Abstract
We examined the spatial structure of Schistosoma mansoni, a parasite of humans, from natural infections at two levels: across the Lake Victoria basin of Kenya and among snail hosts. Using 20 microsatellite markers we examined geographic patterns of relatedness and population structure of cercariae and found weak, but significant structure detected by some, but not all analyses. We hypothesize structure created by aggregations of clonal individuals or adherence of hosts to local transmission sites is eroded by high amounts of gene flow in the region. This finding also supports previous hypotheses concerning the evolution of drug resistance in the region. Intrasnail dynamics were investigated in the context of aggregation and kin selection theory to determine how relatedness and also sex influence host sharing and host exploitation. Cercarial production did not differ significantly between snails with one or two genotypes suggesting that mixed infections resulted in decreased individual fitness and provides a framework for reproductive competition. Coinfection patterns in snails were independent of parasite relatedness indicating that schistosomes were not aggregated according to their relatedness and that kin selection was not influencing host sharing. Additionally, host exploitation in coinfections (measured by cercarial production) was not negatively correlated with relatedness, as predicted by classical models due to increased competition and thus exploitation when parasites are unrelated. Because of the low levels of relatedness within the population, schistosomes may rarely encounter close relatives and kin selection mechanisms that influence the distribution of individuals within snails or the virulence mode of the parasites may simply have not evolved.
Keywords: Schistosoma mansoni, Genetic Structure, Lake Victoria, Kenya, Coinfection, Host Sharing, Competition, Kin Selection
Understanding the transmission patterns of pathogens among host populations is an important goal for the control of infectious diseases. Population genetics can be used to reveal patterns of pathogen transmission. For instance, population subdivision into separate transmission cycles, the amount of gene flow among subpopulations, and the role of hosts and geography in structuring subpopulations can be revealed with these methods (Tibayrenc, 1999; Criscione et al., 2005; Archie et al., 2009). This information also can be used to predict evolutionary dynamics of pathogens such as the evolution of drug resistance alleles and local adaptation or divergence of pathogens (Criscione et al., 2005; Huyse et al., 2005).
Schistosomiasis is a chronic and debilitating disease that is estimated to infect 207 million people world wide, most of whom are in sub-Saharan Africa (King et al., 2005; Steinmann et al., 2006). Schistosoma mansoni is the primary etiological agent of schistosomiasis in the Lake Victoria basin of Africa, a region where schistosomiasis has a serious impact on human health (Kardorff et al., 1997; Handzel et al., 2003; Steinmann et al., 2006; Steinauer et al., 2008c). Previous studies of genetic structure in Kenya have revealed population subdivision across large geographic regions (Agola et al., 2006), but this question has not been investigated within the Lake Victoria basin. This region is of particular interest because of an ongoing longitudinal study that has involved the repeated use of Praziquantel, the primary drug to treat schistosomiasis (Karanja et al., 2002). Although there is no evidence of clinical resistance (Black et al., 2009), drug tolerant parasites have been found in this population (Melman et al., In Review).
Population substructure is formed in large part by the amount and patterns of gene flow of the parasites, which is strongly influenced by the movement of their hosts (Blouin et al., 1995; Blouin et al., 1999; McCoy et al., 2003; Criscione and Blouin, 2004). S. mansoni develops within two hosts, a vertebrate definitive host, typically humans, and a snail intermediate host, snails of the genus Biomphalaria. Also, free living stages, cercariae or miracidia, may have the capability of dispersal through active swimming or via water currents. While dispersal acts to erode genetic structure, spatial aggregation works to create it. In many ways, the life cycles of parasites can create spatial aggregation such as the release of eggs in feces or the release of cercariae from a single snail point source. Furthermore, aggregation can be amplified through the life cycle because it increases the probability that infective stages will be cotransmitted to the same individual hosts (Lotz et al., 1995; Vickery and Poulin, 2002; Keeney et al., 2007). For instance, a fecal sample from a host will contain eggs of related parasites. Because the eggs are spatially aggregated, an individual snail may acquire related worms, and then release the related cercariae, which are likely to encounter the same individual definitive hosts. High aggregation can lead to the formation of local transmission sites containing unique parasite demes (Criscione and Blouin, 2006).
Another consideration is polyembryony within the snail host, resulting in the daily release of hundreds to thousands of cercariae into the environment during patent infection. Particularly if the transmission success of clones is highly variable, so that some clones are overrepresented in the adult population, they will produce most of the offspring which will be related as full or half siblings. If dispersal of the offspring is minimal, such aggregation could create local pedigree structure, which has an effect on the population genetic structure and also could influence intraspecific interactions.
Intraspecific interactions of parasites within natural populations can also be explored using population genetics. The degree of relatedness between individuals that share a host is predicted to play a large role in determining the outcome of their interaction, and the extent to which they harm their host (Reviewed by Buckling and Brockhurst, 2008). In single infections, the optimal strategy is for the parasite to maximize its reproductive rate to maximize fitness without killing its host (Levin and Pimentel, 1981; Bremermann and Pickering, 1983; Buckling and Hodgson, 2007). However, hosts are often exposed to multiple individuals of a parasite species, especially if the infective stages are aggregated in the environment. If resources are limited, a conflict ensues between parasites that share a host. Classically, related pathogens are expected to cooperate resulting in decreased host exploitation, while non-related pathogens are expected to compete resulting in increased reproduction and host exploitation (Hamilton, 1972; Frank, 1992, 1996). If schistosomes can modulate their reproductive rate based on the relatedness of a competitor, the outcome of this interaction may be detected in the population. If coinfections of nonrelatives lead to exclusion or host death, then the parasites that share a host will be more related to each other than expected due to the background relatedness of the parasites. Also, if coinfections of non-relatives lead to increased host exploitation via increased reproduction and relatives moderate exploitation, there will be a negative relationship between relatedness and reproductive rate. This relationship will also give insight into whether this schistosome system fits the predictions of more recent alternative models (Chao et al., 2000; Brown et al., 2002; Buckling and Brockhurst, 2008).
Another factor that could influence host sharing and competition of dioecious parasites is the sex of the participants. The fragmented nature of their habitats, which are individual hosts, can be problematic for dioecious parasites because when a parasite colonizes a host, there is no guarantee that an available mate will be present. Therefore, strategies that increase the probability of co-transmission of mates, like facilitating dual sex infections in snails, could be highly advantageous if the cost of host sharing is not too high.
We examined natural populations of S. mansoni in the Lake Victoria basin in western Kenya to determine the geographic transmission dynamics of this parasite and also investigate intraspecific interactions that may influence transmission among hosts. First, we examined the genetic structure of schistosomes in snails in the Lake Victoria basin using a hierarchical approach. Fine scale structure was determined with spatial patterns of relatedness values. Also, pairwise F-statistics (Wright, 1932) and Bayesian clustering methods were used to determine patterns of subdivision and gene flow. Using these data, we addressed the question of whether schistosomes in the Lake Victoria basin of Kenya function as a single panmictic population or if separate collection sites or water bodies represent subpopulations or separate transmission foci.
We also examined relatedness patterns of individual parasites that shared a snail host. Both spatial aggregation and intraspecific interactions may influence whether related parasites share a host and how relatedness influences their reproductive output. First, we examined the patterns of relatedness of individuals in coinfections to determine if those within a single snail were more related to each other than expected based on background levels of relatedness (predicted by aggregation and kin selection theory). Second, the asexual reproductive output of parasites in single and multiple infections was compared to determine if snails represent a limited resource to the production of infective stages, thus suggesting the potential for interaction among individuals that share a snail host. Third, to determine the association between relatedness and cercarial reproduction, we examined the relatedness of coinfecting individuals and the number of cercariae produced from the infection. We also examined whether or not coinfections were gender biased since bisexual infections may be more favorable if they permit cotransmission of mates. We used a field based approach to address these questions so that a natural population of hosts and parasites and their true relatedness structure could be investigated. This is in contrast to experimental studies using inbred lines of parasites that may not be relevant to what a parasite actually encounters in the natural system. In this regard, a field study places the theory in the context of the natural system and provides a valuable perspective even if all variables cannot be controlled.
Materials and Methods
Collection of Snails and Parasites and Molecular Techniques
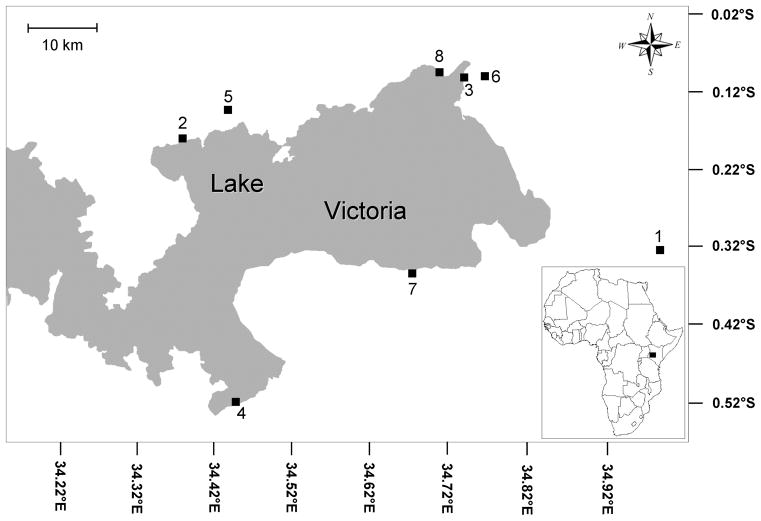
Snails (Biomphalaria sudanica and B. pfeifferi) were collected around the Kenyan portion of Lake Victoria basin (Fig. 1, Table 1) from November 2004–February 2007 (Steinauer et al., 2008c). Snails were examined for infection by bringing them into the laboratory, isolating them in wells of a 24 well plate, and monitoring them for 48 hours for emerging cercariae. Cercariae from infected snails were used to infect mice via skin penetration of the abdomen while the mice were anesthetized with sodium pentobarbital. Infection doses of 10 to 200 cercariae were used depending on the number released by the snail, and 1–8 mice were infected with cercariae for each snail depending on how many cercariae were released and how long the snail survived. Recovery of adult worms from mice 7 weeks post-exposure was accomplished by perfusion (Smithers and Terry, 1965). Gender of the worms was determined by examining adult morphology. The HotSHOT (Truett et al., 2000) method was used to prepare genomic DNA of the worms for PCR. The number of genotypes per snail was previously determined (Steinauer et al., 2008c) using 7 microsatellite loci. The total number of adults genotyped was 4,777, with a mean of 34.1 per snail (2.5 standard error), range of 8–217, and median of 24 adults per snail. Individuals with the same genotypes at all 7 loci that emerged from the same snail were considered to be clones descended from a single miracidium and are referred to as a multilocus genotype. The probability that each multilocus genotype was produced from independent sexual reproduction events rather than clonal reproduction was low and ranged from 1.2 × 10−27 to 7.4 × 10−4 for each multilocus genotype (Steinauer et al., 2008c). No identical multilocus genotypes were found among snails. A total of 182 multilocus genotypes of S. mansoni were collected from over 22,000 snails (158 infected). Each multilocus genotype was further characterized so that a total of 22 loci were amplified (Steinauer et al., 2008b).
Figure 1.
Map of the Lake Victoria region of Kenya and sites sampled for this study. Collection locality numbers correspond to Table 1.
Table 1.
Collection sites for snails of the genus Biomphalaria and their schistosome parasites in Western Kenya. Global Positioning System coordinates are projected in North American Datum 1983 decimal degrees. Sample size of Schistosoma mansoni nsm collected from each site is given.
| Site Name | Map # | Habitat | South | East | nSM |
|---|---|---|---|---|---|
| Asao | 1 | Stream | −0.3325600 | 34.9991440 | 49 |
| Asembo Bay | 2 | Lake | −0.1885080 | 34.3875340 | 25 |
| Kisumu | 3 | Lake | −0.0958667 | 34.7485944 | 50 |
| Homa Bay | 4 | Lake | −0.5226060 | 34.4545590 | 6 |
| Kasabong | 5 | Stream | −0.1519060 | 34.4455280 | 8 |
| Nyabera | 6 | Marsh | −0.1097139 | 34.7746111 | 20 |
| Seka Kagwa | 7 | Lake | −0.3555556 | 34.6827778 | 5 |
| Sandharvester Site | 8 | Lake | −0.1013889 | 34.7147222 | 19 |
Geographic Structure
Hardy-Weinberg equilibrium tests at each locus and genotypic disequilibrium tests for pairs of loci were performed within populations using GENEPOP 4.0 (Rousset, 2008). Significance tests were performed using the Markov chain method (10,000 dememorizations, 10,000 batches, 10,000 iterations per batch) with sequential Bonferroni corrections for multiple comparisons within each sample (Rice, 1989). Because prevalence of infection in snails is often less than 1%, sample sizes at some of the sites were low; therefore, we performed these tests in five of the populations from which more than 18 schistosome individuals were sampled. This dataset included 163 individuals. The software package, MICRO-CHECKER (Van Oosterhout et al., 2004), was used to identify loci that potentially are affected by the presence of null alleles, long allele dropout, or scoring errors due to stuttering. Diversity including the number of alleles (A), allelic richness (Rs, number of alleles rarefied to the smallest sample size), Nei’s estimator of unbiased heterozygosity (Hs) (Nei, 1987), and the Weir and Cockerham estimator f of Fis (Weir and Cockerham, 1984) were calculated for each locus for each population and the population considered as a whole using FSTAT 2.9.3 (Goudet, 2001), and results are shown in Table 2. Because sex specific genetic structure has been reported in a population of schistosomes that infects rats in Guadaloupe (Prugnolle et al., 2002), we grouped the individuals by sex and tested for differentiation with 10,000 permutations of individuals among sex using FSTAT, and calculated values of Hs and Fis including the 95% confidence intervals obtained through bootstrapping procedures. This analysis was performed without regard to geographic location and by only using the largest “lake” population.
Table 2.
Diversity statistics for Schistosoma mansoni in the Lake Victoria Basin by locus and collection sites (water bodies), and averaged over all loci. Sample sizes are given in parentheses after the collection site name. For each of 20 microsatellite loci, and the mean and standard deviation (SD) for all loci, the number of alleles (A), Allelic richness (Rs) rarefied to the smallest sample size, observed heterozygosity (Ho), and gene diversity (Hs), and Weir and Cockerham’s Fis values are given.
| Locus | Asao (49) | Kasabong (8) | Lake (105) | Nyabera (20) | Total | |
|---|---|---|---|---|---|---|
| AF325697 | A | 13 | 9 | 21 | 11 | 22 |
| Rs | 4.14 | 9.000 | 5.106 | 6.678 | 5.197 | |
| Ho | 0.388 | 0.875 | 0.495 | 0.800 | 0.638 | |
| Hs | 0.378 | 0.766 | 0.491 | 0.673 | 0.59 | |
| Fis | −0.016 | −0.077 | −0.001 | −0.165 | −0.022 | |
| AF202966 | A | 3 | 1 | 4 | 2 | 4 |
| Rs | 1.603 | 1.000 | 1.558 | 1.000 | 1.516 | |
| Ho | 0.082 | 0.000 | 0.076 | 0.050 | 0.052 | |
| Hs | 0.079 | 0.000 | 0.074 | 0.049 | 0.052 | |
| Fis | −0.021 | N/A | −0.024 | 0 | −0.025 | |
| AF325698 | A | 28 | 11 | 42 | 17 | 48 |
| Rs | 9.1 | 11 | 9.519 | 9 | 9.519 | |
| Ho | 0.796 | 1.000 | 0.781 | 0.850 | 0.856 | |
| Hs | 0.815 | 0.891 | 0.862 | 0.774 | 0.857 | |
| Fis | 0.033 | −0.057 | 0.099 | −0.073 | 0.059 | |
| AF325695 | A | 11 | 8 | 13 | 12 | 13 |
| Rs | 7.570 | 8.000 | 7.976 | 9.000 | 7.955 | |
| Ho | 0.816 | 0.750 | 0.810 | 0.950 | 0.831 | |
| Hs | 0.872 | 0.844 | 0.884 | 0.886 | 0.896 | |
| Fis | 0.074 | 0.176 | 0.091 | −0.046 | 0.073 | |
| L46951 | A | 17 | 9 | 19 | 17 | 20 |
| Rs | 9.855 | 9.000 | 10.090 | 11.000 | 9.997 | |
| Ho | 0.939 | 1.000 | 0.952 | 1.000 | 0.973 | |
| Hs | 0.919 | 0.852 | 0.928 | 0.913 | 0.925 | |
| Fis | −0.012 | −0.109 | −0.022 | −0.07 | −0.028 | |
| AF202968 | A | 3 | 3 | 6 | 4 | 6 |
| Rs | 2.844 | 3.000 | 3.561 | 3.000 | 3.315 | |
| Ho | 0.469 | 0.500 | 0.648 | 0.500 | 0.531 | |
| Hs | 0.526 | 0.617 | 0.618 | 0.569 | 0.599 | |
| Fis | 0.118 | 0.253 | −0.057 | 0.146 | 0.019 | |
| AF202965 | A | 9 | 6 | 8 | 8 | 11 |
| Rs | 5.791 | 6.000 | 5.248 | 6.000 | 5.583 | |
| Ho | 0.837 | 0.625 | 0.760 | 0.947 | 0.792 | |
| Hs | 0.787 | 0.695 | 0.717 | 0.805 | 0.77 | |
| Fis | −0.052 | 0.167 | −0.056 | −0.151 | −0.036 | |
| BF936409 | A | 10 | 7 | 9 | 7 | 10 |
| Rs | 5.996 | 7.000 | 6.205 | 6.000 | 6.277 | |
| Ho | 0.837 | 1.000 | 0.886 | 1.000 | 0.93 | |
| Hs | 0.784 | 0.828 | 0.822 | 0.816 | 0.832 | |
| Fis | −0.057 | −0.143 | −0.07 | −0.201 | −0.075 | |
| M85305 | A | 13 | 10 | 16 | 8 | 17 |
| Rs | 7.594 | 10.000 | 8.546 | 7.000 | 8.241 | |
| Ho | 0.816 | 1.000 | 0.857 | 0.850 | 0.881 | |
| Hs | 0.865 | 0.875 | 0.885 | 0.835 | 0.888 | |
| Fis | 0.066 | −0.077 | 0.039 | 0.008 | 0.038 | |
| AI395184 | A | 5 | 5 | 7 | 5 | 9 |
| Rs | 4.206 | 5.000 | 4.142 | 4.000 | 4.184 | |
| Ho | 0.592 | 0.750 | 0.533 | 0.500 | 0.593 | |
| Hs | 0.603 | 0.680 | 0.619 | 0.416 | 0.595 | |
| Fis | 0.029 | −0.037 | 0.15 | −0.176 | 0.083 | |
| AI067617 | A | 9 | 4 | 14 | 6 | 15 |
| Rs | 4.509 | 4.000 | 4.724 | 5.000 | 4.619 | |
| Ho | 0.551 | 0.500 | 0.571 | 0.550 | 0.542 | |
| Hs | 0.643 | 0.602 | 0.614 | 0.658 | 0.647 | |
| Fis | 0.154 | 0.233 | 0.073 | 0.188 | 0.111 | |
| R95529 | A | 16 | 7 | 20 | 18 | 26 |
| Rs | 8.508 | 7.000 | 8.055 | 10.000 | 8.793 | |
| Ho | 0.857 | 0.875 | 0.829 | 0.900 | 0.867 | |
| Hs | 0.876 | 0.813 | 0.869 | 0.908 | 0.89 | |
| Fis | 0.031 | −0.01 | 0.042 | 0.034 | 0.044 | |
| BH795456 | A | 14 | 10 | 16 | 13 | 16 |
| Rs | 8.946 | 10.000 | 9.320 | 9.000 | 9.256 | |
| Ho | 0.918 | 0.875 | 0.895 | 1.000 | 0.924 | |
| Hs | 0.902 | 0.875 | 0.915 | 0.899 | 0.921 | |
| Fis | −0.008 | 0.067 | 0.016 | −0.087 | 0.001 | |
| L81235 | A | 8 | 4 | 8 | 6 | 12 |
| Rs | 4.626 | 4.000 | 4.294 | 4.000 | 4.520 | |
| Ho | 0.714 | 0.875 | 0.638 | 0.600 | 0.708 | |
| Hs | 0.746 | 0.750 | 0.704 | 0.710 | 0.747 | |
| Fis | 0.053 | −0.101 | 0.087 | 0.18 | 0.082 | |
| AI068335 | A | 5 | 4 | 7 | 7 | 9 |
| Rs | 4.771 | 4.000 | 5.305 | 5.000 | 5.251 | |
| Ho | 0.813 | 0.625 | 0.829 | 0.850 | 0.779 | |
| Hs | 0.784 | 0.680 | 0.798 | 0.785 | 0.782 | |
| Fis | −0.026 | 0.146 | −0.032 | −0.057 | −0.021 | |
| AI067567 | A | 14 | 8 | 16 | 11 | 19 |
| Rs | 8.003 | 8.000 | 8.052 | 8.000 | 7.912 | |
| Ho | 0.854 | 0.875 | 0.876 | 0.900 | 0.876 | |
| Hs | 0.878 | 0.781 | 0.886 | 0.859 | 0.873 | |
| Fis | 0.038 | −0.054 | 0.017 | −0.022 | 0.014 | |
| AI110905 | A | 13 | 9 | 15 | 11 | 16 |
| Rs | 6.37 | 9.000 | 6.475 | 7.000 | 6.581 | |
| Ho | 0.857 | 0.875 | 0.762 | 0.800 | 0.825 | |
| Hs | 0.806 | 0.867 | 0.778 | 0.770 | 0.826 | |
| Fis | −0.053 | 0.058 | 0.013 | −0.013 | −0.004 | |
| M85304 | A | 11 | 5 | 12 | 8 | 14 |
| Rs | 5.307 | 5000 | 5.755 | 6.000 | 5.656 | |
| Ho | 0.673 | 0.500 | 0.657 | 0.684 | 0.63 | |
| Hs | 0.693 | 0.500 | 0.695 | 0.741 | 0.677 | |
| Fis | 0.039 | 0.067 | 0.056 | 0.103 | 0.056 | |
| AF3256924 | A | 9 | 7 | 10 | 6 | 10 |
| Rs | 5.779 | 7.000 | 5.871 | 5.000 | 5.797 | |
| Ho | 0.857 | 1.000 | 0.838 | 0.789 | 0.871 | |
| Hs | 0.786 | 0.805 | 0.799 | 0.742 | 0.801 | |
| Fis | −0.080 | −0.179 | −0.046 | −0.036 | −0.063 | |
| L25065 | A | 11 | 6 | 11 | 8 | 12 |
| Rs | 6.582 | 600 | 6.857 | 7.000 | 6.781 | |
| Ho | 0.796 | 0.375 | 0.848 | 0.789 | 0.702 | |
| Hs | 0.832 | 0.797 | 0.849 | 0.835 | 0.854 | |
| Fis | 0.053 | 0.576 | 0.006 | 0.082 | 0.052 | |
| All | Ho | 0.723 | 0.744 | 0.727 | 0.765 | 0.740 |
| Hs | 0.729 | 0.726 | 0.740 | 0.732 | 0.751 | |
| Fis | 0.018 | 0.042 | 0.020 | −0.020 | 0.019 |
To test for the presence of population structure of S. mansoni in the Lake Victoria basin indicative of separate local transmission foci, we examined patterns of relatedness among geographic sites (fine scale genetic structure), compared F-statistics among sites, and used a Bayesian clustering method that does not rely on a priori delimitation of populations. For the analyses in which a priori designations were required (all but Bayesian clustering), we designated populations at two spatial scales: by considering only the 5 sites that had samples larger than 18 individuals (sites), and by combining samples based on the boundaries of different water bodies including individuals from the Lake, Asao Stream, Nyabera Marsh, and Kasabong Stream (water bodies).
For the fine scale genetic structure analyses we followed the methodology of Vekemans and Hardy (2004) and examined the patterns of relatedness (r) with regard to site only and with regard to distance among collection localities (analyses were performed on both the sites and water bodies sampling regimes). This type of analysis detects the formation of local pedigree structure. Even though we did not collect adults, this pattern may be detected in the cercariae if transmission is geographically localized, and a few genotypes are responsible for the production of the majority of offspring due to their transmission success. In this case, the parasites within a geographic area will be more related to each other than those from other geographic locations. This will be particularly true if reproductive variance is highly skewed among families (which could be exacerbated by a high number of clones in a host) or the number of reproducing parasites is low (Criscione and Blouin, 2006). This effect may also be enhanced by the longevity of adult worms (6–10 years) (Fulford et al., 1995). Patterns of relatedness were investigated with SPAGeDi 1.2 (Hardy and Vekemans, 2002) and ML-RELATE (Kalinowski et al., 2006). ML-RELATE uses the downhill simplex routine to find the maximum likelihood estimates of r, and SPAGeDi calculates relatedness statistics as described by Loiselle et al. (1995) (rn), Queller and Goodnight (1989) (rqg), Lynch and Ritland (1999) (rlr), and Wang (2002) (rw). Permutation tests (10,000 permutations) were performed on the four relatedness statistics to test the hypothesis that the average relatedness of individuals that inhabit a site was greater than a random distribution of the parasites. To test for the effects of distance, relatedness coefficient values were regressed on the linear spatial distance among sites and also the natural logarithm of distance and tested against 10,000 permutations of individuals among sites using SPAGeDi 1.2.
Bayesian clustering analysis was implemented by STRUCTURE 2.2.3 (Pritchard et al., 2000). This method does not rely on predefined populations and instead delineates clusters of individuals in Hardy-Weinberg equilibrium and assigns each individual a probability of belonging to a cluster (Pritchard et al., 2000); therefore all samples were included. We performed runs with predefined numbers of clusters (K) ranging from 1 to 10, and performed 5 replicates of each using admixture and correlated allele frequency models. Each run was composed of a burnin period of 1 million MCMC, followed by 100,000 iterations. To ensure the length of the run was sufficient, we checked that the likelihood values of the runs had stabilized among replicates. The optimal number of clusters, K, was determined by the maximum values of lnP(D), which is an estimate of the posterior probability of the data for a given K value. Also, the assignment patterns of individuals were investigated to determine if any individuals were strongly associated with a cluster or if they had a relatively equal chance of belonging to any of them.
Population differentiation was also investigated using fixation indices that were calculated using the method of Weir and Cockerham (1984) and the presence of population structure was tested with 10,000 permutations of genotypes among sites using FSTAT 2.9.3 (Goudet, 2001). Pairwise Fst values were calculated using the same software and tested for significant differences between populations with 6,000 permutations. Since subpopulations must be defined a priori for theseanalyses and these parameters were unknown, we tested both the sites and water bodies sampling regimes. All Fst values reported were standardized by calculating the maximum Fst value given the data using RecodeData v. 0.1 (Meirmans, 2006) and dividing the Fst values calculated from the data by the maximum values. This standardization allows comparison of Fst values across studies.
Ideally, population genetics analyses should be performed on a subsample of the adult population. However, it is not possible to obtain adult schistosomes from human transmitted systems because they live within the mesenteric veins and cannot be easily or ethically removed from a living person. Consequently, we sampled cercariae from snails to infer patterns of transmission. Although indirect, the cercariae represent individuals that are being transmitted from snails and should represent the adult population unless very few adults produce the majority of cercariae. In this case, the presence of sibling groups could lead to the conclusion of false genetic structure (Li and Hedgecock, 1998; Waples, 1998). Fortunately, microsatellite data allow the determination of relationship structure so that sibling groups can be detected. Cercariae are less likely to be affected by a skew in adult reproduction than miracidia, which are obtained directly from fecal samples and suggested as an alternative method for schistosome population genetics (e.g. Curtis et al., 2002; Shrivastava et al., 2005; Gower et al., 2007; Steinauer et al., 2008a; Thiele et al., 2008).
Intrasnail Dynamics
To test the hypothesis that the average relatedness of individuals that coinfect the same snail host was different from a random distribution of parasites. Permutation tests were performed with SPAGeDi as described for testing relatedness patterns across geography except individuals were grouped by snail rather than geographic locations.
To determine if cercarial production (as a measure of host exploitation) differed among snails infected with multiple v. single genotypes, an analysis of covariance (ANCOVA) was performed using the natural log transformed values of cercarial abundance as a dependent variable. The model included the natural log of snail size as a covariate, the infection category (single v. multiple infection), and the interaction between these variables as the independent variables. Cercarial production was measured hourly during a 24 hour period and replicated every 4 days as long as the snail lived (Steinauer et al., 2008c). Because the number of replicates varied among snails, cercarial abundance was calculated in two ways: as the mean of all replicates per snail, and as the first observation for each snail. Results did not differ among these methods. To determine if relatedness correlated with the number of cercariae shed as a measure of exploitation, a Pearson correlation was performed on the pairwise relatedness values (rw) and number of cercariae shed for each replicate for each snail and a one tailed significance test was performed. If only one replicate per snail was included, the results did not differ. Snail size was not considered in this analysis as it was not a significant factor in determining cercarial production as determined from the previous analysis (see Results).
We used cercarial production as a measure of host exploitation in terms of the resources used by the parasite to create parasite infective stages and the energetic cost to the snail. Schistosome parasites acquire the majority of their resources directly from snail tissues; therefore greater cercarial production requires a larger amount of resources taken from the snail. We expect this to be a general pattern in a natural population, and it was detected Davies et al. (2002) in a laboratory experiment. However, there may be exceptions due to host-parasite compatibility such as those detected when comparing selected lines of parasites and snails (e.g. Davies et al., 2001).
To determine if dual sex coinfections were more common than single sex coinfections, a χ2 goodness of fit was used to compare the observed and expected ratios. Since sex ratios were not even, permutations were used to determine the expected numbers.
This project has been fully approved for the use of animals by the University of New Mexico Institutional Animal Care and Use Committee (Protocol #07UNM003) and Board of Animal Care and Use of the Kenya Medical Research Institute.
Results
Geographic Dynamics
Marker Validation
After Bonferroni corrections, none of the loci showed statistical deviation from Hardy-Weinberg proportions, although P-values were low, but not significant after correction (0.0009, 0.0017) for the X77211 locus for two populations, Asembo Bay and Kisumu, respectively. The analysis with MICRO-CHECKER also indicated that this locus was likely affected by presence of null alleles, and this locus has been problematic in other studies due to stuttering and long allele dropout (Steinauer et al., 2008a); therefore, this locus was not used in these analyses. Linkage disequilibrium was detected between two loci, which were originally designated as SMC1 (Curtis et al., 2001) and SMMS16 (Silva et al., 2006). Even though they were described as independent loci with different primer sets, they appear to be the same locus since the GenBank accession numbers are identical (AF325694). SMC1 was removed from further analyses.
Sex Biases
Male and female worms were not significantly differentiated regardless if the entire dataset was analyzed or if only the “lake” population was analyzed, therefore only the results using all data are given (P = 0.540 for all populations) and they had similar values of Fis and heterozygosity. For all populations, Fis for females with the 95% confidence interval was 0.010 (0.009–0.016) and for males was 0.019 (−0.005–0.043). Unbiased heterozygosity (Hs) (Nei, 1987) for females and males was 0.744 and 0.749, respectively.
Genetic Structure
The permutation analyses indicated that for 3 of the 4 relatedness statistics the parasites collected from within a site were more related than expected by the background relationship structure when only the five largest populations were considered (sites) and when sites were grouped by water bodies (water bodies) (Table 3). Significant differences were not detected when relatedness was measured with Wang’s estimator. The results of the regression analyses indicated that these patterns were not due to isolation by distance when either a linear or log-linear regression were performed.
Table 3.
Mean pairwise relatedness values for three measures of relatedness, Queller and Goodnight (1989) (rqg), Lynch and Ritland (1999) (rlr), and Wang (2002) (rw), and Nason’s estimator of kinship (Loiselle et al., 1995) (FN). Standard deviations are given in parentheses. Probability values are given from permutation tests that test whether relatedness within the specified category is greater than expected based on the background relatedness structure. Asterisks indicate statistical significance.
| rqg | pqg | rlr | plr | rw | pw | FN | PF | |
|---|---|---|---|---|---|---|---|---|
| All Individuals | −0.005 (0.109) | N/A | −0.005 (0.056) | N/A | −0.004 (0.105) | N/A | 0.0001 (0.052) | N/A |
| Within Coinfections | −0.013 (0.092) | 0.641 | −0.004 (0.060) | 0.442 | −0.015 (0.108) | 0.715 | −0.008 (0.048) | 0.802 |
| Within Sites | −0.001 (0.111) | *0.017 | −0.003 (0.052) | *0.0001 | −0.004 (0.106) | 0.112 | 0.002 (0.052) | *0.006 |
| Within Water Bodies | −0.001 (0.110) | *0.033 | −0.003 (0.059) | *< 0.0001 | −0.002 (0.148) | 0.144 | 0.001 (0.052) | *0.003 |
The analyses with STRUCTURE, the Bayesian clustering analysis that makes no a priori assumptions of population structure, indicated that the most optimal partition of the data consisted of one cluster, with no subdivision, suggesting a single panmictic population. The lnP(D) values for one cluster (K=1) was consistently lower than when multiple clusters were considered. Also, when analyses were performed with more than one cluster, the estimated membership coefficients (Q̂) indicated that each individual was about equally likely to belong to all of the estimated clusters, which supports the presence of only one cluster.
Permutation tests of F statistics indicated the presence of substructure when individual sites were considered (P = 0.001) and when water bodies were considered (P = 0.0001). Overall standardized Fst was 0.012 for sites and 0.016 for water bodies. Pairwise tests indicated that when each site was considered, only the Asao Stream and Nyabera Marsh populations were significantly different. When water bodies were considered, the Fst values among them were low (standardized Fst = 0.010–0.056), but most populations were significantly different from each other except Kasabong Stream and Nyabera Marsh, and Kasabong Stream and the Lake population (Table 4); however, this result should be regarded with caution because of the small sample size of the Kasabong Steam site (n=8).
Table 4.
Pairwise comparisons of assemblages of Schistosoma mansoni collected from sites grouped by lake and non lake sites within the Lake Victoria basin. Standardized pairwise Fst values are in the lower matrix and significance values are in the upper matrix.
| Lake | Asao | Kasabong | Nyabera | |
|---|---|---|---|---|
| Lake | - | *0.0070 | 0.0030 | *0.0020 |
| Asao | 0.0100 | - | *0.0030 | *0.0083 |
| Kasabong | 0.0461 | 0.0564 | - | 0.0153 |
| Nyabera | 0.0150 | 0.0109 | 0.0494 | - |
= statistical significance after Bonferroni correction.
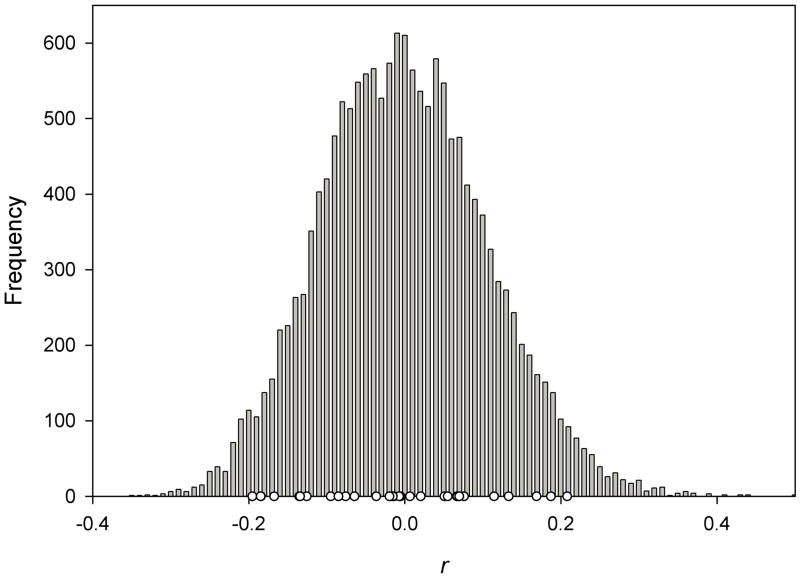
The mean pairwise relatedness values and their standard deviations for the three relatedness test statistics ranged from −0.004 to −0.002 (Table 3). These values along with the frequency histogram of the pairwise values (Fig. 2) indicate that most individuals were unrelated; however, some individuals were related as shown by the values in the right tail of the distribution. ML-RELATE estimated the relationships of 0.03% of the dyads to be full-siblings, 5.52% half-siblings, 0% parent-offspring, and 94.44% unrelated.
Figure 2.
Frequency histogram of pairwise relatedness values (r) of Schistosoma mansoni collected from snails in the Lake Victoria Basin of Kenya. Relatedness was calculated by the method of Wang (2002). Open circles indicated relatedness values of dyads collected from the same individual snail.
Intrasnail Dynamics
Individuals that coinfected the same hosts (21 total coinfections, 18 snails with 2 genotypes and 3 snails with 3 genotypes) were not more related than expected by chance as determined by all three permutation tests that utilized different relationship statistics (Fig. 2, Table 3) (This analysis was also performed within the Lake and Asao populations separately. Results were still insignificant, data not shown). Also, the number of cercariae released per snail over a 24 hour period did not differ between snails that were infected with a single genotype and those multiply infected (First observation only: Size: F1,91 = 0.153, P = 0.153; Infection F1,91 = 0.100, P = 0.753; Interaction: F1,91 = 0.117, P = 0.733), suggesting that cercarial production is reduced per capita in multiple infections. The number of cercariae released from snails with multiple infections was significantly positively correlated with relatedness of the parasites (Pearson Coefficient = 0.409, P = 0.0237, n = 24, using all replicates); however, a single outlier had a large influence, and when removed, the relationship was no longer significant (Pearson Coefficient = 0.223, P = 0.188, n = 23) (Fig. 3). Results using only the first observation were also similar in that a marginal positive relationship was detected (Pearson Coefficient = 0.503, P = 0.0837, n = 9). Interestingly, the positive relationship between these variables, whether considered significant or not due to the outlier, was the opposite of what was predicted by traditional models, which is a negative relationship between relatedness and cercarial production. The distribution of sexes within a coinfection was not significantly different than expected by random association (χ2 = 2.234, df = 2, P = 0.30).
Figure 3.
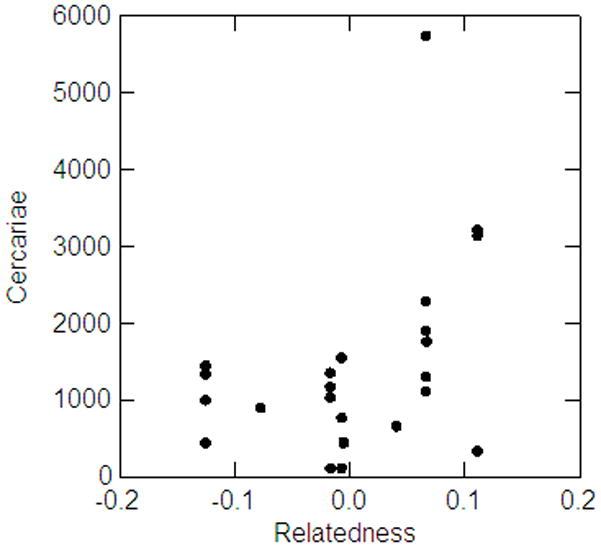
Relationship between the number of cercariae released from multiply infected snails and the relatedness of their parasites.
Discussion
Geographic Dynamics
We examined both fine scale and broad scale population structure of schistosomes in the Kenyan portion of the Lake Victoria basin to investigate their transmission dynamics. There was some evidence of weak substructure at both levels. Bayesian clustering analyses failed to identify unique populations suggesting that schistosomes in this area are better characterized as a single panmictic population. However, the pairwise tests of F-statistics did identify unique populations in the region, although with low Fst values. Because pairwise test have more statistical power to detect differences than the Bayesian analysis, these results suggest the presence of weak, but significant substructure among schistosomes from different water bodies. The differentiation of these populations could be due to watershed characteristics or movement patterns of humans that restrict gene flow among these sites. Watershed characteristics influence parasite gene flow because they restrict movement of both snails and free-living stages of parasites. Stream flow would work to inhibit upstream movement, especially from lake sites into streams, although downstream movement is likely. Also, snails are geographically restricted based on their habitat requirements: B. pfeifferi occurs in the streams and B. sudanica occurs in the lake. Differential use of these host species would work in synergy with microhabitat segregation to increase population subdivision if the parasites at these sites are better adapted to their local intermediate host species. Also, these transmission sites serve as primary sources of water for many humans in the area, and fidelity of humans to these sites would also increase population subdivision.
Fine scale structure was detected using 3 of 4 relatedness estimators, suggesting that individuals collected from a single site or sites grouped into water bodies were more related than expected from a random distribution of parasites. Fine scale structure detects the presence of local patterns of pedigree structure (Vekemans and Hardy, 2004; Born et al., 2008), which in this system could be due to spatial aggregation of siblings in fecal samples or clonal genotypes emerging from a single snail point source that do not get dispersed. The observed structure did not significantly correlate to the geographic distance among sites, which suggests that distance does not necessarily predict dispersal patterns and factors like water currents or wind patterns could be more important determinants of structure at this scale. Although this analysis indicated the presence of fine scale structure, it appears to be weak because collection sites were not characterized by a large number of highly related parasites. In fact, very few closely related individuals were found (Fig. 2).
Schistosoma mansoni has been the focus of similar studies that examine population structure in human transmission foci (e.g. Curtis et al., 2002; Agola et al., 2006; Thiele et al., 2008). Structuring is dependent both on the scale of the study and the particular populations investigated (Thiele et al., 2008). A previous study of schistosome genetic structure in Kenya showed subdivision among three watersheds in Kenya and among sites within two of these watersheds (Agola et al., 2006). Our results show much less differentiation partly due to the smaller scale of the study, but also potentially due to the enormity of the lake and the large number of people who are occupationally tied to it for fishing, harvesting sand, and washing cars, and who rely on it for their primary source of water.
In the Lake Victoria basin, schistosome population structure is of interest because of the possibility for the emergence of drug resistance. In this region, there is an ongoing longitudinal study that has involved the repeated use of Praziquantel to treat a population of approximately 200 men near the city of Kisumu (Karanja et al., 2002; Black et al., 2009). Recently drug tolerant parasites have been isolated from this population (Melman et al., In Review). However, long term epidemiological data suggest that there is no evidence of clinical resistance in this population (Black et al., 2009). One of the primary factors in determining how rapidly drug resistance alleles spread throughout a population is the size of the treated proportion relative to the untreated proportion, termed “refugium” (Geerts and Gryseels, 2000; Sissay et al., 2006; Leathwick et al., 2008; Waghorn et al., 2008). Our findings suggest this refugium is relatively large because of the high rates of gene flow and lack of population substructure throughout the Kenyan portion of the lake. On one hand, high rates of gene flow indicate opportunities for resistance alleles to become distributed widely; however, as long as drug treatment pressure is restricted to a small proportion of the population, such alleles are less likely to increase in frequency to the level of clinical resistance. Our findings are consistent with this theory; however, other factors play a role in the emergence of drug resistance such as the genetic architecture underlying the trait and costs associated with resistance, and these factors have yet to be fully explored (Feng et al., 2001; William et al., 2001).
Intrasnail Dynamics
We also examined the relatedness patterns of parasites within snails. Snails that share a host may be related due to aggregation of relatives within the habitat, or potentially due to intraspecific interactions. We investigated intraspecific interactions in the context of classical kin selection models. These models predict that unrelated individuals should compete and increase reproduction and thus exploitation in the short term to acquire as many resources as possible even at the expense of the host, but related individuals should reduce their levels of competition, and reduce host exploitation (Hamilton, 1972; Frank, 1992; May and Nowak, 1995; Frank, 1996).
First, to determine the potential importance of intraspecific interactions when parasites share a snail host, we compared reproductive success of parasite individuals that shared a host to those that did not. We found that the total reproductive rate of parasites in coinfections did not differ from those in single infections. Therefore, coinfections have severe fitness consequences to the parasites, and there is a context for intraspecific interactions to evolve. Although asexual reproductive rate is not the same as fecundity, it likely is directly associated as the more infective stages that are produced, the greater the probability of transmission to the next host, and thus the higher their total fecundity. However, this prediction has not been validated in natural populations and it is possible that a tradeoff could result in the opposite pattern.
We also found that the parasites that coinfect snails were not more related than expected by a random distribution of parasites. It was predicted that aggregation or kin selection would favor the occurrence of related parasites within a snail. The former would be due to spatial autocorrelation of parasite relatives in fecal samples. The latter could be due to competitive exclusion of unrelated individuals within snails or the death of snails that acquire unrelated parasites due to increased parasite competition. The latter outcome is one prediction of kin selection theory; however, interactions could be manifested differently such as by modifying reproductive rates based on relatedness of coinfecting parasites. However, there was no significant negative relationship between relatedness and the total number of cercariae produced in coinfections, as predicted by classical models. In fact, the trend, which was significant with the inclusion of an outlier, was a positive correlation. These data better fit alternative models that incorporate more details of the interactions between the parasites and their hosts (Chao et al., 2000; Brown et al., 2002; Schjørring and Koella, 2003). For example, if there are costs to competition, then decreased growth or reproductive rates of the parasites is predicted when parasites are less related, creating a positive correlation between relatedness and reproductive rate (Chao et al., 2000). Alternatively, if the parasites only cause sub-lethal damage to the host, and this damage feeds back to affect parasite reproduction, then multiple infections generally lead to reduced reproduction (Schjørring and Koella, 2003). In this case, competition may initially increase reproduction, but damage done to the host limits further parasite reproduction, resulting in no net increase. Further studies are necessary to determine if these models are relevant to this system.
Finally, we determined that parasites that coinfected a host were also randomly distributed with regard for sex. This finding suggests there is no mechanism that would favor the establishment of opposite sex coinfections over same sex coinfections. Opposite sex coinfections were predicted to be beneficial because it is likely that the individuals would be cotransmitted to the final host, thus insuring the presence of a mate.
Contrary to our findings, relatedness of schistosomes in snails has been found by others to be an important factor in determining parasite interactions within snails. Experimental infections of laboratory strains of schistosomes revealed that mixed infections of unrelated individuals led to increased asexual reproductive rates (Davies et al., 2002). It is possible that parasite interactions are context dependent: Davies and colleagues used strains of laboratory bred schistosomes and a different species of snail host than used in our study. Another explanation is that the discrepancies between studies are due to the differences in the degree of relatedness among individuals. The experimental study utilized two highly divergent laboratory strains of parasites and laboratory raised snails while we used individuals from a large, heterogeneous, natural population. It is possible that the relatedness values of the schistosomes in Kenya were not high enough for this theory to be relevant as high levels of relatedness are necessary for kin selection to operate (Buckling and Brockhurst, 2008). For example, interactions may only be detected with full siblings or inbred lines. Finally, competition between parasites could be manifesting itself differently from either complete interference or increased reproductive rates of both competitors. It is possible that one genotype reproduces more rapidly while interfering with the reproduction of the other resulting in no net increase in cercariae (Gower and Webster, 2005). To test this scenario, a detailed examination of the numbers of cercariae produced by each MLG would be necessary.
Field based studies of natural populations such as this one cannot control all possible factors influencing the system and therefore may not be as clear-cut as experimental studies that are necessary to fully understand intraspecific interactions. However, they provide an essential perspective because the data are based on a natural population of hosts and parasites and focus on their true relatedness structure. In populations in Kenya, the fact that schistosomes rarely encounter close relatives in the snail host due to chance alone could make kin selection irrelevant for this system particularly if their current relationship structure is analogous to historical structure. However, relatedness has been shown to positively influence coinfections of tapeworms in sticklebacks even when the relatives were only cousins (r = 0.125) (Jager and Schjørring, 2006).
Summary
Schistosoma mansoni in the Lake Victoria basin is characterized by having very little spatial structure among snail hosts and across geography. Although there is evidence of spatial patterning of related individuals and also spatial substructure based on water bodies, substructure is not strong indicating a significant amount of gene flow across these geographic sites. We also found evidence of local pedigree structure, which may be attributed to the massive production of clonal infective stages in snails; however, the pedigree structure was weak and close relatives were not common. These findings suggest that the variance of success in schistosome cercariae is not highly skewed so that most infections in humans are not derived from a small proportion of snails (i.e. superspreading snails), which would lead to a much more related population of schistosomes. Relatedness or sex of schistosomes did not influence host sharing. Also, relatedness and cercarial production had a positive relationship, the opposite result of our predictions based on classical models. Because of the low levels of relatedness within the population, schistosomes may rarely encounter close relatives and kin selection mechanisms that influence the distribution of individuals within snails or their competitive strategy may simply have not evolved.
Acknowledgments
Primary funding was provided by U.S. National Institutes of Health Grant AI044913. We thank Diana Karanja for providing laboratory space and Ibrahim N. Mwangi, Geoffrey M. Maina, Joseph M. Kinuthia, Martin W. Mutuku, Ben Mungai, John Adero, Sara V. Brant, Elizabeth Hatton, Boniface Mualuko, Esther Mungai, and James Wangunyu for their assistance on this project. Support was also obtained from the University of New Mexico Center for Evolutionary and Theoretical Immunology (CETI), and the UNM Molecular Biology Facility (NIH Grant Number 1P20RR18754 from the Institute Development Award (IDeA) Program of the National Center for Research Resources).
Literature Cited
- Agola LE, Mburu DN, DeJong RJ, Mungai BN, Muluvi GM, Njagi ENM, Loker ES, Mkoji GM. Microsatellite typing reveals strong genetic structure of Schistosoma mansoni from localities in Kenya. Infect Genet Evol. 2006;6:484–490. doi: 10.1016/j.meegid.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Archie EA, Luikart G, Ezenwa VO. Infecting epidemiology with genetics: a new frontier in disease ecology. Trends in Ecology & Evolution. 2009;24:21–30. doi: 10.1016/j.tree.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Black CL, Steinauer ML, Mwinzi PNM, Secor WE, Karanja DMS, Colley DG. Impact of intense, longitudinal retreatment with praziquantel on cure rates of schistosomiasis mansoni in a cohort of occupationally exposed adults in western Kenya. Trop Med Int Health. 2009;14:1–8. doi: 10.1111/j.1365-3156.2009.02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin MS, Yowell CA, Courtney CH, Dame JB. Host movement and the genetic structure of populations of parasitic nematodes. Genetics. 1995;141:1007–1014. doi: 10.1093/genetics/141.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin MS, Liu J, Berry RE. Life cycle variation and the genetic structure of nematode populations. Heredity. 1999;83:253–259. doi: 10.1038/sj.hdy.6885420. [DOI] [PubMed] [Google Scholar]
- Born C, Hardy OJ, Chevallier MH, Ossari S, Atteke C, Wickings EJ, Hosseart-McKey M. Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation. Mol Ecol. 2008;17:2041–2050. doi: 10.1111/j.1365-294X.2007.03685.x. [DOI] [PubMed] [Google Scholar]
- Bremermann HJ, Pickering J. A game-theoretical model of parasite virulence. J Theor Biol. 1983;100:411–426. doi: 10.1016/0022-5193(83)90438-1. [DOI] [PubMed] [Google Scholar]
- Brown SP, Hochberg ME, Grenfell BT. Does multiple infection select for raised virulence? Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. [DOI] [PubMed] [Google Scholar]
- Buckling A, Hodgson DJ. Short-term rates of parasite evolution predict the evolution of host diversity. J Evol Biol. 2007;20:1682–1688. doi: 10.1111/j.1420-9101.2007.01402.x. [DOI] [PubMed] [Google Scholar]
- Buckling A, Brockhurst MA. Kin selection and the evolution of virulence. Heredity. 2008;100:484–488. doi: 10.1038/sj.hdy.6801093. [DOI] [PubMed] [Google Scholar]
- Chao L, Hanley KA, Burch CL, Dahlberg C, Turner PE. Kin selection and parasite evolution: Higher and lower virulence with hard and soft selection. Q Rev Biol. 2000;75:261–275. doi: 10.1086/393499. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Life cycles shape parasite evolution: comparative population genetics of salmon and trematodes. Evolution. 2004;58:198–202. doi: 10.1111/j.0014-3820.2004.tb01587.x. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Poulin R, Blouin MS. Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol Ecol. 2005;14:2247–2257. doi: 10.1111/j.1365-294X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Criscione CD, Blouin MS. Minimal selfing, few clones, and no among-host genetic structure in a hermaphroditic parasite with asexual larval propagation. Evolution. 2006;60:553–562. [PubMed] [Google Scholar]
- Curtis J, Sorensen RE, Page LK, Minchella DJ. Microsatellite loci in the human blood fluke Schistosoma mansoni and their utility for other schistosome species. Mol Ecol Notes. 2001;1:143–145. [Google Scholar]
- Curtis J, Sorensen RE, Minchella DJ. Schistosome genetic diversity: the implications of population structure as detected with microsatellite markers. Parasitology. 2002;125:S51–S59. doi: 10.1017/s0031182002002020. [DOI] [PubMed] [Google Scholar]
- Davies CM, Webster JP, Woolhouse MEJ. Trade-offs in the evolution of virulence in an indirectly transmitted macroparasite. Proc R Soc Lond B Biol Sci. 2001;268:251–257. doi: 10.1098/rspb.2000.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CM, Fairbrother E, Webster JP. Mixed strain schistosome infections of snails and the evolution of parasite virulence. Parasitology. 2002;124:31–38. doi: 10.1017/s0031182001008873. [DOI] [PubMed] [Google Scholar]
- Feng ZL, Curtis J, Minchella DJ. The influence of drug treatment on the maintenance of schistosome genetic diversity. Journal of Mathematical Biology. 2001;43:52–68. doi: 10.1007/s002850100092. [DOI] [PubMed] [Google Scholar]
- Frank SA. A kin selection model for the evolution of virulence. Proc R Soc Lond B Biol Sci. 1992;250:195–197. doi: 10.1098/rspb.1992.0149. [DOI] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Q Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Fulford AJC, Butterworth AE, Ouma JH, Sturrock RF. A statistical approach to schistosome population dynamics and estimation of the life span of Schistosoma mansoni in man. Parasitology. 1995;110:307–316. doi: 10.1017/s0031182000080896. [DOI] [PubMed] [Google Scholar]
- Geerts S, Gryseels B. Drug resistance in human helminths: Current situation and lessons from livestock. Clin Microbiol Rev. 2000;13:207–222. doi: 10.1128/cmr.13.2.207-222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001 posting date. http://www.unil.ch/izea/softwares/fstat.html.
- Gower CM, Webster JP. Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution. 2005;59:544–553. [PubMed] [Google Scholar]
- Gower CM, Shrivastava J, Lamberton PHL, Rollinson D, Webster BL, Emery A, Kabatereine NB, Webster JP. Development and application of an ethically and epidemiologically advantageous assay for the multi-locus microsatellite analysis of Schistosoma mansoni. Parasitology. 2007;134:523–536. doi: 10.1017/S0031182006001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WD. Altruism and related phenomena, mainly in social insects. Annu Rev Ecol Syst. 1972;3:193–232. [Google Scholar]
- Handzel T, Karanja DMS, Addiss DG, Hightower AW, Rosen DH, Colley DG, Andove J, Slutsker L, Secor WE. Geographic distribution of schistosomiasis and soil-transmitted helminths in Western Kenya: Implications for anthelminthic mass treatment. Am J Trop Med Hyg. 2003;69:318–323. [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- Huyse T, Poulin R, Theron A. Speciation in parasites: a population genetics approach. Trends Parasitol. 2005;21:469–475. doi: 10.1016/j.pt.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Jager I, Schjørring S. Multiple infections: Relatedness and time between infections affect the establishment and growth of the cestode Schistocephalus solidus in its stickleback host. Evolution. 2006;60:616–622. doi: 10.1554/05-450.1. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST, Wagner AP, Taper ML. ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes. 2006;6:576–579. [Google Scholar]
- Karanja DMS, Hightower AW, Colley DG, Mwinzi PNM, Galil K, Andove J, Secor WE. Resistance to reinfection with Schistosoma mansoni in occupationally exposed adults and effect of HIV-1 co-infection on susceptibility to schistosomiasis: a longitudinal study. Lancet. 2002;360:592–596. doi: 10.1016/S0140-6736(02)09781-7. [DOI] [PubMed] [Google Scholar]
- Kardorff R, Gabone RM, Mugashe C, Obiga D, Ramarokoto CE, Mahlert C, Spannbrucker N, Lang A, Gunzler V, Gryseels B, Ehrich JHH, Doehring E. Schistosoma mansoni related morbidity on Ukerewe Island, Tanzania: Clinical, ultrasonographical and biochemical parameters. Trop Med Int Health. 1997;2:230–239. doi: 10.1046/j.1365-3156.1997.d01-269.x. [DOI] [PubMed] [Google Scholar]
- Keeney DB, Waters JM, Poulin R. Diversity of trematode genetic clones within amphipods and the timing of same-clone infections. Int J Parasitol. 2007;37:351–357. doi: 10.1016/j.ijpara.2006.11.004. [DOI] [PubMed] [Google Scholar]
- King CH, Dickman K, Tisch DJ. Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet. 2005;365:1561–1569. doi: 10.1016/S0140-6736(05)66457-4. [DOI] [PubMed] [Google Scholar]
- Leathwick DM, Miller CM, Atkinson DS, Haack NA, Waghorn TS, Oliver AM. Managing anthelmintic resistance: Untreated adult ewes as a source of unselected parasites, and their role in reducing parasite populations. New Zealand Veterinary Journal. 2008;56:184–195. doi: 10.1080/00480169.2008.36832. [DOI] [PubMed] [Google Scholar]
- Levin S, Pimentel D. Selection of intermediate rates of increase in parasite-host systems. American Naturalist. 1981;117:308–315. [Google Scholar]
- Li G, Hedgecock D. Genetic heterogeneity, detected by PCR-SSCP, among samples of larval Pacific oysters (Crassostrea gigas) supports the hypothesis of large variance in reproductive success. Canadian Journal of Fisheries and Aquatic Sciences. 1998;55:1025–1033. [Google Scholar]
- Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic-structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae) Am J Bot. 1995;82:1420–1425. [Google Scholar]
- Lotz JM, Bush AO, Font WF. Recruitment-driven, spatially discontinuous communities: A null model for transferred patterns in target communities of intestinal helminths. J Parasitol. 1995;81:12–24. [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May RM, Nowak MA. Coinfection and the evolution of parasite virulence. Proc R Soc Lond B Biol Sci. 1995;261:209–215. doi: 10.1098/rspb.1995.0138. [DOI] [PubMed] [Google Scholar]
- McCoy KD, Boulinier T, Tirard C, Michalakis Y. Host-dependent genetic structure of parasite populations: Differential dispersal of seabird tick host races. Evolution. 2003;57:288–296. doi: 10.1111/j.0014-3820.2003.tb00263.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Melman SD, Steinauer ML, Cunningham C, Salter-Kubatko L, Mwangi IN, Matuku M, Karanja DMS, Colley DG, Black CL, Secor WE, Barker N, Mkoji GM, Loker ES. Reduced susceptibility of Kenyan Schistosoma mansoni to praziquantel following repeated exposures: origin, measurement, and likelihood of persistence. PLos Neg Trop Dis In Review. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, De Meeus T, Durand P, Sire C, Theron A. Sex-specific genetic structure in Schistosoma mansoni: evolutionary and epidemiological implications. Mol Ecol. 2002;11:1231–1238. doi: 10.1046/j.1365-294x.2002.01518.x. [DOI] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistics tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rousset F. genepop™007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schjørring S, Koella JC. Sub-lethal effects of pathogens can lead to the evolution of lower virulence in multiple infections. Proc R Soc Lond B Biol Sci. 2003;270:189–193. doi: 10.1098/rspb.2002.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava J, Gower CM, Balolong E, Wang TP, Qian BZ, Webster JP. Population genetics of multi-host parasites: The case for molecular epidemiological studies of Schistosoma japonicum using larval stages from naturally infected hosts. Parasitology. 2005;131:617–626. doi: 10.1017/S0031182005008413. [DOI] [PubMed] [Google Scholar]
- Silva L, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology. 2006;132:331–338. doi: 10.1017/S0031182005009066. [DOI] [PubMed] [Google Scholar]
- Sissay MM, Asefa A, Uggla A, Waller PJ. Anthelmintic resistance of nematode parasites of small ruminants in eastern Ethiopia: Exploitation of refugia to restore anthelmintic efficacy. Veterinary Parasitology. 2006;135:337–346. doi: 10.1016/j.vetpar.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Smithers SR, Terry RS. The infection of laboratory hosts with cercariae of Schistosoma mansoni and the recovery of the adult worms. Parasitology. 1965;55:695–700. doi: 10.1017/s0031182000086248. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Agola LE, Mwangi IN, Mkoji GM, Loker ES. Molecular epidemiology of Schistosoma mansoni: a robust, high-throughput method to assess multiple microsatellite markers from individual miracidia. Infect Genet Evol. 2008a;8:68–73. doi: 10.1016/j.meegid.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinauer ML, Hanelt B, Mwangi IN, Maina GM, Agola EL, Kinuthia JM, Mutuku MW, Mungai BN, Wilson WD, Mkoji GM, Loker ES. Introgressive hybridization of human and rodent schistosome parasites in western Kenya. Mol Ecol. 2008b;17:5062–5074. doi: 10.1111/j.1365-294X.2008.03957.x. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Mwangi IN, Maina GM, Kinuthia JM, Mutuku MW, Agola LE, Mungai BN, Mkoji GM, Loker ES. Interactions between natural populations of human and rodent schistosomes in the Lake Victoria region of Kenya: a molecular epidemiological approach. PLos Neg Trop Dis. 2008c;2:1–11. doi: 10.1371/journal.pntd.0000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- Thiele EA, Sorensen RE, Gazzinelli A, Minchella DJ. Genetic diversity and population structuring of Schistosoma mansoni in a Brazilian village. Int J Parasitol. 2008;38:389–399. doi: 10.1016/j.ijpara.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M. Toward an integrated genetic epidemiology of parasitic protozoa and other pathogens. Annu Rev Genet. 1999;33:449–477. doi: 10.1146/annurev.genet.33.1.449. [DOI] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) BioTechniques. 2000;29:52–53. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Vickery WL, Poulin R. Can helminth community patterns be amplified when transferred by predation from intermediate to definitive hosts? J Parasitol. 2002;88:650–656. doi: 10.1645/0022-3395(2002)088[0650:CHCPBA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Waghorn TS, Leathwick DM, Miller CM, Atkinson DS. Brave or gullible: Testing the concept that leaving susceptible parasites in refugia will slow the development of anthelmintic resistance. New Zealand Veterinary Journal. 2008;56:158–163. doi: 10.1080/00480169.2008.36828. [DOI] [PubMed] [Google Scholar]
- Wang JL. An estimator for pairwise relatedness using molecular markers. Genetics. 2002;160:1203–1215. doi: 10.1093/genetics/160.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS. Separating the wheat from the chaff: Patterns of genetic differentiation in high gene flow species. Journal of Heredity. 1998;89:438–450. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- William S, Sabra A, Ramzy F, Mousa M, Demerdash Z, Bennett JL, Day TA, Botros S. Stability and reproductive fitness of Schistosoma mansoni isolates with decreased sensitivity to praziquantel. Int J Parasitol. 2001;31:1093–1100. doi: 10.1016/s0020-7519(01)00215-6. [DOI] [PubMed] [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of the VI International Congress of Genetics. 1932;1:356–366. [Google Scholar]