Abstract
Previous studies demonstrate that nitric oxide (NO) promotes p53 transcriptional activity by a classical DNA-damage-responsive mechanism involving activation of ATM/ATR and phosphorylation of p53. These studies intentionally used high doses of NO-donors to achieve the maximum DNA-damage. However, lower concentrations of NO donors also stimulate rapid and unequivocal nuclear retention of p53, but apparently do not require ATM/ATR-dependent p53 phosphorylation or total p53 protein accumulation. To identify possible mechanisms for p53 activation at low NO levels, the role of Tyr nitration in p53 activation was evaluated. Low concentrations of the NO donor, DETA NONOate (<200μM), exclusively nitrate Tyr327 within the tetramerization domain promoting p53 oligomerization, nuclear accumulation and increased DNA-binding activity without p53 Ser15 phosphorylation. Molecular modeling indicates that nitration of one Tyr327 stabilizes the dimer by about 2.67 kcal mol−1. Significant quantitative and qualitative differences in the patterns of p53-target gene modulation by low (50μM), non DNA-damaging and high (500μM), DNA-damaging NO donor concentrations was shown. These results demonstrate a new post-translational mechanism for modulating p53 transcriptional activity responsive to low NO concentrations and independent of DNA damage signaling.
Nitric oxide (NO) activates the tumor suppressor protein p53 through a mechanism involving NO-induced DNA damage and, as a consequence, activation of ATM/ATR-dependent p53 phosphorylation and nuclear uptake (1–4). These studies for the most part employed NO donors at 0.5–1mM concentrations that generate NO levels mimicking a chronic inflammatory state of tissues and that induce maximal levels of p53 Ser15 phosphorylation (2). The downstream transcriptional consequences are similar to those observed after treatment with other DNA damaging agents, e.g. ionizing radiation, ultraviolet light, or adriamycin (2). Some investigations also report results at lower NO donor concentrations that, although stimulating rapid and unequivocal nuclear retention of p53, do so by a mechanism(s) that do not require p53 Ser15 phosphorylation or total cellular p53 protein accumulation (1, 2). These results imply that p53 activity is also responsive to non-DNA damaging physiological stimuli that generate small transient increases in NO.
NO involvement in cellular regulation is usually thought of in terms of activation of soluble guanylate cyclase and protein kinase G (5, 6). However, NO and reactive nitrogen species (RNS) generated as a consequence of the reaction of NO with different reactive oxygen species (ROS) also covalently modify a number of regulatory proteins. The best studied of these covalent modifications is the S-nitrosylation or reversible oxidation of Cys of a number of key regulatory proteins (7–10). RNS can also nitrate Tyrs (11, 12). Although Tyr nitration is usually associated with ischemia-reperfusion conditions, more modest conditions of RNS generation have been shown to nitrate signal transduction proteins. For example, activation of the transcription factor NF-κB by RNS at therapeutic doses of ionizing radiation involves the reversible nitration of a Tyr181 in the inhibitor protein IκBα and, as a consequence, dissociation of IκBα from NF-κB (13). Other examples of Tyr nitration of key regulatory proteins suggest that this post-translational modification has regulatory significance for cellular responses to mild oxidative/nitrosative stresses (12, 14–19).
Tyr nitration of p53 is also observed in vitro and in vivo (20, 21). An increase in nitrated p53 is observed in brain tissue of patients with advanced Alzheimer’s disease (22). Basal levels of p53 Tyr nitration are also reduced in cultured MCF-7 cells by a NOS-inhibitor – consistent with a role for endogenous NOS in modulating p53 function (20). However, identification of the nitrated Tyrs and how their nitration alters p53 function is unexplored. Herein we identify the primary site of p53 nitration at low NO levels as Tyr327 in the tetramerization domain of p53. Nitration of Tyr327 stimulates the oligomerization and nuclear retention of p53. The p53 dependent transcriptional response to nitration of Tyr327 is very different from that observed after DNA-damage induced p53 activation.
Materials and Methods
Cell Culture, Irradiation, NO-donor Treatment
MCF-7 cells were cultured in MEM with 10% FBS, 0.01 mg/ml bovine insulin, and 1xPen/Strep. Saos-2 cells were cultured in McCoy’s 5A Medium with 15% FBS and 1xPen/Strep. Cells were irradiated at a dose rate of 1 Gy/min with a 137Cs source (GammaCell® 40 Exactor, MDS Nordion).
Antibodies and Reagents
Primary antibodies used: beta-actin (Santa Cruz Biotechnology); nitroTyr (Millipore); TATA binding protein (Abcam); p53, phospho-p53 (Ser15), and the 9E10 monoclonal against c-Myc (Cell Signaling). Wild-type pCMV-p53 was purchased from Clontech. Mutants were constructed by QuikChange® PCR technology. Site-Directed mutagenesis Kit (Stratagene) was validated by full-length sequencing. Peroxynitrite was purchased from Calbiochem, and DETA NONOate from Cayman Chemical.
Transfection, Subcellular Fractionation, Mass Spectrometry, Immunoprecipitation and Western Blotting
These procedures have been described (9, 13, 23). Mass spectroscopic analysis of nitrated proteins was performed as detailed previously (13).
Tetramerization assay
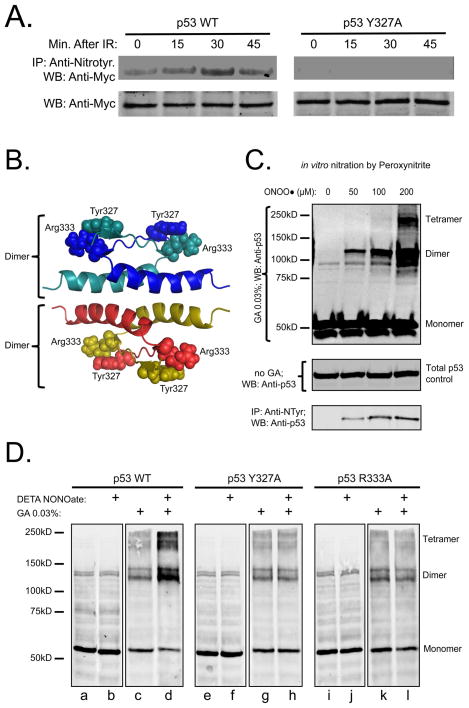
Glutaraldehyde (GI) crosslinking to stabilize p53 oligomers has been extensively described (24–26) Cell lysates prepared in non-denaturing lysis buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 0.5% Triton® x-100, supplemented with protease and phosphatase inhibitors) are incubated with glutaraldehyde on ice for 30 min, boiled with SDS sample buffer, resolved on 8% SDS-PAGE and after transfer to nitrocellulose analyzed for p53 by Western blotting. Western blots obtained with anti-p53 rabbit monoclonal IgG showed a non-specific double band at ~130kD if samples were not incubated with cross-linker (Figure 2D, lanes: a,b,e,f,i,j). These non-specific bands were not observed with anti-p53 mouse monoclonal IgG (see Figure S6, lanes: a,b,e,f,i,j).
Figure 2.
Tyr nitration stimulates p53 tetramerization in vitro and in vivo. (A) MCF-7 cells transfected with equal amounts of Myc-tagged p53 WT or Tyr327Ala were irradiated (5 Gy). Tyr nitration was shown as described above at the indicated time-points (top panels). Both top panels are from the same blot. Total cell lysates were probed with anti-Myc IgG as a loading control (bottom panels). (B) Structure of the p53 tetramerization domain indicating Tyr327/Arg333 inter-monomer interactions in both dimers; (C) Peroxynitrite stimulates endogenous p53 tetramerization (top panel) and Tyr nitration (bottom panel) in MCF-7 cells in a dose dependent manner; (D) Incubation with DETA NONOate stimulates tetramerization of WT p53, but not Tyr327Ala or Arg333Ala mutants. Saos-2 cells were transfected with p53 WT or mutants. 24 h after transfection cells were incubated with 200 μM DETA NONOate for 4 h. Tetramerization of p53 was assessed as described in Methods. All panels are from the same blot. GA – glutaraldehyde.
Electrophoretic Mobility Gel Shift Assay and cDNA Array
p53 DNA binding activity in the nuclear extracts is measured with the Odyssey® EMSA Kit using p53 IRDye® 700 Infrared Dye labeled oligonucleotides (Li-Cor® Biosciences) according to manufacturer’s protocol. Incubation with anti-p53 IgG (DO-1 X, Santa Cruz Biotechnology) (Supershift) and with p53 consensus oligonucleotide (Santa Cruz Biotechnology) was used as a control of specific p53-DNA binding (SI, Figure S2). mRNA was isolated with RNeasy® Mini Kit (QIAGEN) and analyzed for p53-regulated genes with a Human p53-Regulated cDNA Plate Array (Signosis, Inc.).
Molecular Modeling
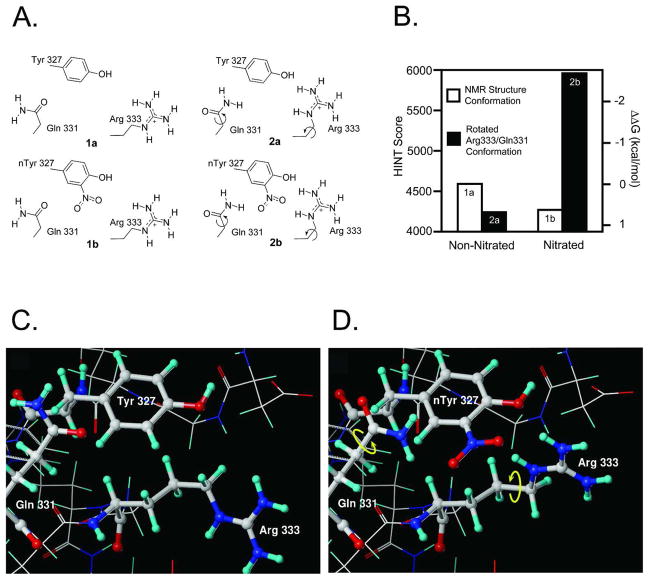
The minimized average NMR structure of the p53 tetramerization domain (PDB 1sak) was obtained from the Protein Data Bank (27, 28). For the structural analysis of nitrated proteins the overall procedure was as described in (13). The geometry of the wild type tetramer structure was further optimized while holding the backbone rigid. Sybyl 8.1 (29) using the Tripos force field, Gasteiger-Hückel charges, a dielectric constant of 6, and the Powell optimization algorithm (gradient of energy = 0.05 kcal Å−1 mol−1). This resulted in structure model 1a (see Figure 6A). Next, this structure was modified by replacing the hydrogen at the 3′ position of Tyr327 on chain A with a nitro group followed by the same geometry optimization protocol to obtain structure 1b. The positioning of the guanidinium group of Arg333 and the amide group on the side chain of Gln331 on chain C was found to be not optimal for the resulting nitrated structure. Thus, the hydrogen bonding was optimized for this structure by first turning the guanidinium group of Arg333 on chain C towards nitroTyr327 (nTyr327) on chain A, allowing the guanidinium hydrogens to form hydrogen bonds with both the hydroxyl and the nitro groups (see Figure 6A). This resulting structure was subjected to geometry optimization, followed by flipping the amide group on the side chain of Gln331 180° to allow a hydrogen bond to form between the amide and the nitro group. The final geometry optimization was performed on this structure (2b in Figure 6A). For comparison, similar manipulations of Arg333 and Gln331 were performed on the non-nitrated optimized structure, resulting in structure 2a.
Figure 6.
Molecular modeling of the interaction changes between two p53 monomers after Tyr327 nitration. (A) The four structures subjected to HINT analysis; (B) Interactions between chains A and C calculated by HINT. The nitrated structure, with the optimized hydrogen bond network has stronger interactions than the conformation derived from the NMR structure. ΔΔG=0 was defined as the interactions in the non-nitrated (NMR experimental) conformation; (C) conformation before Tyr327 nitration; (D) conformation after Tyr327 nitration.
Hydropathic Analysis
The software package HINT (Hydrophatic INTeractions) (30) was used to evaluate the changes in interactions between the chains caused by nitration of Tyr327 on chain A. The HINT score provides an empirical, but quantitative, evaluation of protein-protein interactions as a sum of all single atom-atom interactions using the following equation:
where bij is the score for the interaction between atoms i and j, ai is the hydrophobic atom constant for atom i, Si is the solvent accessible surface area for atom i, and Tij is a logic function that can assume values +1 and −1. The choice of Tij depends on the nature of the interacting atoms. Rij and rij are both functions of the distance between atoms i and j (31). The HINT force field is based on the assumption that each bij is related to a partial δg value and that the HINT score has a linear correlation with the global interaction ΔG°. Higher HINT scores correspond to more favorable binding free energies. The “semi-essential” mode was used: polar hydrogen atoms and hydrogen atoms bonded to unsaturated carbon atoms are treated explicitly. In addition to polar hydrogens, aromatic hydrogens were allowed to act as hydrogen bond donors. For the backbone amide hydrogen atoms the Si terms were corrected by adding a 20 Å2 (32) to improve the relative energetics of inter- and intra-molecular hydrogen bonds involving backbone amides.
Results
Activated endogenous NOS and NO donors stimulate nitration of p53 in vivo
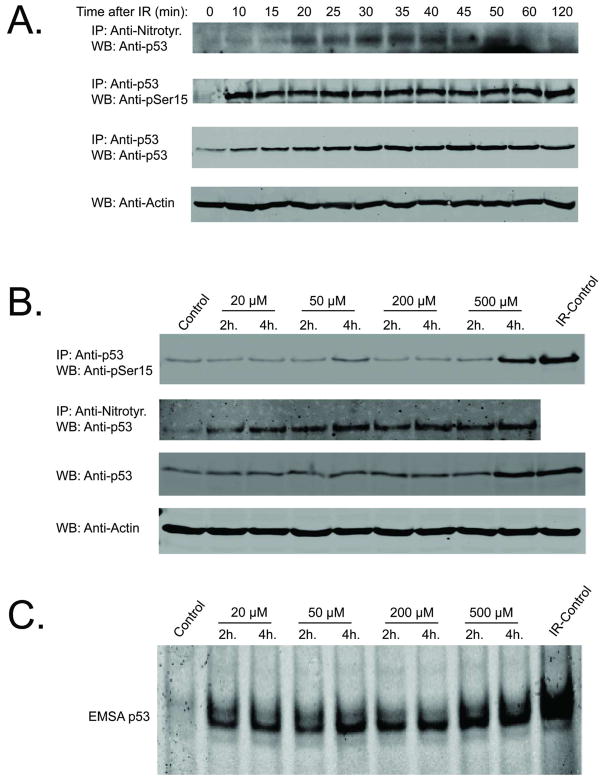
Ionizing radiation at clinically relevant doses (< 8 Gy) activates constitutive NOS in diverse cell types (9, 13, 33). One consequence of this activation is Tyr-nitration of several proteins including Mn-dependent superoxide dismutase and the NF-κB inhibitor protein, IκBα. A single radiation exposure of 5 Gy also stimulates endogenous p53 Tyr nitration (along with Ser15 phosphorylation) in p53 wild type MCF-7 cells (Figure 1A). Tyr nitration of p53 is transient with maximum levels achieved at ~30–40 min after radiation and recovering to baseline levels by 60 mins. Tyr nitration of endogenous p53 was shown in reciprocal immunoprecipitation experiments by: a) immunoprecipitation with anti-nitroTyr IgG followed by blotting with anti-p53 IgG (Figure 1A); and b) immunoprecipitation with anti-p53 IgG followed by blotting with anti-nitroTyr IgG (Supplemental Information (SI): Figure S1). In contrast, radiation induced p53 Ser15 phosphorylation is sustained for a longer period of time (Figure 1A).
Figure 1.
Tyr nitration and phosphorylation of endogenous p53 after radiation. (A) MCF-7 cells were irradiated at 5 Gy and cell lysates were prepared at the indicated time-points for analysis of Tyr nitration, Ser15 phosphorylation, and total p53 accumulation. Cell lysate Actin levels were used to demonstrate equal amounts of starting material prior to immunoprecipitation (IP) (B) MCF-7 cells were with DETA NONOate for the indicated period of time. NO-donor stimulated Ser15 phosphorylation was analyzed by immunoprecipitation with anti-p53 antibody and WB with anti-pSer15 IgG. NO-donor stimulated nitration was analyzed by IP with anti-nitroTyr IgG and WB with anti-p53 IgG. 2 h time-point after a single dose of IR (5 Gy) was used as a positive control for Ser-15 phosphorylation (last lane). (C) DNA-binding activity of endogenous p53 in MCF-7 cells was analyzed by EMSA. 2 h time-point post-radiation (5 Gy) was used as a positive control.
To evaluate the role of NO concentration in p53 activation, cells were incubated with the slow release NO donor, DETA NONOate, at concentrations ranging between 20 and 500 μM to generate steady state levels of 20 to 400 nM (the latter simulating chronic inflammatory conditions (2, 34)). Figure 1B compares the effects of different NO concentrations on p53 Ser15 phosphorylation and p53 Tyr nitration. As observed by previous investigators (1–4) only at the highest NO donor doses (≥ 500μM), is appreciable Ser15 phosphorylation observed. This contrasts with increased Tyr-nitration obtained at the lowest NO donor concentrations. At DETA NONOate concentrations ≤ 200μM, p53 DNA binding is also enhanced to levels not that much different than observed at the highest NO donor concentration examined (500μM, Figure 1C). The 2 hour time-point after 5 Gy of ionizing radiation was used as a positive control for p53 EMSA assay.
To determine what Tyrs are nitrated, cell lysates of MCF-7 cells overexpressing Myc-tagged p53 were prepared under non-denaturing conditions and treated with peroxynitrite to maximally nitrate p53. After immunopurification p53 was processed for mass spectrometry. Seventeen peptides were identified for coverage of 54.7%, included 5 of the 9 Tyrs of p53: Tyr103, Tyr107, Tyr126, Tyr205, Tyr327 (Figure 2A, SI: Table S1). Two of these five, Tyr107 and Tyr327, were identified as nitrated by peroxynitrite (SI: Figure S3A, S4, and Table S1).
A genetic approach was used to test whether these Tyrs are nitrated following treatment of cells with NO donors or gamma-irradiation. MCF-7 cells were transfected with Myc-tagged wild type p53, the single mutants Tyr107Ala and Tyr327Ala, or the double mutant Tyr107Ala/Tyr327Ala. After the indicated treatment immunopurified nitrated proteins were resolved by gel electrophoresis and the resulting Western blots probed with anti-Myc-tag IgG (Figures 2A, 3A, SI: Figure S3B). Mutation of Tyr327 to Ala completely abrogated both radiation-induced and basal p53 nitration (Figure 2A). Similar specificity is observed with DETA NONOate concentrations < 50μM; p53 wild type, but not the Tyr327Ala mutant, is nitrated at these donor concentrations (Figure 3A). At much higher levels of NO donor both wild type and the p53 Tyr327Ala mutant are nitrated, but the level of p53 Tyr327Ala mutant Tyr nitration is significantly lower compared with Tyr nitration of p53 WT (Figure 3A). These observations are in contrast to the relative non-specific nitration obtained by treating cell lysates with high concentrations of peroxynitrite (SI: Figure S3B). We conclude that the transient and low levels of RNS generated metabolically by a modest radiation exposure or with low concentrations of slow release NO donors such as DETA NONOate results in a relatively specific p53 Tyr nitration profile.
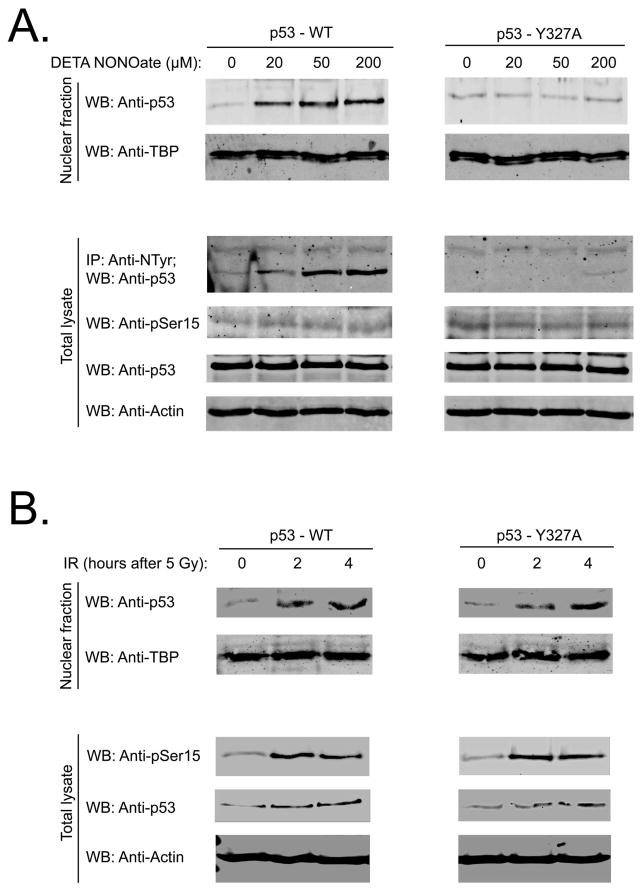
Figure 3.
Low doses of DETA NONOate (20–200 μM) stimulate p53 tetramerization and nuclear accumulation by a Tyr327 nitration dependent mechanism. (A) Saos-2 cells were transfected with p53 WT or Tyr327Ala mutant. 24 h after transfection cells were incubated with different doses of NO-donor for 2 hours. Nuclear fractions were probed with anti-p53 antibody. NO-donor stimulated nitration it the total cell lysates was analyzed by IP with anti-nitroTyr IgG and WB with anti-p53 IgG. Total cell lysates were probed with anti-phospho-p53 (Ser15), anti-p53, and anti-Actin IgG. Anti-TBP (TATA binding protein) antibody was used as a loading control for nuclear extracts; (B) Saos-2 cells were transfected with p53 WT or Tyr327Ala mutant. 24 hour after transfection cells were irradiated (5 Gy). Nuclear fractions were collected at 2 and 4 hour after IR and probed as in Fig. 3A. 0 time-point represents non-radiated controls. Total cell lysates were probed with anti-phospho-p53 (Ser15), anti-p53, and anti-Actin IgG. Anti-TBP (TATA binding protein) IgG was used as a loading control for nuclear extracts.
Nitration of Tyr327 promotes p53 oligomerization in a dose dependent manner
Tyr327 is a proximal member of the β-strand (residues 326–333) of the tetramerization domain whereas Arg333 is the most distal member of the β-strand and forms an inter-monomer hydrogen bond with Tyr327 in an adjacent monomer (35). Hence, a p53 dimer has two Tyr327/Arg333 interacting sites that contribute to the anti-parallel interaction of β-strands of two monomers (Figure 2B). Increased dimer stability promotes p53 tetramerization (dimerization of dimers) through interactions of their α-helices (residues 335–354), resulting in nuclear accumulation of p53 and p53-dependent transcriptional activation (36, 37).
Oligomerization of p53 was examined using previously described methods of glutaraldehyde protein cross-linking (24, 38). Cell lysates from MCF-7 cells with wild type p53 were treated with different doses of peroxynitrite, incubated on ice with 0.03% glutaraldehyde and analyzed by SDS gel electrophoresis. With increasing peroxynitrite concentration there is increasing endogenous p53 Tyr nitration and oligomerization (Figure 2C). The reason for the multiple bands at protein sizes corresponding to dimers and tetramers and observed by all investigators using glutaraldehyde to crosslink p53 is not known (24, 25, 39).
A similar approach was used to examine NO-dependent oligomerization in intact cells at different NO donor concentrations. Saos-2 cells (p53 null) were transfected with wild type p53, or the Tyr327Ala, and Arg333Ala p53 mutants. Twenty-four hrs after transfection cells were incubated for 2 hrs with 200μM of DETA NONOate and lysed in non-denaturing lysis buffer. The cell lysates were divided into equal aliquots: one treated with glutaraldehyde to stabilize the oligomers and the other without. Incubation with the NO-donor promoted both dimerization and tetramerization in cells transfected with wild type p53, but had no effect on p53 oligomerization in the cells expressing either mutant (Figure 2D). In the absence of NO donor the monomer-dimer-tetramer equilibria of wild type p53 and the mutants were identical (compare lanes c,g,k in Figure 2D). These latter results correspond to previous observations that mutation of Tyr327 to Ala or Arg333 to Ala had no effect on unstimulated p53 oligomerization when expressed in yeast or Saos-2 cells (26, 40). On another hand, mutation of Tyr327 to Phe results in a significant decrease of unstimulated p53 oligomerization and cannot be used for our purposes (SI: Figure S5).
Two additional p53 mutants were also evaluated: Arg337Ala and p53 without its tetramerization domain (p53 ΔTD) (SI: Figure S6). Mutation Arg337Ala decreases p53 tetramerization (26, 41). In untreated samples p53 Arg337Ala had significantly lower dimer-tetramer level compared with p53 WT (SI: Figure S6, lanes: c,g), and incubation with 200μM of DETA NONOate did not stimulate tetramerization of this mutant (SI: Figure S6, lanes: g,h). Absence of the tetramerization domain in p53 ΔTD completely abolishes the ability for oligomerization with and without DETA NONOate incubation (SI: Figure S6, lanes: k.l). We conclude that the cross-linking results in Figure 2 support the importance of the Tyr327/Arg333 interaction in the mechanism of NO-dependent oligomerization of p53.
Low doses of NO by nitrating Tyr327 stimulate p53 nuclear retention and p53-dependent gene transcription
We tested whether low doses (≤ 200μM) of DETA NONOate activate p53, not through phosphorylation, but through nitration of Tyr327, protein tetramerization and nuclear retention. Twenty-four hours after transfection with p53 WT or p53 Tyr327Ala mutant, Saos-2 cells were incubated 2 hours with 20–200μM of DETA NONOate. Treatment with these low concentrations of DETA NONOate induced nuclear accumulation of p53 WT and nitration but without increasing Ser15 phosphorylation (Figure 3A). Similar treatment of Saos-2 cells expressing the Tyr327Ala mutant failed to nitrate or enhance the nuclear accumulation of the p53 mutant. In contrast, p53 WT and p53 Tyr327Ala nuclear accumulation and Ser 15 phosphorylation demonstrated identical responses after IR: both stimulate Ser 15 phosphorylation, significant nuclear accumulation and moderate total accumulation of the protein (Figure 3B). Consistent with these results, DETA NONOate at 50 μM stimulated p53 WT but not p53 Tyr327A mutant oligomerization (SI: Figure S7).
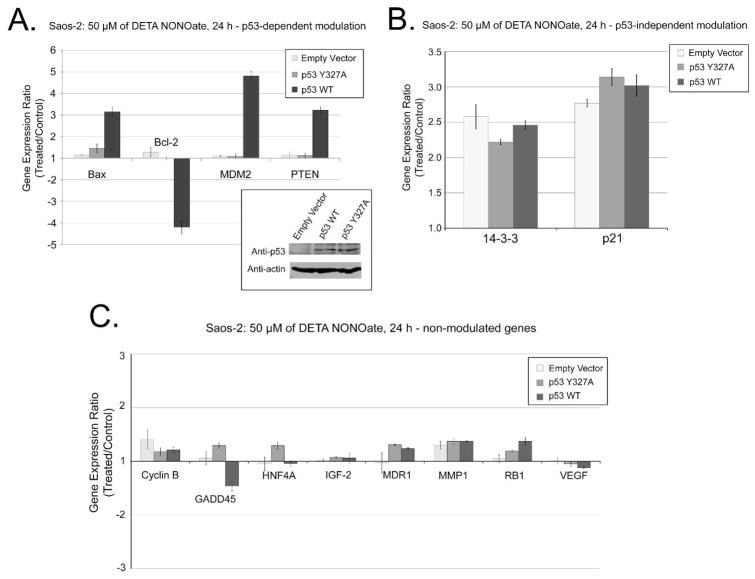
To investigate the role of Tyr327 nitration in p53-dependent gene expression, we treated cells with 50μM of DETA NONOate and analyzed gene expression with a cDNA array specific for p53 target genes. This concentration of NO-donor produces WT p53 nitration (Figure 1B), tetramerization (SI: Figure S7), and nuclear accumulation (Figure 3A) without apparent activation of ATM/ATR signaling pathway (Figures 1B, 3A and references (1, 2)). More than twofold up- or down-regulation of mRNA expression was considered significant. Saos-2 cells expressing WT p53 demonstrated up-regulation of Bax, MDM2, PTEN mRNAs, and down-regulation of Bcl-2 upon treatment with 50μM of DETA NONOate (Figure 4A). In contrast, expressions of these genes were unchanged in Saos-2 cells expressing p53 Tyr327Ala or with empty vector. NO-dependent but p53 independent transcriptional regulation was demonstrated by the comparable up-regulation of 14-3-3 and p21 in Saos-2 cells transfected with empty vector, p53 WT, or Tyr327Ala mutant (Figure 4B). Incubation with 50μM DETA NONOate had no effect on expression of Cyclin B, GADD45, HNF4α, IGF-2, MDR1, MMP1, Rb1, or VEGF in Saos-2 regardless of whether they expressed p53 or not (Figure 4C).
Figure 4.
Analysis of p53-dependent gene expression (Saos-2 cells). The cDNA assay for gene expression was performed with Saos-2 cells transfected with empty Vector, p35 WT, or p53 Tyr327Ala. Panel A: NO-dependent/p53-dependent genes modulation; Panel B: NO-dependent/p53-independent genes modulation; Panel C: genes showing no significant expression changes. Panel D: Comparison of gene expression analysis in MCF-7 cells treated with 50 or 500μM DETA NONOate, or 5 Gy of ionizing radiation at 24 h post-treatment. Genes with less than 2-fold expression changes are not shown. Experimental data are presented as means ± SD for triplicate samples. The embedded panel in panel A shows the level of p53 expression for cells transfected with empty Vector, p53 WT, or p53 Tyr327Ala. Actin expression was shown as a loading control.
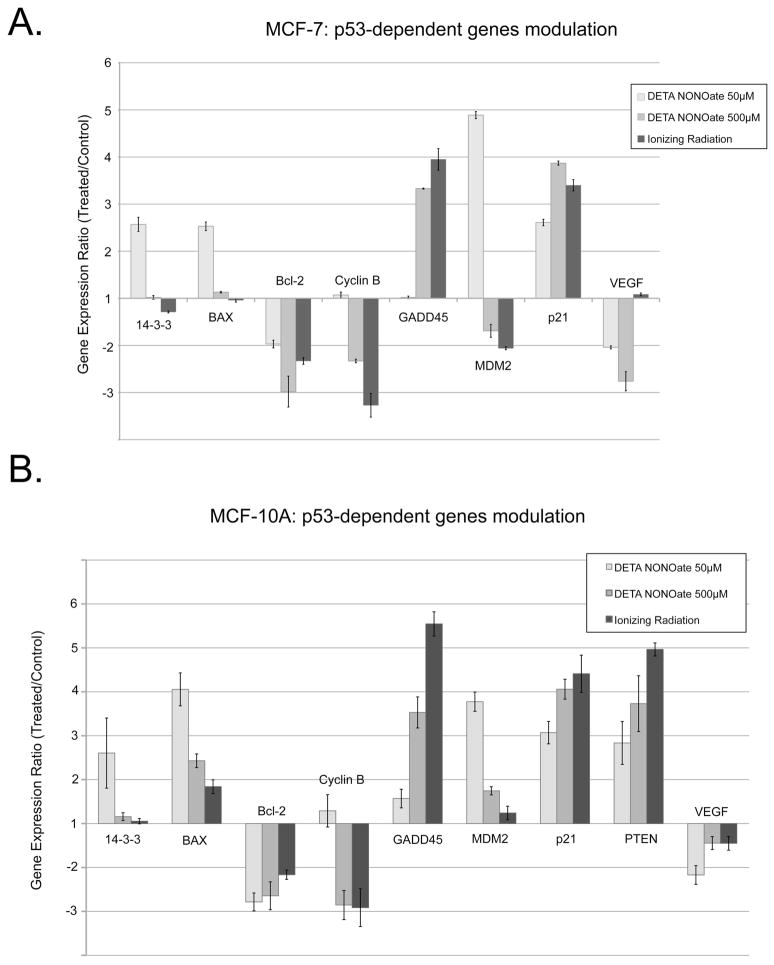
We also compared endogenous p53 gene expression profiles in MCF-7 and MCF- 10A cells treated with 50μM or 500μM of DETA NONOate or irradiated at 5 Gy (Figure 5). At 50μM DETA NONOate, the expression patterns of MCF-7, MCF-10A, and Saos-2 transfected with WT p53 were very similar with expression of MDM2, p21, 14-3-3 and Bax up and expression of Bcl-2 down. The only difference was with PTEN, whose expression was unresponsive in MCF-7 cells irrespective of treatment. Responses to 500μM DETA NONOate or ionizing radiation showed almost the same spectrum of gene modulation that differed significantly from that observed at lower NO donor concentrations, e.g., 14-3-3, Bax, GADD45, Cyclin B, and MDM2. These latter findings parallel the relative effects of these different treatments on p53 Ser15 phosphorylation (Figures 1B, 3 and references (1, 2)) and support the argument that low level NO activates p53 transcriptional activity by mechanisms that differ from that initiated by DNA damage.
Figure 5.
Analysis of p53-dependent gene expression (MCF-7 and MCF-10A cells). Comparison of gene expression analysis in MCF-7 (Panel A) and MCF-10A (Panel B) cells treated with 50 or 500μM DETA NONOate, or 5 Gy of ionizing radiation at 24 h post-treatment. Genes with less than 2-fold expression changes are not shown. Experimental data are presented as means ± SD for triplicate samples.
Molecular modeling of the interaction changes between two p53 monomers after Tyr 327 nitration
The effects of Tyr327 nitration on the p53 subunit interactions were also analyzed with the molecular modeling tool HINT (Hydropathic INTeractions). HINT permits the quantitative analysis of all possible noncovalent atom-atom interactions, including hydrogen bonding, Coulombic, acid-base, and hydrophobic interactions, using X-ray crystal or NMR protein structures (42, 43) as input. HINT uses empirically derived constants based on thermodynamic hydropathy values from solvent partition measurements. In a previous study we used HINT to evaluate the effects of IκBα nitration on its interactions with the p50 subunit of NF-κB (13). HINT calculations have also accurately estimated changes in free energy resulting from site-specific mutations and their effect on hemoglobin dimer-tetramer assembly (44, 45). Calculations were performed on two sets of p53 structures (Figure 6A). Each set consisted of one tetramer structure where Tyr327 on chain A was nitrated (1b and 2b) and one where it was not (1a and 2a). The difference between the structures in these two sets was that for structures 1a and 1b the geometries were built from the optimized NMR structure that has the wild type Tyr327. No experimental structure exists for the nitrated protein and thus H-bonding for 2b with nitrated Tyr327 on chain A, was optimized by rotating the guanidinium of Arg333 and the amide of Gln331 on chain C (see Materials and Methods). 15N NMR relaxation measurements indicate that the side chains on these two residues are flexible (46) and can adopt many orientations (47). In particular, model 24 in 2j0z (47) shows the orientation of Arg333 relative to Tyr327 in an almost identical pose to the one we obtained by optimizing the interactions between this Arg and nitrated Tyr.
This analysis showed that for the non-nitrated (1a) and nitrated (1b) structures, the interactions between chains A and C were 4590 and 4241 HINT units, respectively. Earlier work (42, 43) suggests that 1 kcal mol−1 is equivalent to 515 HINT score units. Thus, the nitrated structure 1b is less stable than 1a by 0.68 kcal mol−1. However, after optimization of the hydrogen bonding network in the nitrated case, as in structure 2b, the interaction score between the two chains is 5961 HINT units, i.e., 2.67 kcal mol−1 more stable than 1a. The non-nitrated structure, 2a, subjected to the same manipulations as 2b, is 0.63 kcal mol−1 less stable than 1a, indicating that it is the nitration and not an optimization artifact that stabilizes the nitrated tetramer structure. Figure 6B summarizes these results. Molecular models of the 1a and 2b conformations are shown in Figures 6C and 6D. A detailed listing of the interactions and HINT/free energy contributions are provided in Table S2 of the SI. As a further note, this analysis only represents the effect of one Tyr nitration. It is possible for the Tyr327 on the opposite strand to be nitrated and interact with its corresponding Arg333. This could as much as double the estimated energy stabilization due to Tyr nitration.
Discussion
Cellular responses ranging from cytoprotective, signaling activity to DNA damaging, cytotoxic effects are initiated at different NO concentrations (reviewed in (48)). With respect to the tumor suppressor protein p53, high NO concentrations (>400 nM) stimulate p53 Ser15 phosphorylation and activation similar to what is observed with DNA damaging agents (1, 2). These earlier studies, however, also demonstrated that the DNA binding activity of p53 is promoted at much lower cellular NO levels without Ser15 phosphorylation. The aim of the present work was to explore this phosphorylation-independent mechanism of p53 activation.
Previously we demonstrated that nitration of Tyr181 of IκBα dissociates IκBα from NF-κB and activates the transcription factor (13). Here we describe how Tyr nitration activates p53 by promoting its oligomerization. A high degree of selectivity in p53 nitration was observed at low NO concentrations generated either by transient activation of endogenous NOS by a single radiation treatment or by incubating cells with 20–50μM of DETA NONOate. Under these conditions only Tyr327 is nitrated (Figures 2A, 3A). At higher DETA NONOate concentrations (≥200μM) or treating cell lysates with peroxynitrite additional Tyrs become nitrated (Figure 3A, SI: Figure S3).
Tyr327 is a proximal member of the β-strand (residues 326–333) of the tetramerization domain and forms an inter-monomer hydrogen bond with Arg333, the most distal member of the β-strand (35). This proximal component of the tetramerization domain (a.a. 325–334) is responsible for the monomer/monomer binding and dimer formation – the rate-limiting step in forming a tetramer (Figure 2B). Mutation of either amino acid prevents NO-induced p53 oligomerization. A molecular modeling suggests that nitration of Tyr327, by changing the monomeric conformation and facilitating the interaction with Arg333 of an adjacent monomer, enhances the stability of the p53 dimer. Dimer stabilization promotes tetramerization (dimerization of dimers) which depends on the distal part of tetramerization domain (a.a. 335–355) (49). Although mutation of Tyr327 or Arg333 to Ala completely blocks the p53 oligomerization in response to a NO-donor, it does not interfere with basal p53 monomer-dimer-tetramer equilibrium (Figure 2D, lanes c,g,k). This latter finding agrees with work showing that mutation of either amino acid does not alter p53 oligomer equilibria when expressed in yeast or Saos-2 cells (26). Control experiments with p53 Arg337Ala and p53 ΔTD provide additional proof that NO-dependent p53 activation depends on the tetramerization domain (SI: Figure S6).
High doses of NO also lead to Tyr327 nitration, but as shown by several investigators DNA-damage is also generated with subsequent activation of ATM/ATR-dependent signaling pathways. These DNA-damage inducible pathways block the binding of mdm2 to p53-tetramer by ATM/ATR-dependent phosphorylation of both proteins thereby stabilizing the p53 tetramer (1–4).
Different patterns of p53-dependent gene expression are also observed at different NO concentrations (Figure 5). Expression modulation of Bax, Cyclin B, GADD45, MDM2 at high NO levels in MCF-7 and MCF-10A cells mimics the radiation-induced pattern and is significantly different from that observed at low doses of NO. Since nitration of p53 occurs at all NO donor concentrations, these differences must reside in the modulation of different co-activators or cooperation factors that enhance or repress p53-induced transcription (see review (50)). We also do not understand whether there is any functional consequence to slow sustained release of NO as obtained with DETA NONOate compared to the relatively rapid, transient increase in NO following NOS activation by radiation. More extensive investigations of p53-dependent gene modulation by different doses of NO and frequency of NO delivery and p53 nitration are planned that address these different mechanisms. Results from such experimentation should provide insight into the physiological function of p53 nitration under acute or chronic inflammatory conditions.
Our results also reveal a NO-dependent, p53-independent activation of two p53-target genes – p21 and 14-3-3. p21 is activated by radiation and both low or high NO-donor concentrations whereas 14-3-3 expression is only activated at low NO doses. p53-independent activation of p21 has been previously reported (51–53), but we are unaware of previous investigations demonstrating p53-independent up-regulation of 14-3-3. The DNA-binding activity of p53 is enhanced by 14-3-3 by stabilization of p53 dimers to form tetramers at low p53 concentrations (54). Hence, NO-dependent up-regulation of 14-3-3 can serve as an additional stabilization mechanism for p53 tetramerization.
Supplementary Material
Acknowledgments
This research was supported by NIH grants P01 CA072955 and R01 CA90881 (RBM), GM071894 (GEK). AB was supported by NIH T32 CA113277.
We thank David C. Williams, Jr. (VCU Department of Pathology) for his assistance in interpreting NMR structures of p53.
ABBREVIATIONS AND TEXTUAL FOOTNOTES
- NO
nitric oxide
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- IgG
immunoglobulin G
- IR
ionizing radiation
- NMR
nuclear magnetic resonance
- FBS
fetal bovine serum
- GA
glutaraldehyde
- IP
immunoprecipitation
Footnotes
Supporting information available: p53 Tyr-nitration after IR (Figure S1); p53 gel mobility shift, supershift, and “cold oligonucleotide” control (Figure S2); p53 Tyr-nitration in vitro (Figure S3); MS/MS spectra of the peptides contained nitroTyr (Figure S4); tetramerization activity of p53 WT, Tyr327Ala and Tyr327Phe mutants (Figure S5); stimulation of p53 WT, Arg337Ala, and ΔTD tetramerization by DETA NONOate (Figure S6); stimulation of p53 WT and Tyr327Ala tetramerization by low dose of DETA NONOate (Figure S7); mass spectroscopy results for p53 protein (Table S1); HINT score calculations for interactions of Tyr327 before and after nitration (Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wang X, Zalcenstein A, Oren M. Nitric oxide promotes p53 nuclear retention and sensitizes neuroblastoma cells to apoptosis by ionizing radiation. Cell Death Differ. 2003;10:468–476. doi: 10.1038/sj.cdd.4401181. [DOI] [PubMed] [Google Scholar]
- 2.Hofseth LJ, Saito S, Hussain SP, Espey MG, Miranda KM, Araki Y, Jhappan C, Higashimoto Y, He P, Linke SP, Quezado MM, Zurer I, Rotter V, Wink DA, Appella E, Harris CC. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc Natl Acad Sci U S A. 2003;100:143–148. doi: 10.1073/pnas.0237083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester K, Ambs S, Lupold SE, Kapust RB, Spillare EA, Weinberg WC, Felley-Bosco E, Wang XW, Geller DA, Tzeng E, Billiar TR, Harris CC. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc Natl Acad Sci U S A. 1996;93:2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneiderhan N, Budde A, Zhang Y, Brune B. Nitric oxide induces phosphorylation of p53 and impairs nuclear export. Oncogene. 2003;22:2857–2868. doi: 10.1038/sj.onc.1206431. [DOI] [PubMed] [Google Scholar]
- 5.Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 6.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamler JS, Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 9.Barrett DM, Black SM, Todor H, Schmidt-Ullrich RK, Dawson KS, Mikkelsen RB. Inhibition of protein-tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J Biol Chem. 2005;280:14453–14461. doi: 10.1074/jbc.M411523200. [DOI] [PubMed] [Google Scholar]
- 10.Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- 11.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration-- functional alteration or just a biomarker? Free Radic Biol Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakovlev VA, Barani IJ, Rabender CS, Black SM, Leach JK, Graves PR, Kellogg GE, Mikkelsen RB. Tyrosine nitration of IkappaBalpha: a novel mechanism for NF-kappaB activation. Biochemistry. 2007;46:11671–11683. doi: 10.1021/bi701107z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balafanova Z, Bolli R, Zhang J, Zheng Y, Pass JM, Bhatnagar A, Tang XL, Wang O, Cardwell E, Ping P. Nitric oxide (NO) induces nitration of protein kinase Cepsilon (PKCepsilon ), facilitating PKCepsilon translocation via enhanced PKCepsilon -RACK2 interactions: a novel mechanism of no-triggered activation of PKCepsilon. J Biol Chem. 2002;277:15021–15027. doi: 10.1074/jbc.M112451200. [DOI] [PubMed] [Google Scholar]
- 15.Elsasser TH, Kahl S, Li CJ, Sartin JL, Garrett WM, Rodrigo J. Caveolae nitration of Janus kinase-2 at the 1007Y–1008Y site: coordinating inflammatory response and metabolic hormone readjustment within the somatotropic axis. Endocrinology. 2007;148:3803–3813. doi: 10.1210/en.2006-1737. [DOI] [PubMed] [Google Scholar]
- 16.Koeck T, Fu X, Hazen SL, Crabb JW, Stuehr DJ, Aulak KS. Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J Biol Chem. 2004;279:27257–27262. doi: 10.1074/jbc.M401586200. [DOI] [PubMed] [Google Scholar]
- 17.Ischiropoulos H. Protein tyrosine nitration--an update. Arch Biochem Biophys. 2009;484:117–121. doi: 10.1016/j.abb.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 18.Wu F, Wilson JX. Peroxynitrite-dependent activation of protein phosphatase type 2A mediates microvascular endothelial barrier dysfunction. Cardiovasc Res. 2009;81:38–45. doi: 10.1093/cvr/cvn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prevotat L, Filomenko R, Solary E, Jeannin JF, Bettaieb A. Nitric oxide-induced down-regulation of beta-catenin in colon cancer cells by a proteasome-independent specific pathway. Gastroenterology. 2006;131:1142–1152. doi: 10.1053/j.gastro.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Chazotte-Aubert L, Hainaut P, Ohshima H. Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem Biophys Res Commun. 2000;267:609–613. doi: 10.1006/bbrc.1999.2003. [DOI] [PubMed] [Google Scholar]
- 21.Cobbs CS, Samanta M, Harkins LE, Gillespie GY, Merrick BA, MacMillan-Crow LA. Evidence for peroxynitrite-mediated modifications to p53 in human gliomas: possible functional consequences. Arch Biochem Biophys. 2001;394:167–172. doi: 10.1006/abbi.2001.2540. [DOI] [PubMed] [Google Scholar]
- 22.Cenini G, Sultana R, Memo M, Butterfield DA. Effects of oxidative and nitrosative stress in brain on p53 proapoptotic protein in amnestic mild cognitive impairment and Alzheimer disease. Free Radic Biol Med. 2008;45:81–85. doi: 10.1016/j.freeradbiomed.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amorino GP, Hamilton VM, Valerie K, Dent P, Lammering G, Schmidt-Ullrich RK. Epidermal growth factor receptor dependence of radiation-induced transcription factor activation in human breast carcinoma cells. Mol Biol Cell. 2002;13:2233–2244. doi: 10.1091/mbc.01-12-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu J, Nie L, Wiederschain D, Yuan ZM. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol Cell Biol. 2001;21:8533–8546. doi: 10.1128/MCB.21.24.8533-8546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foo RS, Nam YJ, Ostreicher MJ, Metzl MD, Whelan RS, Peng CF, Ashton AW, Fu W, Mani K, Chin SF, Provenzano E, Ellis I, Figg N, Pinder S, Bennett MR, Caldas C, Kitsis RN. Regulation of p53 tetramerization and nuclear export by ARC. Proc Natl Acad Sci U S A. 2007;104:20826–20831. doi: 10.1073/pnas.0710017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi T, Kato S, Otsuka K, Watanabe G, Kumabe T, Tominaga T, Yoshimoto T, Ishioka C. The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene. 2005;24:6976–6981. doi: 10.1038/sj.onc.1208839. [DOI] [PubMed] [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clore GM, Ernst J, Clubb R, Omichinski JG, Kennedy WM, Sakaguchi K, Appella E, Gronenborn AM. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 29.www.tripos.com
- 30.Eugene Kellogg G, Abraham DJ. Hydrophobicity: is LogP(o/w) more than the sum of its parts? Eur J Med Chem. 2000;35:651–661. doi: 10.1016/s0223-5234(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 31.Kellogg GE, Burnett JC, Abraham DJ. Very empirical treatment of solvation and entropy: a force field derived from log Po/w. J Comput Aided Mol Des. 2001;15:381–393. doi: 10.1023/a:1011136228678. [DOI] [PubMed] [Google Scholar]
- 32.Porotto M, Fornabaio M, Greengard O, Murrell MT, Kellogg GE, Moscona A. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J Virol. 2006;80:1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leach JK, Black SM, Schmidt-Ullrich RK, Mikkelsen RB. Activation of constitutive nitric-oxide synthase activity is an early signaling event induced by ionizing radiation. J Biol Chem. 2002;277:15400–15406. doi: 10.1074/jbc.M110309200. [DOI] [PubMed] [Google Scholar]
- 34.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1alpha, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proc Natl Acad Sci U S A. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 36.McLure KG, Lee PW. How p53 binds DNA as a tetramer. EMBO J. 1998;17:3342–3350. doi: 10.1093/emboj/17.12.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman PN, Chen X, Bargonetti J, Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci U S A. 1993;90:3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooks CL, Li M, Gu W. Mechanistic studies of MDM2- mediated ubiquitination in p53 regulation. J Biol Chem. 2007;282:22804–22815. doi: 10.1074/jbc.M700961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chene P, Bechter E. Cellular characterisation of p53 mutants with a single missense mutation in the beta-strand 326–333 and correlation of their cellular activities with in vitro properties. J Mol Biol. 1999;288:891–897. doi: 10.1006/jmbi.1999.2738. [DOI] [PubMed] [Google Scholar]
- 41.Mateu MG, Fersht AR. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J. 1998;17:2748–2758. doi: 10.1093/emboj/17.10.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham DJ, Kellogg GE, Holt JM, Ackers GK. Hydropathic analysis of the non-covalent interactions between molecular subunits of structurally characterized hemoglobins. J Mol Biol. 1997;272:613–632. doi: 10.1006/jmbi.1997.1249. [DOI] [PubMed] [Google Scholar]
- 43.Cozzini P, Fornabaio M, Marabotti A, Abraham DJ, Kellogg GE, Mozzarelli A. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J Med Chem. 2002;45:2469–2483. doi: 10.1021/jm0200299. [DOI] [PubMed] [Google Scholar]
- 44.Burnett JC, Botti P, Abraham DJ, Kellogg GE. Computationally accessible method for estimating free energy changes resulting from site-specific mutations of biomolecules: systematic model building and structural/hydropathic analysis of deoxy and oxy hemoglobins. Proteins. 2001;42:355–377. doi: 10.1002/1097-0134(20010215)42:3<355::aid-prot60>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 45.Burnett JC, Kellogg GE, Abraham DJ. Computational methodology for estimating changes in free energies of biomolecular association upon mutation. The importance of bound water in dimer-tetramer assembly for beta 37 mutant hemoglobins. Biochemistry. 2000;39:1622– 1633. doi: 10.1021/bi991724u. [DOI] [PubMed] [Google Scholar]
- 46.Clubb RT, Omichinski JG, Sakaguchi K, Appella E, Gronenborn AM, Clore GM. Backbone dynamics of the oligomerization domain of p53 determined from 15N NMR relaxation measurements. Protein Sci. 1995;4:855–862. doi: 10.1002/pro.5560040505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mora P, Carbajo RJ, Pineda-Lucena A, Sanchez del Pino MM, Perez-Paya E. Solvent-exposed residues located in the beta-sheet modulate the stability of the tetramerization domain of p53--a structural and combinatorial approach. Proteins. 2008;71:1670–1685. doi: 10.1002/prot.21854. [DOI] [PubMed] [Google Scholar]
- 48.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free Radic Biol Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mateu MG, Sanchez Del Pino MM, Fersht AR. Mechanism of folding and assembly of a small tetrameric protein domain from tumor suppressor p53. Nat Struct Biol. 1999;6:191–198. doi: 10.1038/5880. [DOI] [PubMed] [Google Scholar]
- 50.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 51.Zeng YX, el-Deiry WS. Regulation of p21WAF1/CIP1 expression by p53-independent pathways. Oncogene. 1996;12:1557–1564. [PubMed] [Google Scholar]
- 52.Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci U S A. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahyar-Roemer M, Roemer K. p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene. 2001;20:3387–3398. doi: 10.1038/sj.onc.1204440. [DOI] [PubMed] [Google Scholar]
- 54.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36:5983–5991. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.