Abstract
For some time post-translational small protein modifications were found only in eukaryotes; much later, such modifications were identified in some species of bacteria. The recent discovery of ubiquitin-like proteins that form polymeric chains and covalently modify proteins in archaea finally closes the evolutionary gap among the domains of life.
Ubiquitin and related modifiers: elegant chemistry for eukaryotic biology
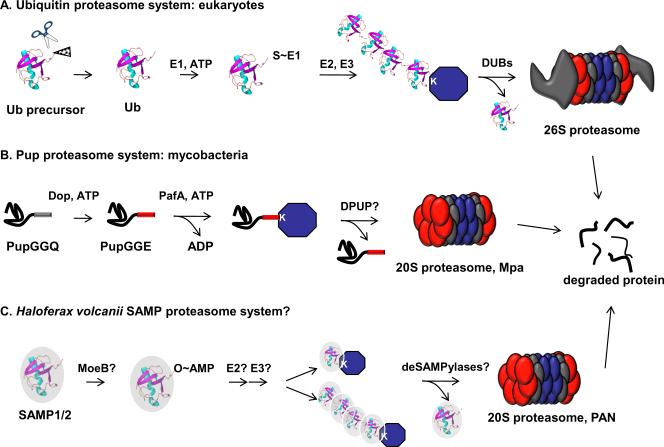
Proteins can be modified in numerous ways, including phosphorylation, acetylation, and glycosylation. In eukaryotes one of the most intriguing areas of investigation focuses on modifications by the small protein ubiquitin (Ub) and its related Ub-like modifiers (Ubl) [reviewed in (1, 2)]. Ub is a 76 amino acid protein that is usually processed from larger polypeptides. The resulting Ub molecules end in Gly-Gly, a motif common to most conjugatable Ubls (reviewed in (1)). The C-terminal Gly is subjected to a series of reactions that result in the conjugation of Ub to lysine (Lys) residues in target proteins. Activating (E1) enzymes utilize ATP to adenylate the C-terminal Gly of Ub, which is then passed to an active-site cysteine in the E1 enzyme. From here, Ub is transferred to a Ub-conjugating enzyme (E2) that delivers Ub to a protein ligase (E3), which catalyses the formation of an isopeptide bond with a Lys on the target substrate. Ub can be further modified with additional Ub molecules on any one of seven Lys or even the N-terminus (3). Proteins doomed for degradation are generally tagged with Lys48-linked Ub chains, and are recognized by the regulatory complex of the 26S proteasome, where deubiquitylating proteases (DUBs) remove and recycle Ub monomers for subsequent ubiquitylation reactions (Fig. 1A).
Fig. 1. Comparison of the eukaryotic and proposed prokaryotic Ub/Ubl-proteasome systems.
(A) Eukaryotic Ub is encoded by four different loci in yeast (33) and is processed by Ub specific proteases that expose a C-terminal GlyGly motif. The C-terminus is the site of adenylation by an E1 enzyme, which via a series of transthioesterification reactions passes Ub to E2 and E3 enzymes. E3 enzymes are numerous (>600 in humans) (1) and determine which proteins are to be ubiquitylated. Ub is recognized by proteins in the regulatory cap of the 26S proteasome and is removed by deubiquitylases (DUBs) prior to destruction of the target substrate. (B) The bacterial Pup conjugation system uses enzymes unrelated to those in the eukaryotic Ub pathway. Pup is synthesized with a C-terminal Gln, which is deamidated by Dop. PafA catalyzes the ligation of Pup to substrates, presumably by a phosphorylated Pup intermediate. Pup is recognized by the Mycobacterial proteasome assocated ATPase, Mpa. It is not know if Pup is removed prior to degradation by a depupylase (DPUP). (C) Proposed H. volcanii SAMP-proteasome pathway. MoeB, a homologue of eukaryotic E1 activating enzymes, may be the E1 for SAMP1 and/or SAMP2. Homologues of eukaryotic E2 or E3 enzymes have not been identified in H. volcanni thus the conjugation of SAMPs to substrates remains to be determined. Furthermore, it is not known if “desampylation” occurs, although H. volcanii encodes two putative JAMM proteases. PAN: proteasome activating nucleotidase is a homologue of proteasome associated ATPases.
In addition to Ub, several Ubls with the canonical β-grasp fold that characterizes Ub have been identified in eukaryotes. These include SUMO (small Ub-like modifier), Nedd8 (neural precursor cell expressed, developmentally downregulated 8), ISG15 (interferon stimulated gene 15), and Urm1 (Ub-related modifier 1). All of these Ubls terminate in GlyGly and several are conjugated to target proteins by enzymes with similar activities [reviewed in (1, 2)]. Unlike Ub, these modifiers, with some exceptions, are not generally associated with proteasomal degradation but instead function to change the interaction properties of proteins, transfer sulfur, or target proteins to specific sub-cellular locations. Taken together, there is little question that Ub and its related modifiers are essential for eukaryotic life.
A bacterial small protein modifier and ubiquitin-like proteins
Despite the existence of proteasomes (4) and proteins with Ub folds (5) in prokaryotes, post-translational modifications by small proteins had not been identified in archaea or bacteria until recently. Thus, a puzzle of the prokaryotic proteasome systems was to understand how proteins were targeted for degradation. Eventually, a small protein modifier, prokaryotic ubiquitin-like protein (Pup), was identified in mycobacteria as a signal for proteasomal degradation [reviewed in (6)]. Despite its name, Pup does not have a Ub-fold, but is an intrinsically disordered protein (7–9). Furthermore, although Pup has a penultimate C-terminal Gly-Gly motif, it does not attach to substrates via Gly, but uses glutamate (Glu) (10, 11). Although there is a lack of sequence and structural homology with Ub, Pup functions to target proteins for degradation (10). It appears that Pup targets substrates to the mycobacterial proteasome (Fig. 1B), however this activity has not been directly demonstrated in vitro. Interestingly, monopupylation seems to be sufficient to doom proteins for degradation in vivo because polypupylation has not been observed (12, 13); however, we cannot rule out the possibility of polypup-dependent degradation.
Open reading frames encoding Pup homologues are only found in a limited number of organisms, namely, the bacterial order of Actinomycetales (6) and the Nitrospirales (14). By contrast, numerous Ub or β-grasp domain proteins are present in both bacteria and archaea (15). For example, ThiS and MoaD are proteins with a β-grasp fold, conserved in most bacteria, and are involved in thiamine and molybdopterin co-factor biosynthesis, respectively (16). ThiS and MoaD have C-terminal Gly-Gly motifs that undergo a chemistry that resembles Ub activation, however, these proteins transfer sulfur rather than conjugate to proteins. Urm1, which is similar to ThiS and MoaD, acts both as a protein modifier (17) and as a sulfur carrier in a tRNA modification pathway (18, 19). The nature and function of the chemical bond between Urm1 and its single protein target, Ahp1p (alkyl hydroperoxide reductase), remains a matter of debate (20), and the majority of the observed Urm1 deletion phenotypes can be rescued by supplementing the modified tRNA. Despite these findings, it was proposed that bacterial Ub-fold proteins could possibly covalently modify other proteins (6, 15).
Perhaps inspired by the presence of small protein modifiers in bacteria, Humbard and co-workers recently tested the ability of five small proteins to form conjugates in Haloferax volcanii, a halophilic (“salt-loving”) archaeon (21). The criteria used to predict potential modifiers were that the proteins be small and terminate in Gly-Gly. Although five candidates ended in Gly-Gly, only three of the selected genes (HVO_2619, HVO_0202, HVO_2177) were predicted to encode Ub-like, β-grasp fold proteins. Ectopic expression of these genes in H. volcanii revealed that HVO_2619 and HVO_0202 conjugated to other proteins. Based on this result, HVO_2619 and HVO_0202 were named “small archaeal modifier protein” (SAMP) 1 and SAMP2.
The pattern of protein modification with either SAMP1 or SAMP2 (“SAMPylation”) varied depending on the growth conditions used, as well as whether or not certain proteasomal subunits were present. H. volcanii encodes two types of proteasomal alpha subunits (α, α) that form 20S core particles with a single type of β-subunit (22, 23), and also interact with one or both proteasome activating nucleotidases, PAN-A and PAN-B (24). The α1 proteasome and PAN-A subunits are highly abundant in H. volcanii during all phases of growth (24) and mutants lacking these proteins display deficiencies in stress response (25). Humbard and co-workers examined proteasomal mutants for accumulated SAMPylated proteins. SAMP1ylation increased only when both panA and psmA (encoding the α1 subunit) were deleted (21). This suggested that SAMP1 conjugates might be proteasome substrates. In striking contrast, SAMP2 conjugates were less abundant in the double mutant strain, as well as in the single psmA, panA, and panB mutants. This result might suggest some form of feedback inhibition of SAMP2ylation or SAMP2 removal (“deSAMPylation”) in the absence of a full repertoire of proteasome function. In addition to defects in proteasome function, nitrogen limitation dramatically increased both SAMP1ylation and SAMP2ylation, although the reasons for this are not clear. It is possible that nitrogen limitation represents a stress that requires proteomic adaptation by this archaeal species.
A single step immunopurification of the SAMP1 and SAMP2 proteomes (“SAMPylomes”) identified 34 proteins that are potentially modified with either or both SAMPs. Deletion of Gly-Gly from SAMP1 or SAMP2 abolished SAMPylation. Although it was presumed that SAMP1 conjugates to target Lys via a C-terminal Gly, SAMP1 attachment sites were not identified; SAMP1 lacks tryptic sites [arginine (Arg) or Lys] that would allow for the facile identification of modified peptides. The introduction of Arg or Lys near the C-terminus of SAMP1 might allow investigators to eventually map SAMP1 sites. Nonetheless, it seems likely that Lys is the targeted amino acid in light of how related modifiers conjugate to proteins, although it is possible that SAMP1 forms conjugates to amino acids other than Lys.
The method used to purify SAMP1 conjugated proteins could present a potential caveat to the SAMP1ylome analysis. Although high salt is required for optimal H. volcanii protein folding or activity, immunoprecipitation using the anti-FLAG antibody was performed under low salt, non-denaturing conditions (21), which could preserve some non-covalent interactions. Thus several of the proteins reported in the proteomic analysis could have co-purified with truly SAMPylated proteins, rather than be targets of SAMPylation.
In contrast to SAMP1, SAMP2 has a Lys immediately before the Gly-Gly motif; thus trypic digestion and mass spectrometry (MS) analysis was able to identify nine proteins with a SAMP2 modification. Two proteins had two different SAMP2ylated sites, suggesting that either or both could be modified on a single molecule, although no peptide was identified with both Lys modified simultaneously. Perhaps most intriguingly, SAMP2 could form Lys58 conjugates, a feature resembling polyubiquitylation, although the position of SAMP2 Lys58 does not have a corresponding Lys in Ub. It was not clear how many SAMP2 monomers could polymerize into a single chain, but the possibility of polySAMP2ylation is exciting, and could suggest roles for SAMP2 other than, or in addition to, proteasomal degradation (Fig. 1C).
A motif for SAMPylation has not been identified, although this is also largely true for Ub and Pup. Interestingly, Kirkland and colleagues had previously tested the possibility that phosphorylation might play a role in targeting proteins for proteasomal degradation in H. volcanii because protein phosphorylation can signal the ubiquitylation of eukaryotic proteasome substrates (26). The phosphoproteomes of wild type and panA H. volcanii were compared, and a dramatic increase in phosphoproteins in the proteasome mutants was observed (27). However, comparison of the phosphoproteome and SAMPylomes identified little overlap with the exception of two proteins [OsmC-like protein (HVO_1289); 2-oxoglutarate oxidoreductase, HVO_0887)]. Still, it will be interesting to see if any SAMPylated proteins are also phosphorylated. Additional proteins that were previously reported to accumulate in the proteomes of proteasome-defective H. volcanii also appeared to be targets of SAMPylation, consistent with a potential role for SAMPylation with proteasome function.
SAMP enzymology: more like us than bacteria?
SAMPylome analysis might also have revealed some insight into the enzymology of SAMPylation. Both SAMP1 and SAMP2 co-purified with MoeB (HVO_0558, molybdopterin biosynthesis protein), a homologue of the E1 enzyme of the Urm1 pathway, Uba4p (18, 28, 29). Interestingly, SAMP2 forms an isopeptide bond with MoeB at Lys113, a finding that might suggest that SAMP2 regulates MoeB activity or stability. It remains to be determined if MoeB is the E1-like activating enzyme for either SAMP1 or SAMP2, but it seems to be a likely candidate based on its presumed adenylating activity.
Although a potential E1 enzyme was identified, no potential E2 or E3 candidates were identified in the H. volcanii genome. This suggests that either H. volcanii has dispensed for the need of such enzymes or, more likely, has a Ubl system that is a progenitor to the more complex eukaryotic Ub and Ubl pathways. Alternatively, H. volcanii conjugation enzymes might have highly divergent sequence or structural similarity with eukaryotic conjugating enzymes. Nonetheless, because SAMPs conjugate to proteins via Gly and not Glu, and because it appears a eukaryotic E1 homologue could be involved in SAMPylation, it preliminarily looks as if the archaeal system is more like the eukaryotic Ub system than the Pup system of bacteria (Fig. 1C).
Although no homologues of the eukaryotic conjugation enzymes were identified, H. volcanii could have deconjugating proteases. Indeed, it seems likely that a SAMP deconjugation system is present as both the conjugation and removal of Ubls are critical to the function of these modifiers in eukaryotes. Several sequenced archaeal species are known to encode JAB1/MPN/Mov34 metalloenzyme motif (JAMM) proteases (30), proteins that have been associated with the deconjugation of Ubls in eukaryotes, and have homologues in bacteria as well as archaea (31, 32). The H. volcanii genome contains two putative JAMM proteases (HVO_1016 and HVO_2505) that could be possible “deSAMPylases”.
Prospects
The discovery of Ubls in an archaeal species finally gives investigators new leads into how proteins are targeted to a proteasome, but much work is needed to be done to link the identification of these modifiers with protein degradation. There is no direct evidence to show that SAMP1 or SAMP2 are implicated in the targeting of proteins to the proteasome like their eukaryotic and bacterial counterparts, Ub and Pup, respectively. Although the number or amount of certain proteins increased in the proteasome defective strains, this could be due to transcriptional, translational or other effects. In order to begin to pinpoint the role of proteasome function on SAMPylation, investigators will need to check for transcriptional changes in proteasome mutants. For example, are genes encoding proteins identified in the SAMPylome transcriptionally upregulated in proteasome mutants? If not, this would begin to support the notion that proteins of interest post-translationally accumulate in proteasome mutants. Another possibility is that defective proteasome function increases the translation of specific transcripts, which would also result in protein accumulation. Ectopic expression of putative proteasome substrate genes from a heterologous promoter in H. volcanii could rule out this possibility.
SAMP2 appears to form conjugates, but it is not yet clear if SAMP1 can do so as well. As previously suggested, improved proteomic analysis of the SAMP1 proteome will hopefully reveal more information on the nature of SAMP1 conjugation. An exciting future prospect will be to learn if polySAMPylation will have roles independent of proteasome function in the physiology of H. volcanii.
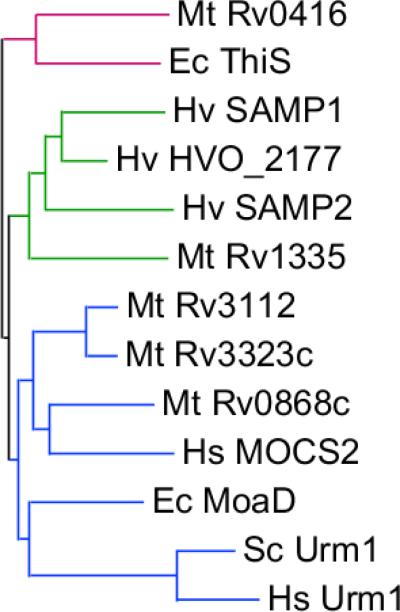
Finally, researchers have noted the presence of Ubl proteins in all prokaryotes, not just archaea (6, 15). The archaeal SAMP proteins form a subfamily of β-grasp proteins that is distinct from the MoaD/Urm1 and ThiS families of sulfur-carrier proteins (Fig. 2). The clustering of the non-conjugated HVO_2177 protein with the SAMPs suggests functions of the SAMP subfamily beyond protein modification. It will be interesting to elucidate the biological role of other members of the subfamily, which are found in both archaea and bacteria. For example, do any covalently attach to other proteins in these prokaryotes? Although post-translational small protein modifiers were once considered unique to the eukaryotic domain of life, it is now clear that these proteins will most likely have widespread roles in the biology of all organisms.
Fig. 2. Sequence relationship between archaeal, bacterial and eukaryotic β-grasp proteins.
Neighbor-joining dendrogram visualizing the evolutionary relationship between the β-grasp proteome of Haloferax volcanii (Hv), Escherichia coli (Ec), and M. tuberculosis (Mt). Eukaryotic Urm1-like proteins from human (Hs) and budding yeast (Sc) are included for reference. The proteins can be classified into three major subfamilies groups: ThiS-like (red), SAMP-like (green) and MoaD/Urm1-like (blue).
Acknowledgements
We are grateful to Kristin Burns, Andrew Darwin and Sebastien Leidel for helpful suggestions and critical review of this manuscript. K.H.D. is supported by NIH grant HL092774 and is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease. K.H. is supported by DFG grant SPP1365.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7(8):750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin KH. Prokaryotic Ubiquitin-Like Protein, Proteasomes, and Pathogenesis. Nat. Rev. Microbiol. 2009;7(7):485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, et al. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392(1):208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao S, et al. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422(2):207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- 9.Striebel F, et al. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16(6):647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 10.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322(5904):1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284(5):3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Festa RA, et al. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5(1):e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watrous J, et al. Expansion of the mycobacterial “PUPylome”. Mol Biosyst. 2010;6(2):376–385. doi: 10.1039/b916104j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Mot R. Actinomycete-like proteasomes in a Gram-negative bacterium. Trends Microbiol. 2007;15(8):335–338. doi: 10.1016/j.tim.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7(7):R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev. 2006;30(6):825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 17.Goehring AS, Rivers DM, Sprague GF., Jr. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell. 2003;14(11):4329–4341. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leidel S, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458(7235):228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 19.Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci U S A. 2008;105(47):18255–18260. doi: 10.1073/pnas.0808756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedrioli PG, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 2008;9(12):1196–1202. doi: 10.1038/embor.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humbard MA, et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463(7277):54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaczowka SJ, Maupin-Furlow JA. Subunit topology of two 20S proteasomes from Haloferax volcanii. J Bacteriol. 2003;185(1):165–174. doi: 10.1128/JB.185.1.165-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome alpha1 influences its N(alpha) acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J Bacteriol. 2009;191(12):3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186(22):7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190(24):8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28(5):730–738. doi: 10.1016/j.molcel.2007.11.019. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 27.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190(1):193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37(4):1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goehring AS, Rivers DM, Sprague GF., Jr. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell. 2003;2(5):930–936. doi: 10.1128/EC.2.5.930-936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 31.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419(6905):403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 32.Burns KE, et al. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J Am Chem Soc. 2005;127(33):11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. Embo J. 1987;6(5):1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]