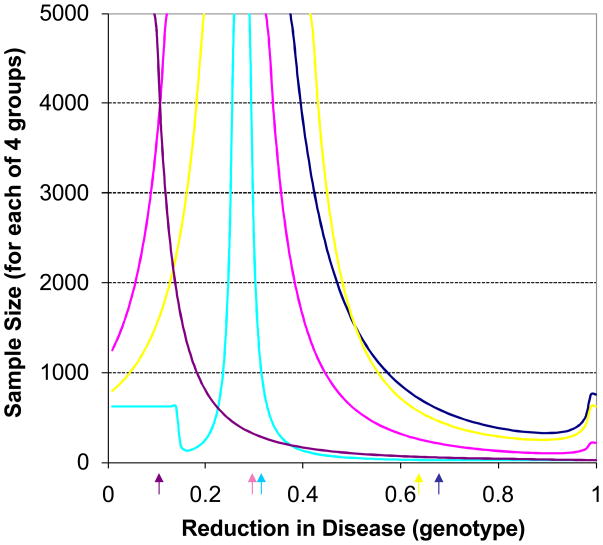
We extended to study considered above to situations in which different assumptions were made concerning the reduction in disease outcome rates among treated individuals with the susceptibility genotype. Figure 2 displays the results graphically. The y-axis reflects the sample size necessary for a study testing an interaction effect, as before, with a power of 0.8 and a type I error rate of 0.05. The fact that the lines in Figure 2 would go beyond a sample of 5000 if it was extended reflect the fact that as the reductions in disease outcomes approach those observed in the actual trial then no pharmacogenetic effect would be detected, given the way in which we constrained the overall rates to match those in the actual trials considered. Note that for some of the situations we considered, lines converge on the reductions observed in the actual trial from reductions less than and greater than this observed reduction. The arrows underneath the x-axis reflect points beyond which the calculated efficacy rates for the treated group without the genotype would be higher than the placebo group without the genotypes and hence suggest toxicity of the drug for individuals with the genotype, as in Figure 1.
Figure 2.
Sample size requirements to detect an interaction effect between genotype and intervention on the rates of disease outcomes assuming that equal numbers of subjects with and without the genotype were assigned to the intervention and control groups for overall rates of the outcome associated with the trials listed in Table 2 and the genetic factors outlined in Table 3. Color coding is the same as that used in Figure 1. The sample size provides the size of each of the four groups needed for the trial. The figure relates the sample size needed to detect an interaction effect with a power level of 0.80 and type I error rate of 0.05 as a function of the assumed decrease in the rate of the disease outcome among individuals with the genotype and on the intervention. Constraints on this rate, as well as the rates for those on placebo and without the genotype, were based on the observed rates and genotype effects in Tables 2 and 3 and the algebraic identities discussed in the text. The arrows underneath the figure are as described in Figure 1.
DISCUSSION
GWA studies pursued in the last 2–3 years have yielded a number of unequivocal, replicable, statistical associations between inherited genetic variations and traits and diseases across all medical disciplines (Manolio et al. 2008). As impressive as these findings are, they raise many questions. For example, many of the genetic variations identified in these studies are not in known functional regions of the genome, raising questions about their ultimate biological significance. In addition, the genetic variations identified to date for any one disease typically explain only a small fraction of the risk of developing that disease, raising questions about not only the missing heritability accounted for by other genetic variations contributing to the disease, but also the potential clinical utility of the current genetic associations.
The use of genetic variations to restrict entry into clinical trials has been proposed and evaluated theoretically by Simon and colleagues (Simon and Maitournam 2006; Maitournam and Simon 2005). The motivation for the studies by Simon and colleagues was to assess potential efficiency gains in clinical trials limited to participants possessing a genetic profile that is known to respond better to the drug in question. These calculations assumed a pharmacogenetic effect; i.e., individuals with the genotype respond better to the treatment than individuals without the genotype. We have considered trials designed to make use of genotype-restricted entry criteria on two fronts: 1. leveraging genotype-based sampling in order to simply enrich the trial for individuals predisposed to the condition or outcome the drug/intervention was designed to prevent; and 2. in order to test pharmacogenetic effects. We did this in the context of genetic variations identified from recent GWA studies and in the context of the results from actual historical clinical trials. In this way we could evaluate potential utility of recently identified genetic variations in genotype-restricted clinical trials that are realistic in terms of the efficacy of actual drugs/interventions. Our results suggest that, despite the low effect sizes of recently identified genetic variations from GWA studies, realistic clinical trials making use of these genetic variations for restricting entry into the trial can lead to efficiency gains. However, there are a few issues that should be discussed.
First, clinical trials leveraging genotype-restricted entry must identify individuals with the genotypes in question. This would require screening. If the necessary genotypes are rare, then a large number of individuals would have to be screened (e.g., Table 3–5). Depending on the costs of screening individuals relative to the conduct of the trial, a genotype-restricted trial may be more expensive. In addition, most high-throughput and cost effective genotyping technologies work in batch, where costs are minimized by processing a large number of samples at once. Sampling individuals for a clinical trial would likely be more akin to screening and involve point-of-care diagnostics whereby subjects are tested sequentially to gain immediate feedback on their eligibility in the trial over some period of time. Second, we assumed that the participants in the historical trials considered were not already enriched for the susceptibility genotypes also considered. Clinical trials designed to study a drug/intervention to prevent a disease outcome often recruit individuals that are at risk for the disease outcome of interest. Susceptibility genotypes are likely to predispose individuals to the traditional factors used to recruit patients into a trial (e.g., obesity, elevated cholesterol level, etc. or a study investigating a drug/intervention used to prevent heart attack). Therefore, the clinical trials investigated here may have been enriched for individuals with the susceptibility genotypes. However, it has been shown to be the case that for many disease conditions the genetic variations associated with that condition are often associated independently of other risk factors (Talmud et al. 2008; Aulechenko et al. 2009; Bouatia-Naji et al. 2009; Lyssenko et al. 2008). The degree to which a possible ‘overlap’ or redundancy in genotypic and traditional risk factor-based restricted sampling for a trial affects the results of our calculations is unknown. Therefore we regard our calculations as ultimately providing upper bounds to the efficiency gains that could have been obtained through the use of genotype-restricted sampling for the trials that we considered.
Acknowledgments
The authors are supported in part by the following research grants: The National Institute on Aging Longevity Consortium [grant number U19 AG023122-01]; The NIMH-funded Genetic Association Information Network Study of Bipolar Disorder National [grant number 1 R01 MH078151-01A1]; National Institutes of Health grants: grant numbers N01 MH22005, U01 DA024417-01, P50 MH081755-01; the Scripps Translational Sciences Institute Clinical Translational Science Award [grant number U54 RR0252204-01], the Price Foundation and Scripps Genomic Medicine.
References
- Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, Martin NG, Pedersen NL, Kyvik KO, Kaprio J, Hofman A, Freimer NB, Jarvelin MR, Gyllensten U, Campbell H, Rudan I, Johansson A, Marroni F, Hayward C, Vitart V, Jonasson I, Pattaro C, Wright A, Hastie N, Pichler I, Hicks AA, Falchi M, Willemsen G, Hottenga JJ, de Geus EJ, Montgomery GW, Whitfield J, Magnusson P, Saharinen J, Perola M, Silander K, Isaacs A, Sijbrands EJ, Uitterlinden AG, Witteman JC, Oostra BA, Elliott P, Ruokonen A, Sabatti C, Gieger C, Meitinger T, Kronenberg F, Döring A, Wichmann HE, Smit JH, McCarthy MI, van Duijn CM, Peltonen L ENGAGE Consortium. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009 Jan;41(1):47–55. doi: 10.1038/ng.269. Epub 2008 Dec 7. PMID: 19060911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiplatelet Trialists’ Collaboration. Collaborative overview of randomised trials of antiplatelet therapy--I: Prevention of death, myocardial infarction, and stroke by prolonged antiplatelet therapy in various categories of patients. Antiplatelet Trialists’ Collaboration. BMJ. 1994 Jan 8;308(6921):81–106. Erratum in: BMJ. 1994, Jun 11;308(6943):1540. [PMC free article] [PubMed] [Google Scholar]
- Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes R Cholesterol Treatment Trialists’ (CTT) Collaborators. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005 Oct 8;366(9493):1267–78. doi: 10.1016/S0140-6736(05)67394-1. Epub 2005 Sep 27. Erratum in: Lancet. 2005 Oct 15–21;366(9494):1358. [DOI] [PubMed] [Google Scholar]
- Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, Sparsø T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, De Graeve F, Chèvre JC, Borch-Johnsen K, Hartikainen AL, Ruokonen A, Tichet J, Marre M, Weill J, Heude B, Tauber M, Lemaire K, Schuit F, Elliott P, Jørgensen T, Charpentier G, Hadjadj S, Cauchi S, Vaxillaire M, Sladek R, Visvikis-Siest S, Balkau B, Lévy-Marchal C, Pattou F, Meyre D, Blakemore AI, Jarvelin MR, Walley AJ, Hansen T, Dina C, Pedersen O, Froguel P. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009 Jan;41(1):89–94. doi: 10.1038/ng.277. Epub 2008 Dec 7. PMID: 19060909. [DOI] [PubMed] [Google Scholar]
- Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, 2nd, Lachno DR, Salazar D, Winters KJ. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007 Dec;5(12):2429–36. doi: 10.1111/j.1538-7836.2007.02775.x. Epub 2007 Sep 26. PMID: 17900275. [DOI] [PubMed] [Google Scholar]
- Butler D. Translational research: crossing the valley of death. Nature. 2008 Jun 12;453(7197):840–2. doi: 10.1038/453840a. No abstract available. PMID: 18548043. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Idury RM, Harris TJ, Witte JS, Elston RC. Testing drug response in the presence of genetic information: sampling issues for clinical trials. Pharmacogenetics. 2000 Aug;10(6):503–10. doi: 10.1097/00008571-200008000-00003. [DOI] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007 Apr;68(4):613–8. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Boström K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Råstam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjögren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007 Jun 1;316(5829):1331–6. doi: 10.1126/science.1142358. Epub 2007 Apr 26. [DOI] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougnères P, Kovacs P, Marre M, Balkau B, Cauchi S, Chèvre JC, Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007 Jun;39(6):724–6. doi: 10.1038/ng2048. Epub 2007 May 13. [DOI] [PubMed] [Google Scholar]
- Elston RC, Idury RM, Cardon LR, Lichter JB. The study of candidate genes in drug trials: sample size considerations. Stat Med. 1999 Mar 30;18(6):741–51. doi: 10.1002/(sici)1097-0258(19990330)18:6<741::aid-sim64>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- The EPIC Investigation. Use of a monoclonal antibody directed against the platelet glycoprotein IIb/IIIa receptor in high-risk coronary angioplasty. The EPIC Investigation. N Engl J Med. 1994 Apr 7;330(14):956–61. doi: 10.1056/NEJM199404073301402. [DOI] [PubMed] [Google Scholar]
- Fibrinolytic Therapy Trialists’ (FTT) Collaborative Group. Indications for fibrinolytic therapy in suspected acute myocardial infarction: collaborative overview of early mortality and major morbidity results from all randomised trials of more than 1000 patients. Fibrinolytic Therapy Trialists’ (FTT) Collaborative Group. Lancet. 1994 Feb 5;343(8893):311–22. Review. Erratum in: Lancet. 1994 Mar 19;343(8899):742. [PubMed] [Google Scholar]
- Fijal BA, Hall JM, Witte JS. Clinical trials in the genomic era: effects of protective genotypes on sample size and duration of trial. Control Clin Trials. 2000 Feb;21(1):7–20. doi: 10.1016/s0197-2456(99)00039-2. [DOI] [PubMed] [Google Scholar]
- Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. New Jersey: John Wiley & Sons; 2003. [Google Scholar]
- Gortmaker SL, Peterson K, Wiecha J, Sobol AM, Dixit S, Fox MK, Laird N. Reducing obesity via a school-based interdisciplinary intervention among youth: Planet Health. Arch Pediatr Adolesc Med. 1999 Apr;153(4):409–18. doi: 10.1001/archpedi.153.4.409. [DOI] [PubMed] [Google Scholar]
- Gurtsman B. Epidemiology Kept Simple. New York: Wiley-Liss; 1998. [Google Scholar]
- Heart Protection Study Collaborative Group. MRC/BHF Heart Protection Study of antioxidant vitamin supplementation in 20,536 high-risk individuals: a randomised placebo-controlled trial. Lancet. 2002 Jul 6;360(9326):23–33. doi: 10.1016/S0140-6736(02)09328-5.Summary for patients in: J Fam Pract. 2002 Oct;51(10):810.
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007 Jun 8;316(5830):1491–3. doi: 10.1126/science.1142842. Epub 2007 May 3. PMID: 17478679. [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008 Oct 23;359(17):1757–65. doi: 10.1056/NEJMoa0804385. PMID: 18946061. [DOI] [PubMed] [Google Scholar]
- Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007 Jun 8;316(5830):1491–3. doi: 10.1126/science.1142842. Epub 2007 May 3. [DOI] [PubMed] [Google Scholar]
- LaRosa JC, Grundy SM, Waters DD, Shear C, Barter P, Fruchart JC, Gotto AM, Greten H, Kastelein JJ, Shepherd J, Wenger NK Treating to New Targets (TNT) Investigators. Intensive lipid lowering with atorvastatin in patients with stable coronary disease. N Engl J Med. 2005 Apr 7;352(14):1425–35. doi: 10.1056/NEJMoa050461. Epub 2005 Mar 8. [DOI] [PubMed] [Google Scholar]
- Law MR, Wald NJ, Rudnicka AR. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ. 2003 Jun 28;326(7404):1423. doi: 10.1136/bmj.326.7404.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Zhang X, Clark DJ, Peltz A genomic “roadmap” to “better” drugs. Drug Metab Rev. 2008;40(2):225–39. doi: 10.1080/03602530801952815. Review. PMID: 18464044. [DOI] [PubMed] [Google Scholar]
- Liggett SB, Cresci S, Kelly RJ, Syed FM, Matkovich SJ, Hahn HS, Diwan A, Martini JS, Sparks L, Parekh RR, Spertus JA, Koch WJ, Kardia SL, Dorn GW., 2nd A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008 May;14(5):510–7. doi: 10.1038/nm1750. Epub 2008 Apr 20. PMID: 18425130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Obuchowski N, Won S, Zhu X, Elston RC. Using the Optimal Robust Receiver Operating Characteristic (ROC) Curve for Predictive Genetic Tests. Biometrics. 2009 Jun 8; doi: 10.1111/j.1541-0420.2009.01278.x. [Epub ahead of print] PMID: 19508241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, Berglund G, Altshuler D, Nilsson P, Groop L. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008 Nov 20;359(21):2220–32. doi: 10.1056/NEJMoa0801869. PMID: 19020324. [DOI] [PubMed] [Google Scholar]
- Maitournam A, Simon R. On the efficiency of targeted clinical trials. Stat Med. 2005 Feb 15;24(3):329–39. doi: 10.1002/sim.1975. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008 May;118(5):1590–605. doi: 10.1172/JCI34772. Review. PMID: 18451988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Topol EJ. Pharmacogenomics in cardiovascular diseases. Prog Cardiovasc Dis. 2002 May-Jun;44(6):479–98. doi: 10.1053/pcad.2002.123467. Review. [DOI] [PubMed] [Google Scholar]
- Salpeter SR, Buckley NS, Kahn JA, Salpeter EE. Meta-analysis: metformin treatment in persons at risk for diabetes mellitus. Am J Med. 2008 Feb;121(2):149–157.e2. doi: 10.1016/j.amjmed.2007.09.016. Review. [DOI] [PubMed] [Google Scholar]
- Schlesselman JJ. Case-Control Studies: Design, Conduct, Analysis. New York: Oxford University Press; 1982. [Google Scholar]
- Schork NJ, Weder AB. The use of genetic information in large-scale clinical trials: applications to Alzheimer research. Alzheimer Dis Assoc Disord. 1996 Fall;10( Suppl 1):22–6. doi: 10.1097/00002093-199601031-00007. [DOI] [PubMed] [Google Scholar]
- SEARCH Collaborative Group. Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008 Aug 21;359(8):789–99. doi: 10.1056/NEJMoa0801936. Epub 2008 Jul 23. PMID: 18650507. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007 Apr 25;297(16):1793–800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- Simon R. New challenges for 21st century clinical trials. Clin Trials. 2007;4(2):167–9. doi: 10.1177/1740774507076800. No abstract available. [DOI] [PubMed] [Google Scholar]
- Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clin Cancer Res. 2004 Oct 15;10(20):6759–63. doi: 10.1158/1078-0432.CCR-04-0496. Erratum in: Clin Cancer Res. 2006 May 15; 12(10):3229. [DOI] [PubMed] [Google Scholar]
- Simon R. Validation of pharmacogenomic biomarker classifiers for treatment selection. Cancer Biomark. 2006;2(3–4):89–96. doi: 10.3233/cbm-2006-23-402. Review. [DOI] [PubMed] [Google Scholar]
- Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE. Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008 Mar;54(3):467–74. doi: 10.1373/clinchem.2007.095489. Epub 2008 Feb 4. PMID: 18250146. [DOI] [PubMed] [Google Scholar]
- Thompson IM, Goodman PJ, Tangen CM, Lucia MS, Miller GJ, Ford LG, Lieber MM, Cespedes RD, Atkins JN, Lippman SM, Carlin SM, Ryan A, Szczepanek CM, Crowley JJ, Coltman CA., Jr The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003 Jul 17;349(3):215–24. doi: 10.1056/NEJMoa030660. Epub 2003 Jun 24. [DOI] [PubMed] [Google Scholar]
- Trepicchio WL, Essayan D, Hall ST, Schechter G, Tezak Z, Wang SJ, Weinreich D, Simon R. Designing prospective clinical pharmacogenomic (PG) trials: meeting report on drug development strategies to enhance therapeutic decision making. Pharmacogenomics J. 2006 Mar-Apr;6(2):89–94. doi: 10.1038/sj.tpj.6500344. No abstract available. [DOI] [PubMed] [Google Scholar]
- Yusuf S, Sleight P, Pogue J, Bosch J, Davies R, Dagenais G. Effects of an angiotensin-converting-enzyme inhibitor, ramipril, on cardiovascular events in high-risk patients: The Heart Outcomes Prevention Evaluation Study Investigators. N Engl J Med. 2000 Jan 20;342(3):145–53. doi: 10.1056/NEJM200001203420301. [DOI] [PubMed] [Google Scholar]
- Yusuf S, Zhao F, Mehta SR, Chrolavicius S, Tognoni G, Fox KK Clopidogrel in Unstable Angina to Prevent Recurrent Events Trial Investigators. Effects of clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med. 2001 Aug 16;345(7):494–502. doi: 10.1056/NEJMoa010746. Erratum in: N Engl J Med 2001 Dec 6;345(23):1716. N Engl J Med 2001 Nov 15;345(20):1506. [DOI] [PubMed] [Google Scholar]
- Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Bälter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Grönberg H. Cumulative Association of Five Genetic Variants with Prostate Cancer. N Engl J Med. 2008 Jan 16; doi: 10.1056/NEJMoa075819. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]