Abstract
Cells living in a complex environment must constantly detect, process, and appropriately respond to changing signals. Therefore, all cellular information processing is dynamic in nature. As a consequence, understanding the process of signal transduction often requires detailed quantitative analysis of dynamical behaviors. Here we focus on the oscillatory dynamics of the tumor suppressor protein p53 as a model for studying protein dynamics in single cells to better understand its regulation and function.
Introduction
How do signals received by a cell get translated into decisions such as growth, death, or movement? In the past several decades there has been a great deal of success in identifying the proteins and genes that are activated or repressed in response to specific inputs and in assembling them into signal transduction pathways. However, even though we now have maps of many signaling pathways, new questions have arisen due to the complexity of the pathways they represent. How can we move beyond describing the structure of biological networks to developing a detailed, quantitative understanding of their function and behavior? One promising approach is to investigate the dynamics of key proteins within the network (Figure 1). In this context, dynamics is defined as the change of any variable that can be quantitatively measured over time, such as protein concentration, activity, modification state, or localization. These data are complementary to the information originally used to describe the network, and have great potential to provide new insight into the relationship between network structure and function. For example, if the activity of a signaling molecule is measured at only a single point in time, the signal could be interpreted as binary – being either “on” or “off.” If, however, the signaling activity is quantitatively measured with high temporal resolution over a long period it could show a large number of distinct behaviors. Detailed analysis of dynamical behaviors in diverse systems and under various conditions will likely provide new levels of understanding of how cells detect inputs and translate them into outputs.
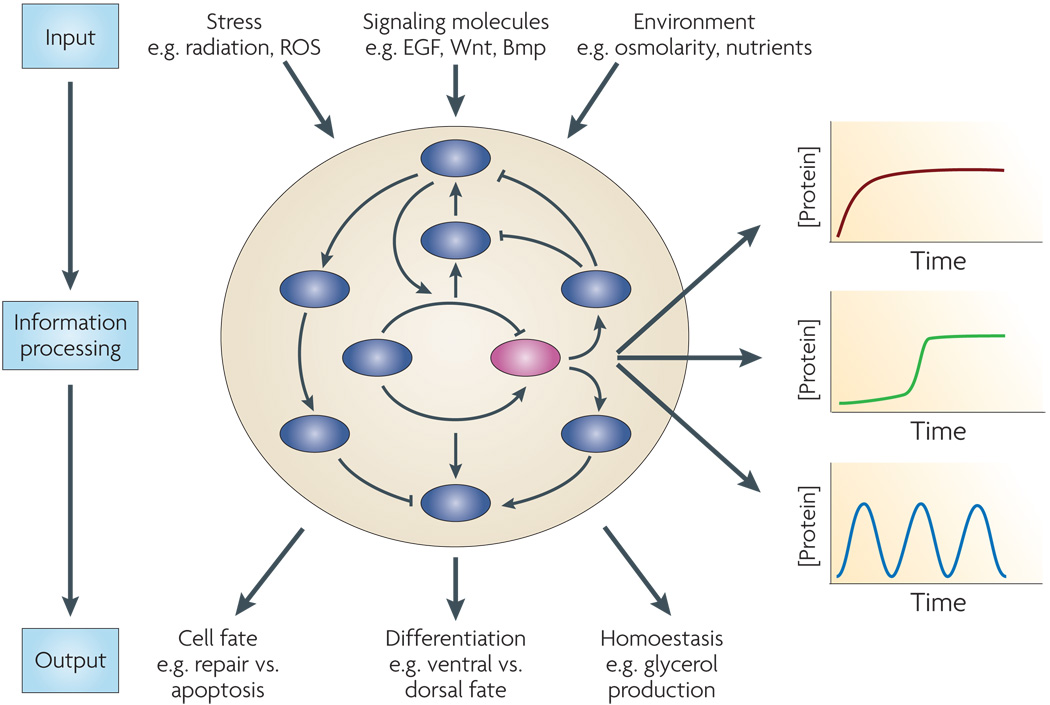
Figure 1. Dynamics in signal transduction pathways.
A complex protein network senses information about the intra- and extra-cellular environment (input), processes the information, and triggers a response (output). Currently, the information processing step is usually represented as a static drawing of binary (inhibitory or activating) arrows connecting different components of the network. An important aspect that is missing from such diagrams is the dynamical behavior of key members of the network. For example, the purple protein might show different dynamical behaviors in response to different inputs, in different cell types, or even between genetically identical cells. These dynamics can provide new insights about the specific interactions that are functional in each condition and the role of these interactions in triggering the right outcome.
The analysis of cellular dynamics often requires measurements in single cells since measurements of averaged dynamics in a population of cells can be misleading. For example, in response to certain doses of antibiotics, some cells live while others die1. These different outcomes might reflect differences in the initial state of the cell (such as its cell cycle state, basal level of network components, or local environment) which in turn lead to differences in the quantitative behavior of the information-processing network. By visualizing the dynamical behavior and identifying how it varies among cells (or cell types), we might be able to explain varying behaviors both within cell populations and in different cell types.
Single cell analyses of signaling systems have already revealed important information about the role of dynamics in regulating various cellular responses. For example, the transcription factor nuclear factor kappa B (NF-κB) in mammalian cells shows pulses of nuclear localization upon stimulation2, 3. Single cell analysis of luciferase expression from a synthetic NF-κB responsive promoter suggested that the pulses play a role in maintaining target gene expression3, 4. In S. cerevisiae, the mitogen-activated protein kinase Fus3 shows oscillations in activity in response to mating pheromone5. The Fus3 oscillations correlate with oscillations in mating gene expression and the formation of new mating projections, as determined by fluorescence microscopy and flow cytometry using cells expressing fluorescent fusion proteins5.
In this Perspective, we focus on the p53 network, as a model for studying the dynamics of a signal transduction pathway in single cells to better understand its structure and function. This is a new area of p53 research, which is still in its early stages of development, and therefore the data covered in this article should be viewed in this context. We will discuss the proper characterization of p53 dynamics in single cells, especially as it relates to the structure of the network that shapes this dynamical response. We will also describe possible functions of p53 dynamics, in terms of both the fate of individual cells and the survival of the entire organism.
p53 Dynamics in Single Cells
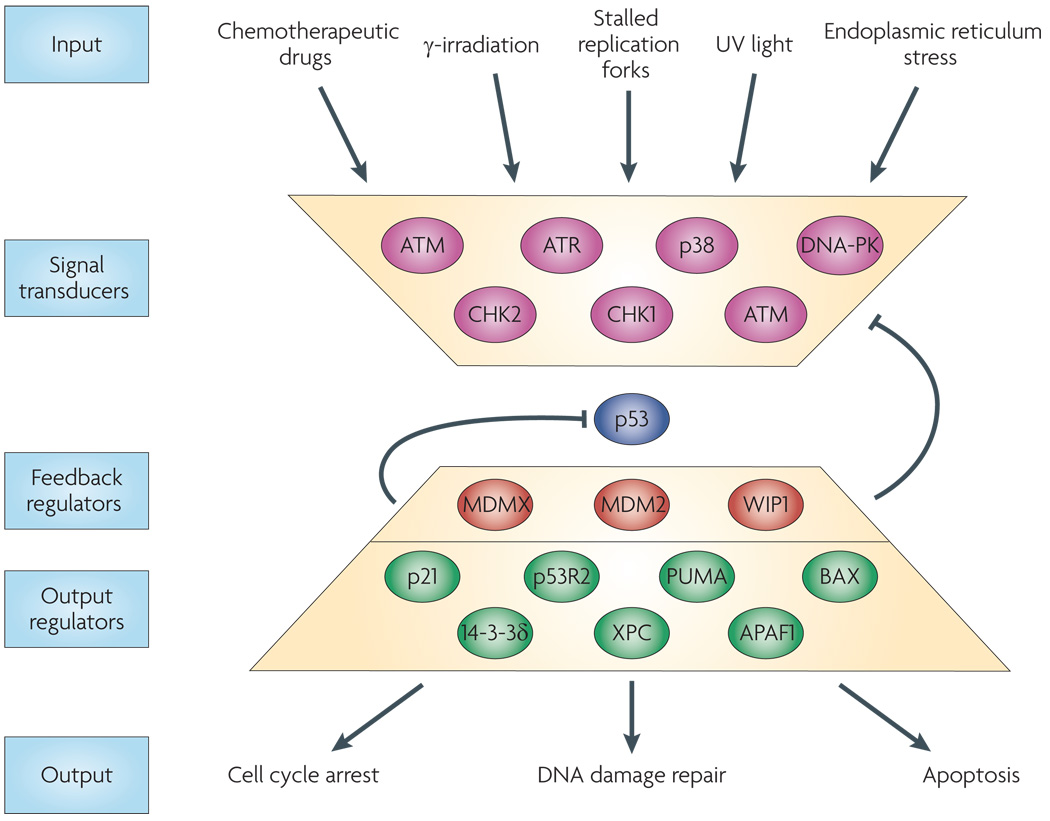
Due to its important role in maintaining genomic integrity6–8, the p53 network has been studied extensively over the past three decades (Figure 2). p53 is upregulated in response to many forms of cellular stress, including various types of DNA damage9. The presence of stress is detected and transmitted to p53 by the action of signal transducers which post-translationally modify p53 and affect its stability and activity. Upon activation, p53 regulates the transcription of hundreds of genes7, 10. These genes code for proteins with a wide range of functions including inhibition of cell cycle progression, activation of apoptosis, and regulation of p53 itself directly or through the upstream signal transducers7, 10 (Figure 2).
Figure 2. The p53 signaling network.
Stress signals (inputs) are detected and transduced to p53 through several kinases (signal trasducers). Upon activation, p53 upregulates the transcription of numerous genes. Some p53 targets act as “feedback regulators,” altering the activity of the kinases or the stability of p53. Other p53 targets are “output regulators” that trigger specific cellular outcomes, including cell cycle arrest and apoptosis.
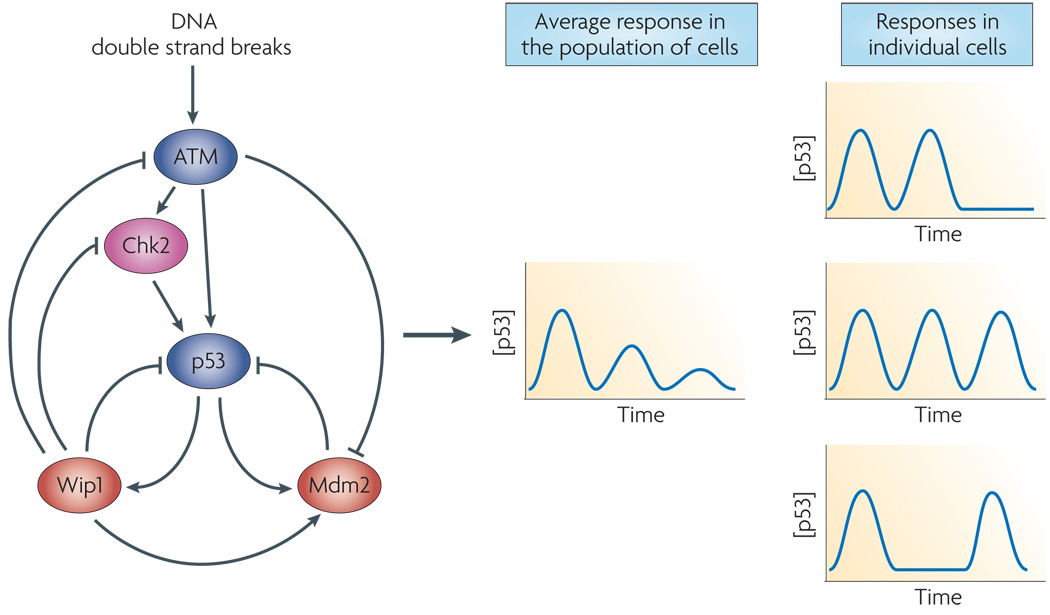
Previous studies revealed that p53 undergoes a complex dynamical response to DNA damage. Work by Lev Bar-Or et al.11 indicated that, in response to double strand breaks (DSBs) caused by γ-irradiation, p53 levels increased dramatically, then declined in a series of damped oscillations, in which the amplitude of the oscillations decreases in time (Figure 3). Single live cell analyses using fluorescently-tagged p53 and higher temporal resolution revealed that these population studies masked the true behavior of the network. Instead of damped oscillations, individual cells show series of undamped p53 pulses with fixed amplitude and duration, independent of the amount of γ-irradiation12, 13 (Figure 3). The initial characterization of the pulses as damped oscillations was a result of averaging across a population of cells. The apparently lower amplitude of p53 in later pulses as observed in western blots results from several factors, including a reduction in the number of cells pulsing at later times12 and loss of synchronization between individual cells13, 14.
Figure 3. p53’s response to DNA double strand breaks.
In response to DSBs ataxia telangiectasia mutated (ATM) kinase is activated64, and activates checkpoint kinase 2 (Chk2)65, 66. Both of these kinases upregulate p53 by disrupting its interaction with one of its target genes, the E3 ubiquitin ligase Mdm252, 53, 67–69. p53 also upregulates the transcription of the phosphatase Wip1 which negatively feeds back on the entire circuit by dephosphorylating ATM, Chk2, p53, and Mdm2. Solid lines represent protein-protein interactions, dashed lines represent transcriptional activation. Middle column; measurements averaged over populations of cells show damped oscillations of p53. Right column; single cell measurements show a series of undamped pulses with different cells showing different numbers of pulses.
Oscillations of p53 activity have also been observed in vivo using a mouse model. Hamstra et al.15 constructed a transgenic mouse line in which firefly luciferase was expressed from a p53-responsive promoter. Upon irradiation, oscillations of luciferase were observed in a p53-dependent manner in the intestinal tissue. The timing of the oscillations were consistent with those observed in cultured cancer cells11, 12, indicating that oscillatory dynamics in the p53 network is not limited to cultured human cancer cells. Interestingly, the response in vivo was tissue-specific, suggesting that cell-type specific differences in the p53 network might give rise to differences in p53 dynamical responses to the same stimulus.
Understanding p53 Dynamics: “Oscillations” Versus “Pulses”
The frequency of p53 pulses in response to γ-irradiation appears to be tightly controlled. Do these pulses truly constitute “oscillations” or are they actually a series of independent pulses? Is this distinction biologically relevant in a sense that it would provide mechanistic insight into the function of the network, or is it merely a matter of semantics? We believe it is crucial to distinguish between “oscillations” and “repeated independent pulses” since the functions of the two behaviors are generally different, and they are used to “solve” two different problems. Oscillators tend to be autonomous, and are often important for setting a well-regulated time-scale or sequence to biological events. In contrast, pulse generators respond to stimulation with a single burst of activity, which can be repeatedly triggered. In addition, the network structures and kinetic parameters that are required for regulating these behaviors are different. For example, oscillators can arise from a single negative feedback loop, whereas pulse generators often exhibit excitability as a result of a fast and strong positive feedback that is crucial in setting an activation threshold16, 17.
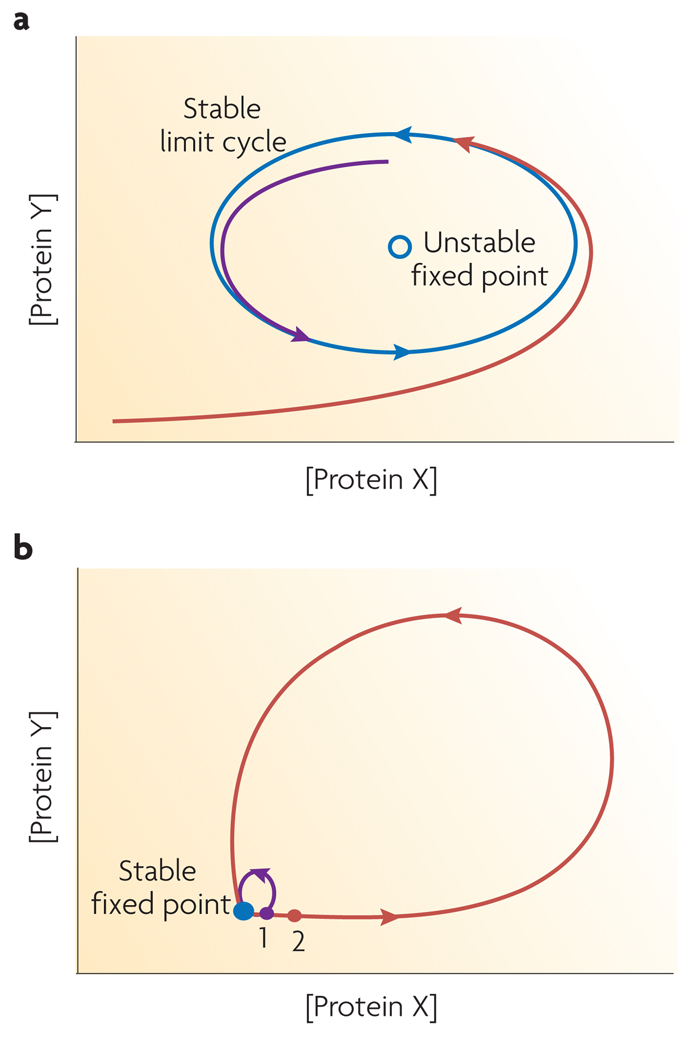
The p53 system shares several features with oscillating systems and specifically with a class of dynamical systems called “limit-cycle oscillators”16–18. The components of limit-cycle oscillators change in concentration or location in a regular, repetitive pattern. In general, the changes are resistant to small fluctuations from a basal temporal trajectory. In mathematical terms, this trajectory is referred to as a “stable limit cycle”16–18 (Figure 4a). When operating in the stable limit cycle, oscillators tend to have characteristic frequencies and amplitudes. Specific examples of biological limit cycle oscillators include the regulation of sustained cAMP oscillations in the amoeba Dictyostelium19, 20, regulation of circadian rhythms such as the PER-TIM system in Drosophila21 or the KaiABC system in cyanobacteria22, 23, and the eukaryotic mitotic clock24, 25.
Figure 4. Phase plane trajectories of oscillators and pulse generators.
(a) The concentration of proteins composing an oscillator follow a cyclic path in phase space called a limit cycle (blue). The limit cycle is stable in that fluctuations of protein concentrations away from the limit cycle are suppressed. If the concentrations of the proteins in a system are initially different from values on the limit cycle, the system will relax away from unstable fixed points and will gradually approach the limit cycle. Two sample trajectories with different initial protein concentrations are shown in purple and red. (b) The concentration of proteins in a pulse generator will remain at a stable fixed point as long as the system is unperturbed. If the system is perturbed slightly, as indicated in point 1, it relaxes back to the stable point with relatively small changes in the protein concentrations (purple trajectory). However, if the system receives a large enough perturbation, such as to point 2, the concentrations of the proteins in the system change greatly, making a large excursion in phase space before relaxing back to the stable fixed point (red trajectory).
An important (but not sufficient) component of oscillators is a negative feedback26. We have recently shown14 that p53 dynamics in response to DSBs are shaped by a combination of two negative feedbacks including the negative feedback between p53 and Mdm2, and the negative feedback between p53, ATM and Chk2 mediated by the phosphatase Wip127–30 (Figure 3). Careful analysis of the dynamics of these feedbacks revealed that the upstream regulators ATM and Chk2 show oscillations that are both required for and shaped by p53 dynamics14. Computational work on p53 dynamics has suggested that additional negative feedbacks on p53, such as the feedback mediated by ARF31, may also play a role in regulating p53’s dynamical response to DNA damage. Additional work is required to determine whether this and other known negative feedbacks on p5332 are required for p53 oscillations.
The p53 system also shares features with excitable “pulse generator” systems. In these systems, the basal state is an “off” state and it is referred to as a “stable steady state.” Minor fluctuations from the stable steady state can immediately return back to the stable point (Figure 4b). However, fluctuations from the steady state that are of sufficient size are amplified, usually through a strong positive feedback. The system is then excited and undergoes a large change in component concentrations, relative to the initial perturbation, before it returns to the stable steady state (Figure 4b). Examples of pulse generator systems include protein networks regulating action potentials in neurons33–36 and the switch from the vegetal growth state to the competent state in B. subtilis37. Recently, we showed that transient activation of ATM and Chk2 results in a full pulse of p5314 away from its basal state, suggesting that there may be an excitable mechanism controlling p53 pulses. The fact that a simple negative feedback loop can generate sustained oscillations but not excitability suggests the existence of a positive feedback in the network controlling p53 levels. Several positive feedbacks have been identified in the p53 network32, but additional studies are required to determine if these, or as yet unidentified positive feedbacks, are important in shaping the p53 response to DSBs.
So, does the p53 system behave as a limit cycle oscillator or as a pulse generator? Is it possible that the p53 system switches between both kinds of dynamical behaviors? In general, biological systems can generate different dynamical behaviors, switching between qualitatively distinct regimes depending on the concentrations of network components and values of kinetic parameters for a given condition. For example, in the absence of growth factors, mammalian cells are in a stable, non-oscillatory steady state in which the concentrations of cell cycle factors remain fixed. In the presence of growth factors, the cell cycle regulators are forced out of the stable steady state into a limit-cycle and continue to cycle as long as growth factors are present38, 39. Theoretical analysis of the developmental decision in B. subtilis showed that multiple regimes of qualitatively distinct behavior exist depending on key parameters in the network of interactions governing the system37. These regimes include both oscillatory and excitable pulse generating regions37. Another example is the behavior of giant squid axons. Experimental and mathematical analysis showed that stimulation away from a stable steady state above a threshold voltage leads to nerve signal propagation in an excitable manner33, 35. However, applying an additional biasing current to the nerve leads to the formation of an unstable state surrounded by a stable limit cycle, converting the system from a pulse generator to oscillations34. A similar case may hold for the p53 network; depending on cellular conditions, such as the amount of DNA damage or initial concentrations of regulatory proteins at the time of damage, the network may shift between pulse-generating and oscillatory behavior. One could even speculate that in the absence of severe DNA damage p53 levels are low and excitable. Any brief damage elicits one pulse of p53. Sustained external damage might destabilize the system’s steady state and lead to a series of oscillations until damage is repaired.
Based on our current understanding of the structure and function of the p53 network, we (at this point) favor the hypothesis that the network acts as a pulse generator in response to DSBs. It seems reasonable that, like a pulse generator, p53 is maintained in an “off” state until a stimulus is present. When a stimulus such as DNA damage is present, p53 shows a pulse only if the stimulus is large enough to push p53 over an activation threshold. After the completion of the pulse, if the stimulus is still present, and of sufficient magnitude, a subsequent pulse of p53 occurs. In this scenario, repeated activation would not require the formation of a stable limit cycle, but could arise due to repeated perturbation from a stable steady state (Fig. 4). We therefore choose the word “pulses” for describing p53 dynamics throughout the second half of this article while keeping in mind that additional research is required to determine the proper classification and characterization of p53’s dynamical behavior.
Possible Functions of the Pulses in the p53 Network
Even before we develop a complete understanding of the exact mechanism controlling p53 dynamics, we can begin to address the role of its dynamics in responding to DNA damage. One possible explanation is that ATM pulses14 are the ones that are important for proper repair of DNA DSBs, and the p53 pulses are only an epiphenomenon of this behavior (Figure 5). In this scenario, when a DSB occurs, break recognition proteins including the Mre11-Rad50-Nbs1 (MRN) complex40, 41 localize to the site of damage. ATM is recruited to sites of DSBs through interactions with the MRN complex, which is believed to be required for kinase activation42. Following activation of ATM, a pulse of p53 occurs14, leading to the transcription of several target genes including DNA repair proteins and negative feedback regulators. The negative regulator Wip1 plays an important role in inhibiting ATM27, resulting in a pulse of ATM activity14. We hypothesize that the inhibition of ATM may lead to dissociation of ATM from damage sites, thereby allowing repair proteins access to the damage. If a repair attempt is unsuccessful, a new round of ATM and p53 activation is initiated. In support of this idea, mutually exclusive binding of ATM and the repair cofactor XRCC4 to a site-specific DSB has been recently reported41. In contrast, many other proteins that associate with DSBs, including MDC1 and 53BP1, appear to remain stably associated with damage sites for prolonged times43. This suggests that if there is shuttling of proteins on and off damage sites, it may be specific to a subset of the proteins involved in the DSB response. Further studies, together with the development and refinement of technology for measuring kinase activity in single cells, are required to quantify ATM’s binding to the breaks over time and to determine how this affects the repair rate and the dynamics of many additional ATM substrates44.
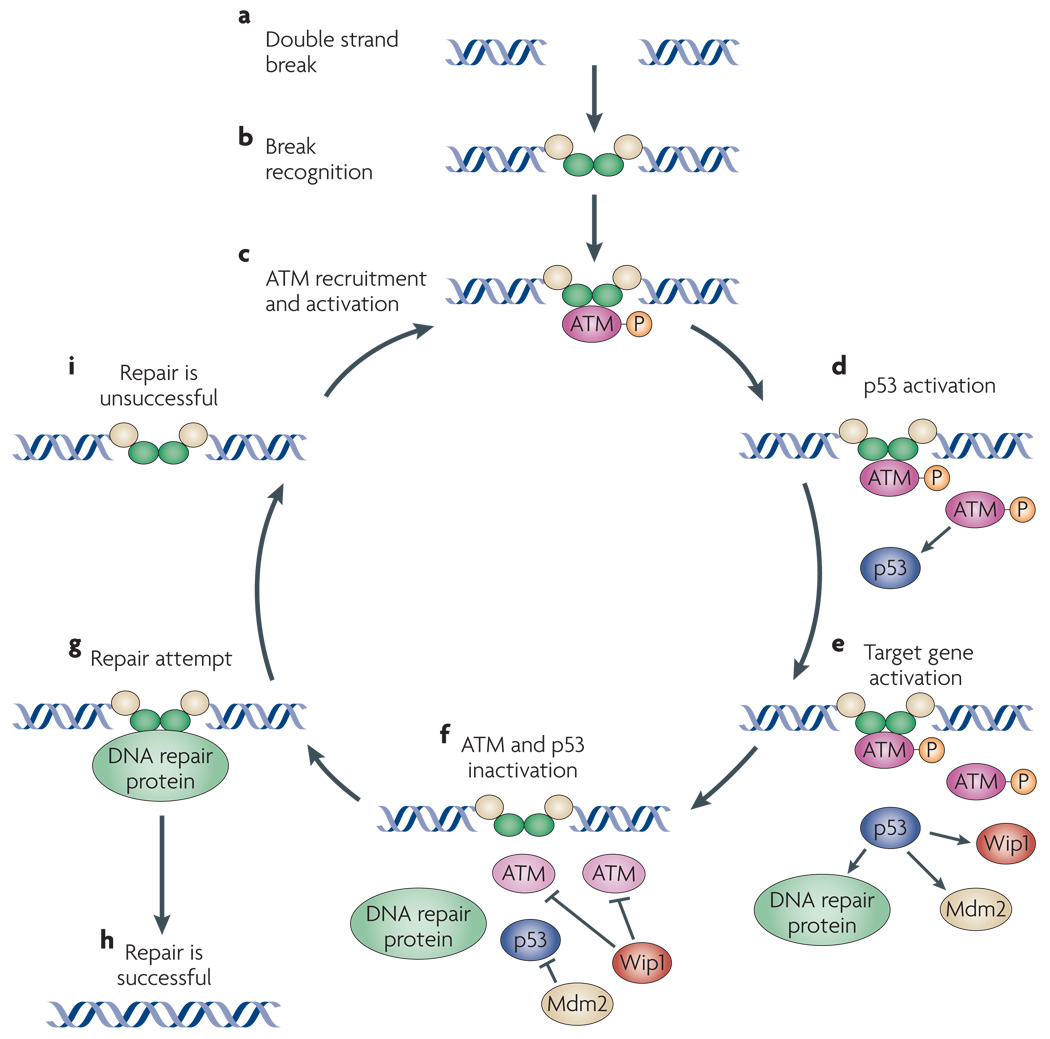
Figure 5. Model for the potential role of ATM pulses in the DNA repair process.
Upon occurrence of a DNA DSB (a), several proteins form a complex at the break site (b). ATM is recruited and activated by break recognition proteins (c). Once activated, ATM stabilizes p53 and triggers its accumulation. (d). p53 then transcriptionally upregulates the expression of several genes, including DNA repair genes and the inhibitor of ATM, Wip1 (e). Inhibition of ATM by Wip1 might lead to dissociation of ATM from the break (f), potentially allowing DNA repair proteins access to the DSB (g). If the DSB is repaired, ATM would not be recruited again and the signaling to p53 would be halted (h). If the DSB is not repaired (or if new breaks are detected), a new round of ATM recruitment would begin, resulting in a subsequent pulse of ATM and p53 (i).
On the other hand, it is possible that ATM pulses serve a purely regulatory function for driving p53 pulses in response to DSBs. The question then becomes what is the function of p53 pulses in regard to its activity as a transcription factor? One of the simplest explanations is that the p53 pulses function to keep p53 below a threshold concentration to prevent premature activation of certain target genes, for example pro-apoptotic genes. While this is certainly plausible, it is unlikely to be the entire reason for p53’s pulsatile behavior since simpler regulatory mechanisms, such as maintaining a constant low level of p53, could be used with similar results.
Are there any cellular benefits that could arise from p53 pulses? One possibility is that p53 pulses may coordinate regulation of p53’s target genes. Recent work in the yeast S. cerevisiae showed that the transcription factor Crz1 undergoes bursts of nuclear localization in response to calcium45. Using single cell time lapse microscopy, it was shown that the frequency, but not the amplitude, of Crz1 pulses increased with greater concentrations of calcium. This behavior led to expression from various Crz1 target promoters in fixed ratios across a wide range of calcium concentrations; while the absolute level of activity of any given promoter depended on the concentration of calcium, the ratio of the activity of one promoter to that of another promoter was independent of the calcium concentration. Such fixed ratios would not occur if the amplitude of Crz1 bursts changes with calcium concentration. These results suggest that frequency modulated pulses might coordinate the expression levels of multiple target genes without the need to specifically tune the activity of individual promoters. The fact that the amplitude of p53 pulses is independent of stimulus strength (γ-irradiation dose) suggests that the ratio of p53 target genes may be fixed in a manner similar to the targets of Crz1. It might be beneficial for cells to coordinate upregulation of large sets of cell cycle arrest or DNA damage repair genes using such a regulatory mechanism. However, as opposed to the Crz1 system, the frequency of p53 pulses appears to be tightly regulated and fixed13. Frequency modulation of p53 pulses, if it indeed exists and plays a role in its function, has yet to be observed.
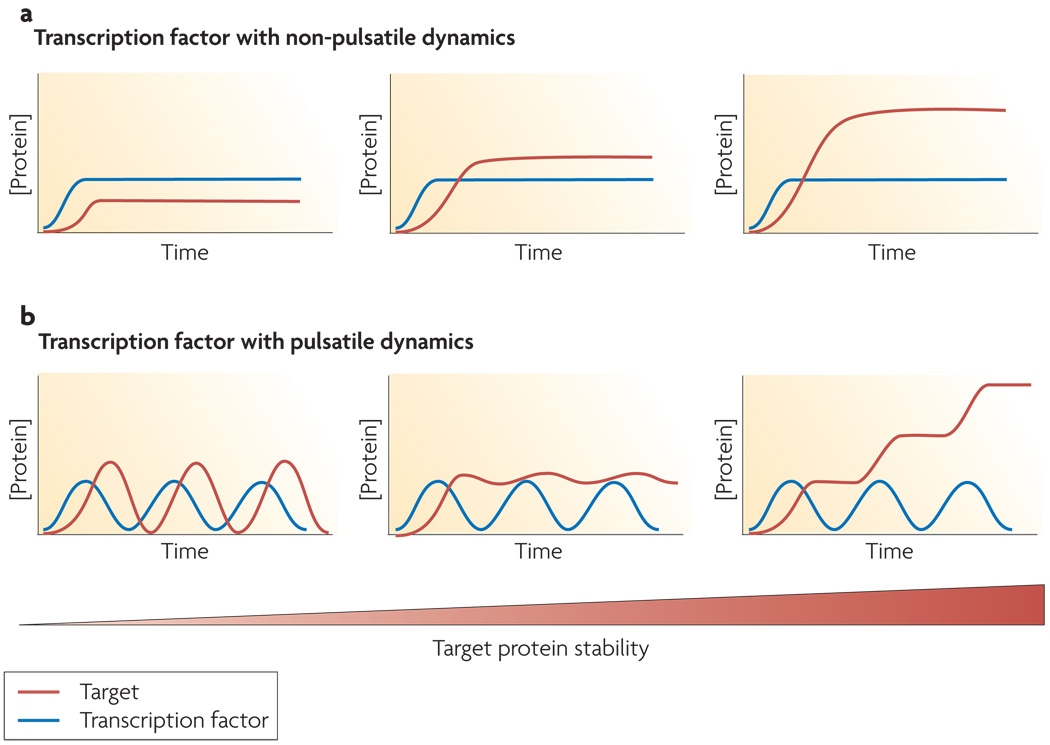
The widely varying functions of p53 targets pointed us recently to an alternative hypothesis for the function of its pulses. We suspect that the pulsatile dynamics of p53 increases the range of possible target gene dynamics compared with constant p53 levels. p53 regulates hundreds of genes46, which are involved in diverse functions such as cell cycle regulation and apoptosis. It is likely that cells would benefit from having distinct temporal expression patterns for genes in different programs. Constant p53 levels do not allow this wide range of behaviors since target genes with similar promoter activity but different protein degradation rates result in qualitatively similar temporal expression patterns (Figure 6a). Pulses of p53, however, allow a broader range of temporal patterns depending on the stability of the target’s mRNA and protein (Figure 6b). One simple example of an expression pattern that can result from a pulsing transcription factor is a pulsatile pattern. This pattern arises when the degradation rates of the target’s mRNA and protein are faster than the frequency of the transcription factor pulses. We observed such a pattern for the p53-regulated cell cycle regulator p2114. On the other hand, if the mRNA and protein half-life of the target gene are comparable to the frequency of the transcription factor pulse, the expression pattern remains constant following induction. If, however, the mRNA is rapidly degraded but the resulting protein is relatively stable, the target gene will show a step-like expression pattern. Further studies of the dynamics of different p53 target genes in response to DSBs are required to determine whether p53 pulses do translate to various dynamical patterns of its target genes.
Figure 6. Pulsing transcription factors can give rise to multiple dynamical patterns of their target genes.
(a) If upon activation a transcription factor becomes elevated to a new concentration that remains constant in time, the level of its target genes will also remain constant in time with a steady state level that depends on the target protein’s stability (b) If a transcription factor undergoes pulses upon activation, the dynamics of its target genes can take on a wider range of qualitatively distinct behaviors. For example, targets can show pulsatile, steady, or step-like expression patterns depending on the stability of the target protein.
Alternatively, it has recently been suggested that p53 pulses may be a fast and effective method to regulate p53’s post-translational modifications to effect an orderly temporal pattern of cellular responses47, 48. There are over 30 currently known sites of post-translational modification in p53, including numerous sites of phosphorylation, acetylation, and ubiquitination49–51. While some of the sites (such as serine-15, serine-18, and C-terminal lysines) are important for regulating p53 stability52–54, other sites alter p53’s gene regulatory functions (such as lysine-373 and lysine-382)55. It has been speculated that specific patterns of post-translational modification on p53 might act as a molecular barcode, indicating specific cellular response programs such as DNA repair or apoptosis56, 57. If this is true, orderly progression from one stage to the next might require eliminating a large number of current modifications and establishing a new barcode. Instead of reversing a large number of modifications on the current set of p53 molecules, it might be more efficient to degrade the existing p53 and synthesize new p53 with a different barcode.
Potential Connections between p53 Pulses and Cancer
As it is often noted, p53 is estimated to be mutated in half of all cancers, and mutations in the p53 network are estimated to occur in nearly all cancers. An interesting possibility is that some mutations in the p53 network could affect the regulation of p53 dynamics, which might then contribute to the transformation of cells and the development of cancer. Recent work has suggested a correlation between a particular polymorphism in the p53 network, changes in p53 dynamics, and predisposition to cancer. A single nucleotide polymorphism (SNP309G) in the mdm2 promoter increases expression of Mdm258. Hu et al.59 showed that cultured cells containing the wild-type SNP309T/T or heterozygous SNP309T/G sequences showed pulsatile p53 responses to DSBs caused by γ-irradiation. In contrast, cells carrying the homozygous SNP309G/G sequences responded to γ-irradiation by increasing the concentration of p53 and maintaining it at high levels without pulses59. Evidence has linked the SNP309G/G allele to earlier onset of several cancers, including breast carcinoma and colon carcinoma, in certain groups of patients60. However, contradictory data has also been presented that failed to find a statistical significance between SNP309G and tumor onset in other groups61–63. Clearly, more work is required to determine whether there is a connection between SNP309G, the alteration of p53 dynamics, and tumor progression. Yet, understanding the role of p53 dynamics in healthy cells, and developing new ways to manipulate p53 dynamics in transformed cells, may one day prove invaluable for cancer treatment.
Concluding Remarks
Our ability to monitor and interpret dynamical changes is an important step for understanding a wide range of signaling events in both healthy cells and in the context of disease. As was clearly the case for the p53 network, proper identification of network dynamics depends strongly on our ability to monitor cellular events at the single cell level. It is likely that we will identify additional systems exhibiting pulsatile or oscillatory dynamics as more analyses are performed at the single cell level with the appropriate temporal resolution. Combining our current knowledge of the interactions that exist within various networks with new information regarding the dynamics of network components will likely prove valuable in attempts to manipulate and control signaling systems for therapeutic purposes.
Acknowledgments
We thank all the members of our laboratory for useful discussions. This work was supported by National Institutes of Health Grant GM083303. E.B. was supported by the American Cancer Society, California Division, Pamela and Edward Taft Postdoctoral Fellowship.
References
- 1.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 3.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DE, See V, Nelson G, White MR. Oscillations in transcription factor dynamics: a new way to control gene expression. Biochem Soc Trans. 2004;32:1090–1092. doi: 10.1042/BST0321090. [DOI] [PubMed] [Google Scholar]
- 5.Hilioti Z, et al. Oscillatory phosphorylation of yeast Fus3 MAP kinase controls periodic gene expression and morphogenesis. Curr Biol. 2008;18:1700–1706. doi: 10.1016/j.cub.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 8.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 9.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 10.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 11.Lev Bar-Or R, et al. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lahav G, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 13.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100068. 2006 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamstra DA, et al. Real-time evaluation of p53 oscillatory behavior in vivo using bioluminescent imaging. Cancer Res. 2006;66:7482–7489. doi: 10.1158/0008-5472.CAN-06-1405. [DOI] [PubMed] [Google Scholar]
- 16.Murray JD. Mathematical Biology. In: Antman SS, Marsden JE, Sirovich L, Wiggins S, editors. New York, NY: Springer; 2002. [Google Scholar]
- 17.Edelstein-Keshet L. Philadelphia, PA: Society for Industrial and Applied Mathematics; 2005. Mathematical Models in Biology. [Google Scholar]
- 18.Strogatz SH. Cambridge, MA: Perseus Books Publishing, LLC; 1994. Nonlinear Dynamics and Chaos. [Google Scholar]
- 19.Halloy J, Lauzeral J, Goldbeter A. Modeling oscillations and waves of cAMP in Dictyostelium discoideum cells. Biophys Chem. 1998;72:9–19. doi: 10.1016/s0301-4622(98)00119-7. [DOI] [PubMed] [Google Scholar]
- 20.Tyson JJ, Murray JD. Cyclic AMP waves during aggregation of Dictyostelium amoebae. Development. 1989;106:421–426. doi: 10.1242/dev.106.3.421. [DOI] [PubMed] [Google Scholar]
- 21.Leloup JC, Gonze D, Goldbeter A. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J Biol Rhythms. 1999;14:433–448. doi: 10.1177/074873099129000948. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pomerening JR, Sontag ED, Ferrell JE., Jr. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 25.Tyson JJ, Csikasz-Nagy A, Novak B. The dynamics of cell cycle regulation. Bioessays. 2002;24:1095–1109. doi: 10.1002/bies.10191. [DOI] [PubMed] [Google Scholar]
- 26.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shreeram S, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto H, et al. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2005 doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-Mediated DNA Damage Response by the Oncogenic Phosphatase PPM1D. Cell Cycle. 2005;4 [PubMed] [Google Scholar]
- 30.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proctor CJ, Gray DA. Explaining oscillations and variability in the p53-Mdm2 system. BMC Syst Biol. 2008;2:75. doi: 10.1186/1752-0509-2-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 33.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzhugh R. Impulses and physiological states in theoretical models of nerve membrane. Biophys. J. 1961;1:445–466. doi: 10.1016/s0006-3495(61)86902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzhugh R. Thresholds and plateaus in the Hodgkin-Huxley nerve equations. J Gen Physiol. 1960;43:867–896. doi: 10.1085/jgp.43.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagumo J, Arimoto S, Yoshizawa S. An active pulse transmission line simulating nerve axon. Proc. IRE. 1962;50 [Google Scholar]
- 37.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 38.Murray AW, Kirschner MW. Dominoes and clocks: the union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- 39.Gonze D, Goldbeter A. A model for a network of phosphorylation-dephosphorylation cycles displaying the dynamics of dominoes and clocks. J Theor Biol. 2001;210:167–186. doi: 10.1006/jtbi.2000.2294. [DOI] [PubMed] [Google Scholar]
- 40.Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 43.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 45.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 47.Lahav G. The strength of indecisiveness: oscillatory behavior for better cell fate determination. Sci STKE. 2004;2004:pe55. doi: 10.1126/stke.2642004pe55. [DOI] [PubMed] [Google Scholar]
- 48.Zhang T, Brazhnik P, Tyson JJ. Exploring mechanisms of the DNA-damage response: p53 pulses and their possible relevance to apoptosis. Cell Cycle. 2007;6:85–94. doi: 10.4161/cc.6.1.3705. [DOI] [PubMed] [Google Scholar]
- 49.Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930–930. doi: 10.1016/j.cell.2008.05.020. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 51.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 52.Banin S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 53.Canman CE, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 54.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 55.Li AG, et al. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 57.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 58.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 59.Hu W, et al. A Single Nucleotide Polymorphism in the MDM2 Gene Disrupts the Oscillation of p53 and MDM2 Levels in Cells. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-06-2656. in press. [DOI] [PubMed] [Google Scholar]
- 60.Bond GL, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 61.Petenkaya A, et al. Lack of association between the MDM2-SNP309 polymorphism and breast cancer risk. Anticancer Res. 2006;26:4975–4977. [PubMed] [Google Scholar]
- 62.Campbell IG, Eccles DM, Choong DY. No association of the MDM2 SNP309 polymorphism with risk of breast or ovarian cancer. Cancer Lett. 2006;240:195–197. doi: 10.1016/j.canlet.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Krekac D, et al. MDM2SNP309 does not associate with elevated MDM2 protein expression or breast cancer risk. Oncology. 2008;74:84–87. doi: 10.1159/000139135. [DOI] [PubMed] [Google Scholar]
- 64.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 65.Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- 66.Matsuoka S, et al. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khosravi R, et al. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]