The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms
Mutations in the mitochondrial pro-fusion dynamin Opa1 cause optic nerve degeneration and autosomal dominant optic atrophy. The study by Landes et al reports that Opa1 and the mitochondrial pro-apoptotic Bnip3 interact and that both proteins together modulate mitochondrial morphology and cell death. This interaction may regulate the adaptive response of retinal ganglion cells to hypoxia, a potent inductor of Bnip3.
Keywords: ADOA, apoptosis, Bnip3, mitochondrial dynamics, Opa1
Abstract
Opa1 modulates mitochondrial fusion, cristae structure and apoptosis. The relationships between these functions and autosomal dominant optic atrophy, caused by mutations in Opa1, are poorly defined. We show that Bnip3 interacts with Opa1, leading to mitochondrial fragmentation and apoptosis. Fission is due to inhibition of Opa1-mediated fusion and is counteracted by Opa1 in an Mfn1-dependent manner. Bnip3–Opa1 interaction is necessary to trigger Opa1 complex disruption in a Bax- and/or Bak-dependent manner, ultimately leading to apoptosis. Our results uncover a direct link between Opa1 on the inner mitochondrial membrane and the apoptotic machinery on the outer membrane that modulates fusion and cristae structure by separate mechanisms. These findings might help to unravel optic atrophy aetiology as retinal ganglion cells are particularly prone to hypoxia, an inductor of Bnip3 expression.
Introduction
Mitochondria are highly dynamic organelles that continually fuse and divide to yield mitochondrial morphologies ranging from an interconnected filamentous network to small spherical grains in adaptation to cell needs. Principal actors of mitochondrial dynamics have been reviewed by Detmer & Chan (2007). On the mitochondrial outer membrane (MOM), Drp1 and Fis1 control fission whereas mitofusins (Mfn1 and Mfn2) are involved in fusion. We have identified Opa1, a dynamin of the mitochondrial inner membrane (MIM) that regulates fusion, cristae structure and apoptosis (Landes et al, 2010). The importance of mitochondrial dynamics in cellular homeostasis is highlighted by the involvement of several of these effectors in neurodegenerative disorders, such as Charcot–Marie–Tooth disease and autosomal dominant optic atrophy (ADOA; Detmer & Chan, 2007).
Reciprocal links between mitochondrial dynamics and apoptosis have been discussed by Suen et al (2008). Apoptosis is often associated with mitochondrial fission, and displacement of the dynamics equilibrium towards fission or fusion, respectively, enhances or decreases apoptosis. Members of the Bcl2 family that regulate apoptogenic protein mobilization and MOM permeabilization modulate mitochondrial dynamics both during apoptosis and in healthy cells.
Mitochondrial dynamics and apoptosis involve coordinated reorganization of MIM and MOM to achieve faithful fission and fusion of the organelle, or mobilization and release of cytochrome c. Although distinguishable, machineries executing the two processes might use common elements to promote such rearrangements of the two membranes. Opa1, which regulates both mitochondrial dynamics and apoptosis, has been suggested to be a crucial organizer of MIM. In this study, we identify Bnip3, a mitochondrial proapoptotic BH3-only protein of the Bcl2 family (Burton & Gibson, 2009), as a partner of Opa1. We show that Bnip3 links two distinct Opa1-regulated machineries involved either in mitochondrial dynamics or in apoptosis, and operating at fusion sites or cristae junctions, respectively.
Results And Discussion
By using a yeast two-hybrid screen, we identified Bnip3 as an interacting partner of Opa1 (supplementary Fig S1A online). This was substantiated by glutathione-S-transferase (GST) pulldown assays (supplementary Fig S1B online) and confirmed by reciprocal co-immunoprecipitation of Opa1 and haemagglutinin-tagged Bnip3 (HA–Bnip3) on crosslinked mitochondria from HeLa cells ectopically expressing both proteins (Fig 1A). Co-immunoprecipitation of Opa1 and Bnip3 did not arise with protein A–sepharose alone nor did it carry along major proteins of various mitochondrial compartments (Fig 1A; supplementary Fig S2A online, left). It also occurred in non-crosslinked mitochondria (supplementary Fig S2C online) and after in cellulo crosslinking (supplementary Fig S2B online), showing that it was not due to artefactual crosslinking or to mitochondrial compartments mixing on cells lysis. Physiological interaction was evidenced between endogenous Opa1 and Bnip3, the latter being induced by exposure of cells to severe hypoxia (Fig 1C; supplementary Fig S2A online, right) or CoCl2 (supplementary Fig S2D online).
Figure 1.
Bnip3 interacts with Opa1. (A) HeLa cells were co-transfected with plasmids for Opa1 and HA–Bnip3, or (B) HA–Bnip3 mutants, as indicated. (C) Endogenous interactions were observed in HeLa cells exposed (+) or not (−) to hypoxia to induce Bnip3 expression. Immunoprecipitations (IPs) were performed on crosslinked mitochondria (mito input, 1/20 of IP) using anti-Opa1, anti-HA or no antibodies (−), and analysed by western blots using the indicated antibodies. Migration of molecular weight markers is shown on the right. The batch of affinity media used in (C) released a major contaminant (indicated by the asterisk), probably immunoglobulin light chains (MW∼25 kDa). Note in (C) that low basal levels of endogenous Bnip3 expressed under normoxia (IP Opa1, hypoxia−) are associated with Opa1. HA, haemagglutinin; HA–Bnip3, haemagglutinin-tagged Bnip3.
Bnip3 integrates into the MOM through its carboxy-terminal transmembrane domain (TMD) leaving the last 10 residues into the intermembrane space (IMS; Vande Velde et al, 2000). As the bulk of Opa1, both regions are thus proximal to, or within, the IMS. Mutants of Bnip3 (supplementary Table S1 online), in which the TMD and IMS tail or TMD alone were replaced by the equivalent parts of Omp25 (Bnip3TMOmp25) or Bcl2 (Bnip3TMBcl2), respectively, to ensure MOM localization no longer interacted with Opa1 (Fig 1B). Bnip3ΔC that conserves the TMD but lacks the IMS tail had a reduced interaction with Opa1 (35%), whereas mutants with a deletion of, or point mutations in, the cytosol-exposed BH3 domain (Bnip3ΔBH3 and Bnip3mutBH3, respectively) interacted fully with Opa1 (Fig 1B). These observations suggest that parts of Opa1 might penetrate into the MOM inner leaflet to contact the TMD of Bnip3, the IMS tail of which might stabilize the association.
As the proapoptotic Bnip3 interacts with a pro-fusion protein, we assessed the resulting effects on mitochondrial dynamics. HeLa cells had a tubular mitochondrial network that fragmented in 58% of the cells expressing HA–Bnip3 (Fig 2A,B). Bnip3-induced fission depended on Drp1 activity as it occurred in only 30% of cells that co-expressed dominant-negative Drp1K38A (Fig 2B). The Bnip3–Opa1 interaction might thus have relieved Drp1-driven fission by inhibiting antagonistic Opa1 fusion activity. This was investigated by a polyethylene glycol fusion assay using a mixture of HeLa cells separately harbouring green fluorescent protein (GFP)- or DsRed-labelled mitochondria (Fig 2C). Fused control cells not expressing HA–Bnip3 had filamentous mitochondria with superimposed colours showing mitochondrial fusion. Conversely, those expressing HA–Bnip3 had fragmented green or red mitochondria, implying a lack of fusion. This was verified by using the photoconvertible fluorescent protein Dendra targeted to the mitochondrial matrix (mito–Dendra). In control HeLa cells, 40% of the laser-converted red fluorescence disappeared within 40 min after illumination, indicating its redistribution in the matrix of fused mitochondria (Fig 2D). In cells transfected with Bnip3, redistribution was much slower and fluorescence decreased very slightly, similar to that in Opa1 short interfering RNA (siRNA)-transfected cells (data not shown), showing that Bnip3 indeed inhibited fusion.
Figure 2.
Bnip3 induces mitochondrial fragmentation by inhibiting fusion. HeLa cells were transfected with GFP and either empty (control), HA–Bnip3 or Drp1K38A vectors during 24 h. Mitochondria were observed after MitoTracker staining. Typical morphologies are exemplified (A) and were quantified (B). Data represent mean±s.d. of three independent experiments; n⩾200 GFP-expressing cells per condition. (C) HeLa cells co-transfected separately with mtDsRed or mtGFP, and HA–Bnip3 or empty (control) vectors were fused with polyethylene glycol and observed by fluorescence microscopy. (D) HeLa cells were transfected with mito-Dendra and either HA–Bnip3 or empty vectors (control). Images were acquired in GFP and TRITC channels before photoconversion, and in TRITC channel immediately after (t=0 min) and every 10 min subsequently. The graph depicts the time course of fluorescence. Data represent mean±s.d. of four separate experiments; n⩾20 single time lapse per condition. (A,C,D) Scale bars, 10 μm. GFP, green fluorescent protein; HA–Bnip3, haemagglutinin-tagged Bnip3; TRITC, tetramethyl rhodamine isothiocyanate.
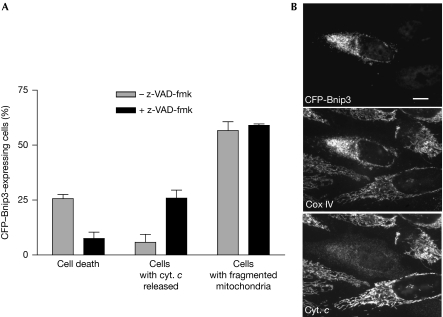
According to cell type and stimuli, Bnip3 might induce cell death by apoptosis or autophagy. Twenty-five per cent of HeLa cells overexpressing cerulean fluorescent protein-tagged Bnip3 (CFP–Bnip3) had condensed nuclei (Fig 3A), often associated with decreased MitoTracker staining indicative of a low membrane potential (data not shown). The inhibitor of late apoptotic events, z-VAD-fmk, effectively blocked cell death and allowed observation of early cytochrome-c release, showing that Bnip3 induced a caspase-dependent apoptosis (Fig 3). Interestingly, such inhibition did not block mitochondrial fragmentation, which still occurred in 56% of CFP–Bnip3-expressing cells (Fig 3A) and half of these cells with punctuated mitochondria had mitochondrially localized cytochrome-c (Fig 3A). These results suggest that Bnip3 triggered mitochondrial fragmentation before cytochrome-c release and loss of membrane potential that were ultimately followed by apoptosis. Bnip3 overexpression in HeLa cells was not associated to autophagy as the LC3-II-specific marker was not induced (data not shown).
Figure 3.
Bnip3 induces apoptotic cell death. HeLa cells were transfected with CFP–Bnip3 for 24 h with or without z-VAD-fmk. The cells were observed by immunofluorescence using antibodies against cytochrome-c (cyt. c) or subunit IV of the cytochrome-c oxidase (Cox IV) and stained by Hoescht 3325. (A) The data represent the percentage of CFP–Bnip3-transfected cells with apoptotic nuclei, cytosolic cytochrome-c or fragmented mitochondria and are the mean±s.d. of three independent experiments; n⩾200 cells per condition. (B) Representative images are shown. Scale bar, 10 μm. CFP–Bnip3, cerulean fluorescent protein-tagged Bnip3.
Mechanisms by which the Bnip3–Opa1 interaction influences mitochondrial morphology and cell death were then studied. A stable Tet-Off HeLa cell line expressing inducible and moderate levels of Opa1 (supplementary Fig S3A online) that did not alter mitochondrial morphology (Fig 4A) was derived. Overexpression of HA–Bnip3 without induction of Opa1 led to 36% of cells with punctuated mitochondria and to 15% of cell death (Fig 4A; supplementary Fig S3A online). Concomitant expression of Opa1 and Bnip3 returned fragmentation and cell death levels to control values. Opa1 thus clearly antagonized both effects of Bnip3. The interaction-less mutants Bnip3TMOmp25 and Bnip3TMBcl2 induced neither fragmentation nor cell death (Fig 4B) while being expressed at comparable levels as Bnip3 (supplementary Fig S3B online). Bnip3ΔC was similarly ineffective although it interacted slightly with Opa1 (Fig 1B), suggesting that the IMS tail, by stabilizing the interaction, might also functionalize it. Conversely, both Bnip3ΔBH3 and Bnip3mutBH3 induced mitochondrial fragmentation but failed to trigger apoptosis. These observations imply that the interaction of Opa1 with Bnip3 is required to induce mitochondrial fission and cell death, the latter also requiring the BH3 domain of Bnip3.
Figure 4.
Interactions of Bnip3 and Opa1 induce mitochondrial fragmentation and cell death by distinct mechanisms. (A) Established HeLa Tet-Off cells were induced (+) or not (−) for Opa1 expression during 48 h. For the last 24 h, cells were co-transfected with GFP and either HA–Bnip3 (+) or empty (−) vectors. (B) HeLa cells were transfected for 24 h with GFP and either empty (−) or the indicated wild-type or mutant HA–Bnip3 vectors. (C) Cells were as in (A) and cultured with siRNA to Mfn1 (Mfn1) or the off-target luciferase (luc) during the Opa1 induction period. (D) HeLa cells were cultured under hypoxia (+) or normoxia (−) in the presence of siRNA to Bnip3 or luciferase. Fragmentation occurred rapidly (24 h) and was later (72 h) followed by apoptosis. For each panel, data (mean±s.d. of ⩾3 independent experiments; n⩾200 cells per condition) show quantification of mitochondrial fragmentation (left axis) and of apoptosis (right axis) on observation by fluorescence microscopy after MitoTracker and Hoescht 33258 staining. GFP, green fluorescent protein; HA–Bnip3, haemagglutinin-tagged Bnip3; Mfn, mitofusin; siRNA, short interfering RNA.
We then assessed whether the counteracting effects of Opa1 relied on its Mfn1-dependent fusogenic function. Mfn1 extinction slightly shortened mitochondrial filaments (data not shown) but neither modified Bnip3-induced mitochondrial fragmentation nor apoptosis (Fig 4C; supplementary Fig S3C online). In cells expressing both Opa1 and Bnip3, Mfn1 extinction prevented Opa1 from blocking the effect of Bnip3 on mitochondrial fragmentation but not on cell death. Thus, Bnip3-induced inhibition of Opa1 fusion activity indeed triggered mitochondrial fragmentation but did not constitute a death signal. This suggested that the apoptotic effects of Bnip3 depend on an Opa1 function distinct from its fusion activity.
Opa1 is thought to control cytochrome-c mobilization by forming oligomers at cristae junctions (Frezza et al, 2006; Yamaguchi et al, 2008). When added to HeLa mitochondria or to mouse embryonic fibroblast (MEF) mitochondria, Bnip3, similarly to t-Bid, led to the disassembly of Opa1 complexes (Fig 5A; supplementary Fig S4A online), as observed recently with heart mitochondria (Quinsay et al, 2009). This effect relied on Bnip3–Opa1 interaction as it was not induced by Bnip3TMOmp25 or Bnip3TMBcl2 (Fig 5A). Opa1 complex disassembly was neither observed with Bnip3mutBH3 (Fig 5A) nor in mitochondria from Bax/Bak-knockout MEFs (supplementary Fig S4A online) and thus also needed a functional Bnip3 BH3 domain and Bax and/or Bak.
Figure 5.
Bnip3 promotes Opa1 oligomer disassembly. (A) Mitochondria isolated from HeLa cells were incubated without (−) or with t-Bid, or with thrombin cleavage products from GST (mock), GST-Bnip3, GST-Bnip3ΔC, GST-Bnip3TMBcl2, GST-Bnip3TMOmp25 or GST-Bnip3mutBH3 beads. (B) HeLa cells were treated with siRNA to luciferase (luc) or Bnip3 for 24 h and cultured (+) or not (−) under hypoxia for an additional 24 h. For (A) and (B), mitochondria were then crosslinked and analysed by western blots using the indicated antibodies. Quantification of Opa1 complex-to-monomer ratios is shown for each figure. Data represent the mean±s.e.m. of ⩾3 independent experiments; *P<0.05. GST, glutathione-S-transferase; siRNA, short interfering RNA.
Exposure of HeLa cells to severe hypoxia confirmed the physiological bearings of the effects of Bnip3 detailed here. Under hypoxia, upregulated Bnip3 that interacted with Opa1 (Fig 1C) induced mitochondrial fragmentation and caspase-dependent cell death (Fig 4D; supplementary Fig S3D online). Each effect depended on Bnip3 as it was not produced when Bnip3 was not expressed or suppressed by specific siRNA. Exposure of HeLa cells to hypoxia (Fig 5B) or CoCl2 (supplementary Fig S4B online) elevated the levels of Bnip3 and induced a Bnip3-dependent Opa1 complex rupture accompanied by cytochrome-c and Opa1 release. MOM permeabilization was also induced on isolated mitochondria by recombinant t-Bid but not by purified Bnip3 (Fig 5A; supplementary Fig S4A online), suggesting either that recombinant Bnip3 was not fully functional or that unlike t-Bid functioning with Bax to induce MOM permeabilization, Bnip3 requires factors either lacking or inactive in the isolated mitochondria. Together, our findings suggest that elevated levels of Bnip3 induce a prerequisite interaction of its TMD and IMS tail with Opa1. Bax and/or Bak might be ‘recruited' by reorganization of MOM on conformational changes of the TMD-proximal BH3 domain or by modulation of Bcl2-family member(s) activity on Bnip3 induction. These initial events would in turn disrupt Opa1 complexes and trigger cytochrome-c mobilization.
Speculation
Evidence is presented for a direct and causative link between the apoptotic machinery on the MOM and Opa1 on the inner membrane. This dual functioning could rely on distinct Opa1-containing complexes specific either for fusion or for cristae structure depending on the presence of particular isoforms and partners of Opa1. Entry of Bnip3 into Opa1 complexes could modulate both their composition by molecular competition and their GTPase activity as it does by interacting with Rheb, a Ras GTPase (Li et al, 2007).
Bnip3 was recently involved in mitochondrial autophagy, or mitophagy, an adaptive response to hypoxia (Zhang et al, 2008). It was also suggested that fragmented mitochondria are more amenable to engulfment into autophagosomes (Twig et al, 2008). Bnip3-induced mitochondrial fragmentation might thus favour mitophagy. Retinal ganglion cells are often exposed to stresses as hypoxia (Kergoat et al, 2006). Under controlled stress exposure, Bnip3–Opa1 interaction might favour mitophagy by inhibiting Opa1-mediated fusion, whereas more severe conditions might lead to disassembly of Opa1 complexes and death of retinal ganglion cells. By impairing the survival response, mutations in Opa1 might lower the tolerance threshold to such stresses, leading to progressive optic nerve degeneration and to ADOA.
Methods
Cell biology. The Tet-Off Expression System (Clontech, Saint Germain en Laye, France) was used to derive stable HeLa cells with inducible expression of Opa1. Lipofectin (Invitrogen, Cergy Pontoise, France) and Amaxa Nucleofector kits (Amaxa Biosystems, Köln, Germany) were used for transfections. In co-transfection experiments, the GFP vector for transfected cells visualization and the Bnip3 vector of interest were in 1:5 ratio, respectively. z-VAD-fmk (100 μM) was added 4 h after transfection. Cells were cultivated under hypoxia (0.1% O2) or in serum-free medium containing 1 mM CoCl2 to induce endogenous Bnip3 expression.
To quantify the mitochondrial phenotypes, cells were stained directly in the cultures using 100 nM Mitotracker Red (Molecular Probes, Invitrogen) for 30 min, then fixed and stained with Hoescht 33258.
Mitochondrial fusion was assessed by the polyethylene glycol assay (Legros et al, 2002) or by using mito-Dendra, a mitochondrially targeted photoactivatable fluorescent protein Dendra (Gurskaya et al, 2006). After photoconversion (10 ms, 405 nm diode laser), z-stacks sections (0.3 μm) were acquired (FITC and TRITC filter sets, DMI6000B microscope; Leica Microsystems) onto a depth of 3 μm and analysed (Karbowski et al, 2004).
For immunofluorescence, antibodies to cytochrome-c (6H2.B4; Promega, Charbonnières-les Bains, France) and subunit IV of cytochrome-c oxidase (Cox IV; 3E11 Cell Signaling, Ozyme, Saint Quentin en Yvelines, France) were used, and imaging was performed with the DM5000 microscope (Leica Microsystems, Nanterre, France).
Molecular biology and biochemistry. Opa1 (residues 260–960) was used in a yeast two-hybrid screen of a human retinal library (a gift from C. Petit) according to the manufacturer's instructions (Matchmaker System; Clontech). The pCCEY-Opa1 plasmid was described by Olichon et al (2007). Full-length Bnip3 was isolated by PCR. Bnip3ΔC, Bnip3mutBH3 and Bnip3ΔBH3 were obtained by inverse PCR and Bnip3TMBcl2 and Bnip3TMOmp25 by synthetic oligonucleotide replacement (supplementary Table S1 online). Constructs were sequenced before subcloning into pCDNA3-HA, pmCerulean and pGEX2T'6. Luciferase, Mfn1 and Bnip3 siRNA (Dharmacon Research, Abgene Ltd, Epsom, UK) were used at 100 nM.
GST and GST-Bnip3 wild-type and mutants were produced in Escherichia coli BL21 and purified by thrombin cleavage (Smith & Johnson, 1988). For pulldown experiments, GST–sepharose beads were incubated in 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.1% Triton X-100 and a cocktail of proteases inhibitors for 6 h at 4°C in the presence of supernatants (30,000g, 15 min, 4°C) of S35methionine-labelled in vitro-translated Opa1, or of solubilized HeLa cells mitochondria (900 μg) prepared as described (Olichon et al, 2002). Beads were washed three times in the same buffer and processed for sodium dodecyl sulphate–polacrylamide gel electrophoresis (SDS–PAGE) and autoradiography or immunoblots.
For co-immunoprecipitation, mitochondrial extracts (6 mg/ml) were incubated for 2 h at 4°C with 1 mM dithio-bis (succinimidyl propionate) (DSP; Pierce, ThermoFisher Scientific, Brebières, France). Reactions were quenched with 125 mM glycine and mitochondria were disrupted by solubilization for 30 min in lysis buffer (50 mM Tris–HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% Triton X-100, protease inhibitors) and sonication. After centrifugation (25,000g, 15 min, 4°C) solubilized proteins (500 μg) were incubated for 4 h at 4°C with antibodies and protein A–sepharose in lysis buffer adjusted to 0.25% Triton X-100. Beads were washed in lysis buffer containing 0.1% Triton X-100 and processed for SDS–PAGE. For in cellulo crosslinking, transfected cells were harvested with trypsin and incubated (2 h, 4°C) in phosphate-buffered saline with 2 mM DSP. Reactions were stopped with 20 mM Tris–HCl (pH 7.5) and solubilized as above. After centrifugation (100,000g, 30 min, 4°C) solubilized proteins (7 mg) were used for immunoprecipitation.
For Opa1 oligomers assessment, mitochondria were incubated (30 min, 30°C) either alone or with 50 nM t-Bid (R&D Systems, Lille, France), or thrombin cleavage products from GST and GST-Bnip3 beads in 15 mM Hepes–KOH (pH 7.4), 125 mM KCl, 4 mM MgCl2, 210 mM mannitol, 5 mM NaH2PO4, 0.5 mM EGTA, 5 mM succinate, 2 mM ATP, 10 mM phosphocreatine, 10 μg/ml creatine kinase and proteases inhibitors. Samples were centrifuged (12,000g, 5 min, 4°C), harvested in phosphate-buffered saline containing 10 mM 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC; Pierce) and incubated (30 min, 37°C) before centrifugation and analysis by SDS–PAGE (4–12% Bis–Tris; Invitrogen).
For western blots, antibodies to the following proteins were used: actin (C4; Chemicon, Millipore, Molsheim, France), Bnip3 (ANa40; Novus Biologicals, Cambridge, UK), Hsp60 and Smac/Diablo (LK2 and S0941; Sigma-Aldrich, Saint Quentin Fallavier, France), cytochrome c and PARP (7H8.2C12 and 4C10-5; BD Pharmingen, BD Biosciences, Le Pont en Claix, France), Opa1 (clone-18; BD Biosciences), HA (12CA5; Roche), VDAC (89-173/016; Calbiochem), CIII (13G12-AF12-BB11; Molecular Probes), Cox IV (3E11; Cell Signaling) and Mfn1 (a gift from M. Rojo). Antibodies to Bnip3 (559690; BD Pharmingen), HA (12CA5; Roche) and Opa1 (Olichon et al, 2002) were used for immunoprecipitations.
The ImageJ software was used for quantification of protein expression levels in western blots, and unpaired Student's t-test was used for statistical analysis.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank C. Petit for the human retina two-hybrid library and D. Arnoult for MEF cells. This work was supported by grants from the Centre National de la Recherche Scientifique, Université Paul Sabatier, Rétina France, Association Française contre les Myopathies, Association pour la Recherche sur le Cancer, Ligue Contre le Cancer Haute-Garonne and Association contre les Maladies Mitochondriales. T.L. is the recipient of a Ligue Nationale contre le Cancer fellowship.
Footnotes
The authors declare that they have no conflict of interest.
References
- Burton TR, Gibson SB (2009) The role of Bcl-2 family member BNIP3 in cell death and disease: NIPping at the heels of cell death. Cell Death Differ 16: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer SA, Chan DC (2007) Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8: 870–879 [DOI] [PubMed] [Google Scholar]
- Frezza C et al. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189 [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA (2006) Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24: 461–465 [DOI] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ (2004) Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol 164: 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kergoat H, Herard ME, Lemay M (2006) RGC sensitivity to mild systemic hypoxia. Invest Ophthalmol Vis Sci 47: 5423–5427 [DOI] [PubMed] [Google Scholar]
- Landes T et al. (2010) OPA1 (dys)functions. Semin Cell Dev Biol (in press); doi:10.1016/j.semcdb.2009.12.012 [DOI] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M (2002) Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13: 4343–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, You M, Guan KL (2007) Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem 282: 35803–35813 [DOI] [PubMed] [Google Scholar]
- Olichon A et al. (2002) The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett 523: 171–176 [DOI] [PubMed] [Google Scholar]
- Olichon A et al. (2007) Effects of OPA1 mutations on mitochondrial morphology and apoptosis: relevance to ADOA pathogenesis. J Cell Physiol 211: 423–430 [DOI] [PubMed] [Google Scholar]
- Quinsay MN, Lee Y, Rikka S, Sayen MR, Molkentin JD, Gottlieb RA, Gustafsson AB (2009) Bnip3 mediates permeabilization of mitochondria and release of cytochrome c via a novel mechanism. J Mol Cell Cardiol (in press); doi:10.1016/j.yjmcc.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Johnson K (1988) Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ (2008) Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G et al. (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, Cizeau J, Dubik D, Alimonti J, Brown T, Israels S, Hakem R, Greenberg AH (2000) BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol Cell Biol 20: 5454–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi R, Lartigue L, Perkins G, Scott RT, Dixit A, Kushnareva Y, Kuwana T, Ellisman MH, Newmeyer DD (2008) Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol Cell 31: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283: 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.