Abstract
Orotate phosphoribosyltransferases from Plasmodium falciparum and human sources (PfOPRT and HsOPRT) use orotidine as a slow substrate in the pyrophosphorolysis reaction. With orotidine, intrinsic kinetic isotope effects (KIEs) can be measured for pyrophosphorolysis, providing the first use of pyrophosphate (PPi) in solving an enzymatic transition state. Transition state structures of PfOPRT and HsOPRT were solved through quantum chemical matching of computed and experimental intrinsic KIEs and can be compared to transition states solved with pyrophosphate analogues as slow substrates. PfOPRT and HsOPRT are characterized by late transition states with fully dissociated orotate, well-developed ribocations and weakly bonded PPi nucleophiles. The leaving orotates are 2.8 Å distant from the anomeric carbons at the transition states. Weak participation of the PPi nucleophiles give C1'-OPPi bond distances of approximately 2.3 Å. These transition states are characterized by C2'-endo ribosyl pucker, based on the β-secondary [2'-3H] KIEs. The geometry at the 5'-region is similar for both enzymes, with C3'-C4'-C5'-O5' dihedral angles near −170°. These novel phosphoribosyltransferase transition states are similar to but occur earlier in the reaction coordinate than those previously determined with orotidine 5'-monophosphate and phosphonoacetic acid as substrates. The similarity between the transition states with different substrate analogues supports similar transition state structures imposed by PfOPRT and HsOPRT even with distinct reactants. We propose that the transition state similarity with different nucleophiles is determined, in part, by the geometric constraints imposed by the catalytic sites.
Introduction
Orotate phosphoribosyltransferase (OPRT; EC 2.4.2.10) is the fifth enzyme in the de novo pyrimidine biosynthesis pathway. It catalyzes the formation of orotidine 5'-monophosphate (OMP) from α-D-phosphoribosylpyrophosphate (PRPP) and orotate. The OMP product is subsequently converted to uridine 5'-monophosphate (UMP) by OMP decarboxylase and is the precursor for all pyrimidines used in cellular DNA and RNA synthesis. Plasmodium falciparum, the most virulent parasite causing malaria, is responsible for the annual deaths of over a million people, with a majority under the age of five.1 Currently, increasing resistance of P. falciparum parasites to drug treatment brings the development of novel antimalarial drugs to the top-priority. P. falciparum requires de novo pyrimidine synthesis as it does not encode enzymes for pyrimidine salvage.2–6 PfOPRT-specific inhibitors shows antimalarial activity at low micromolar concentration.7,8 5'-Fluoroorotate, an alternative substrate of PfOPRT, inhibits the in vitro growth of P. falciparum with an IC50 less than 10 nM, through the inactivation of thymidylate synthase.7,9 Inhibitors blocking pyrimidine synthesis at other steps in the pathway have shown antimalarial potency in P. falciparum.10–13 Unlike the P. falciparum parasites, host cells are complemented by pyrimidine salvage pathways.14 However, rapidly growing cells, including cancer cells, place a high demand on de novo pyrimidine synthesis.15 Inhibitors of the pyrimidine pathway, including the antifolates, have a long history in the treatment of malignant neoplastic and autoimmune diseases.16–20 OPRT from P. falciparum and human sources are therefore potential targets for antimalarial and anticancer drugs.
Intrinsic kinetic isotope effects (KIEs) provide access to information about enzyme transition states and have been used to establish the transition state structures of N-ribosyltransferases.21–25 In enzymatic reactions, competitive V/K KIEs are contributed by the isotope effects from all kinetically significant steps between free substrate and the first irreversible step.26 When bond-breaking is the first irreversible step, the V/K KIEs can be corrected for commitment factors to give intrinsic KIEs. The intrinsic KIEs reflect the bond vibrational differences between the free substrate in solution and the transition state. With the experimental intrinsic KIEs as boundary conditions, enzymatic transition states can be estimated through quantum chemical calculations. The geometrical and electrostatic characteristics of the transition state have been used for the design of transition state analogues.27,28 By converting the forces involved in catalysis into thermodynamic forces, faithful mimics of enzymatic transition states are bound to their cognate enzymes with limits of the rate enhancements imposed by the enzyme. In practice, inhibitors with Kd values as low as 10−14 M have been reported.29–32 Most N-ribosyltransferases are characterized by SN1 reaction mechanisms with ribocationic transition states.23–25,33 Immucillin-H, a first generation transition state analogue of human purine nucleoside phosphorylase (PNP), has a dissociation constant of 58 pM for human PNP and is currently in clinical development.34,35
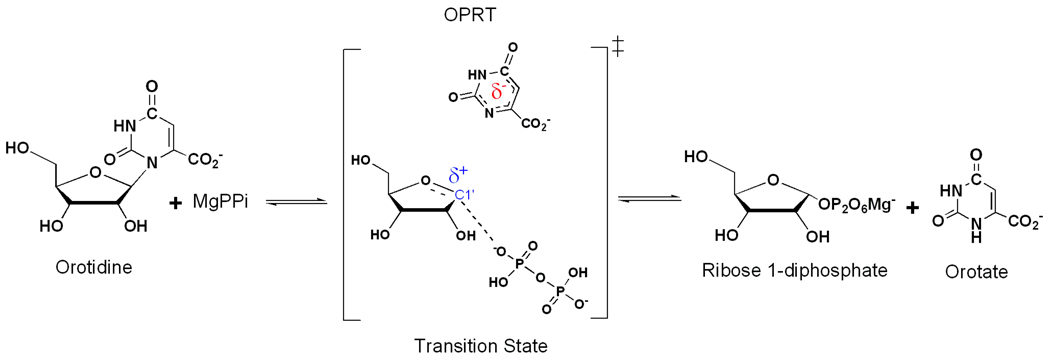
Similar development with PRPP-related phosphoribosyltransferases has been problematic because of suppressed KIE values. Phosphonoacetic acid has been used as a slow substrate analogue of pyrophosphate for OPRT and has been used for transition state analysis of Salmonella typhimurium, Plasmodium falciparum and human OPRT enzymes (Figure 1).36,37 However, replacing PPi with phosphonoacetic acid raises questions about the role of the physiological nucleophile in transition state formation and there are no literature reports of any enzymatic transition state structure with pyrophosphate as the nucleophile, presumably because of slow catalytic site exchange of the Mg-pyrophosphate complex.
Figure 1.
Pyrophosphorolysis of orotidine catalyzed by PfOPRT and HsOPRT. The full characterization of α-D-ribose 1-pyrophosphate is provided in the Supporting Information. This pyrophospho-sugar has not been previously reported or characterized.
The discovery that orotidine is a slow substrate of both PfOPRT and HsOPRT permits intrinsic KIE measurement with pyrophosphate (PPi) and with isotopic labels in the nucleoside (Supporting Information).36 Both enzymes display late associative DN*AN‡ transition states, with dissociation of leaving group followed by nucleophilic attack on the ribocation intermediate.38 Both transition states exhibit fully dissociated orotate, ribooxacarbenium ion character, partial participation of the PPi nucleophile and C2'-endo ribosyl pucker. Having solved this problem, we find that the PfOPRT and HsOPRT transition states with PPi and orotidine are closely related to the previously determined transition states with phosphonoacetic acid and OMP as reactants.36 The transition states for the pyrophosphorolysis reactions catalyzed by PfOPRT and HsOPRT provide two new examples of ribosyltransferase transition states where the transition state occurs beyond the point of ribocation formation with partial nucleophile participation. Similarities in transition state features for OPRTs with chemically distinct nucleophiles support development of transition state features independent of the nucleophile. These transition states provide new insight into the OPRT catalytic mechanism and are anticipated to assist in the design of potent OPRT transition state analogue inhibitors.
Experimental Methods
Reagents and Materials
D-[1-14C]Ribose, D-[1-3H]ribose, D-[5-3H]glucose, D-[6-3H2]glucose 6-phosphate and D-[6-14C]glucose were purchased from American Radiolabeled Chemicals Inc. [1, 3-15N2]Orotic acid and [15N2]urea were purchased from Cambridge Isotope Laboratories. [2-3H]Ribose was prepared as described previously.39 Hexokinase (HK), glucose 6-phosphate dehydrogenase (G6PDH), phosphogluconic acid dehydrogenase (PGDH), L-glutamic acid dehydrogenase (GDH), phosphoriboisomerase (PRI), adenylate kinase (AK) and pyruvate kinase (PK) were purchased from Sigma-Aldrich. Phospho-D-ribosyl-α-1-pyrophosphate synthase (PRPPase) and ribokinase (RK) were prepared as described before.24,40 Alkaline phosphatase (AP) was purchased from Roche Applied Science. PfOPRT and HsOPRT were expressed in E. coli cells and purified.36 All other reagents were purchased from readily available commercial sources and used without further purification. Products of the orotidine pyrophosphorolysis reaction were characterized by NMR and mass spectral analysis (see Supporting Information).
Synthesis of Isotopically Labeled Orotidines
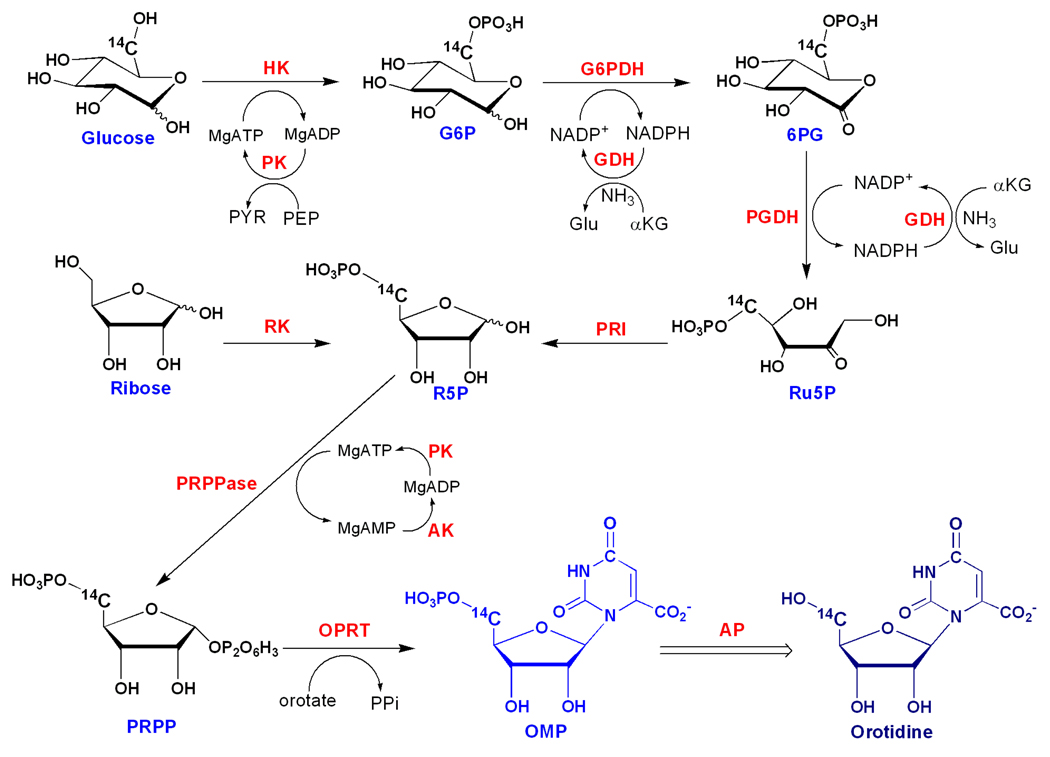
Orotidines were synthesized enzymatically from OMPs, prepared as previously described (Figure 2).36 The isotopically labeled OMPs were treated with alkaline phosphatase (60 units per µmol) (Roche Applied Science) in reaction buffer overnight at room temperature. The resulting orotidines were purified by reverse phase HPLC (C-18 Deltapak column, 20 mM KH2PO4, 8 mM tetrabutylammonium, pH = 6.0 as eluting buffer). The isotopically labeled orotidines were concentrated and re-purified by HPLC, eluted with a solution containing 100 mM acetic acid and 100 mM triethylamine, pH = 6.0. The purified orotidines were lyophilized, dissolved in H2O, and stored at −80 °C for further use. The overall yields of orotidine were 80–90% from starting OMP.
Figure 2.
Enzymatic synthesis of isotopically labeled orotidines. [5'-14C]Orotidine is used as an example. [1'-14C]-, [1'-3H]-, [2'-3H]-, [4'-3H]-, [5'-14C]-, [5'-3H2]-, [1, 3-15N2, 5'-14C]- and [3-15N, 5'-14C]Orotidine were prepared enzymatically from isotopically labeled riboses, glucoses and orotates through two-step procedure (see Experimental Methods for details). The enzymes used for the synthesis include ribokinase (RK), hexokinase (HK), glucose-6-phosphate dehydrogenase (G6PDH), phosphogluconic acid dehydrogenase (PGDH), L-glutamic dehydrogenase (GDH), phosphoriboisomerase (PRI), adenylate kinase (AK), pyruvate kinase (PK), phospho-D-ribosyl-1-pyrophosphate synthase (PRPPase), orotate phosphoribosyltransferase (OPRT) and alkaline phosphatase (AP).
Determination of Kinetic Isotope Effects
Experimental KIEs were measured by analyzing isotope ratios of products from partial and complete reactant conversions while using pairs of isotopically labeled substrates.41,42 Briefly, assays were performed at 25 °C in a total volume of 300 µL containing isotopically labeled orotidines (cpm of 3H:14C = 4:1 and at least 5 × 104 cpm for 14C). The reaction was initiated by adding 2.5 µM PfOPRT or 7.2 µM HsOPRT to a solution with 50 mM Tris-HCl (pH = 8.0), 5 mM MgCl2, 5 mM PPi and 500 µM orotidine (labeled and unlabeled). The reaction mixture was divided into two portions (240 µL and 60 µL). The 240 µL portion was loaded onto a charcoal-cellulose column (activated charcoal and cellulose powder in a 1:4 w/w ratio, mixed with H2O into slurry and settled into disposable Pasteur pipets) at 20–30% conversion to product. The 60 µL portion was converted completely to product (100%) by adding an additional 16.5 µM PfOPRT and incubating for an additional 1 h. Reaction mixture of the 100% conversion was then loaded onto a charcoal-cellulose column (1:4 w/w) pre-equilibrated with H2O. The columns were washed with 1 mL of H2O, followed by elution with 3 mL 3.3% (v/v) NH4OH/H2O (pH = 6.6 with acetic acid). The 3 mL fractions containing the isotopically labeled products were dried by speedvac, dissolved in 200 µL of H2O and mixed with 10 mL scintillation fluid (Ultima Gold). The samples were counted for at least 8 cycles (20 min/cycle, Wallac 1414 LSC, PerkinElmer) to determine the 3H:14C ratios. At least 5 replicates were performed for each KIE measurement.
The ratios of 3H to 14C in products were calculated using equations 1 and 2. The 14C channel parameters were determined by counting a [1-14C]ribose standard to calculate the ratio of counts appearing in channels A and B. The channels were set to keep all 3H counts in channel A and only 14C counts in channel B with a fraction of 14C counts appearing in channel A.
| (1) |
| (2) |
After determining the 3H:14C ratio for the partial and complete conversion, the KIEs were corrected to 0% reaction based on equation 3, where f is the fraction of reactants converted to products and Rf and R0 are the ratios of the heavy isotope to the light isotope at the partial and complete conversion, respectively.
| (3) |
Measurement of Forward Commitment Factors
Isotope trapping was used to measure the forward commitment factors for orotidine pyrophosphorolysis by PfOPRT and HsOPRT.43 Briefly, 60 µM PfOPRT or HsOPRT was pre-incubated with 500 µM [1'-3H]orotidine for 10 s at 25 °C in a total volume of 1.5 µL containing 50 mM Tris-HCl (pH = 8.0) and 5 mM MgCl2. The mixture was then rapidly diluted with 148.5 µL of chase solution containing a large excess of unlabeled orotidine (5 mM) in 50 mM Tris-HCl (pH = 8.0), 5 mM MgCl2 and various concentrations of PPi. After five catalytic turnovers (2 min for PfOPRT and 3.5 min for HsOPRT), the reaction mixtures were loaded onto a charcoal-cellulose column (1:4 w/w) pre-equilibrated with H2O. The column was washed with 1 mL of H2O and isotopically labeled products were eluted with 3 mL 3.3% (v/v) NH4OH/H2O (pH = 6.6 with acetic acid). The radioactivity of products was measured by liquid scintillation counting. Control experiments were performed in the absence of PfOPRT and HsOPRT to give background corrections. The forward commitment factors were calculated from the fractions of enzyme-bound orotidine converted to product in the presence of excess unlabeled orotidine.
Computational Modeling of the Transition State Structures
The transition state structures were calculated in vacuo using hybrid density functional theory implemented in Gaussian 03.44 At the B3LYP/6-31G (d,p) level, geometry optimizations were performed for the structures of orotidine and PPi substrates and transition state candidates which include the ribocation with partially or fully dissociated orotate in different ionic states and varied extents of PPi nucleophilic participation. Bond frequencies of the optimized structures were calculated at the same level of theory and basis set. KIEs were calculated from the computed frequencies of substrate and transition states at 298 K using ISOEFF98.45 All 3N - 6 vibrational modes were used to calculate the KIEs. The frequencies of substrate, transition states, and reaction-coordinate imaginary frequencies of 52.3i cm−1 (PfOPRT) and 52.1i cm−1 (HsOPRT) were used as inputs. On the basis of experimental intrinsic KIEs, the transition state structures were iteratively altered to generate models with the best matches.
Calculation of Molecular Electrostatic Potential Surfaces
The CUBE subprogram from Gaussian 03 was used to generate the molecular electrostatic potential (MEP) surfaces of transition state structures. The formatted checkpoint files from the geometry optimization processes were used as inputs. The MEP surfaces were visualized at an isovalue of 0.068 using GaussView 3.09.44
Results and Discussion
Experimental KIEs and Commitment to Catalysis
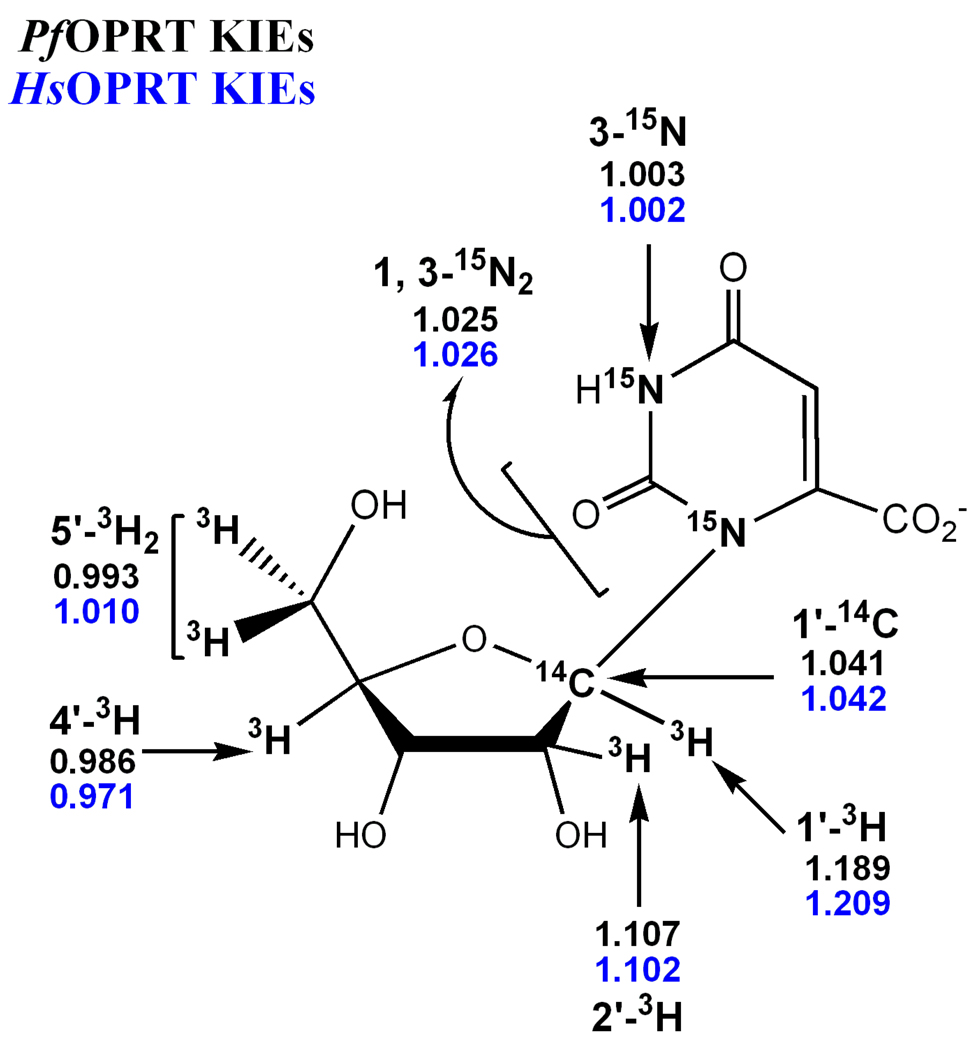
The experimental KIEs were measured using 3H and 14C internal competition methods. The [5'-14C] KIE is assumed to be unity, since the 5'-14C of orotidine is four bonds away from the center of reaction and 14C atoms have been shown to give negligible binding isotope effects.46 The [1'-3H]-, [2'-3H]-, [4'-3H]- and [5'-3H2]orotidine KIEs were measured using [5'-14C]orotidine as the remote label. [4'-3H]Orotidine was used as the remote label for the KIEs with [1'-14C]-, [1, 3-15N2]- and [3-15N]orotidine. The experimental KIEs were obtained by correcting for the remote [4'-3H]orotidine KIE.
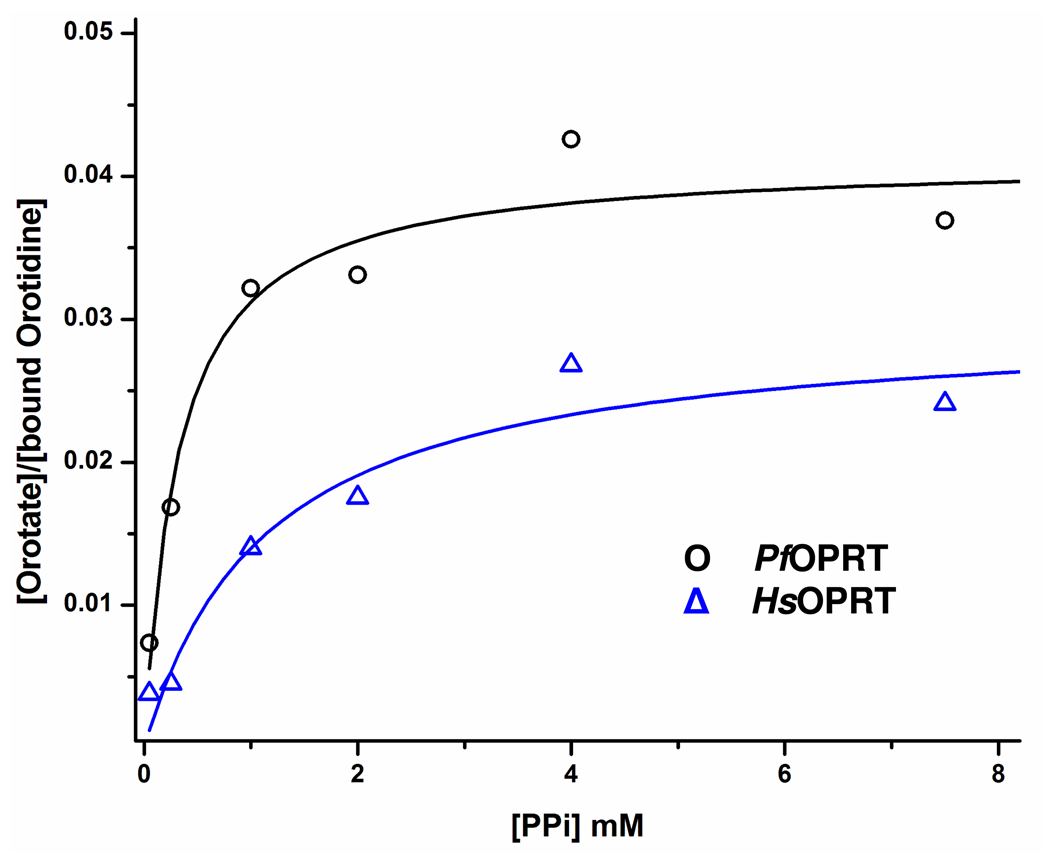
The intrinsic KIEs, reflecting the bond-breaking step, were obtained by correcting the experimental KIEs for commitment factors. The forward commitment factors for PfOPRT and HsOPRT were determined to be 0.043 and 0.031, respectively (Figure 3). By applying the forward commitment factors to the equation x(V/K) = (xk + Cf)/(1 + Cf),47 where x(V/K) is the experimental KIE, xk is the intrinsic KIE and Cf is the forward commitment factor, the intrinsic KIEs are within experimental error of the experimental KIEs. The negligible forward commitments result from the substrate properties of orotidine. With Km values of 96 µM for PfOPRT and 91 µM for HsOPRT and kcat values of 0.042 s−1 for PfOPRT and 0.024 s−1 for HsOPRT, the kcat/Km values are 4.4 × 102 M−1 s−1 and 2.6 × 102 M−1 s−1 for PfOPRT and HsOPRT, respectively. Weaker nucleoside binding and a higher transition state barrier than with OMP (Km: 3.7 µM for PfOPRT and 1.5 µM for HsOPRT) reduces the forward commitments normally observed for ribosyl pyrophosphorolysis reactions. The large [1'-3H]- and [1, 3-15N2]orotidine KIEs are near theoretical maxima for these isotopic substitutions (Table 1) and therefore support the proposal that the KIEs are not obscured by reverse commitment factors, defined as the probability of bound products reforming substrate relative to product release.
Figure 3.
Measurements of forward commitment factors of PfOPRT and HsOPRT. [1'-3H]Orotidine was pre-incubated with OPRTs. The mixture was diluted with a large excess of cold orotidine and various concentrations of PPi. The fractions of formed product from bound substrate were plotted versus the corresponding PPi concentrations. The forward commitment factors (Cf) were calculated from the equation
, where Ymax is the y axial intercept upon extrapolating the plateau region. The values of Cf are 0.043 ± 0.002 for PfOPRT ( ) and 0.031 ± 0.004 for HsOPRT (
) and 0.031 ± 0.004 for HsOPRT ( ).
).
Table 1.
| PfOPRT | HsOPRT | ||||
|---|---|---|---|---|---|
| Orotidine pairs | Type of KIEs | Intrinsic | Calcd. | Intrinsic | Calcd. |
| [1'-14C] vs [4'-3H] | primary | 1.041 ± 0.007 | 1.041 | 1.042 ± 0.008 | 1.042 |
| [1, 3-15N2, 5'-14C] vs [4'-3H] | primary | 1.025 ± 0.008 | 1.026 | 1.026 ± 0.006 | 1.027 |
| [3-15N, 5' 14C] vs [4'-3H] | β-secondary | 1.003 ± 0.008 | 1.004 | 1.002 ± 0.006 | 1.004 |
| [1'-3H] vs [5'-14C] | α-secondary | 1.189 ± 0.006 | 1.405 | 1.209 ± 0.004 | 1.411 |
| [2'-3H] vs [5'-14C] | β-secondary | 1.107 ± 0.004 | 1.111 | 1.102 ± 0.005 | 1.113 |
| [4'-3H] vs [5'-14C] | γ-secondary | 0.986 ± 0.005 | 0.973 | 0.971 ± 0.003 | 0.967 |
| [5'-3H2] vs [5'-14C] | δ-secondary | 0.993 ± 0.005 | 1.002 | 1.010 ± 0.005 | 1.016 |
Intrinsic KIEs were obtained after correcting for the remote [4'-3H] KIEs and the forward commitment factors. At least 5 measurements were made for each experimental KIE. Due to the small commitment factors, experimental KIEs are the intrinsic KIEs.
Computationally matched KIEs were established by comparing the calculated KIEs from the structures of transition state candidates with the intrinsic KIEs. The calculated KIEs were determined from the bond frequencies of the substrate and the transition states using the ISOEFF98 program.
Computation of Transition States for PfOPRT and HsOPRT
The transition state structures of PfOPRT and HsOPRT were determined computationally in vacuo with the experimental intrinsic KIEs as boundary conditions. All calculations were performed at the B3LYP/6-31G (d,p) level of theory. The transition state structures were modeled by matching the calculated and experimental intrinsic KIEs (Figure 4). The computational KIEs were obtained from the vibrational differences between reactants and the transition state. Optimized orotidine with the lowest zero-point energy was selected as the reactant geometry for the KIEs calculation. It represents the ground state of free substrate in solution. The experimental KIEs for [1'-3H]orotidine of approximately 1.2 and that for [1'-14C]orotidine near 1.04 are characteristic of SN1 transition states with well-developed ribocations but with partial bonding to leaving group or attacking nucleophile. SN2 transition states were ruled out on the basis of unmatched intrinsic and calculated [1'-14C] and [1'-3H] KIEs (see below). Consequently, the transition state calculations were initiated with the ribooxacarbenium ion with varied degrees of nucleophilic participation by orotate and/or PPi. A dianionic PPi nucleophile was assumed, considering the experimental pH and the Mg2+ coordination at the active site. Bond distances of C1'-N1 and C1'-OPPi were varied systematically between 1.9 Å and 2.8 Å to match the experimental intrinsic KIEs. Structures with a C1'-N1 distance of 2.8 Å and C1'-OPPi distances of 2.34 Å gave the computational KIEs of [1'-14C], [1, 3-15N2], [1'-3H] and [2'-3H] consistent with the experimental intrinsic KIEs. Optimization of the geometries at 5'-hydroxyl group provided structures with optimal matches between the [4'-3H] and [5'-3H2] intrinsic KIEs.
Figure 4.
Intrinsic KIEs for orotidine pyrophosphorolysis catalyzed by PfOPRT and HsOPRT. The orotidine structure is shown with a summary of KIEs indicated.
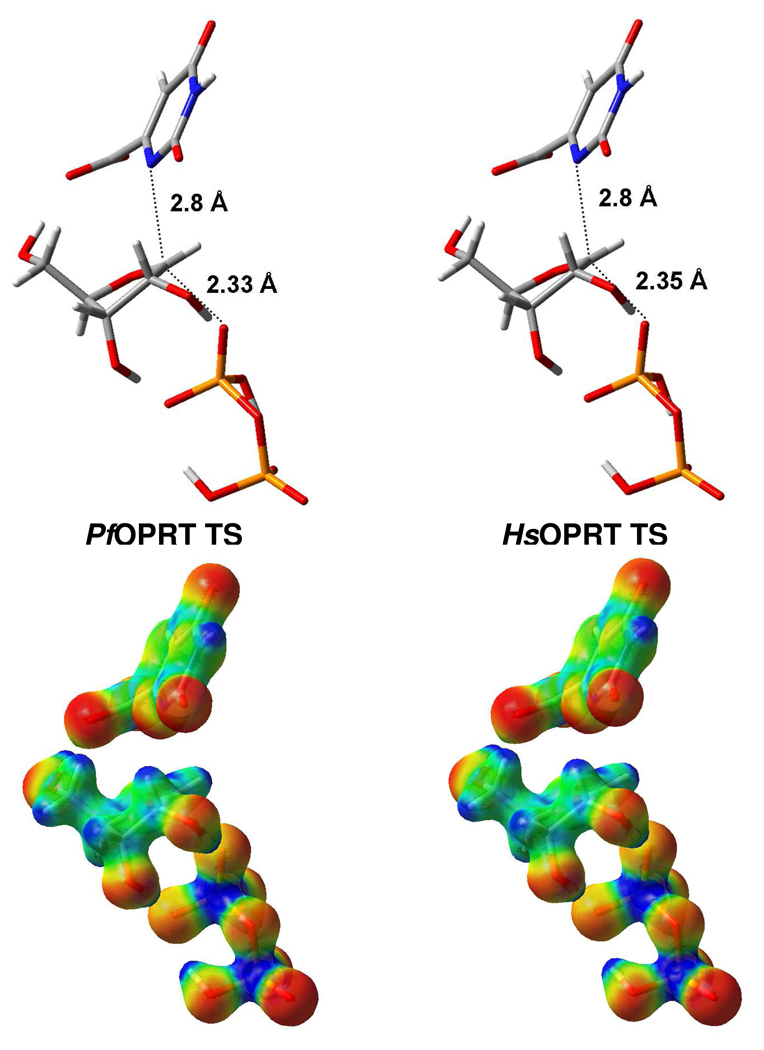
PfOPRT and HsOPRT display late associative DN*AN‡ characteristics (Figure 5). The ribocations at the transition states exhibit full loss of C1'-N1 bond order and weak nucleophilic participation by the PPi nucleophiles. The fully dissociated orotate is 2.8 Å away from the anomeric carbon (C1'-N1 bond order: 0.01). Nucleophilic attack by PPi on the ribooxacarbenium ion gives a C1'-OPPi bond distance of 2.33 Å for the PfOPRT transition state and 2.35 Å for the HsOPRT transition state (C1'-OPPi bond order: 0.05). The ribosyl rings in these transition states adopt C2'-endo conformations with O4'-C1'-C2'-C3' dihedral angles around 16.5° at the transition states to give calculated [2'-3H]orotidine KIEs of 11% consistent with the experimental KIEs. The ribosyl geometries at the 5'-region are also similar with a C3'-C4'-C5'-O5' dihedral angle of −171° at the PfOPRT transition state and −175° at the HsOPRT transition state. This small difference in geometry variation at the 5’-hydroxyl group gives rise to slight differences in the [4'-3H]- and [5'-3H2]orotidine KIEs for PfOPRT and HsOPRT. These differences are proposed to reflect distinct bond distortions imposed by PfOPRT and HsOPRT, since sequence homology from the crystal structure of S. typhimurium OPRT suggests distinct contacts (see below).
Figure 5.
Structures and molecular electrostatic potential (MEP) surfaces of PfOPRT and HsOPRT transition state. The distances of C1'–N1 and C1'–OPPi in the reaction coordinate are shown in the structures of PfOPRT and HsOPRT transition state. The transition state structures were calculated in vacuo using Gaussian 03 at the B3LYP/6-31G (d,p) level of theory. The MEP surfaces of PfOPRT and HsOPRT transition states were generated by CUBE subprogram from Gaussian 03 and visualized using GaussView 3.09 at an isovalue of 0.068. Electron rich and deficient are represented by red and blue, respectively.
[1'-14C]- and [1, 3-15N2]Orotine Primary KIEs
The [1'-14C]orotidine primary KIE is the most useful parameter to establish nucleophilic substitution at the transition state (SN1 or SN2) of N-ribosyltransferases. It reports on the interactions of the anomeric carbon with adjacent nucleophiles and is sensitive to motion in the reaction coordinate. A [1'-14C] KIE of 1.01–1.08 indicates a transition state with weak to partial participation of nucleophiles at the transition state.48,49 A unity or slightly inverse [1'-14C] KIE indicates a fully dissociated transition state structure with fully developed ribocation and no nucleophilic participation. A [1'-14C] KIE above 1.08 indicates SN2 character at the transition state.24,48
The intrinsic KIE of 1.041 for [1'-14C]orotidine with PfOPRT and 1.042 with HsOPRT, indicates SN1 transition states with significant bond order to the leaving group (early dissociative) or the attacking nucleophile (late dissociative). The C1'-N1 and C1'-OPPi bond distances were altered systematically to search for matches in [1'-14C] KIEs. SN2 transition states were ruled out due to the large calculated [1'-14C] KIEs (>1.08). Early dissociative transition state candidates were excluded on the basis of the [1, 3-15N2]- and [2'-3H]orotidine KIE values. Transition states matching both the primary [1'-14C]- and [1, 3-15N2]orotidine and the secondary [1'-3H]- and [2'-3H]orotidine KIEs are characterized by late dissociative transition states with weak nucleophilic participation. The C1'-OPPi bond distance is 2.33 Å for PfOPRT and 2.35 Å for HsOPRT transition states. The calculated [1'-14C] and [1'-3H] KIEs with these distances show optimal agreement with the intrinsic KIEs. The partial bond orders of C1'-OPPi from PPi nucleophilic participations give rise to the modest intrinsic [1'-14C]orotidine KIEs of 1.041 and 1.042. The large [1'-3H]orotidine KIEs of 1.189 and 1.209 arise from the well-developed ribocations at these transition states.
The [1, 3-15N2]- and [3-15N]orotidine KIEs reflect the extent of orotate dissociation at the transition state and the nature of the enzymatic interactions with orotate. For example, when unprotonated orotate is the leaving group, the calculated [1, 3-15N2]orotidine KIE increases with the C1'-N1 bond distance to a maximum of 1.027, with a 1.022 KIE contribution from [1-15N]orotidine and a 1.005 KIE contribution from [3-15N]orotidine. Intrinsic KIEs from [1, 3-15N2]- and [3-15N]orotidine were 1.025 and 1.003 for PfOPRT and 1.026 and 1.002 for HsOPRT, consistent with transition states characterized by fully dissociated dianionic orotate. The C1'-N1 bond distances of 2.8 Å for PfOPRT and HsOPRT transition states matched the [1, 3-15N2]- and [3-15N]orotidine KIEs. Protonated orotates as leaving groups were ruled out as these gave calculated [1, 3-15N2]- and [3-15N]orotidine KIEs inconsistent with the intrinsic KIEs. Thus, PfOPRT and HsOPRT show late dissociative transition states with full loss of C1'-N1 bond order to dianionic orotate and weak PPi nucleophilic participation. Matching the calculated and experimental primary KIEs of [1'-14C]- and [1, 3-15N2]orotidine, the bond distances of C1'-N1 and C1'-OPPi at transition states were determined and this transition state is additionally supported by the secondary [1'-3H]- and [2'-3H]orotidine KIEs (see below).
Full dissociation of dianionic orotate at the transition states of PfOPRT and HsOPRT distinguishes these transition states from the N-ribosyltransferase transition states where the leaving group is protonated to form a neutral species.24,25,33 Although dianionic orotate is implicated from KIEs, it is certain to be stabilized by multiple hydrogen bond and ionic interactions at the active site. In crystal structures of OPRT homologues, interaction networks involving bound nucleotide, ion-pair formation to the OMP carboxylate, hydrogen bonds to both carbonyl oxygens and to N3 or the pyrimidine are conserved at these catalytic sites.50,51 Candidates for leaving group activation, based on the S. cerevisiae crystal structure, include the hydroxyl of Thr135 and backbone NH of Lys29 interacting with the 6-carboxyl group, the cationic NH2 of Arg163, the backbone NH of Phe38 and two water molecules in favorable H-bonds to the 2- and 4-carbonyl oxygens and a backbone C=O of Phe38 hydrogen bonded to N3 of the pyrimidine.
Along the reaction coordinate, bound OMP is sufficiently activated through these interactions to permit formation of the transition state with relatively weak participation of the PPi nucleophile. This transition state can be contrasted with the earlier transition states found in the purine nucleoside hydrolases and phosphorylases where the leaving group is more strongly activated by protonation and transition states are often found earlier in the reaction coordinate.28,33,50
[1'-3H]Orotidine α-Secondary KIEs
The [1'-3H]- and [2'-3H]orotidine KIEs result from C1' rehybridization from sp3 toward sp2 following C1'-N1 bond dissociation at the transition state. The [1'-3H] KIE arises mainly from the out-of-plane bending mode of C1'-H1' due to increased steric freedom. This isotope effect is reduced by restricted stretching modes but this effect is subordinate to the out-of-plane mode. The [1'-3H]orotidine KIE is sensitive to reaction coordinate motion. Small [1'-3H]orotidine KIEs indicate SN2-like transition states. Large [1'-3H]orotidine KIEs are characteristic of fully or partly dissociative transition states. The magnitude of the [1'-3H]orotidine KIE is dependent on the bond order of C1'-N1 and C1'-OPPi at the transition state. The experimental intrinsic KIE of [1'-3H]orotidine is 1.189 for PfOPRT and 1.209 for HsOPRT. Calculations for SN2 transition state candidates gave [1'-3H]orotidine KIEs far below these intrinsic values. Combined with the [1'-14C]- and [1, 3-15N2]orotidine KIEs, the large [1'-3H]orotidine KIEs support ribocationic transition states with fully dissociated orotate and weak PPi nucleophilic participation. With full loss of the C1'-N1 bond and weak bonding to PPi, the out-of-plane bending modes of the ribocation dominate the observed intrinsic [1'-3H]orotidine KIEs. The calculated [1'-3H]orotidine KIEs of 1.405 for PfOPRT and 1.411 for HsOPRT are significantly larger than the experimental values. However, this overestimation of α-secondary [1'-3H] KIEs from in vacuo calculation has been well documented.25
The [1'-3H]orotidine secondary KIE is sensitive to non-covalent and remote interactions.48 Catalytic site contacts stabilizing the ribocationic transition states are implicated to hinder the out-of-plane bending motion of C1'-H1' bond, causing smaller than calculated [1'-3H] KIEs. Compared to the [1'-3H] KIEs from in vacuo calculations, the smaller intrinsic [1'-3H] KIEs suggest a more constrained environment near the 1’-H at the OPRT transition states. The lack of structural information for catalytic site contacts and dynamic excursions associated with barrier passage prevents accurate modeling of the calculated and experimental [1'-3H] KIEs. Computational overestimation of [1'-3H] KIEs can also result from distorted C1'-H1' geometry in the orotidine reactant, giving a larger difference in the out-of-plane bending modes between reactant and transition state. The anionic 6-carboxyl group of orotidine causes calculations of the reactant state to overestimate intramolecular interactions between the 6-carboxylate and 2'-hydroxyl groups, resulting in distorted geometry for adjacent the C1'-H1'. Fortunately, transition state features in the OPRTs are over-determined with the intrinsic KIE sets reported here and are therefore not dependent on exact matching of the [1'-3H] KIEs. However, the measured [1'-3H] orotidine KIEs are consistent with other [1'-3H] KIEs in transition states known to have ribooxacarbenium ion character.28,49
[2'-3H]Orotidine β-Secondary KIEs
The β-secondary [2'-3H]orotidine KIE originates from the hyperconjugation of a σ bond (C2'-H2') to the partially vacant 2pz orbital of the anomeric carbon at the transition state. Its magnitude depends on the occupancy of the 2pz orbital and the orbital overlap of the 2pz orbital with the C2'-H2' σ bond. The occupancy of the 2pz orbital is dependent on the extent of leaving group dissociation and the nucleophilic participation. The degree of orbital overlap between the 2pz orbital and the C2'-H2' σ bond is established by the ribose pucker. Thus, the [2'-3H]orotidine KIE includes information from leaving group dissociation, nucleophile association and ribosyl geometry at the transition state. The intrinsic [2'-3H]orotidine KIE is 1.107 for PfOPRT and 1.102 for HsOPRT, consistent with C2'-endo ribosyl pucker at the PfOPRT and HsOPRT transition states. Early dissociative transition state structures gave rise to smaller [2'-3H]orotidine KIEs inconsistent with the intrinsic KIE values. The late dissociative transition states of PfOPRT and HsOPRT adopt C2'-endo geometries and matched the [2'-3H]orotidine KIEs. At both transition states, the H2'-C2'-C1'-H1' dihedral angles of 76° contribute strongly to the C2'-H2'→C1'-2pz hyperconjugation giving rise to the [2'-3H]orotidine KIEs. The experimental [2'-3H]orotidine KIEs support the C1'-N1 and C1'-OPPi bond distances of the transition states derived from the primary [1'-14C]- and [1, 3-15N2]orotidine KIEs. Structures with C2'-exo conformations showed smaller [2'-3H]orotidine outside experimental KIEs because of compromised hyperconjugation.
[4'-3H]- and [5'-3H2]Orotidine Remote KIEs
The [4'-3H]orotidine KIE results from the hyperconjugation of the σ * (C4'-H4') antibonding orbital to the O4' lone pair (np). At the ribocationic transition state, altered hyperconjugation from the O4' gives an inverse [4'-3H]orotidine KIE. Some N-ribosyltransferase transition states show polarization of the 3'-OH to give normal KIEs.52 PfOPRT and HsOPRT have slightly inverse [4'-3H] KIEs of 0.986 and 0.971, respectively, indicating little polarization of the 3'-OH and a dominant O4’ effect at these transition states. These late dissociative transition states give calculated [4'-3H]orotidine KIEs of 0.973 for PfOPRT and 0.967 for HsOPRT, consistent with the intrinsic [4'-3H] KIEs. The C4'-H4' bond distances decrease from 1.099 Å at the reactant state to 1.090 Å at the transition states to generate these isotope effects (Figure 6).
Figure 6.
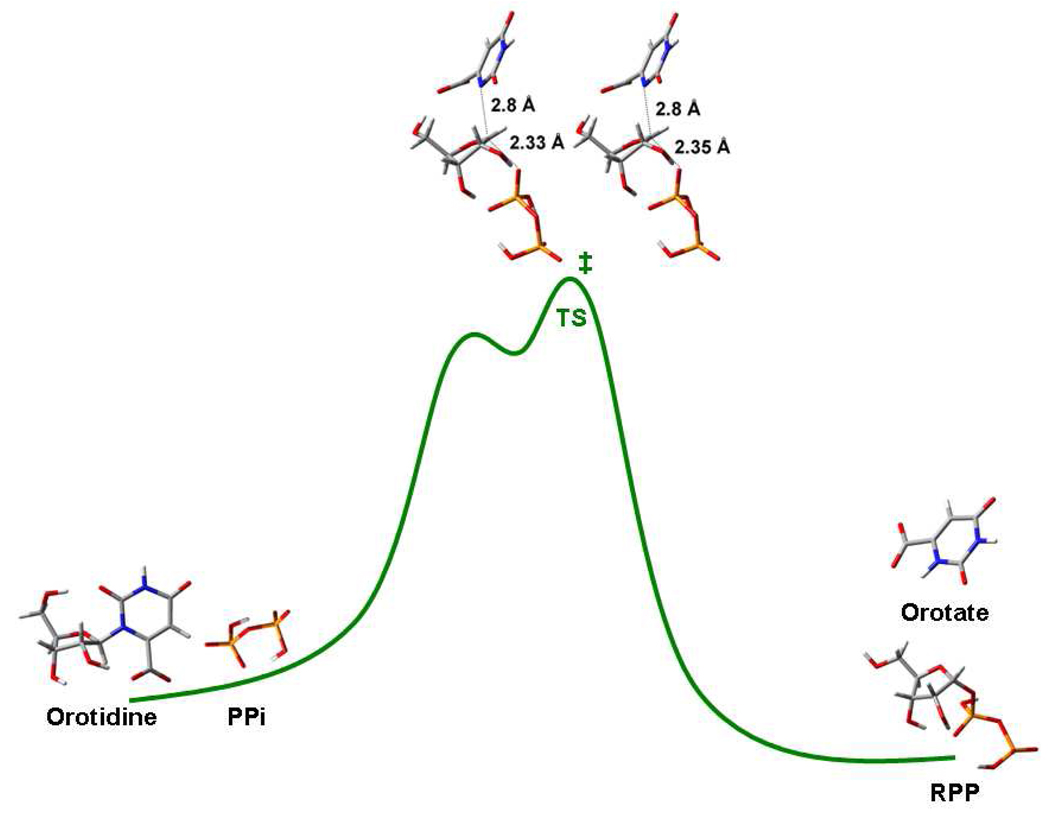
Reaction coordinate of pyrophosphorolysis of orotidine by PfOPRT and HsOPRT. The relative energy along the reaction coordinate is arbitrary with the highest energetic point as the transition state. The first reaction barrier represents the dissociation of dianionic orotate. The first energetic local minimum represents the fully developed ribocation expected to form prior to the weak attack of the nucleophile to form the transition state. Ribocations are not expected to be intermediates with significant lifetimes.53 However, intermediacy for the ribocation is neither proved or excluded by the present data. The structures of reactants (orotidine and PPi), products (orotate and RPP) and transition states are shown in stick mode and were optimized at the same level of theory and basis set.
The [5'-3H2]orotidine KIE reflects the 5’-hydroxyl group bond distortion imposed by the catalytic site and includes information from binding isotope effects and from the transition state.46 PfOPRT and HsOPRT have intrinsic [5'-3H2] KIEs of 0.993 and 1.010, respectively. The varied [5'-3H2] KIEs indicate geometry difference at the remote regions of the transition states. By restricting the O4'-C4'-C5'-O5' dihedral angle of 72.6° for PfOPRT and 68.8° for HsOPRT, the transition state structures were optimized to match the [5'-3H2] KIEs. Differences in the [4'-3H]- and [5'-3H]orotidine KIE values are proposed to arise from distinct catalytic site contacts. The crystal structure for S. typhimirium OPRT shows the carbonyl oxygen of Val126 and OH of Thr128 as the nearest 5'-OMP contacts. Sequence alignments of human and PfOPRTs indicate Val125 and Thr127 in the human sequence and Lys191 and Thr193 in the PfOPRT sequence as predicted contacts in the 5’-region, thus accounting for different remote isotope effects.
Transition State Analysis with Substrate Analogues
The transition state structures of PfOPRT and HsOPRT solved with phosphonoacetic acid (PA) and isotopically labeled OMPs gave C1'-OPA bond distances of 2.14 to 2.15 Å.36 The C1'-OPPi bond distances of approximately 2.34 Å at the transition states indicate an earlier transition state with PPi. The variation in nucleophilic participation and the slow catalytic rate with PA established different nucleophilic efficiency at the transition states. The pKa values for PA are 2.0, 5.1 and 8.7, while those for PPi are 0.91, 2.10, 6.7 and 9.32. After formation of a ribocation in the reaction coordinate, enzyme-bound PPi forms an earlier transition state than does the PA nucleophile.
Transition state structures for OPRTs determined with the PA + OMP or PPi + orotidine are similar in regard to the extent of orotate dissociation, ribosyl geometry, and the nature of ribooxacarbenium ion at the transition states. KIEs measurements using orotidine and PPi indicate dianionic orotates at the OPRT transition states, the same as found for the transition states with OMP and PA.36 Thus, protonation of oratate prior to the transition state is not a feature of OPRT and suggests a reaction coordinate with activation of the leaving group by multiple hydrogen bond interactions. The extent of nucleophilic participation differs with PPi favoring earlier transition states than PA. This difference is small, and it can be concluded that these substrate analogues give rise to similar late dissociative transition states. KIE measurements with OMP and PA showed almost same [4'-3H]- and [5'-3H2] KIEs for PfOPRT and HsOPRT. With orotidine and PPi, the differences are seen. Comparing transition states with OMP and orotidine, remote KIEs for PfOPRT and HsOPRT are the same with the 5'-phosphate but differ for the nucleoside. Without the 5'-phosphate, geometric constraints at the 5'-region are relaxed to permit distinct result in the [4'-3H] and [5'-3H2] KIEs. The similarity of all four transition states suggest but do not establish that PfOPRT and HsOPRT may also have similar transition states with the native substrates (OMP and PPi). Similar transition states with different substrate analogues imply that the determinants of the nature of enzymatic transition states include catalytic site structure and geometric constraints imposed by the catalytic site.
This is the first report of a phosphoribosyltransferase transition state with PPi as the nucleophile. Natural bond analysis of the nucleophilic oxygens of pyrophosphate (−3) and phosphonoacetic acid (−2) shows natural charges of −1.232 and −1.189 respectively. Thus the earlier transition states found with PPi are consistent with its increased nucleophilicity. Attempts to measure KIEs for PfOPRT and HsOPRT using PPi and isotopically labeled OMPs under several conditions revealed small KIE consistent with high forward and/or internal commitment to catalysis. Slowly reacting substrate analogues permit transition state analysis of PfOPRT and HsOPR and their similarity are encouraging for the interpretation of the natural transition state structure. Transition states solved with orotidine and PPi provide a new approach for the phosphoribosyltransferases, otherwise intractable for transition state analysis.
Conclusion
The PfOPRT and HsOPRT transition state structures were determined through quantum chemistry computation with experimental intrinsic KIEs as constraints. The PfOPRT and HsOPRT transition states are characterized by well-developed ribocations, full dissociation of orotate, partial participation of the PPi nucleophile and C2'-endo ribosyl puckers. This is the first report of phosphoribosyltransferase transition states with the PPi nucleophile. These transition states are similar to the previously solved transition states with the PA and OMP. The extent of nucleophilic participation is the only significant difference in the bond-breaking reaction coordinate and is in agreement with oxygen nucleophilicity. Similar transition states with different substrate analogues suggest that OPRTs may have a similar transition states with its native substrates. Similar transition states with different slow substrates suggests that the nature of the transition states is determined by the catalytic site architecture.
Supplementary Material
Acknowledgement
We thank Dr. Jennifer S. Hirschi (Department of Biochemistry, Albert Einstein College of Medicine) for help on transition state calculations, Dr. Sean M. Cahill (Department of Biochemistry, Albert Einstein College of Medicine) for NMR measurement and Dr. Hui Xiao (The Laboratory for Macromolecular Analysis & Proteomics, Albert Einstein College of Medicine) for the mass spectra measurements. We also thank Prof. Jiali Gao of the University of Minnesota Supercomputer Institute for computer time. This work was supported by grants from the NIH (AI049512 and GM041916).
Footnotes
Supporting Information Available: NMR and mass spectral results, full author list of Ref. 44 and complete calculation results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Guerin PJ, Olliaro P, Nosten F, Druilhe P, Laxminarayan R, Binka F, Kilama WL, Ford N, White NJ. Lancet. Infect. Dis. 2002;2:564–573. doi: 10.1016/s1473-3099(02)00372-9. [DOI] [PubMed] [Google Scholar]
- 2.Jones ME. Annu. Rev. Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 3.Reyes P, Rathod PK, Sanchez DJ, Mrema JE, Rieckmann KH, Heidrich HG. Mol. Biochem. Parasitol. 1982;5:275–290. doi: 10.1016/0166-6851(82)90035-4. [DOI] [PubMed] [Google Scholar]
- 4.Gero AM, O'Sullivan WJ. Blood Cells. 1990;16:467–484. [PubMed] [Google Scholar]
- 5.Baldwin J, Michnoff CH, Malmquist NA, White J, Roth MG, Rathod PK, Phillips MA. J. Biol. Chem. 2005;280:21847–21853. doi: 10.1074/jbc.M501100200. [DOI] [PubMed] [Google Scholar]
- 6.Krungkrai SR, DelFraino BJ, Smiley JA, Prapunwattana P, Mitamura T, Horii T, Krungkrai J. Biochemistry. 2005;44:1643–1652. doi: 10.1021/bi048439h. [DOI] [PubMed] [Google Scholar]
- 7.Queen SA, Jagt DL, Reyes P. Antimicrob. Agents Chemother. 1990;34:1393–1398. doi: 10.1128/aac.34.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott HV, Gero AM, O'Sullivan WJ. Mol. Biochem. Parasitol. 1986;18:3–15. doi: 10.1016/0166-6851(86)90045-9. [DOI] [PubMed] [Google Scholar]
- 9.Rathod PK, Khatri A, Hubbert T, Milhous WK. Antimicrob. Agents Chemother. 1989;33:1090–1094. doi: 10.1128/aac.33.7.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krungkrai SR, Aoki S, Palacpac NM, Sato D, Mitamura T, Krungkrai J, Horii T. Mol. Biochem. Parasitol. 2004;134:245–255. doi: 10.1016/j.molbiopara.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Seymour KK, Lyons SD, Phillips L, Rieckmann KH, Christopherson RI. Biochemistry. 1994;33:5268–5274. doi: 10.1021/bi00183a033. [DOI] [PubMed] [Google Scholar]
- 12.Phillips MA, Gujjar R, Malmquist NA, White J, El Mazouni F, Baldwin J, Rathod PK. J. Med. Chem. 2008;51:3649–3653. doi: 10.1021/jm8001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krungkrai J, Krungkrai SR, Phakanont K. Biochem. Pharmacol. 1992;43:1295–1301. doi: 10.1016/0006-2952(92)90506-e. [DOI] [PubMed] [Google Scholar]
- 14.Krungkrai J. Biochim. Biophys. Acta. 1995;1243:351–360. doi: 10.1016/0304-4165(94)00158-t. [DOI] [PubMed] [Google Scholar]
- 15.Quemeneur L, Beloeil L, Michallet MC, Angelov G, Tomkowiak M, Revillard JP, Marvel J. J. Immunol. 2004;173:4945–4952. doi: 10.4049/jimmunol.173.8.4945. [DOI] [PubMed] [Google Scholar]
- 16.Chu E, Callender MA, Farrell MP, Schmitz JC. Cancer Chemother. Pharmacol. 2003;52 Suppl 1:80–89. doi: 10.1007/s00280-003-0625-9. [DOI] [PubMed] [Google Scholar]
- 17.Allison AC. Immunopharmacology. 2000;47:63–83. doi: 10.1016/s0162-3109(00)00186-7. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann ML, Schleyerbach R, Kirschbaum B. J. Immunopharmacology. 2000;47:273–289. doi: 10.1016/s0162-3109(00)00191-0. [DOI] [PubMed] [Google Scholar]
- 19.Christopherson RI, Lyons SD, Wilson PK. Acc. Chem. Res. 2002;35:961–971. doi: 10.1021/ar0000509. [DOI] [PubMed] [Google Scholar]
- 20.Weber G. Biochemistry (Mosc) 2001;66:1164–1173. doi: 10.1023/a:1012493232344. [DOI] [PubMed] [Google Scholar]
- 21.Schramm VL. Meth. Enzymol. 1999;308:301–355. doi: 10.1016/s0076-6879(99)08015-5. [DOI] [PubMed] [Google Scholar]
- 22.Cleland WW. Arch. Biochem. Biophys. 2005;433:2–12. doi: 10.1016/j.abb.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 23.Singh V, Schramm VL. J. Am. Chem. Soc. 2006;128:14691–14696. doi: 10.1021/ja065419p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 25.Matsson O, Westaway KC. Adv. Phys. Org. Chem. 1998;31:143–248. [Google Scholar]
- 26.Schramm VL. Arch. Biochem. Biophys. 2005;433:13–26. doi: 10.1016/j.abb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 27.Schramm VL. Curr. Opin. Struct. Biol. 2005;15:604–613. doi: 10.1016/j.sbi.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Schramm VL. J. Biol. Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 29.Wolfenden R, Snider MJ. Acc. Chem. Res. 2001;34:938–945. doi: 10.1021/ar000058i. [DOI] [PubMed] [Google Scholar]
- 30.Wolfenden R. Nature. 1969;223:704–705. doi: 10.1038/223704a0. [DOI] [PubMed] [Google Scholar]
- 31.Schramm VL. Nucleic Acids Res. Suppl. 2003:107–108. doi: 10.1093/nass/3.1.107. [DOI] [PubMed] [Google Scholar]
- 32.Schramm VL. Acc. Chem. Res. 2003;36:588–596. doi: 10.1021/ar0200495. [DOI] [PubMed] [Google Scholar]
- 33.Singh V, Luo M, Brown RL, Norris GE, Schramm VL. J. Am. Chem. Soc. 2007;129:13831–13833. doi: 10.1021/ja0754204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murkin AS, Birck MR, Rinaldo-Matthis A, Shi W, Taylor EA, Almo SC, Schramm VL. Biochemistry. 2007;46:5038–5049. doi: 10.1021/bi700147b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.BioCryst Pharmaceuticals; Inc.; http://www.biocryst.com. [Google Scholar]
- 36.Zhang Y, Luo M, Schramm VL. J. Am. Chem. Soc. 2009;131:4685–4694. doi: 10.1021/ja808346y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao W, Grubmeyer C, Blanchard JS. Biochemistry. 1996;35:14–21. doi: 10.1021/bi951898l. [DOI] [PubMed] [Google Scholar]
- 38.Guthrie RD, Jencks WP. Acc. Chem. Res. 1989;22:343–349. [Google Scholar]
- 39.Rising KA, Schramm VL. J. Am. Chem. Soc. 1994;116:6531–6536. [Google Scholar]
- 40.Parkin DW, Leung HB, Schramm VL. J. Biol. Chem. 1984;259:9411–9417. [PubMed] [Google Scholar]
- 41.Luo M, Singh V, Taylor EA, Schramm VL. J. Am. Chem. Soc. 2007;129:8008–8017. doi: 10.1021/ja072122y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo M, Schramm VL. J. Am. Chem. Soc. 2008;130:2649–2655. doi: 10.1021/ja078008x. [DOI] [PubMed] [Google Scholar]
- 43.Rose IA. Meth. Enzymol. 1980;64:47–59. doi: 10.1016/s0076-6879(80)64004-x. [DOI] [PubMed] [Google Scholar]
- 44.Frisch MJ, et al. Wallingford, CT: Gaussian, Inc.; 2004. [Google Scholar]
- 45.Anisimov V, Paneth P. J. Math. Chem. 1999;26:75–86. [Google Scholar]
- 46.Lewis BE, Schramm VL. In: Isotope Effects in Chemistry and Biology. Kohen A, Limbach H-H, editors. CRC Press; 2006. pp. 1019–1053. [Google Scholar]
- 47.Northrop DB. Biochemistry. 1975;14:2644–2651. doi: 10.1021/bi00683a013. [DOI] [PubMed] [Google Scholar]
- 48.Berti PJ, Tanaka KSE. Adv. Phys. Org. Chem. 2002;37:239–314. [Google Scholar]
- 49.Luo M, Li L, Schramm VL. Biochemistry. 2008;47:2565–2576. doi: 10.1021/bi702133x. [DOI] [PubMed] [Google Scholar]
- 50.Scapin G, Grubmeyer C, Sacchettini JC. Biochemistry. 1994;33:1287–1294. doi: 10.1021/bi00172a001. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez-Segura L, Witte JF, McClard RW, Hurley TD. Biochemistry. 2007;46:14075–14086. doi: 10.1021/bi701023z. [DOI] [PubMed] [Google Scholar]
- 52.McCann JA, Berti PJ. J. Am. Chem. Soc. 2007;129:7055–7064. doi: 10.1021/ja067371l. [DOI] [PubMed] [Google Scholar]
- 53.Saen-oon S, Quaytman-Machleder S, Schramm VL, Schwartz SD. Proc. Natl. Acad. Sci. USA. 2008;105:16543–16548. doi: 10.1073/pnas.0808413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.